Background: Calmodulin (CaM) binds the calmodulin binding domain (CMBD) of Orai1 causing Ca2+·CaM-dependent inhibition (CDI).
Results: Orai1-CMBD binds the C-lobe of CaM in crystal, whereas in solution Orai1-CMBD interacts with both N- and C-lobes of CaM.
Conclusion: One CaM, in an extended conformation, binds to two Orai1-CMBDs.
Significance: This expands our knowledge of the interaction of CaM with its targets, and provides important information for understanding CDI.
Keywords: Calcium Channels, Calcium Signaling, Calmodulin, Crystal Structure, Structural Biology, Orai1, Store-operated Calcium Entry, Calcium-dependent Inactivation, Calmodulin-binding Domain
Abstract
Orai1 is a plasma membrane protein that in its tetrameric form is responsible for calcium influx from the extracellular environment into the cytosol in response to interaction with the Ca2+-depletion sensor STIM1. This is followed by a fast Ca2+·calmodulin (CaM)-dependent inhibition, resulting from CaM binding to an Orai1 region called the calmodulin binding domain (CMBD). The interaction between Orai1 and CaM at the atomic level remains unknown. Here, we report the crystal structure of a CaM·Orai1-CMBD complex showing one CMBD bound to the C-terminal lobe of CaM, differing from other CaM-target protein complexes, in which both N- and C-terminal lobes of CaM (CaM-N and CaM-C) are involved in target binding. Orai1-CMBD binds CaM-C mainly through hydrophobic interactions, primarily involving residue Trp76 of Orai1-CMBD, which interacts with the hydrophobic pocket of CaM-C. However, NMR data, isothermal titration calorimetry data, and pulldown assays indicated that CaM-N and CaM-C both can bind Orai1-CMBD, with CaM-N having ∼4 times weaker affinity than CaM-C. Pulldown assays of a Orai1-CMBD(W76E) mutant, gel filtration chromatography data, and NOE signals indicated that CaM-N and CaM-C can each bind one Orai1-CMBD. Thus our studies support an unusual, extended 1:2 binding mode of CaM to Orai1-CMBDs, and quantify the affinity of Orai1 for CaM. We propose a two-step mechanism for CaM-dependent Orai1 inactivation initiated by binding of the C-lobe of CaM to the CMBD of one Orai1 followed by the binding of the N-lobe of CaM to the CMBD of a neighboring Orai1.
Introduction
Calcium is an important second messenger involved in many cell processes such as cell proliferation, egg fertilization, T cell activation, muscle contraction, apoptosis, and vesicular fusion. One way for a cell to regulate calcium concentration in the cytosol is through store-operated calcium entry (1). STIM1 and Orai1 are the two major components involved in store-operated calcium entry (2–7). Upon calcium depletion from the cisternae delimited by the endoplasmic reticulum membrane, STIM1, which resides in this membrane forms oligomers and migrates to be adjacent to the plasma membrane (8–10), where it activates Orai1, resulting in calcium influx (11, 12). In resting cells, the free calcium concentration in the cytosol is maintained at an extremely low level, near 100 nm. This requires prompt inactivation of Orai1 leading to cessation of Ca2+ entry, a process called calcium-dependent fast inactivation (CDI).3 Lewis and colleagues (13) showed that calmodulin (CaM) and a cytosolic domain of STIM1 are important for CDI. They further showed that binding of CaM to an N-terminal fragment of Orai1 adjacent to its first transmembrane helix is calcium-dependent and critical for CDI. It remains unknown, however, how CaM interacts with Orai1 to inactivate it.
CaM interacts with a large number of proteins to regulate their biological functions. It is composed of N- and C-terminal lobes (CaM-N and CaM-C) that are connected by a central linker. Structures of Ca2+-free and Ca2+-bound CaM show that upon calcium binding, CaM-N and CaM-C undergo conformational changes to expose a hydrophobic pocket in each lobe for the binding of target proteins (14–16). In the absence of target proteins, the central linker of CaM is flexible in solution, and CaM-N and CaM-C do not interact with each other. In the presence of target proteins, CaM-N and CaM-C of a CaM molecule typically wrap around the CaM binding domains (CMBDs) of target proteins (usually an α helix) in a collapsed 1:1 binding mode. Exceptions, however, have been reported, in which CaM binds its target CMBDs in a 2:2 or 1:2 binding mode. For example, in the case of the small conductance Ca2+-activated potassium channel SKca, three helices are bound between CaM-N and CaM-C in an extended 2:2 binding mode (17). In the case of glutamate decarboxylase, each lobe of CaM binds one peptide of glutamate decarboxylase, with the two peptides interacting with each other, resulting in a collapsed 1:2 binding mode (18). The different structures of CaM·CMBD complexes indicate that CaM is very flexible allowing it to adopt different conformations. This conformational flexibility is consistent with the lack of sequence homology among CMBDs of its target proteins. The conformational flexibility of CaM and the lack of sequence homology among CMBDs make it difficult to use existing structural information to accurately predict the interaction between CaM and the CMBD of Orai1. We therefore carried out structural studies on the CaM·Orai1-CMBD complex, hoping to 1) understand how CaM facilitates CDI of Orai1; 2) add to our knowledge of the interaction modes of CaM with its target proteins.
