Background: The mechanisms that regulate intracellular trafficking of the SUMOylation enzymes are not well understood.
Results: SUMO modification of SAE2 at and near the NLS, in addition to its NLS, is required for its nuclear localization.
Conclusion: A mechanism regulating SUMOylation activity in different cellular compartments was identified.
Significance: This SUMOylation-dependent mechanism of regulating intracellular localization may occur widely.
Keywords: Intracellular Trafficking, Post-translational Modification, Protein Export, Sumo, Sumoylation, NLS, SAE, SAE2, SIM, SUMO-interacting Motif
Abstract
SUMOylation occurs predominantly in the nucleus, but non-nuclear proteins can also be SUMOylated. It is unclear how intracellular trafficking of the SUMOylation enzymes is regulated to catalyze SUMOylation in different cellular compartments. Here we report that the SAE2 subunit of human SUMO activation enzyme (SAE) underwent rapid nucleocytoplasmic shuttling and its nuclear accumulation depended on SUMO modification at the C terminus. The SUMOylation sites included three Lys residues on the bipartite nuclear localization sequence (NLS) and two Lys residues outside of but adjacent to the NLS, and their SUMOylation was catalyzed by Ubc9. Because SAE2 forms a tight heterodimer with SAE1 and it controls the trafficking of the heterodimer, this study has identified the mechanism used to localize SAE to the nucleus. Similar mechanisms are likely to exist for other proteins that depend on SUMOylation for nuclear localization.
Introduction
Reversible attachments of the small ubiquitin-like modifiers (SUMO)2 to other cellular proteins are essential mechanisms that regulate important functions, including gene expression, DNA damage response, and the cell cycle progression (1–3). The SUMO activation enzyme (SAE), which catalyzes the first step of SUMOylation, is predominantly located in the nucleus. Some SAE is also present in the cytoplasm, consistent with a role for SAE in SUMOylating cytoplasmic proteins, such as the plasma membrane protein K2P, and proteins known to require SUMOylation to enter the nucleus, including NEMO and PAP (4, 5). However, the mechanism that regulates the nucleocytoplasmic trafficking of SAE itself is not well understood.
SAE is a tight heterodimer of two polypeptides: SAE1 (also known as Aos1) and SAE2 (also known as Uba2), and both SAE subunits contain nuclear localization sequences (NLS) (6–11). SAE1 resembles the N terminus of the ubiquitin E1 enzyme with a classical NLS in its mid region defined by the short sequence motif of K(K/R)X(K/R), where K is Lys, R is Arg and X can be any residue. SAE2 is homologous to the C terminus of ubiquitin E1, but has an 80 amino acid extension that contains a bipartite NLS consisting of two clusters of basic amino acids separated by a 9–10 amino acid segment (6, 11). The C-terminal extension is unstructured and its deletion causes SAE2 to localize to the cytoplasm, but otherwise has no effect on the in vitro enzymatic activity (6, 13). Each component of the SAE can localize to the nucleus separately using its own NLS. However, although the SAE1 NLS is blocked in the heterodimer, the SAE2 NLS can still transport the SAE1/2 heterodimer to the nucleus (11). For proteins with the classical NLS, nuclear import is mediated by the importin β/α complex, with importin α acting as an adaptor (14–16). Proteins with a non-classical NLS (17, 18), or an Arg/Ser (RS)-rich domain (19, 20) can interact directly with distinct members of the importin β family. For SAE2, the two basic regions that form the bipartite NLS are at residues 610–613 (NLS1) and 623–626 (NLS2) (11). However, mutations that destroy NLS1 have little effect on the location of SAE2, and the elimination of NLS2 does not abrogate SAE nuclear localization (11). These results indicate that there are other undefined factors that regulate the nuclear localization of SAE2.
Previous studies have shown that SUMOylation occurs at the NLS or sites adjacent to the NLS in several proteins, including Rad52 and the bovine papilloma virus E1 protein (21, 22). Furthermore, SUMO modification of these proteins at their NLS or SUMOylation of other proteins at non-NLS sites, including actin, SENP1, ErB4, and Smad4, promotes their localization to the nucleus (23–26). However, it is unclear whether this is because their SUMOylation leads to nuclear localization or if SUMOylation prevents these proteins from being exported to the cytoplasm.
In this study, we show that SAE2 was SUMOylated at several C-terminal residues, including three K residues in the bipartite NLS and two K residues adjacent to the NLS regions. SUMOylation of these residues were not required for the nuclear import of SAE2, but were important for its nuclear retention. Without SUMOylation, SAE2 was rapidly exported to the cytoplasm. Because SAE2 forms a tight heterodimer with SAE1 and controls the trafficking of the SUMO SAE heterodimer, this study has identified a post-translational modification that regulates the intracellular localization of SAE, and hence SUMOylation activities in different cellular compartments.
EXPERIMENTAL PROCEDURES
Plasmid, Mutations, and Protein Expression
The His6-SUMO-1, His6-SUMO-3, His6-Ubc9, GST, or GST-Ubc9, RanGAP1, GST-Sp100, SAE1, His6-SAE2, and His6-SAE2 Δ575–640 were cloned, then expressed in Escherichia coli and purified as previously described (27). The mammalian expression plasmids of SAE1-Myc-DKK and SAE2-GFP were obtained from Origene. Point mutations were introduced using a QuickChange Lightning mutagenesis kit (Agilent Technology) according to the manufacturer's protocol.
SUMOylation of SAE
The SUMOylation of SAE in vitro was performed by incubating a reaction mixture (500 μl) containing 3.6 μm SAE, 63 μm SUMO-1, 5 mm ATP, in buffer (5 mm MgCl2, 20 mm Hepes pH 7.5, and 50 mm NaCl) with 20 μm Ubc9 for 40 min at 37 °C to obtain SUMOylated SAE. The reactions were quenched by adding 2× protein loading buffer containing 360 mm DTT, and were separated by SDS-PAGE.
Western Blots
Western blots were performed as previously described (27). SAE2, actin, and SUMOylated proteins were detected by probing with rabbit anti-SAE2 antibody (Abcam), mouse anti-actin antibody (Sigma), mouse anti-SUMO-1 antibody (Abgen), and rabbit anti-SUMO-2/3 antibody (Abcam), respectively, followed by donkey anti-mouse or anti-rabbit secondary antibodies. Signals were detected using an Odyssey fluorescent scanner (Li-Cor Biosciences, Nebraska).
