Background: LBR is an inner nuclear membrane protein that participates in heterochromatin organization.
Results: LBR recognizes specific histone modifications and induces chromatin compaction and transcriptional repression.
Conclusion: LBR tethers epigenetically marked chromatin to the NE to repress transcription.
Significance: This finding provides an implication of how transcriptional activities are repressed beneath the NE.
Keywords: Chromatin, Chromatin Histone Modification, Heterochromatin, Nuclear Membrane, Nuclear Organization, Chromatin Organization, Histone Code, Lamin B Receptor, Nuclear Envelope, Tudor Domain
Abstract
Inner nuclear membrane proteins provide a structural framework for chromatin, modulating transcription beneath the nuclear envelope. Lamin B receptor (LBR) is a classical inner nuclear membrane protein that associates with heterochromatin, and its mutations are known to cause Pelger-Huët anomaly in humans. However, the mechanisms by which LBR organizes heterochromatin remain to be elucidated. Here, we show that LBR represses transcription by binding to chromatin regions that are marked by specific histone modifications. The tudor domain (residues 1–62) of LBR primarily recognizes histone H4 lysine 20 dimethylation and is essential for chromatin compaction, whereas the whole nucleoplasmic region (residues 1–211) is required for transcriptional repression. We propose a model in which the nucleoplasmic domain of LBR tethers epigenetically marked chromatin to the nuclear envelope and transcriptional repressors are loaded onto the chromatin through their interaction with LBR.
Introduction
The eukaryotic genome is organized within the nucleus, and functional organization of the nucleus is crucial for its activities (1–3). It is known that heterochromatin is formed beneath the nuclear envelope (NE)2 (4, 5) and that INM proteins can play a key role in heterochromatin formation (6). LBR is a classical member of the conserved INM protein family and binds to various nuclear components via its nucleoplasmic region (Fig. 1A) (7–11). Because chromatin pulled down by LBR is enriched for heterochromatin marks such as histone H3 lysine 9 tri-methylation (H3K9me3) and lysine 27 tri-methylation (H3K27me3), it is thought that LBR is likely to bind to heterochromatin (6). Dominant mutations in LBR are known to cause human Pelger-Huët anomaly which is characterized by an aberrant neutrophil nuclear shape: in contrast to the characteristic hyperlobulated nucleus of normal neutrophils, the neutrophils of patients with Pelger-Huët anomaly have a bi-lobed nucleus (12). Chromatin clumping and disorganization of pericentric heterochromatin (13, 14) are observed in a related LBR-associated disease in mice, ichthyosis. These studies strongly suggest that LBR is one of the key proteins involved in organizing chromatin beneath the NE. However, the mechanism by which LBR binds to chromatin is unclear.
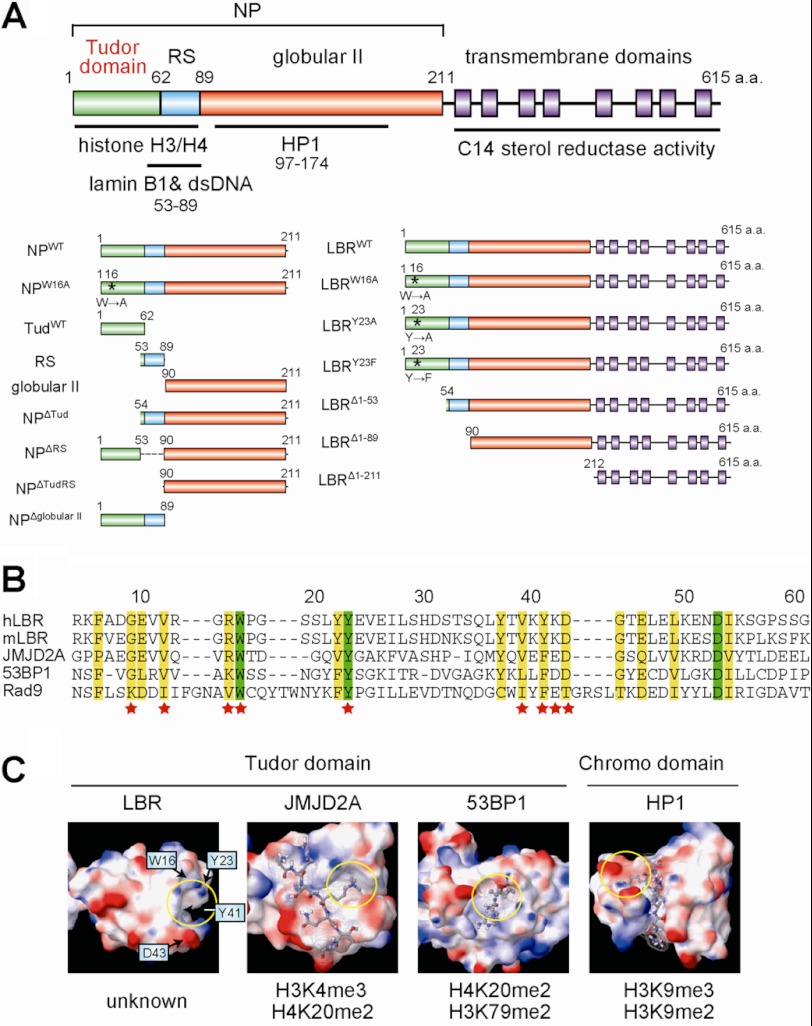
FIGURE 1.
LBR possesses a tudor domain in the N-terminal region. A, schematic illustration of the full-length LBR. The amino acids 1–62 correspond to the tudor domain (green). Blue, red, and purple squares correspond to RS, globular II, and transmembrane regions, respectively. NP represents the whole nucleoplasmic region of LBR. Asterisks indicate the positions of mutated amino acid residues. Numbers indicate the number of amino acid residues from the N terminus. B, alignment of the tudor domain sequences. The amino acid residues of LBR are numbered above the sequences. Green and yellow boxes indicate identical and highly conserved amino acids, respectively. The amino acid residues substituted in this study are indicated by stars. C, comparison of the three-dimensional structure of the tudor and chromo domains. Structural information was obtained from the NCBI Data Bank (Protein Data Bank ID codes for LBR, JMJD2A, 53BP1, and HP1 are 2DIG, 2GFA, 2IG0, and 1Q3L, respectively). The electrostatic potential of the molecular surface was calculated by MacroModel software. Red, blue, and white regions indicate negative, positive, and neutral charges, respectively. The histone peptides recognized by each domain are shown as stick models, and the specific histone codes recognized by the domains are shown at the bottom. Histone code-reading pockets are indicated by yellow circles.