Here, we report the crystal structure of Ca2+·CaM in complex with Orai1-CMBD at 1.9-Å resolution. The structure shows that in the crystal Orai1-CMBD is bound only to CaM-C, with residue Trp76 of Orai1-CMBD fitting into the hydrophobic pocket of CaM-C and making extensive interactions with hydrophobic residues of CaM. Further biochemical analyses and mutational studies indicate that in solution CaM interacts with Orai1-CMBDs in a 1:2 binding mode, with CaM-N and CaM-C each binding one Orai1-CMBD.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma unless indicated elsewhere. All restriction enzymes were purchased from New England Biolabs. Crystallization chemicals were purchased from Hampton Research. The Orai1-CMBD peptide, (sequence HSMQALSWRKLYLSRAKLKA), corresponding to residues His69-Ala88 of Orai1, was obtained from GenScript USA Inc.
Cloning, Expression, and Purification of CaM and CaM(Ca2+-Nmut)
The rat CaM cDNA was amplified by PCR using Supermix solution (Invitrogen) and cloned into pET21b vector using NdeI and BamHI restriction enzymes. CaM(Ca2+-Nmut), a CaM mutant with abolished Ca2+ binding capacity at its N-lobe, was constructed by mutating Asp20, Asp22, Asp24, Glu31, Asp56, Asp58, Asn60, and Glu67 to histidines. Primers containing the mutations were synthesized by Sigma. The mutant gene was amplified using multistep PCR, and cloned into pET21b as wild type CaM. The plasmids CaM-pET21 and CaM(Ca2+-Nmut)-pET21 were sequenced to confirm that the gene was correctly inserted. The plasmids were transformed into BL21(DE3) Gold (Agilent Technologies) for protein expression. The bacteria were grown in LB medium at 37 °C, 220 rpm until cell density reached A600 nm = 0.6. The bacteria were then cultured at 23 °C for 30 min before induction with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside (Invitrogen). The culture was continued at 23 °C, 170 rpm for another 4 h. The bacteria were harvested by centrifugation at 4,000 × g for 10 min. Cell pellets were kept at −80 °C until further use. CaM and CaM(Ca2+-Nmut) were purified according to a reported method (19). Briefly, 1-g portions of frozen cell pellets were suspended in 20 ml of lysis buffer (50 mm Tris, pH 7.5, 2 mm EDTA, 0.2 mm PMSF). The bacteria were lysed by sonication. The lysate was centrifuged at 16,000 × g for 20 min to remove cell debris. The supernatant was supplemented with 5 mm CaCl2 before it was loaded onto a 5-ml HiTrap Phenyl HP column (GE Healthcare). The column was then washed with 50 ml of phenyl wash buffer 1 (50 mm Tris, pH 7.5, 0.1 mm CaCl2, 100 mm NaCl), followed by a wash with 50 ml of phenyl wash buffer 2 (50 mm Tris, pH 7.5, 5 mm CaCl2, 500 mm NaCl). The column was eluted with phenyl elute buffer (50 mm Tris, pH 7.5, 1 mm EGTA). The eluate was concentrated and further purified on a Superdex 200 column (GE Healthcare) equilibrated with sizing buffer (10 mm Tris, pH 7.5, 2 mm CaCl2, 100 mm NaCl).