Identification of SUMOylated SAE2 in Cells
Anti-GFP antibody (Clontech) was attached to AL20 beads (Applied Biosystems) following manufacturer's protocol with slight modifications. The final product was stored in 100 μl of 50 mm Tris buffer, pH 7.5. Under native conditions, 400 μl of cell lysate was diluted to 2 ml in 1× PBS buffer contained 25 mm N-ethylmaleimide (NEM) and 1× protease inhibitor, then incubated with the beads at 4 °C overnight. The beads were washed five times with 500 μl of 1.5 m NaCl in 50 mm Tris buffer, pH 7.5. After immunoprecipitation (IP), beads were resuspended in 2× protein sample buffer for separation by SDS-PAGE and Western blot analysis.
To pull-down the His6-SUMOylated proteins, 100 μl of Ni2+ beads (Qiagen) was added to 900 μl of cell lysate prepared under denaturing conditions. The samples were incubated at room temperature with shaking for 1 h and 40 min. The beads were spun down then washed three times with 2 ml of wash buffer containing 1% SDS and 1 m NaCl in 50 mm Tris buffer, pH 7.6. Bound proteins were eluted by 200 mm imidazole, separated by SDS-PAGE, and detected by Western blot.
Cell Culture and Transfection
HEK293T cells were grown in Dulbecco modified Eagle's medium (DMEM) (CellGro) supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 100 units/ml streptomycin.
DNA transfections were performed using Lipofectamin following the manufacturer's protocol (Invitrogen). After 25 h, cells were washed twice with 3 ml of PBS and lysed with 1 ml of RIPA buffer containing 1% SDS. Lysates were stored at −80 °C for further use. For Leptomycin B (LMB) treatment, 25 h after transfection, cells were transferred to DMEM without FBS and grown for 20 h then treated with LMB (10 ng/μl).
Cells were lysed under native or denaturing conditions. For native lysis, cells at 90% confluence were collected from 15-cm plates, then lysed in 500 μl of Passive Lysis Buffer (Promega) that contained 25 mm NEM and 1× protease inhibitor (Roche) on ice for 30 min. Supernatants were separated by centrifugation at 12,000 rpm for 10 min. Both the supernatants and precipitate of the whole cell lysates (WCE) were stored at −80 °C for future use.
For denaturing lysis, cells at 90% confluence on 15 cm plates were directly lysed on the plate by the addition of 1× PBS buffer containing 1 ml of 1% SDS, 25 mm NEM, and 1× protease inhibitor (Roche). The whole mixture was incubated at 95 °C for 10 min and then stored at −80 °C for future use.
Mass Spectrometry
SUMOylated SAE2 bands were excised from an SDS-PAGE gel and subjected to in-gel reduction, alkylation, and digestion using Glu-C (Roche), Trypsin, or a mixture of Glu-C and Trypsin (27). The LC-MS/MS data were acquired using an Eksigent nanoLC-2D equipped with a self-packed C18 column connected to a hybrid linear ion trap (LTQ-FT) mass spectrometer (Thermo Electron, San Jose). MS/MS spectra were matched to a database of generated FASTA files created by ChopNSpice (chopnspice.gwdg.de) using GPM's X!Tandem database search engine. All identified SUMOylated MS spectra were confirmed manually.
Fluorescence Microscopy
HEK293T cells were grown in 8-well glass cover slips for 30 h prior to transfection with 0.1 μg of SAE2-GFP DNA using the Lipofectamin reagent (Invitrogen) according to the protocol provided by the manufacturer. After 30 h of transfections, cells were fixed for 15 min in 3.7% formaldehyde, and stained with DAPI. The GFP proteins were visualized by Z-stack confocal microscopy using a Zeiss confocal microscope (LSM 510meta) with an Achroplan 40x/0.6 Korr or a Zeiss LD Plan-Neufluar 20×/0.5 objective.
Statistical Analysis
The average ratios of cells with cytoplasmic or nuclear localization divided by the total number of cells (in multiple experiments) were used to calculate the probability of localization. The calculation was done using the Student's t test function (two tail, unequal variance) in Excel. The probability that the localization of the Cterm-5K/R mutant without LMB treatment versus Cterm-5K/R mutant treated with LMB was calculated using the average ratios of cells with localization of SAE2 to the cytoplasm divided by the total number of cells. Differences were considered to be significant if the calculated p value was less than 0.01.
RESULTS
Identification of SAE2 C-terminal Lys Residues That Are SUMOylated
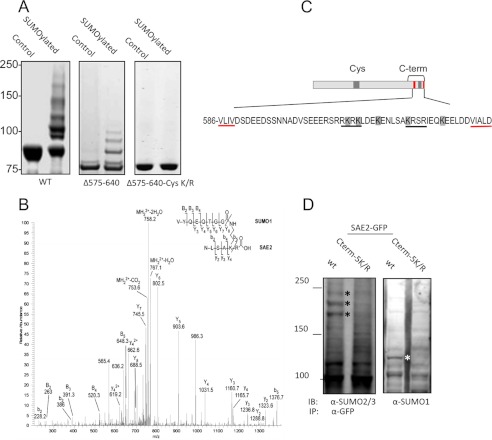
We have previously shown SUMOylation of SAE2 in vitro and in cells (27). Briefly, Ubc9 catalyzed SUMOylation of SAE2 was obtained by incubation of E1 and E2 with SUMO, ATP, and Mg2+, as previously described (27, 28) (Fig. 1A, left panel). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to analyze SUMOylated purified recombinant SAE1/SAE2. The SAE2 SUMOylation sites were clustered in two regions: the Cys domain, which included residues K190, K236, K257, K271, and K275, as previously described (27), and the C-terminal residues of SAE2, K611, K613, K617, K623, and K630 (Fig. 1B and highlighted by gray boxes in 1C). The modification at the C terminus was not observed in SAE2 auto-SUMOylation, i.e. without the catalysis of Ubc9 (27). The C-terminal SUMOylation sites of SAE2 were within or adjacent to the bipartite NLS consensus sequence, indicated by black underlining in the expanded C terminus (Fig. 1C).
FIGURE 1.