It has been reported that LBR binds to histones and DNA via its tudor domain (amino acid residues 1–62) and RS domain (amino acid residues 53–89), respectively (11, 15). Tudor domains are methylation-specific “histone code-reading” domains, as are chromo, MBT (malignant brain tumor) and WD-repeat domains (16–18). Indeed, the tudor domains of 53BP1 and JMJD2A bind specifically to H4K20me2/H3K79me2 and H3K4me3/H4K20me2, respectively (17, 19). Several aromatic amino acid residues in those tudor domains, which make a structure called a “histone code-reading pocket,” are known to be needed for recognition of specific histone modifications (17). These amino acid residues are also conserved in the tudor domain of LBR (Fig. 1, B and C). Thus, LBR may also recognize some specific histone modifications. However, recent studies show that the tudor domain of LBR does not appear to bind methylated lysine or arginine residues (15). They show that not only the tudor domain but also the RS domain binds to histone and that the binding of LBR to chromatin is affected by the globular II domain in vivo (20) (for the domain structures of LBR, see Fig. 1A), suggesting a role for the nucleoplasmic region, outside the tudor domain, of LBR. Here we demonstrate that the whole nucleoplasmic domain of LBR is required for transcriptional repression beneath the NE, whereas the tudor domain of LBR primarily recognizes histone H4 lysine 20 dimethylation (H4K20me2) to recruit epigenetically marked chromatin to the NE.
EXPERIMENTAL PROCEDURES
Materials
Primary antibodies for Western blotting were purchased and used at the indicated dilutions: rabbit polyclonal anti-LBR (1:500, E398L; Epitomics, Burlingame, CA), rabbit polyclonal anti-lamin B1 (1:300; Abcam, Cambridge, MA), rabbit anti-lamin B2 (1:1,000; Epitomics), mouse anti-lamin A/C (1:1,000; 4C11, Sigma-Aldrich), rabbit anti-GAL4 DNA-BD (1:2,000; Sigma-Aldrich), mouse monoclonal anti-GST (1:1,000; GS019, Nacalai Tesque, Kyoto, Japan), monoclonal anti-FLAG M2 (1:2,000; Sigma-Aldrich), anti-panH4 (0.2 μg/ml) and anti-H4K20me2 (0.2 μg/ml) antibodies.3 Anti-mouse HRP-conjugated IgG and anti-rabbit HRP-conjugated IgG were purchased from GE Healthcare.
Plasmids encoding the full-length and the whole nucleoplasmic region of the LBR protein were constructed as reported previously (7, 21) and used as a template for further plasmid constructions as described below. Specific regions of interest were amplified by PCR using either one of these DNA plasmids as a template. The amplified DNA was inserted into the pGEX-5X-1 vector at the EcoRI and XhoI sites, or the pCMX-GAL4 vector at the EcoRI and BamHI sites. The LBR mutants (NPW16A, LBRW16A, LBRY23A, LBRY23F, and others listed in Fig. 3B) were generated using the GeneTailorTM Site-Directed Mutagenesis System (Invitrogen) according to the manufacturer's protocol. The DNA sequences of all plasmids used in this study were confirmed using a CEQ2000 DNA sequencer (Beckman Coulter). The 26-kbp DNA plasmids used for chromatin reconstitution experiments were gifts from Dr. W. de Laat at Erasmus University Medical Center, The Netherlands (22). Stealth siRNAs for luciferase, lamin B1 (HSS106098: AAUUGUAACAGUCUGGCCUGCCUUC) and lamin B2 (HSS189215: UUCUCAUUCUCACGCAUCACCGAGG) were purchased from Invitrogen.
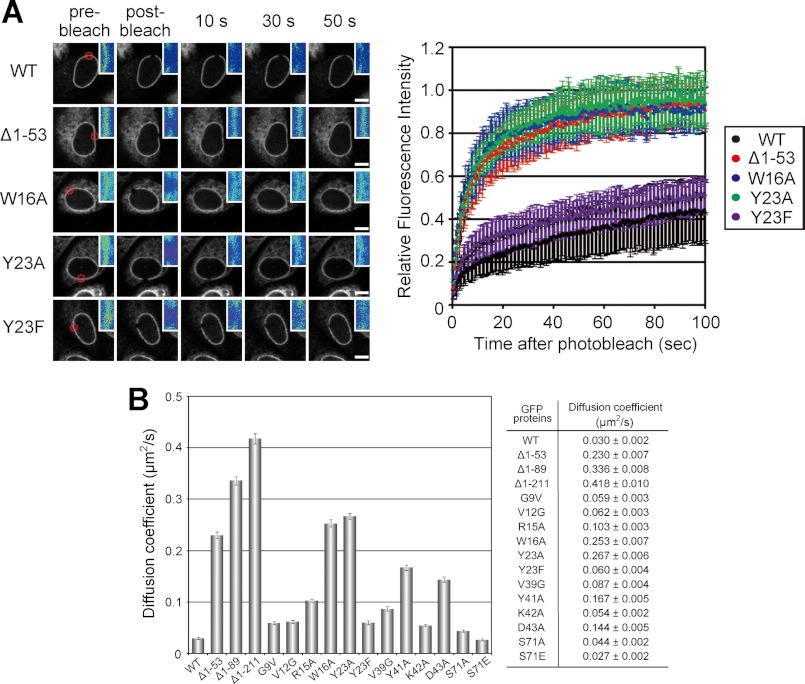
FIGURE 3.
The histone modification-specific binding activity of LBR is necessary to restrict its mobility in the NE. A, GFP-fused LBRWT (WT) and its mutants (Δ1–53, W16A, Y23A, and Y23F) expressed in HeLa cells. Mobility was analyzed by fluorescence recovery after bleaching a 2-μm spot (left panels, red circle). The means of the relative intensity in the bleached area are indicated with the S.D. (right panel, n ≥ 7). Enlarged images around the bleached region in the left panels are shown in pseudo-color at the top right. Scale bars, 10 μm. B, summary of FRAP experiments. Calculated diffusion coefficients of LBR and its mutants are shown.
Glutathione S-transferase (GST) fused with LBR fragment proteins (GST, GST-fused NPWT, and GST-fused NPW16A) were expressed in Escherichia coli and purified with glutathione-Sepharose 4B beads (GE Healthcare) according to the manufacturer's methods with the exception that the beads were washed with high salt washing buffer (phosphate-buffered saline (PBS) containing 1 m NaCl, 1% Triton X-100) for 30 min at 4 °C before elution: the proteins bound to the beads were eluted with GSH buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 50 mm reduced glutathione, 0.1% Triton X-100). The purified proteins were subjected to SDS-PAGE, stained with Coomassie Brilliant Blue, and then quantified by measuring the staining intensity using a LAS-3000 mini image analyzer (Fujifilm, Tokyo, Japan) and ImageQuant 5.0 software (GE Healthcare). His-tagged LBR fragment protein (His-NPWT) was expressed in E. coli and purified with Ni2+-nitrilotriacetic acid beads (Qiagen) according to the manufacturer's method with the exception that the beads were washed with high salt washing buffer as described above. The proteins were eluted with PBS containing 200 mm imidazole and the purified proteins quantified as described above. Core histone proteins for chromatin reconstitution experiments were prepared from HeLa cells by a standard salt extraction method (23). Tail-less histone proteins were prepared according to Hayes et al. (24). Recombinant histone H4 proteins with a single modification (unmodified, K20me1 or K20me2) were purchased from Active Motif (Carlsbad, CA).