Expression and Purification of CaM-N, CaM-C, Orai1-CMBD, and Orai1-CMBD(W76E)
CaM was also cloned into pGEX-4T1 vector using BamHI and XhoI restriction enzymes. To obtain CaM-N (aa 1–75), a stop codon was added after Lys75 in the CaM-pGEX plasmid using the QuikChange mutagenesis kit (Agilent Technologies). The same strategy was used to make the CaM-N-pET21 plasmid from CaM-pET21. The gene of CaM-C (aa 79–148) was amplified by PCR from CaM-pET21 and cloned into pET28b (Novagen) with a His tag at its N terminus using NheI and BamHI restriction enzymes. Orai1-CMBD (aa 69–91) was cloned into pET32b vector (Novagen) using NcoI and EcoRI restriction enzymes as a thioredoxin fusion protein (Orai1-CMBD-Trx) with a His tag. Orai1-CMBD(W76E) was made from Orai1-CMBD-pET32 plasmid using the QuikChange mutagenesis kit. All constructs were expressed in Escherichia coli BL21(DE3) Gold (Agilent Technologies) in the same way as described above for the expression of CaM. CaM-N-GST fusion protein was loaded onto a GSTrap HP column (GE Healthcare), which was then washed with 10 column volumes of GSH wash buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 2 mm CaCl2), and eluted with GSH elution buffer (100 mm Tris-HCl, pH 7.5, 50 mm NaCl, 2 mm CaCl2, 10 mm GSH). CaM-C and Orai1-CMBD-Trx were purified on a HisTrap FF column (GE Healthcare), which was washed with Ni wash buffer (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 2 mm CaCl2, 20 mm imidazole), and eluted with Ni elution buffer (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 100 mm EDTA). All eluates were concentrated using Amicon concentrators and further purified on a Superdex 200 column equilibrated with sizing buffer.
Crystallization of CaM·Orai1-CMBD Complex
Peak fractions of CaM from the Superdex 200 column were pooled and concentrated to 10–20 mg/ml using an Amicon concentrator. The Orai1-CMBD peptide powder was dissolved in sizing buffer, and mixed with CaM at a 2:1 molar ratio. Initial crystallization trials were screened around the reported conditions for CaM and CaM·target peptide complexes. Clusters of long needles were initially obtained in 0.1 m BisTris-HCl (pH 6.0), 45% polypropylene glycol P400 (PPG P400) at 4 °C. Optimization of crystallization was carried out using an Additive Screen (Hampton Research) with a Mosquito liquid handling system (TTP Labtech). Microseeding significantly facilitated the crystal growth of CaM·Orai1-CMBD. Single needle crystals were obtained using 1 μl of calmodulin (20 mg/ml) + synthetic Orai1-CMBD at a 1:4 CaM:peptide molar ratio in 10 mm Tris-HCl (pH 7.5), 100 mm NaCl, 5 mm CaCl2, mixed with 1 μl of reservior solution (0.1 m BisTris-HCl (pH 6.0), 40% PPG P400, 14% γ-butyrolactone, 1% n-octyl-β-d-glucopyranoside), and equilibrated against 500 μl of reservoir solution at 20 °C by the hanging drop vapor diffusion method.
Data Collection and Processing
Crystals of CaM·Orai1-CMBD were flash frozen in liquid nitrogen without additional cryo-protectant. X-ray diffraction data were collected using in-house x-ray generator MicroMax-007 HF (Rigaku). The crystals diffracted to 1.9 Å in space group C2, with cell dimensions of a = 100.07 Å, b = 24.71 Å, c = 60.90Å, and β = 100.1°. The data were processed and scaled with DENZO/SCALEPACK (20). The statistics of the diffraction data are summarized in Table 1.
TABLE 1.
Diffraction data and refinement statistics
| Diffraction data statistics | |
| Space group | C2 |
| Cell dimensions | a = 100.07 Å, b = 24.71 Å, c = 60.9 Å, β = 100.1° |
| Solvent content (%) | 36.95 |
| No. of molecules in ASU | 1 |
| Resolution range (Å) | 50–1.9 |
| No. of unique reflections | 11497 |
| Overall Rsym (%) | 8.4 (23.1)a |
| Completeness (%) | 96.3 (74)a |
| I/σ | 12.9 (3.9)a |
| Refinement statistics | |
| Resolution range ( Å) | 20–1.9 |
| σ cutoff for refinement | > 0 |
| Rcrystal (Rfree) | 0.18 (0.22) |
| No. of reflections in work set | 10594 |
| No. of reflections in test set | 524 |
| No. of nonhydrogen protein atoms | 1364 (B factor: 21.5) |
| No. of calcium atoms | 4 (B factor: 16.6) |
| No. of water molecules | 88 (B factor: 28.7) |
| No. of solvent molecules | 4 (B factor: 38.6) |
| Root mean square deviation bond length (Å) | 0.014 |
| Root mean square deviation bond angle (°) | 0.86 |
a Numbers in parentheses are for the highest resolution shell.