SAE is SUMOylated at the SAE2 C terminus. A, SAE2 is SUMOylated at its C terminus as shown in Coomassie Blue-stained gels of the in vitro SUMOylated SAE2 samples. Deletion from residue 575 to the C terminus (Δ575–640) showed a marked reduction of SAE SUMOylation (middle panel) in comparison to the WT enzyme (left panel). SAE2 was also SUMOylated at its Cys domain (27), and mutations that remove SUMOylation sites within the Cys domain and with Δ575–640 deletion completely eliminated SUMOylation (right panel). B, representative MS/MS spectrum for the identification of the SUMOylation sites. Shown here is the spectrum for SUMOylation at K623. The b and y ion series were numbered from the N terminus. Capital letters were used for SUMO ion series and the lowercase letters for the substrate peptide ion series. C, schematic diagram of SAE2 showing SUMOylation sites clustered at the Cys domain and C terminus (gray-shaded segments), and the SIMs (red segments) (29, 30). In the expanded C-terminal regions, all SUMOylation sites, K611, K613, K617, K623, and K630, are highlighted, the bipartite NLS is underlined in black, and the SIMs are underlined in red. D, SUMOylation sites were confirmed in cells. Plasmids expressing the SAE2-GFP fusion protein with all C-terminal SUMOylation sites mutated (Cterm-5K/R) or the wild type (WT) were transiently transfected into HEK293T. The GFP fusion proteins were immunoprecipitated and SUMOylated SAE2 was identified by immunoblotting with SUMO-2/3 or SUMO-1 antibodies. The asterisks indicate the bands found in the WT protein but not in the mutant.
To confirm the identified SUMOylation sites, we used site-directed mutagenesis and deletion, combined with Ubc9-catalyzed SUMOylation. The complete deletion of the C-terminal region of SAE2 from residue 575 (Δ575–640) resulted in a marked loss of Ubc9-catalyzed SUMOylation in vitro, confirming that the SAE2 C-terminal region was SUMOylated (Fig. 1A, middle panel). SAE2 can also be SUMOylated at its Cys domain (27). Therefore, we tested a mutant that eliminated the SUMOylation sites in the SAE2 Cys domain (Cys-K/R) and also contained the Δ575–640 C-terminal deletion. SUMOylation of this mutant was completely abolished (Fig. 1A, right panel). This result confirmed that all of the SAE2 SUMOylation sites that can be modified in vitro are located in the Cys and C-terminal domains (illustrated in Fig. 1C). The C-terminal SUMOylation sites are flanked by two conserved SUMO-interaction motifs (SIMs) (29–31), indicated by red segments in the schematic diagram and underlined in red in the expanded C-terminal region shown in Fig. 1C.
To confirm SUMOylation occurs on the C-terminal sites in cells, we expressed a mutant of SAE2 as a SAE2-GFP fusion protein. We mutated all of the C-terminal SUMOylation sites (K611, K613, K617, K623, and K630) from K to R (Cterm-5K/R) and then transfected the mutant construct, or WT SAE2-GFP as the control, into HEK293T cells. After 25 h, the cells were lysed, immunoprecipitated with anti-GFP antibody followed by Western blotting with anti-SUMO-1 or anti-SUMO2/3 antibodies. Removal of the SUMOylation sites removed some bands (starred) identified by SUMO-1 or SUMO-2/3 antibodies, indicating the disappearance of some SUMOylated SAE2 species. Therefore, this result indicates that the SUMOylation sites identified in vitro are utilized for modification in cells by both SUMO-1 and SUMO-2/3 (Fig. 1D). The patterns of SUMOylation bands observed in vitro and in cells are different, likely due to one or more of the following reasons. First, in vitro systems lack other factors involved in promoting SUMOylation or the formation of poly-SUMO chains (i.e. E3 ligases). Second, deSUMOylation enzymes (SENP) that remove or edit poly-SUMO chain lengths are present in cells but absent in in vitro assays. Third, cross-talk between post-translational modifications, such as SUMOylation-dependent ubiquitination, is absent in vitro but can affect the pattern of SUMOylated species in cells (32–34). Taken together, our results indicate that SAE2 is SUMOylated at the SAE2 C terminus in vitro and in cells.
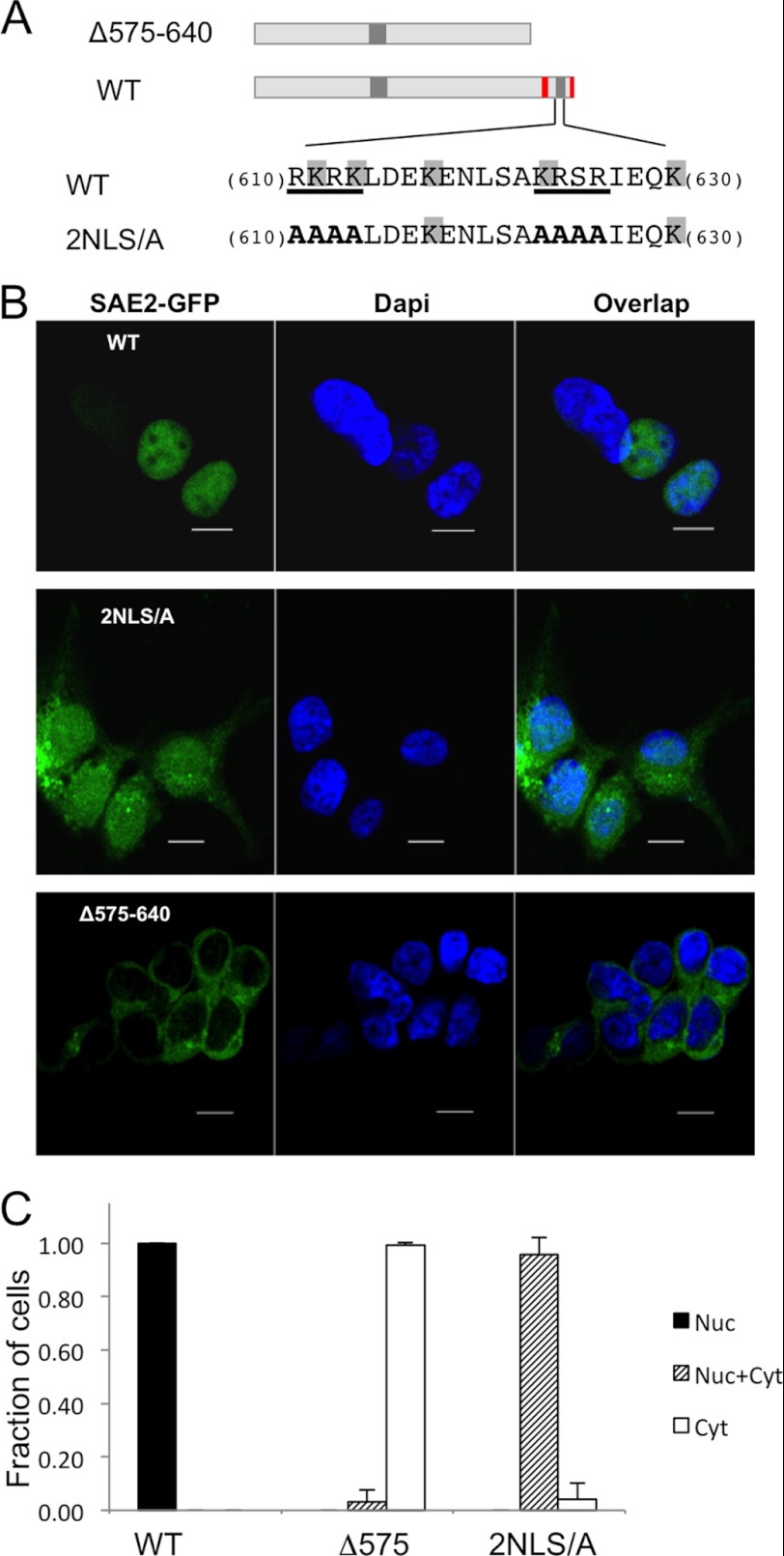
The NLS Is Not Solely Responsible for SAE2 Nuclear Localization