Cells
HeLa cells were obtained from Riken Cell Bank (Tsukuba, Japan). HEK293T and PANC1 cells were kind gifts from Drs. H. Ogawa and M. Tsuchiya, and N. Matsuura, respectively, at Osaka University. These cells were cultured in DMEM containing 10% fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 atmosphere.
Histone Modification-Recognition Assay Using a Histone Peptide Array
CelluspotTM, comprising 384 histone tail peptides with various combinations of histone modifications, was purchased from Active Motif and used to identify what combination of histone modifications bound to the target protein of interest (the NP domain of LBR in this study). Details of the complete matrix of peptides are provided in supplemental Table 1. Celluspot was first treated with Blocking One (Nacalai Tesque) for 1 h at room temperature to block nonspecific binding, and then incubated with 100 nm GST-NPWT, GST-NPW16A, or GST-TudWT protein at 4 °C for 1.5 h in binding buffer (20 mm Tris-HCl (pH 7.5), 300 mm NaCl, 250 mm sucrose, 2 mm MgCl2, 0.1 mm EDTA) with 1 mg/ml BSA. After washing five times with PBS containing 0.05% Tween 20 (PBS-T), Celluspot was incubated with anti-GST antibody as the primary antibody for 1 h at room temperature, washed with PBS-T five times, and then incubated with anti-mouse HRP-conjugated IgG as the secondary antibody. Spots were stained with Chemi-Lumi One L (Nacalai Tesque), and the positive spots were detected by chemiluminescence LAS1000 (Fujifilm).
Pulldown Assay of Histone H4 with LBR-conjugated Beads
LBR fragment protein-conjugated beads were generated as described above. The GST-fused protein-conjugated beads (GST, GST-NPWT, and GST-NPW16A) were incubated with 200 ng each of a recombinant histone H4 protein (unmodified, K20me1 or K20me2) at 4 °C for 1.5 h in binding buffer with 1 mg/ml BSA. Then, the beads were washed five times with the binding buffer and twice with washing buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 250 mm sucrose, 2 mm MgCl2, 0.1 mm EDTA). The proteins were eluted from the beads with SDS-sample buffer, subjected to SDS-PAGE, and stained with Coomassie Brilliant Blue or analyzed by Western blotting using modification-independent anti-histone H4 monoclonal antibody (anti-panH4 antibody).
Cross-linked Chromatin Immunoprecipitation (ChIP)
A ChIP experiment was performed as described previously with modifications (25). PANC1 cells (5 × 106) were cross-linked with 1% formaldehyde in medium for 10 min at room temperature and then incubated in 200 mm glycine in medium for 5 min. After washing cells with PBS and addition of lysis buffer, cells were harvested using a cell scraper, collected by centrifugation, and then resuspended in 1 ml Tx-lysis buffer (5 mm Hepes-NaOH (pH 8.0), 200 mm KCl, 1.5 mm MgCl2, 1 mm CaCl2, 5% sucrose, 0.5% Triton X-100, 1 mm PMSF, protease inhibitor mixture (Nacalai Tesque)). Chromatin was fragmented by sonication (Branson Sonifier 250 with microtip; six times for 12 s; output level 1.2) and further digested with 200 gel units of micrococcal nuclease (New England BioLabs) for 1 h at 37 °C. After centrifugation to remove insoluble materials, the supernatants were incubated with 10 μg of anti-LBR and normal rabbit IgG, and Dynabeads (protein A; Invitrogen; 100-μl original suspension) overnight at 4 °C with rotation. Beads were washed five times with 1 ml of Tx-lysis buffer and then suspended in SDS-sample buffer. After incubation at 95 °C for 1 h to reverse cross-linking, the supernatants were subjected to Western blotting.
siRNA Knockdown
HeLa cells (5 × 104) were added to a 35-mm dish on day 0. Stealth siRNAs for luciferase, lamin B1, and lamin B2 (20 nm) were transfected into HeLa cells using Lipofectamine RNAiMAX transfection reagents according to the manufacturer's instructions on day 1. The plasmid encoding LBR-GFP (2 μg) and the siRNAs (20 nm) were co-transfected into HeLa cells using Lipofectamine 2000 transfection reagent on day 2. Determination of knockdown efficiency by Western blotting and immunostaining, and fluorescence recovery after photobleaching (FRAP) experiments were performed on day 3.
FRAP Analysis
FRAP analyses were performed as described previously with slight modifications (26). The mobility of GFP-tagged LBR and its mutants was analyzed using a confocal microscope (LSM510META; Zeiss; operated by the built-in software) with a Plan-Neofluar 40× NA 1.30 oil immersion lens. Twenty images were collected using 488-nm argon ion laser excitation (1% transmission; 494 ms/frame; 1.6 μs/pixel; 512 × 512 pixels; pinhole 160 μm; 6× zoom) before bleaching, then a 2-μm diameter spot was bleached (100% 488-nm laser transmission; eight iterations), and a further 200 images were collected using the original settings. Photobleaching during imaging was monitored in all experiments and was normalized before drawing the recovery curve. The decrease in fluorescence intensity during imaging was <15%. Diffusion coefficients of LBRs were calculated as a single diffusion model according to Axelrod et al. (27). Sprague et al. have indicated that slowly diffused molecules including the binding/dissociation reactions in the cells can be described as an effective diffusion (28). In the case of LBR, the diffusion coefficient of the LBR-chromatin complex could be approximated to zero during the observation time. Therefore, the LBR dynamics could be considered a single population diffusion coefficient, and we were able to use the Axelrod model.
Co-immunoprecipitation Experiment
FLAG-tagged LBRWT or FLAG-tagged LBRW16A was transiently expressed in HeLa cells. The cells (6.0 × 104) were treated at 4 °C for 2 h with lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 250 mm sucrose, 2 mm MgCl2, 0.1 mm EDTA, 0.1% Triton X-100, 1 mm PMSF and protease inhibitor mixture (Nacalai Tesque)) containing 1 unit/ml DNase (Roche Applied Science) and 100 μg/ml RNase (Sigma-Aldrich). The cell lysate was centrifuged at 15,000 × g for 10 min, and the supernatant was collected. The supernatant was mixed with 10 μl of anti-FLAG antibody-conjugated Sepharose beads (Sigma) and incubated overnight at 4 °C. After washing, the beads were treated with SDS-sample buffer to elute the proteins. The fraction containing the eluted protein was subjected to SDS-PAGE and transferred to PVDF membrane and analyzed by Western blotting using anti-LBR, anti-lamin B1, and anti-lamin B2 antibodies.