Structure Determination and Refinement
The structure was solved by molecular replacement using PHASER molecular replacement from the PHENIX program (21). Initial search with CaM from the CaM·MARCKS structure (PDB code 1IWQ) as a search model did not yield a solution. When CaM-N (aa 4–75) and CaM-C (aa 85–146) were used as individual search models, we were able to find rotation and translation solutions for molecular replacement. After several rounds of refinement using PHENIX and manual model building with COOT (22), the central linker of CaM and Orai1-CMBD were successfully built into the electron density map. Four γ-butyrolactone molecules were also built and refined. The statistics of refinement are shown in Table 1. The interaction interfaces were analyzed with PISA program in the CCP4 Molecular Graphics Documentation (23). All the graphic presentations of structures were made with PyMOL.
Pulldown Assays
In pulldown assays for synthetic Orai1-CMBD, 50 μl of 50 μm CaM-N-GST or CaM-C-His6 were mixed with 50 μl of 50 μm Orai1-CMBD. All samples were in sizing buffer. 50 μl of GSH resin or nickel resin was added to mixtures of CaM-N-GST + Orai1-CMBD or CaM-C-His6 + Orai1-CMBD, respectively. The mixtures were incubated on ice for 20 min, and centrifuged at 2,000 × g for 3 min. The supernatants were removed and the resins were washed three times with 1 ml of GSH wash buffer or Ni wash buffer. 100 μl of 2× SDS sample buffer was added to each resin. The resins were spun at 16,000 × g for 1 min and the supernatants were loaded onto a SDS-PAGE gel.
The pulldown assays testing for interaction of CaM and CaM-N with recombinantly expressed wild type (WT) Orai1-CMBD-Trx or Orai1-CMBD(W76E)-Trx mutants were carried out in a similar manner as described above. Orai1-CMBDs were expressed as His6-tagged thioredoxin fusion proteins. WT and W76E Orai1-CMBD were incubated with GSH and nickel resins as a control to test the nonspecific binding.
Isothermal Titration Calorimetry (ITC) Experiments
ITC measurements were carried out in sizing buffer using an iTC200 MicroCalorimeter (GE Healthcare) at 25 °C. Substrate solutions containing synthetic or recombinantly expressed Orai1-CMBD at 0.3–1 mm were injected into a reaction cell containing 30–60 μm CaM, CaM-N, or CaM-C. Fifty injections of 0.4–0.7 μl at 120-s intervals were performed. Data acquisition and analysis were performed using the Origin Scientific Graphing and Analysis software package (OriginLab). Data analysis was performed by generating a binding isotherm and best fit using the following fitting parameters: N (number of sites), ΔH (cal/mol), ΔS (cal/mol/deg), and K (binding constant in m−1) and the standard Levenberg-Marquardt methods (24). Following data analysis, K (m−1) is then converted to Kd (μm).
Determination of Protein Concentrations
CaM has a low extinction coefficient due to the lack of tryptophan, the main contributor for UV absorbance of a protein at 280 nm. Therefore, the CaM concentration calculated with the extinction coefficient is susceptible to influence of contaminating proteins or DNA. Accuracy of the protein concentration is critical for ITC data analysis. Therefore, we determined the CaM concentration using a bicinchoninic acid protein assay kit (ThermoScientific). BSA at different concentrations determined by weight over volume was used as standard. The concentrations of His-tagged CaM-C, CaM-N-GST, and synthetic Orai1-CMBD were also determined using the same bicinchoninic acid method.
Calculation of Molecular Weights by Size Exclusion Chromatography
Bio-Rad gel filtration standard containing bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1350 Da) were run on a Superdex 200 column eluted with sizing buffer. The log10(MW) were plotted against the elution volumes to obtain a standard curve. The molecular weights of protein samples were calculated by interpolation based on their elution volumes.
NMR Assignments
Calmodulin and CaM-N in pET21 vector were expressed in BL21(DE3) Gold in minimal media containing [U-13C]glucose and 15NH4Cl, and purified as described above. Prior to the NMR experiments the protein was concentrated to 60 μm. NMR spectra were acquired with Orai1-CMBD at 0, 30, 60, 90, and 120 μm. The experiments to assign the protein resonances were acquired in the presence of Orai1-CMBD at 120 μm. The buffer was: 10 mm NaPi (pH 6.5), 2 mm CaCl2, 5% D2O, 50 μm 5,5-dimethylsilapentanesulfonate to serve as a chemical shift reference. NMR spectroscopy was performed at 25 °C on Varian UNITY INOVA 500 MHz spectrometer equipped with a 5-mm triple resonance cold probe with actively shielded z axis gradients. The NMR data were processed with NMRPipe (25) and the spectra were analyzed with NMRView (26). The backbone resonance assignments were made from analysis of the HNCACB and CBCA(CO)NH experiments acquired using the Varian BioPack pulse sequences. Assignment of the methyl resonances was completed with the HCCH-TOCSY, H(CCO)NH-, and C(CO)NH-TOCSY experiments using the standard Varian sequences. The filtered NOE experiments were performed for the mixtures of 700 μm CaM and 700 μm Orai1-CMBD or for 250 μm CaM-N and 300 μm Orai-CMBD with a 200-ms mixing time (27).