Previous studies have indicated that the bipartite NLS of SAE2 is important for the nuclear localization of both SAE2 alone and the SAE1/SAE2 heterodimer. However, mutating both NLS segments of the bipartite NLS does not completely remove SAE2 from the nucleus (11). This would seem to indicate that, in addition to the SAE2 bipartite NLS, another mechanism regulates SAE2 localization to the nucleus (11). To test this hypothesis, SAE2 mutants were constructed and expressed in human cells (Fig. 2A). For the 2NLS/A mutant, both groups of the bipartite NLS were destroyed by mutating residues 610–613 and 623–626 to Ala. The 2NLS/A expression plasmid was transiently transfected into HEK293T cells and the same expression vector containing the WT SAE2 was transfected as the control. Intracellular localization of the WT and mutant proteins was compared using Z-stack confocal microscopy. Our results showed that WT SAE2-GFP was located in the nucleus (Fig. 2, B, top, and C), which is consistent with other reports in the literature. For 2NLS/A, the majority of the mutant protein was also in the nucleus even though both the SAE2 NLS had been eliminated (Fig. 2, B, middle, and C). This result indicated that additional nuclear localization mechanisms exist for SAE2. To determine if additional factors required for the nuclear localization of SAE2 were within the C terminus of the protein, we examined cells expressing the SAE2 C-terminal deletion construct (Δ575–640), in which we deleted the residues from 575 to the C terminus. This region contains the NLS but is not required for the enzymatic activity of the SAE (13). In contrast to the 2NLS/A mutant, the Δ575–640 protein localized exclusively to the cytoplasm confirming that in addition to the NLS, other mechanisms involving the C-terminal region are required to localize SAE2 in the nucleus (Fig. 2, B, bottom, and C).
FIGURE 2.
A mechanism in addition to NLS is required for nuclear localization of SAE2. A, schematic illustration of the Δ575–640 deletion and the site-directed mutations (WT construct shown for comparison). B, plasmids shown in A were transiently transfected into HEK239T with or without SAE1, which produced similar results, followed by confocal imaging. C, statistical analysis of localization of the SAE2 variants as shown in B. The y axis is the ratio of the cell with the phenotype divided by the total number of cells. Nuc: found in nucleus only, Nuc+Cyt: found in both nucleus and cytoplasm, and Cyt: found in cytoplasm only. We counted 74–213 cells for each construct from multiple experiments to obtain the averages and standard deviations.
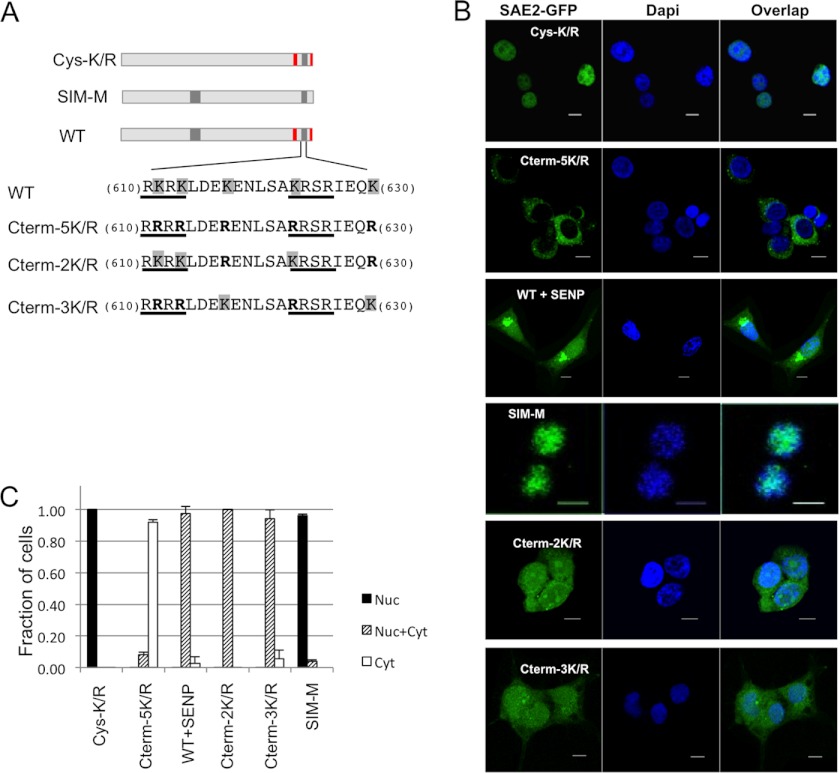
The Role of SUMOylation on the SAE2 C Terminus in Nuclear Localization
Because we found a cluster of SUMOylation sites within (K611/613/623) and adjacent to (K617, K630) the bipartite NLS, we investigated if SUMOylation played a role in the nuclear localization of SAE2. We created conservative mutations for the SUMOylation sites that would abolish SUMOylation but preserve the positive charge of the residues (Fig. 3A). To examine the role of Cys domain SUMOylation in intracellular trafficking, we made the Cys-5K/R mutant, which mutates K to R at all SUMOylation sites on the Cys domain (K190, K236, K257, K271, and K275). To remove the SUMO-binding ability of the two SIMs, we mutated V589, I590, V637, and V638 to Ala (SIM-M; Fig. 1C). In addition, site-specific mutations of the SUMOylation sites were constructed to investigate the contribution of two C-terminal SUMOylation regions to nuclear localization. The 3K/R construct mutated the three SUMOylation sites within the two NLS segments (K611, K613, and K623) to R, whereas 2K/R mutated the two SUMOylation sites adjacent to the NLS (K617 and K630) to R. These SAE2-GFP fusion variants were transiently transfected into HEK293T cells, without or with the SAE1 plasmid. After 30 h SAE2 localization was examined by Z-stack confocal microscopy. In all cases, the absence or presence of SAE1 did not affect the localization of SAE2, which is consistent with the previous report that SAE1 does not play a role in directing the localization of either SAE2 or the SAE1/SAE2 heterodimer (11).