Atomic Force Microscopy (AFM) Imaging
The DNA plasmid, pBlueScript II KS(−), was digested with ScaI and XhoI restriction enzymes. The digested 1.8-kbp linear dsDNA was purified by gel extraction and used for chromatin reconstitution experiments as described previously (23). Briefly, the DNA was mixed with core histone proteins in high salt buffer and dialyzed against low salt buffer. The reconstituted chromatin was incubated with LBR fragments for 30 min at 4 °C, fixed for 30 min at room temperature with fixation buffer (10 mm Hepes-KOH (pH 7.4), 50 mm NaCl, 0.1% glutaraldehyde) and then observed using a Nanoscope IIIa (Veeco, Plainview, NY) as described previously (23). The calculation of the molar ratio of LBR to histone is discussed in the supplemental Experimental Procedures. Nucleosome sizes were measured as described by Ohniwa et al. (29). The radius of curvature of the cantilever was determined using a DNA width of 2 nm in the images.
Reporter Assay
Reporter assays were performed as described previously with slight modifications (30). Briefly, we used 200 ng of the MH100×4-tk-Luc plasmid as a reporter gene. This plasmid contains four copies of an MH100 gene and a basal promoter, thymidine kinase (tk), upstream of luciferase. Luciferase activity was measured 48 h after transfection. The expressed GAL4-LBR protein fragments were detected with anti-GAL4 DNA-BD antibody.
RESULTS
LBR Specifically Binds to H4K20me2
We expressed the whole nucleoplasmic region of LBR, corresponding to amino acid residues 1–211 (designated the NP domain; see Fig. 1 for LBR domain nomenclature), to investigate whether the LBR NP domain recognizes a specific histone modification (16–18) (Fig. 1, B and C). First, we used a Celluspots peptide array comprising 384 histone tail peptides with various combinations of modifications; this array is a powerful tool for determining the specificity of histone modification-recognition domains (31). The array was incubated with the GST-fused wild-type NP domain (NPWT) and then probed with anti-GST antibody. NPWT specifically bound to peptides containing the single modifications H4K20me2 and H4K20ac and exhibited partial binding to H4K20me1 (Fig. 2A; see supplemental Fig. S1 and Table S1 for the complete data). Interestingly, these binding specificities were altered by additional modifications around H4K20 (Fig. 2A, H4R19me2s, H4R19me2a, H4R23me2s, and H4R23me2a; and see Fig. 8A). To confirm the histone modification-binding specificity of LBR, we next performed a pulldown assay by using recombinant histone H4 proteins with different modifications (i.e. unmodified, K20me1-modified, and K20me2-modified), and K20me2-containing H4 was selectively pulled down with NPWT (Fig. 2B, WT). To elucidate the role of the tudor domain, we generated a point mutant in which tryptophan 16 was substituted with alanine (NPW16A), because aromatic residues such as tryptophan and tyrosine in the histone code-reading pocket of tudor domains are essential for recognizing specific histone modifications (17) (Fig. 1C). NPW16A did not bind to any of the recombinant H4 proteins tested (Fig. 2B, W16A). Also, in experiments using the array, NPW16A lost specificity (supplemental Fig. S2), indicating tryptophan 16 is a key residue for determining binding specificity.
FIGURE 2.
LBR specifically bind to H4K20me2. A, histone peptide array. Celluspot was incubated with 100 nm of GST-NPWT and probed with anti-GST antibody. The spots were detected by chemiluminescence. Detected spots related to H4K20 modifications are shown (see supplemental Fig. S1 and Table S1 for complete data). B, GST-NPWT and -NPW16A beads incubated with recombinant histone H4, either unmodified (Un), monomethylated at Lys20 (me1) or dimethylated at Lys20 (me2), washed, and then subjected to SDS-PAGE. Bound H4 was detected by modification-independent anti-H4 antibody. GST and GST-NP fragments on beads are shown in the lower panel. C, ChIP. PANC1 cells (5.0 × 106) were cross-linked with paraformaldehyde, and the chromatin was digested by sonication and micrococcal nuclease until most of the chromatin became a mononucleosome. After immunoprecipitation by the indicated antibodies (normal IgG and LBR), the beads were washed and reverse cross-linked by heating, and then the histone modification profile was analyzed by Western blotting (WB), indicated on the right. Inputs were 0.5, 0.25, and 0.05%, respectively. D, H4K20me2 modification enriched beneath the NE. HeLa cells were fixed and immunostained with anti-H4K20me2 and -LBR antibodies. The boxed area is enlarged at the bottom right. Arrows indicate the NE. Scale bar, 20 μm.
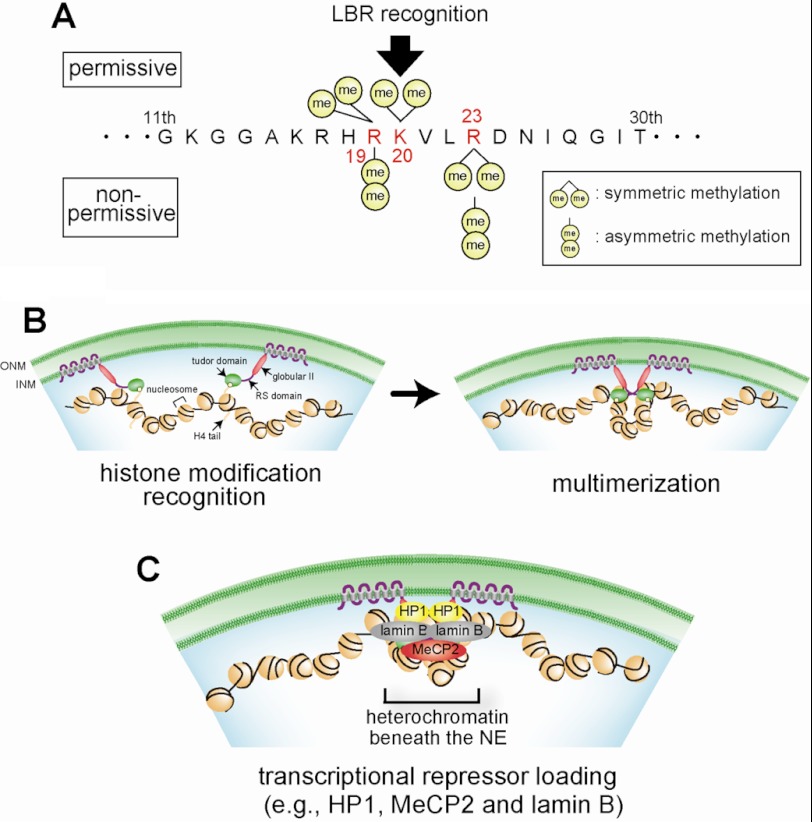
FIGURE 8.