FIGURE 3.
SUMOylation of SAE2 C terminus is required for nuclear accumulation of SAE2. A, schematic illustrations of the mutants used for the study (WT construct shown for comparison). B, plasmids shown in A were transiently transfected into HEK239T with or without SAE1, which produced similar results, followed by confocal imaging. SUMOylation of the Cys domain (Cys-K/R) had no effect on SAE2 (top). Conservative mutations that abolished all of the C-terminal SUMOylation sites (Cterm-5K/R) resulted in SAE2 localizing almost completely to the cytoplasm, similar to the deletion of the SAE2 C terminus (Δ575–640, Fig. 2). Transfection and expression of the de-SUMOylation enzyme SENP1 prior to transfection with WT SAE2 induced partial localization of SAE2 to the cytoplasm. Mutations that eliminated the SUMOylation sites outside of NLS (K617/K630 to R; Cterm-2K/R), caused partial localization of SAE2 to the cytoplasm. Mutations that eliminated the SUMOylation sites within the SAE2 NLS (K611,/K613/K623 to R; Cterm-3K/R), also caused a partial localization of SAE2 to the cytoplasm. C, statistical analysis of the localization of SAE2 mutants. The y axis is the ratio of cell with the phenotype divided by the total number of cells. Nuc: found in nucleus only, Nuc+Cyt: found in both nucleus and cytoplasm, and Cyt: found in cytoplasm only. We counted 74–300 cells for each construct from multiple experiments to obtain the averages and standard deviations.
Our results showed that mutating all the SUMOylation sites of the SAE2 Cys domain did not affect SAE2 localization because, similar to WT, the Cys-K/R mutant protein localized in the nucleus (Fig. 3, B and C). Interestingly, the Cterm-5K/R mutant protein, in which all SAE2 C-term SUMOylation sites were eliminated but the charge of the bipartite NLS was preserved, localized predominantly in the cytoplasm (Fig. 3, B and C). This result was in contrast to the partial mis-localization of the 2NLS/A mutant protein, which had lost the bipartite NLS but retained the adjacent SUMOylation sites (K617 and K630), and still partially localized in the nucleus (Fig. 3, B and C). The results indicate that SUMOylation is important for regulating the localization of SAE2. Furthermore, although the Cterm-5K/R mutant protein had two intact SIMs on its C terminus (Fig. 3A), they were not able to direct SAE2 to the nucleus (Fig. 3B). Thus, the potential non-covalent interaction of the SAE2 SIMs with free SUMO or SUMO conjugated to the adjacent Lys residues could not direct SAE2 to the nucleus. The result was confirmed by the fact that the SIM-M mutant protein, in which both SIM motifs were eliminated, also localized to the nucleus (Fig. 3, B and C).
There remained the possibility that the cytoplasmic localization of SAE2 was caused by the substitution of R for K at the SUMOylation sites rather than the loss of SUMOylation. To eliminate this possibility, we overexpressed the deSUMOylation enzyme SENP1 together with WT SAE2-GFP in HEK293T cells to deSUMOylate the SAE2 without altering its bipartite NLS. Our images showed that in the cells overexpressing SENP1, increased levels of WT SAE2-GFP were localized in the cytoplasm (Fig. 3, B and C), confirming the importance of SUMOylation for the nuclear localization of SAE2. The portion of WT SAE2 that remained in the nucleus in the SENP expressing cells was probably due to incomplete de-SUMOylation of the SAE2-GFP protein. It is unlikely that SENP1 plays a general role in the export of nuclear proteins, because its overexpression does not impair the nuclear localization of the androgen receptor (36), which appears to localize to the nucleus independently of SUMOylation. Altogether, the results indicate that the SUMOylation on the C terminus of SAE2 regulates SAE localization.
To determine if the SUMOylation sites that are within the NLS and those adjacent to it play different roles, we examined the localization of the mutations Cterm-2K/R (K617/630R) and Cterm-3K/R (K611/613/623R). We transiently transfected the plasmids into 293T cells and observed the localization of SAE2 using confocal imaging after 30 h. We found that both of the mutant proteins partially mis-localized to the cytoplasm (Fig. 3, B and C). The Cterm-2K/R protein had an intact NLS, but it could not localize SAE completely in the nucleus, whereas the Cterm-3K/R mutant had intact neighboring SUMOylation sites and also partially mislocalized to the cytoplasm. The results indicate that both the SUMOylation sites within the NLS and those adjacent to it play important roles in SAE2 nuclear localization. The finding further suggests that SUMOylation regulates SAE localization. In addition, because both mutants by themselves could not completely redirect SAE to the cytoplasm as could the Cterm-5K/R mutant, each SUMOylation group could only partially regulate SAE localization. In summary, our results indicate that the covalent SUMOylation of the SAE2 C terminus is required to direct SAE to the nucleus.
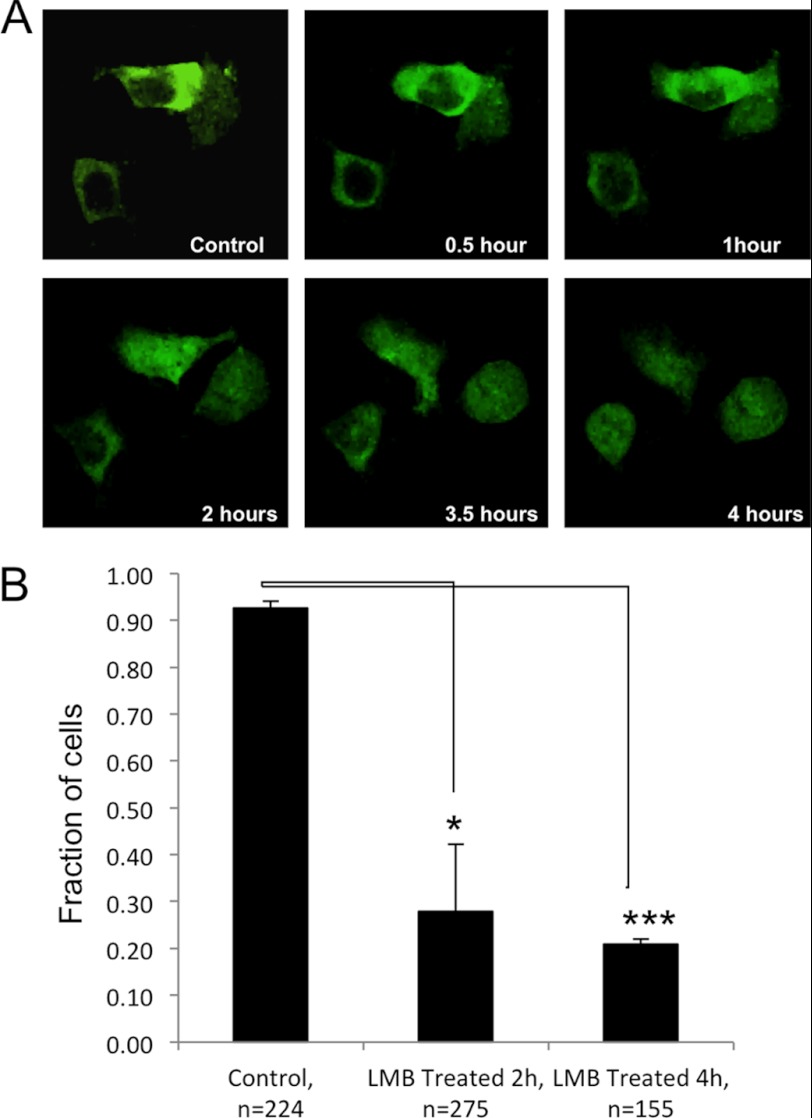
Nucleocytoplasmic Shuttling of SAE2
We used the leptomycin B (LMB) nuclear export inhibitor to investigate if SUMOylation plays a role in nuclear import or nuclear retention. LMB is a widely used nuclear export inhibitor. It inhibits CRM1/Exportin-1, a protein required for nuclear export of proteins containing a nuclear export sequence (NES), by glycosylating a cysteine residue (37–39). The Cterm-5K/R SAE2 mutant that completely abolished C-terminal SUMOylation was transiently expressed in 293T cells together with a WT SAE1 plasmid. After 25 h of expression, the cells were serum starved for 20 h to synchronize them in the G0/G1 phase, so that any observed differences in SAE localization would not be due to differences in cell cycle stage. The cells were then treated with LMB and images of live cells were captured at different time points using confocal microscopy to observe the location of SAE2.
This experiment unexpectedly revealed that SAE is rapidly shuttled into and out of the nucleus. Although the Cterm-5K/R mutant almost completely localized to the cytoplasm in the absence of LMB (Fig. 3), LMB treatment lead to an accumulation of the mutant in the nucleus that increased as time progressed (Fig. 4). This finding suggests that the Cterm-5K/R protein was localized in the cytoplasm not because of an inability of the mutant protein to enter the nucleus, but rather due to its rapid export back to the cytoplasm because it is not SUMOylated. The amount of translocated SAE varied among different cells at any given time, which is consistent with the findings about the nuclear transport process of other proteins (40, 41). To confirm the result, we repeated the experiment multiple times and fixed cells at 2 and 4 h after treatment with LMB, then stained with DAPI to determine with certainty whether SAE2 florescence was present in the nucleus using Z-stack confocal microscopy. We found that treatment of the cells with LMB increased the number of cells that had the SAE2 Cterm-5K/R mutant protein located in the nucleus over time when compared with the untreated control (2 h, p < 0.01; 4 h, p < 0.001) (Fig. 4B). Taken together, the results indicate that SAE2 is rapidly shuttled in and out of the nucleus, but SUMOylation at the C terminus of SAE2 sequestered the enzyme in the nucleus.
FIGURE 4.
Nucleocytoplasmic shuttling of SAE2. A, nucleocytoplasmic trafficking of the SAE2 Cterm-5K/R mutant, which almost completely localized to the cytoplasm. The construct was expressed in HEK239T for 25 h, followed by serum starvation for 20 h to synchronize cell cycle stage, then addition of 10 ng/ml of the LMB nuclear export inhibitor. Live cells were examined by confocal microscopy beginning 20 min after adding LMB and images were captured at the indicated time points. As time increased, the nuclear accumulation of the Cterm5K/R SAE2 mutant protein was observed. B, statistical analysis of cells with exclusive cytoplasmic localization of the Cterm-5K/R mutant after treatment for 2 h and 4 h. At 2 and 4 h after the treatment, the disappearance of cytoplasmic SAE2 and accumulation of nuclear SAE2 was significant. * indicates p < 0.01, and ***, p < 0.0001.