Model of transcriptional repression beneath the NE by LBR. A, permissive (top) or nonpermissive (bottom) modifications of histone H4 for binding with LBR. B and C, heterochromatin formation by LBR. B, chromatin compaction. The tudor domain of LBR binds to chromatin-bearing histone H4 with permissive modifications. The RS domain of LBR tethers chromatin through its multimerization. C, transcriptional repressor loading. The NP domain of LBR recruits transcriptional repressors such as HP1, MeCP2, and lamin B.
To verify the binding of LBR and H4 modifications in vivo, we carried out a ChIP assay. The ChIP assay revealed that H4K20me2 was enriched with LBR (Fig. 2C). On the other hand, it was difficult to detect the enrichment of H4K20ac because it was significantly less abundant in the cells (data not shown). In addition, we investigated the H4K20me2 distribution in the nucleus by immunostaining. As shown in Fig. 2D, H4K20me2 was enriched beneath the INM (Fig. 2D, arrows). Taken together, we concluded that LBR binds to at least H4K20me2.
The Histone Modification-specific Binding Activity of LBR Is Necessary to Restrict Its Mobility in the NE
To verify the LBR-histone H4 interaction in vivo, we applied FRAP to map a residue that affects the mobility of LBR in the NE (Fig. 3). GFP-tagged full-length LBR (LBRWT) moved slowly in the NE (diffusion coefficient, 0.030 ± 0.002 μm2/s), comparable with a previous report (32). In contrast, a tudor domain-deletion mutant, LBRΔ1–53, and the mutants in which aromatic residues were substituted with alanine, LBRW16A and LBRY23A, moved ∼7 times faster than the LBRWT (diffusion coefficients were 0.230 ± 0.007, 0.253 ± 0.007, and 0.267 ± 0.006 μm2/s, respectively). A similar result showing increased mobility of a tudor domain-deletion LBR mutant by FRAP analysis was also reported previously (15, 33). Further experiments showed that alanine substitutions of tyrosine 41 and aspartic acid 43 in the histone code-reading pocket of the tudor domain (Fig. 1C) increased the mobility of the LBR relative to the LBRWT (Fig. 3B), consistent with an essential role for the corresponding aromatic residues in the 53BP1 tudor domain in binding to histone modifications (17). On the other hand, a point mutant, in which tyrosine 23 was substituted with another aromatic residue, phenylalanine (LBRY23F), behaved similarly to the LBRWT. Taken together, binding of the LBR tudor domain to histone is necessary to restrict its mobility in the NE, and these findings suggest that LBR forms a stable complex with peripheral chromatin.
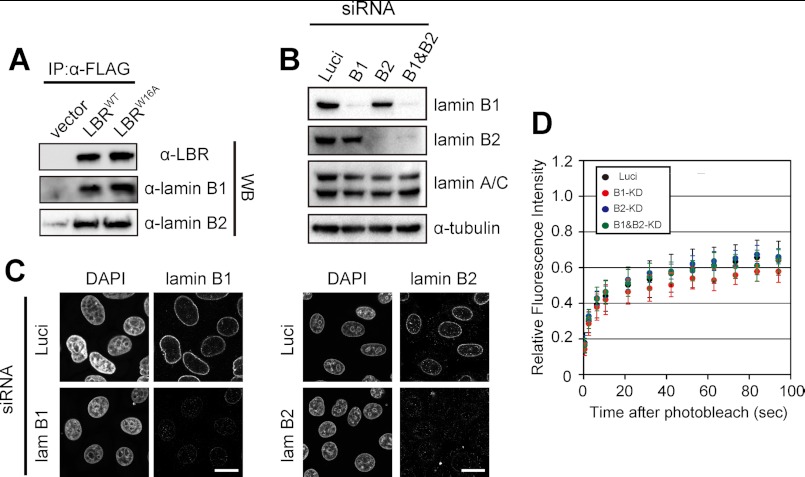
Because LBR also binds to lamin B (11), mobility of LBR in the NE can be restricted by its binding to lamin B (34). If so, it is expected that the mutant LBRs could move faster if the mutations disrupted the binding of LBR to lamin B. Thus, we investigated the effect of lamin binding on LBR dynamics. Lamin B1 and B2 were co-immunoprecipitated with both LBRWT and LBRW16A (Fig. 4A), suggesting that the W16A mutation did not affect the binding of LBR to lamin B. Moreover, siRNA knockdown of lamin B1 and B2 separately or together did not affect LBR dynamics (Fig. 4, B–D). These results indicate that the low mobility of LBR is not attributable to lamin binding.
FIGURE 4.
Lamin B does not affect LBR dynamics in the NE. A, the point mutation W16A in LBR does not affect LBR binding to lamin B. HeLa cells were transfected with plasmids encoding FLAG-LBRWT (LBRWT) or FLAG-LBRW16A (LBRW16A) or with empty vector (vector). The cells (6.0 × 104) were lysed as described under “Experimental Procedures.” The cell lysates were mixed with anti-FLAG antibody-conjugated beads at 4 °C overnight, and the beads were collected after washing. Proteins that bound to the beads were eluted and analyzed by Western blotting with anti-LBR (upper panel), anti-lamin B1 (middle panel), or anti-lamin B2 (lower panel) antibody (WB). B and C, siRNA knockdown of lamin B1 and B2 is shown. siRNA oligonucleotides for luciferase (Luci), lamin B1 (lam B1), and lamin B2 (lam B2) were transfected twice into HeLa cells. The cells were subjected to Western blotting (B) and immunostaining (C). Scale bars, 20 μm. D, FRAP of LBRs in lamin B-knocked down cells. FRAP was performed as described in Fig. 3.
LBR Induces Chromatin Compaction through Its Histone Modification-specific Interaction
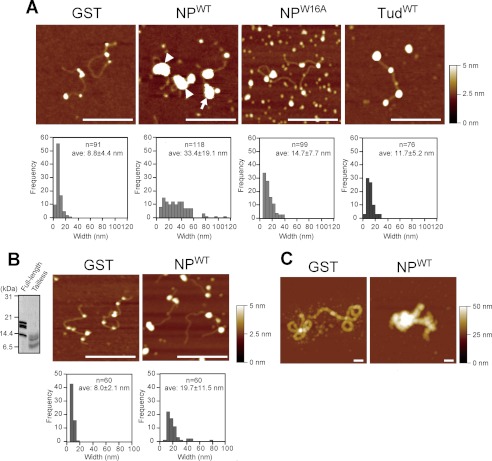
The results also give rise to the possibility that LBR per se participates in chromatin organization beneath the NE. Thus, we examined whether LBR affected chromatin structures by using AFM. We reconstituted chromatin with 1.8-kbp linear dsDNA and human core histones by salt dialysis. In this preparation (Fig. 5A, upper panel, GST), beads-on-a-string chromatin, which had 3–4 nucleosomes on the DNA with an average nucleosome diameter of ∼9 nm, was observed (Fig. 5A, bottom panel, GST; see supplemental Fig. S4 for a low power view). This nucleosome density is lower than the physiological density, but this assay system is useful for observing chromatin compaction (23, 35). With the higher nucleosome density, chromatin tended to be compact by itself, and it was difficult to assess the effect of LBR. We then incubated this reconstituted chromatin with GST-fused LBR fragments (Fig. 5A). In the presence of NPWT, highly aggregated chromatin (typically >30 nm in diameter) was formed (Fig. 5A, NPWT, arrow and arrowheads), and beads-on-a-string structures as observed in the control were virtually absent (Fig. 5A, compare GST and NPWT). In these experiments, we used a 4:1 molar ratio of LBRs to histones, based on the calculated molar ratio of these molecules beneath the NE (supplemental Fig. S3); similar results were obtained using a 1:1 molar ratio. A DNA loop was sometimes observed with the aggregated chromatin (Fig. 5A, NPWT, arrowheads), suggesting that NPWT bridges nucleosomes to induce chromatin compaction. For NPW16A and TudWT (Fig. 5A, NPW16A and TudWT), the size of the nucleosome-like structures appeared bigger than the control (∼15- and 12-nm diameters, respectively), but aggregated chromatin (>30-nm diameter) was rarely observed. These results indicate that NPW16A and TudWT bound to nucleosomes but that these proteins were less able to induce chromatin compaction than NPWT. Similar results were obtained when chromatin was reconstituted using a 26-kbp plasmid dsDNA instead of 1.8-kbp linear dsDNA (supplemental Fig. S5). However, NPWT-induced chromatin compaction was not observed when chromatin was reconstituted with tail-less histones (Fig. 5B). These results indicate that chromatin compaction requires modification-specific interaction of histone H4 with the tudor domain of LBR. Importantly, the histone modification-specific binding activity of the LBR tudor domain is not sufficient to induce chromatin compaction because TudWT alone failed to induce chromatin compaction (Fig. 5A, TudWT). A DNA loop observed in compacted chromatin suggests that chromatin compaction may involve multimerization of LBR as suggested previously (6). Our bead binding assay experiments indicated that the RS domain of LBR was sufficient for its multimerization (Fig. 6). This result also shows that the multimerization activity of LBR is independent of the histone-modification-binding activity because the bead binding assay does not include histones.
FIGURE 5.
LBR induces chromatin compaction. A, chromatin compaction was induced by LBR. Chromatin was reconstituted with 1.8-kbp linear dsDNA and core histones by salt dialysis. The chromatin was incubated with GST-LBR fragments, bound to mica, and then observed using AFM (upper panel). The arrows and arrowhead indicate aggregated chromatin without and with a DNA loop, respectively. Histograms of the nucleosome width are shown at the bottom. B, chromatin reconstituted with tail-less histones is not aggregated by LBR. Tail-less histones were prepared by partial digestion with trypsin (left panel, Coomassie Brilliant Blue stain). Chromatin was reconstituted with 1.8-kbp linear dsDNA and tail-less histones, and observed using AFM. C, chromatin compaction induced by LBRs is independent of histone H1. A 30-nm chromatin fiber on the 26-kbp plasmid was reconstituted according to a previous report (23). After incubation with GST or GST-NPWT, the chromatin was observed using AFM. Scale bars, 200 nm. Scales to indicate height are shown on the right.
FIGURE 6.
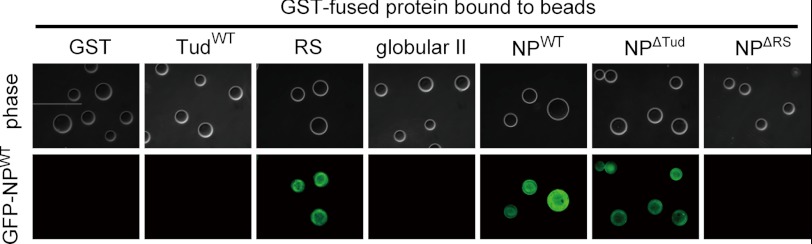
LBR multimerizes by its RS region. Proteins indicated on the panel were conjugated to glutathione-Sepharose beads. Purified GFP-fused NPWT was incubated with the beads. Phase contrast (upper panel) and fluorescence (lower panel) images of the beads are indicated.
Chromatin compaction by LBR is distinct from H1-induced higher order chromatin formation. When we reconstituted a 30-nm chromatin fiber with histone H1 on the 26-kbp reconstituted chromatin and then incubated it with NPWT, the 30-nm chromatin fiber was further compacted as well as the beads-on-a-string chromatin (Fig. 5C).
Histone Modification-specific Binding Activity Is Required for Transcriptional Repression by LBR
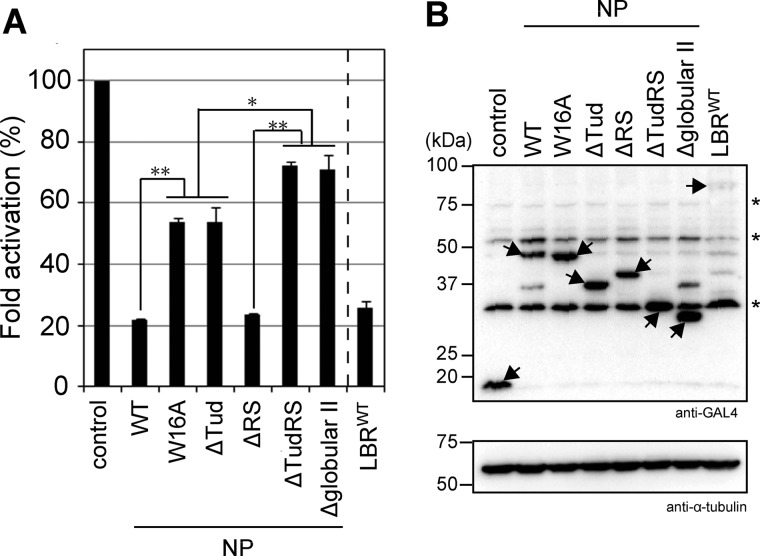
Next, using a luciferase reporter assay, we tested whether LBR modulates transcription (Fig. 7A). GAL4-fused LBR and its fragments were co-transfected with a reporter plasmid, MH100×4-tk-Luc, into HEK293T cells and the luciferase activity driven by the tk promoter was quantified. The cells expressed similar levels of LBR proteins (Fig. 7B, arrows). First, GAL4-LBRWT repressed transcription of the reporter plasmid (Fig. 7A, LBRWT), indicating that LBR was able to repress transcription beneath the NE. Because GAL4-NPWT showed similar transcriptional repression activity to LBRWT (Fig. 7A, WT), we attempted to determine the region responsible for the transcriptional repression using various truncates of the NP fragment. NPΔTudRS and NPΔglobular II (see Fig. 1A) significantly decreased transcriptional repression activity whereas NPΔRS showed similar effects to NPWT. On the other hand, NPΔTud showed a slight decrease in transcriptional repression activity (Fig. 7A, ΔTud, ΔRS, ΔTudRS, and Δglobular II). Histone modification-specific binding activity seems to be crucial for the tudor domain-mediated transcriptional repression because NPW16A showed transcriptional repression activity similar to NPΔTud (Fig. 7A, W16A and ΔTud). These findings suggest that LBR plays at least two important roles in transcriptional repression: chromatin compaction and transcriptional repressor recruitment.