DISCUSSION
Our results indicate that SAE2 is rapidly shuttled in and out of the nucleus, and is sequestered inside of nucleus by SUMOylation of the SAE2 C terminus (Fig. 5). SAE2 protein with mutations in the bipartite NLS still partially localized in the nucleus (11), because SUMOylation could still occur at the adjacent Lys residues (K617 and K630). When all of the SAE2 C terminus SUMOylation sites were removed by conservative K to R mutations, SAE2 almost completely localized in the cytoplasm. However, it did accumulate in the nucleus in the presence of the LMB nuclear export inhibitor, suggesting that SAE2 can enter the nucleus without SUMOylation, but the SUMO moiety is necessary for the protein to remain there. Because many nuclear proteins contain SIMs (29–31), it is likely that the covalent SUMO moiety promotes nuclear retention by interacting with SUMO-binding nuclear proteins. The availability of multiple SUMOylation sites could ensure that a higher percentage of SAE2 is SUMOylated than would be possible with a single SUMOylation site, and/or ensures that SAE2 has a higher binding affinity for SIM-containing proteins due to the multivalency. Overexpression of SENP1 provided further evidence to supporting a role for SUMOylation in the nuclear localization of SAE2. SENP1 overexpression caused a partial localization of WT SAE2 in the cytoplasm and this result eliminated the possibility that mis-localization of the mutant proteins was due to the conservative substitutions of R for K in the C terminus, rather than the loss of SUMOylation. Thus, we have identified a mechanism that regulates nuclear localization of SAE in addition to the NLS, and SUMOylation at the C terminus most likely serves as an “on-off” switch for the export of SAE to the cytoplasm.
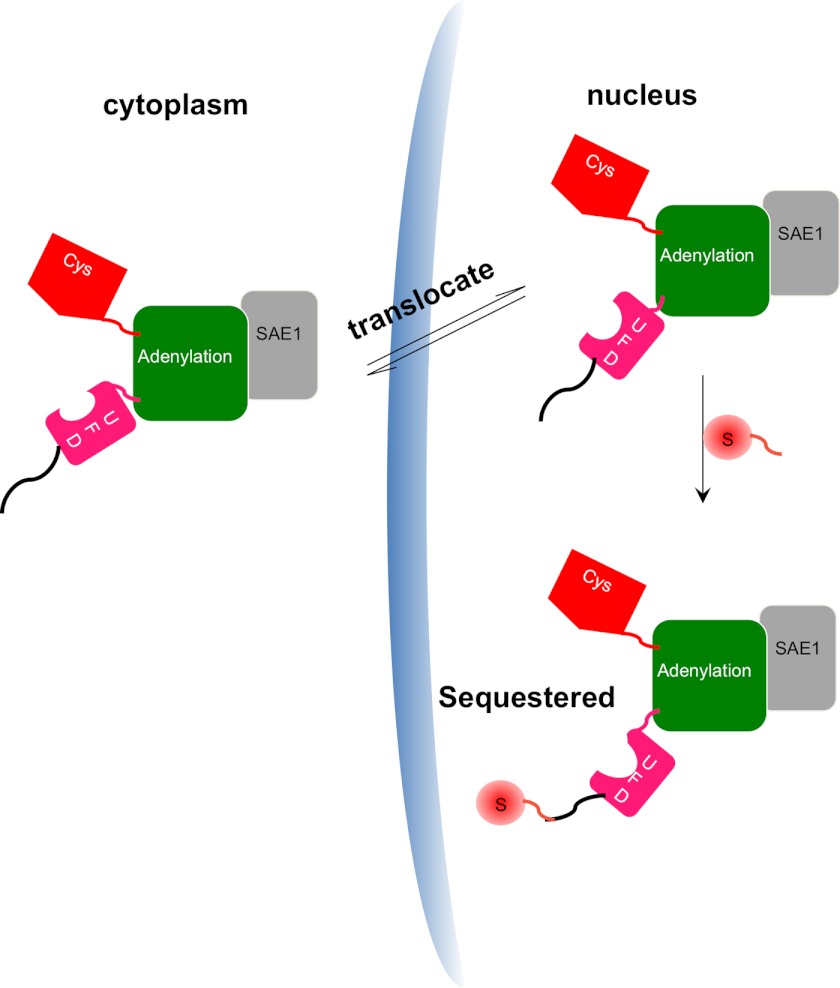
FIGURE 5.
A model for the role of SUMOylation in regulating SAE localization. The SAE1 subunit is shown in light gray, and the different domains of SAE2 are shown in color: the green domain contains the adenylation catalytic center; the red domain contains the catalytic Cys; and the pink region represents the ubiquitin-fold domain (UFD). SAE that is not SUMOylated at the SAE2 C terminus is rapidly shuttled in and out of nucleus, but SUMOylation of SAE2 at the C terminus sequesters the SAE in the nucleus.
None of the SUMOylation sites at the C terminus of SAE2 have the consensus motif of ΨKX(E/D), where Ψ is a bulky hydrophobic residue and X is any residue. Neither do they have its variants such as the inverted motif, hydrophobic cluster, phosphorylation-dependent SUMOylation (PDSM), or the negatively charged amino acid-dependent SUMOylation (NDSM) motif (42). Although SIMs can promote SUMOylation of some proteins (43), the two conserved SIMs that flank the C terminus SUMOylation sites of SAE2 are unlikely to be required for C-terminal SUMOylation. Mutations of both SIMs did not alter the nuclear localization of SAE2, indicating that SIMs do not affect SUMOylation on the C terminus (Fig. 3). It is likely that an interaction between Ubc9 and the UFD domain of SAE2, which is adjacent to its C-terminal extension (44) (Fig. 5), promotes Ubc9-catalyzed SUMOylation of the C-terminal Lys residues of SAE2.
Cross talk between SUMOylation and other post-translational modifications at the C terminus of SAE2 are also likely to contribute to the regulation of SAE2 localization. Two SUMOylation sites of the SAE2 C terminus are within seven residues of either a casein kinase II consensus phosphorylation site, [S/T]XX[E/D] or its inverted motif [E/D]XX[S/T] (sites K611 and K617). In fact, S592 was previously reported to be phosphorylated (45). In addition, the K617 SUMOylation site was reported to be acetylated (46). Therefore, the cross-regulation between SUMOylation and phosphorylation, or SUMOylation and acetylation is likely to occur at the C terminus of SAE2.
The important role we identified here for SUMOylation in the nuclear localization of SAE2 is likely to exist for other proteins that have been reported to depend on SUMOylation for nuclear localization (5, 21, 23, 24, 47). In particular, overexpression of WT SENP1, but not the C603A catalytically inactive SENP1 mutant, was reported to enhance cytoplasmic localization of HIPK1 and poly (A) polymerase (5, 35). In addition, SENP2 contains a bipartite NLS similar to that of SAE2, and is known to shuttle between nucleus and cytoplasm in a de-SUMOylation activity-dependent fashion (12). SUMOylation, as revealed by this study, may serve as the mechanism for the nuclear accumulation of many proteins.