FIGURE 7.
LBR represses transcription. HEK293T cells were transiently transfected with the reporter gene (MH100×4-tk-Luc) and GAL4-fused NP fragments and full-length LBR (see Fig. 1). A, the relative -fold changes in luciferase activities are shown: luciferase activity in the presence of empty vector is set at 100 (control). Values are the mean ± S.D. (error bars) of three independent experiments. Statistical analyses were performed using Student's t test. Asterisks and double asterisks indicate p < 0.05 and 0.001, respectively. B, the GAL4-LBR protein fragments were detected with anti-GAL4 antibody (top). α-Tubulin levels were detected as a loading control (bottom). Arrows and asterisks indicate GAL4-LBR fragments and nonspecific bands, respectively.
DISCUSSION
Histone Modification Binding Specificity of LBR
The LBR-mediated reorganization of chromatin beneath the NE possibly leads to transcriptional repression of developmentally regulated genes because the loss of LBR causes abnormal chromatin organization in mouse granulocyte differentiation (13). On the other hand, H4K20me2, which is the main histone modification recognized by LBR, is broadly spread throughout the genome and is the most abundant H4K20 modification (∼80%) in HeLa cells (36). Thus, why LBR recognizes the broadly spread H4K20me2 modification and whether this binding regulates any biological pathways remain to be elucidated. Although H4K20me2 is the most abundant and broadly spread, it causes specific biological responses such as the recruitment of 53BP1 onto DNA double-strand break sites (17, 37, 38). For this specific binding, a histone methyltransferase MMSET detects and then accumulates at double-strand break sites and results in an increase in the local level of H4 methylation, such that 53BP1 is concentrated. This result suggests the local concentration, not overall amount, of H4K20me2 is important in determining the specificity and the formation of stable interactions with 53BP1. This may be applicable to the LBR-H4K20me2 interaction because H4K20me2 is relatively concentrated beneath the NE (Fig. 2D). Another possible determinant of specificity might be modification patterns surrounding H4K20. Here we have presented evidence that LBR recognizes H4K20me2 in combination with certain surrounding modified residues, and this is distinct from the binding specificity of 53BP1 (31). Although the roles of H4R19 and H4R23 methylation have not been identified, the combination appears to be important for LBR functions; that is, for chromatin compaction and transcriptional repression.
H4K20ac was a candidate modification for LBR recognition, but it was significantly less abundant in several of the cancer cell lines examined (data not shown). Thus, it was difficult to evaluate the significance of the LBR-H4K20ac interaction and to determine a biological role for this interaction in vivo.
Model of LBR-mediated Heterochromatin Formation beneath the NE
Based on our data and published data, we propose a model for the induction of transcriptional repression by LBR (Fig. 8). LBR perhaps possesses at least two important roles in transcriptional repression: chromatin compaction and transcriptional repressor recruitment.
In chromatin compaction, LBR binds to histone H4 containing a specific pattern of modifications (Figs. 2 and 8A), and form a complex with epigenetically marked chromatin (Fig. 8B). A previous report showed that the LBR tudor domain binds to histone H3 but not to H4. However, the study also demonstrated that the RS domain of LBR can bind to histone H4 in vitro (15). We argue that the tudor domain is essential for recognition of the H4 modifications because NPW16A lacks histone modification binding specificity. However, that is not sufficient because our Celluspot peptide array experiments showed that the tudor domain alone did not bind to modified histone H4 peptides (data not shown). The globular II domain affects the binding of LBR to chromatin in vivo (20), suggesting that the binding of LBR to chromatin can be regulated by the whole nucleoplasmic region. Thus, we conclude that the whole nucleoplasmic region is necessary for its binding to histone H4 and that the tudor domain determines its histone modification specificity. At the next step, LBR tethers those chromatin regions together to form a stable LBR-chromatin complex, termed primitive heterochromatin (Fig. 8B). Because the tudor domain alone did not induce chromatin compaction (Fig. 5A, TudWT), another factor(s), e.g. the DNA-binding or multimerization activity of LBR (6, 11, 39), may be required. Our finding that LBR multimerizes via the RS domain implies that this domain participates in primitive heterochromatin formation.
For full transcriptional repression, transcriptional repressor loading at the position recognized by LBR is probably needed because the transcriptional repression activity of LBR is almost completely eliminated by the deletion of the globular II domain (Fig. 7A, Δglobular II). It has been reported that this domain binds to HP1 which is a strong transcriptional repressor (40). As described above, however, HP1 binding to chromatin is not sufficient to repress transcription because ΔTudRS did not repress transcription. Other transcriptional repressors, MeCP2 and lamin B, also bind to LBR and may cooperatively induce transcriptional repression (41, 42). Thus, we speculate that primitive heterochromatin provides a structural framework to recruit transcriptional repressors to form mature heterochromatin (Fig. 8C). It is likely that LBR is a unique INM protein that plays a role in both chromatin organization and transcriptional repression.
Supplementary Material
Acknowledgments
We thank Y. Suzuki and H. Takahashi for helping prepare samples for AFM observation and T. Horigome for critical discussion.
This work was supported by a grant-in-aid for scientific research on innovative areas (to Y. Hiraoka, H. K., and T. H.) and a grant-in-aid for scientific research on priority areas (to K. T.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a grant-in-aid for research activity start-up (to Y. Hirano) from the Japan Society for the Promotion of Science.

This article contains supplemental Experimental Procedures and additional references, Figs. S1–S5, and Table S1.
Y. Hayashi-Takanaka and H. Kimura, unpublished data.
- NE
- nuclear envelope
- AFM
- atomic force microscopy
- FRAP
- fluorescence recovery after photobleaching
- H3K9me3
- histone H3 lysine 9 tri-methylation
- H3K27me3
- histone H3 lysine 27 tri-methylation
- H4K20me2
- histone H4 lysine 20 di-methylation
- INM
- inner nuclear membrane
- LBR
- lamin B receptor
- NP
- nucleoplasmic region of LBR
- tk
- thymidine kinase.
REFERENCES
- 1. Akhtar A., Gasser S. M. (2007) The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8, 507–517 [DOI] [PubMed] [Google Scholar]
- 2. Shaklai S., Amariglio N., Rechavi G., Simon A. J. (2007) Gene silencing at the nuclear periphery. FEBS J. 274, 1383–1392 [DOI] [PubMed] [Google Scholar]
- 3. Towbin B. D., Meister P., Gasser S. M. (2009) The nuclear envelope: a scaffold for silencing? Curr. Opin. Genet. Dev. 19, 180–186 [DOI] [PubMed] [Google Scholar]
- 4. Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 5. Reddy K. L., Zullo J. M., Bertolino E., Singh H. (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 [DOI] [PubMed] [Google Scholar]
- 6. Makatsori D., Kourmouli N., Polioudaki H., Shultz L. D., McLean K., Theodoropoulos P. A., Singh P. B., Georgatos S. D. (2004) The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J. Biol. Chem. 279, 25567–25573 [DOI] [PubMed] [Google Scholar]
- 7. Takano M., Takeuchi M., Ito H., Furukawa K., Sugimoto K., Omata S., Horigome T. (2002) The binding of lamin B receptor to chromatin is regulated by phosphorylation in the RS region. Eur. J. Biochem. 269, 943–953 [DOI] [PubMed] [Google Scholar]
- 8. Martins S. B., Eide T., Steen R. L., Jahnsen T., Skålhegg B. S., Collas P. (2000) HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J. Cell Sci. 113, 3703–3713 [DOI] [PubMed] [Google Scholar]
- 9. Polioudaki H., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P. A., Singh P. B., Giannakouros T., Georgatos S. D. (2001) Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2, 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ye Q., Callebaut I., Pezhman A., Courvalin J. C., Worman H. J. (1997) Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272, 14983–14989 [DOI] [PubMed] [Google Scholar]
- 11. Ye Q., Worman H. J. (1994) Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem. 269, 11306–11311 [PubMed] [Google Scholar]
- 12. Hoffmann K., Dreger C. K., Olins A. L., Olins D. E., Shultz L. D., Lucke B., Karl H., Kaps R., Müller D., Vayá A., Aznar J., Ware R. E., Sotelo Cruz N., Lindner T. H., Herrmann H., Reis A., Sperling K. (2002) Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huët anomaly). Nat. Genet. 31, 410–414 [DOI] [PubMed] [Google Scholar]
- 13. Zwerger M., Herrmann H., Gaines P., Olins A. L., Olins D. E. (2008) Granulocytic nuclear differentiation of lamin B receptor-deficient mouse EPRO cells. Exp. Hematol. 36, 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann K., Sperling K., Olins A. L., Olins D. E. (2007) The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma 116, 227–235 [DOI] [PubMed] [Google Scholar]
- 15. Liokatis S., Edlich C., Soupsana K., Giannios I., Panagiotidou P., Tripsianes K., Sattler M., Georgatos S. D., Politou A. S. (2012) Solution structure and molecular interactions of lamin B receptor tudor domain. J. Biol. Chem. 287, 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y., Fang J., Bedford M. T., Zhang Y., Xu R. M. (2006) Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 [DOI] [PubMed] [Google Scholar]
- 17. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 19. Lee J., Thompson J. R., Botuyan M. V., Mer G. (2008) Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma Y., Cai S., Lv Q., Jiang Q., Zhang Q., Sodmergen, Zhai Z., Zhang C. (2007) Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin β. J. Cell Sci. 120, 520–530 [DOI] [PubMed] [Google Scholar]
- 21. Haraguchi T., Koujin T., Hayakawa T., Kaneda T., Tsutsumi C., Imamoto N., Akazawa C., Sukegawa J., Yoneda Y., Hiraoka Y. (2000) Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113, 779–794 [DOI] [PubMed] [Google Scholar]
- 22. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 23. Hizume K., Yoshimura S. H., Takeyasu K. (2005) Linker histone H1 per se can induce three-dimensional folding of chromatin fiber. Biochemistry 44, 12978–12989 [DOI] [PubMed] [Google Scholar]
- 24. Hayes J. J., Clark D. J., Wolffe A. P. (1991) Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 88, 6829–6833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. (2008) The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33, 61–73 [DOI] [PubMed] [Google Scholar]
- 26. Hirano Y., Ishii K., Kumeta M., Furukawa K., Takeyasu K., Horigome T. (2009) Proteomic and targeted analytical identification of BXDC1 and EBNA1BP2 as dynamic scaffold proteins in the nucleolus. Genes Cells 14, 155–166 [DOI] [PubMed] [Google Scholar]
- 27. Axelrod D., Koppel D. E., Schlessinger J., Elson E., Webb W. W. (1976) Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16, 1055–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sprague B. L., Pego R. L., Stavreva D. A., McNally J. G. (2004) Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86, 3473–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohniwa R. L., Morikawa K., Takeshita S. L., Kim J., Ohta T., Wada C., Takeyasu K. (2007) Transcription-coupled nucleoid architecture in bacteria. Genes Cells 12, 1141–1152 [DOI] [PubMed] [Google Scholar]
- 30. Tsuchiya M., Ogawa H., Suzuki T., Sugiyama N., Haraguchi T., Hiraoka Y. (2011) Exportin 4 interacts with Sox9 through the HMG box and inhibits the DNA binding of Sox9. PLoS One 6, e25694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bock I., Kudithipudi S., Tamas R., Kungulovski G., Dhayalan A., Jeltsch A. (2011) Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC Biochem. 12, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ostlund C., Sullivan T., Stewart C. L., Worman H. J. (2006) Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry 45, 1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirano Y., Takahashi H., Kumeta M., Hizume K., Hirai Y., Otsuka S., Yoshimura S. H., Takeyasu K. (2008) Nuclear architecture and chromatin dynamics revealed by atomic force microscopy in combination with biochemistry and cell biology. Pflugers Arch. 456, 139–153 [DOI] [PubMed] [Google Scholar]
- 34. Moir R. D., Yoon M., Khuon S., Goldman R. D. (2000) Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151, 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hizume K., Araki S., Yoshikawa K., Takeyasu K. (2007) Topoisomerase II, scaffold component, promotes chromatin compaction in vitro in a linker-histone H1-dependent manner. Nucleic Acids Res. 35, 2787–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pesavento J. J., Yang H., Kelleher N. L., Mizzen C. A. (2008) Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 28, 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greeson N. T., Sengupta R., Arida A. R., Jenuwein T., Sanders S. L. (2008) Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J. Biol. Chem. 283, 33168–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P. L., Wang L., You Z., Lou Z. (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duband-Goulet I., Courvalin J. C. (2000) Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry 39, 6483–6488 [DOI] [PubMed] [Google Scholar]
- 40. Ye Q., Worman H. J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 271, 14653–14656 [DOI] [PubMed] [Google Scholar]
- 41. Guarda A., Bolognese F., Bonapace I. M., Badaracco G. (2009) Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp. Cell Res. 315, 1895–1903 [DOI] [PubMed] [Google Scholar]
- 42. Worman H. J., Yuan J., Blobel G., Georgatos S. D. (1988) A lamin B receptor in the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 85, 8531–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.