Acknowledgment
We thank the City of Hope, Beckman Research Institute Mass Spectrometry Core.
This work was supported, in whole or in part, by Grant R01 GM086171 and R01 GM102538 (to Y. C.) from the NIGMS, National Institutes of Health and Grant P30 CA033572 from the NCI, National Institutes of Health.
- SUMO
- small ubiquitin-like modifier
- SAE
- SUMO activation enzyme
- NLS
- nuclear localization sequence
- NEM
- N-ethylmaleimide
- LMB
- Leptomycin B
- SIM
- SUMO-interaction motif
- NES
- nuclear export sequence
- PDSM
- phosphorylation-dependent SUMOylation.
REFERENCES
- 1. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 2. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Yeh E. T. (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hay R. T. (2004) Modifying NEMO. Nat. Cell Biol. 6, 89–91 [DOI] [PubMed] [Google Scholar]
- 5. Vethantham V., Rao N., Manley J. L. (2008) Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 22, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dohmen R. J., Stappen R., McGrath J. P., Forrová H., Kolarov J., Goffeau A., Varshavsky A. (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270, 18099–18109 [DOI] [PubMed] [Google Scholar]
- 7. Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gong L., Li B., Millas S., Yeh E. T. (1999) Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448, 185–189 [DOI] [PubMed] [Google Scholar]
- 9. Okuma T., Honda R., Ichikawa G., Tsumagari N., Yasuda H. (1999) In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254, 693–698 [DOI] [PubMed] [Google Scholar]
- 10. Rodriguez M. S., Dargemont C., Hay R. T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 11. Moutty M. C., Sakin V., Melchior F. (2011) Importin α/β mediates nuclear import of individual SUMO E1 subunits and of the holo-enzyme. Mol. Biol.Cell 22, 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itahana Y., Yeh E. T., Zhang Y. (2006) Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell. Biol. 26, 4675–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lois L. M., Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adam S. A., Gerace L. (1991) Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66, 837–847 [DOI] [PubMed] [Google Scholar]
- 15. Moroianu J., Hijikata M., Blobel G., Radu A. (1995) Mammalian karyopherin α1β and α2 β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. U.S.A. 92, 6532–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weis K., Mattaj I. W., Lamond A. I. (1995) Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science 268, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 17. Siomi H., Dreyfuss G. (1995) A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakel S., Gorlich D. (1998) Importin β, transportin, RanBP5, and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kataoka N., Bachorik J. L., Dreyfuss G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai M. C., Lin R. I., Tarn W. Y. (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rangasamy D., Woytek K., Khan S. A., Wilson V. G. (2000) SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275, 37999–38004 [DOI] [PubMed] [Google Scholar]
- 22. Saito K., Kagawa W., Suzuki T., Suzuki H., Yokoyama S., Saitoh H., Tashiro S., Dohmae N., Kurumizaka H. (2010) The putative nuclear localization signal of the human RAD52 protein is a potential sumoylation site. J. Biochem. 147, 833–842 [DOI] [PubMed] [Google Scholar]
- 23. Lin X., Liang M., Liang Y. Y., Brunicardi F. C., Feng X. H. (2003) SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 278, 31043–31048 [DOI] [PubMed] [Google Scholar]
- 24. Hofmann W. A., Arduini A., Nicol S. M., Camacho C. J., Lessard J. L., Fuller-Pace F. V., de Lanerolle P. (2009) SUMOylation of nuclear actin. J. Cell Biol. 186, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bailey D., O'Hare P. (2004) Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279, 692–703 [DOI] [PubMed] [Google Scholar]
- 26. Sundvall M., Korhonen A., Vaparanta K., Anckar J., Halkilahti K., Salah Z., Aqeilan R. I., Palvimo J. J., Sistonen L., Elenius K. (2012) Protein Inhibitor of Activated STAT3 (PIAS3) Protein Promotes SUMOylation and Nuclear Sequestration of the Intracellular Domain of ErbB4 Protein. J. Biol. Chem. 287, 23216–23226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truong K., Lee T. D., Chen Y. (2012) Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J. Biol. Chem. 287, 15154–15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramaniam S., Mealer R. G., Sixt K. M., Barrow R. K., Usiello A., Snyder S. H. (2010) Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285, 20428–20432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song J., Zhang Z., Hu W., Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 31. Namanja A. T., Li Y. J., Su Y., Wong S., Lu J., Colson L. T., Wu C., Li S. S., Chen Y. (2012) Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis. J. Biol. Chem. 287, 3231–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun H., Leverson J. D., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 34. Weisshaar S. R., Keusekotten K., Krause A., Horst C., Springer H. M., Göttsche K., Dohmen R. J., Praefcke G. J. (2008) Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582, 3174–3178 [DOI] [PubMed] [Google Scholar]
- 35. Li X., Luo Y., Yu L., Lin Y., Luo D., Zhang H., He Y., Kim Y. O., Kim Y., Tang S., Min W. (2008) SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 15, 739–750 [DOI] [PubMed] [Google Scholar]
- 36. Kaikkonen S., Jääskeläinen T., Karvonen U., Rytinki M. M., Makkonen H., Gioeli D., Paschal B. M., Palvimo J. J. (2009) SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol. Endocrinol. 23, 292–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kudo N., Wolff B., Sekimoto T., Schreiner E. P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540–547 [DOI] [PubMed] [Google Scholar]
- 39. Wolff B., Sanglier J. J., Wang Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 40. Reszka A. A., Seger R., Diltz C. D., Krebs E. G., Fischer E. H. (1995) Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 92, 8881–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao Z., Flash I., Raviv Z., Yung Y., Asscher Y., Pleban S., Seger R. (2001) Non-regulated and stimulated mechanisms cooperate in the nuclear accumulation of MEK1. Oncogene 20, 7588–7596 [DOI] [PubMed] [Google Scholar]
- 42. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 43. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 44. Wang J., Lee B., Cai S., Fukui L., Hu W., Chen Y. (2009) Conformational transition associated with E1-E2 interaction in small ubiquitin-like modifications. J. Biol. Chem. 284, 20340–20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 46. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 47. Stade K., Vogel F., Schwienhorst I., Meusser B., Volkwein C., Nentwig B., Dohmen R. J., Sommer T. (2002) A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J. Biol. Chem. 277, 49554–49561 [DOI] [PubMed] [Google Scholar]