RESULTS
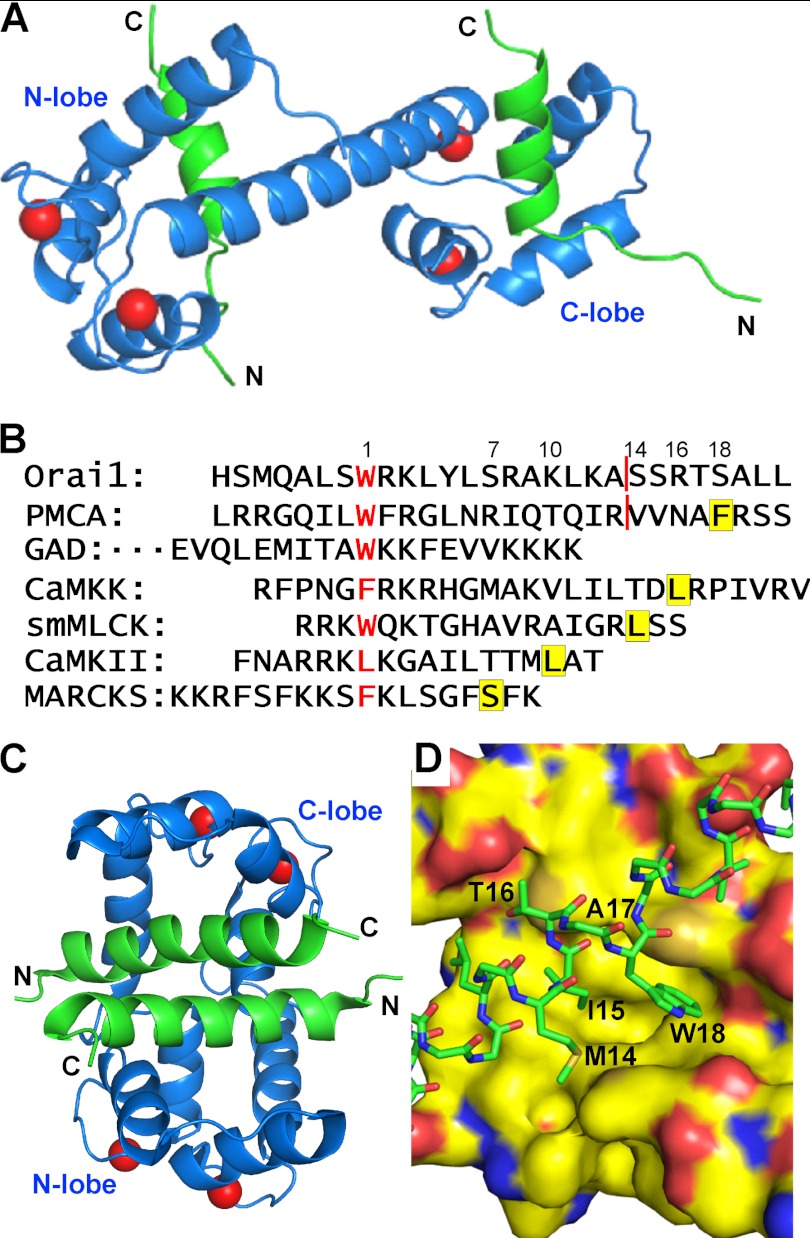
Orai1-CMBD Interacts Only with CaM-C in Crystal
We have determined the structure of CaM·Orai1-CMBD in the presence of Ca2+ in space group C2 at 1.9 Å. There is one CaM·Orai1-CMBD complex in one asymmetric unit. The final R/Rfree factors were 0.18/0.22 (Table 1). There are four calcium ions bound to CaM, two to each lobe. Residues 3–146 of CaM are well defined by the electron density map, and all residues from Orai1-CMBD fit well to the electron density map. The overall quality of the electron density is good (supplemental Fig. S1).
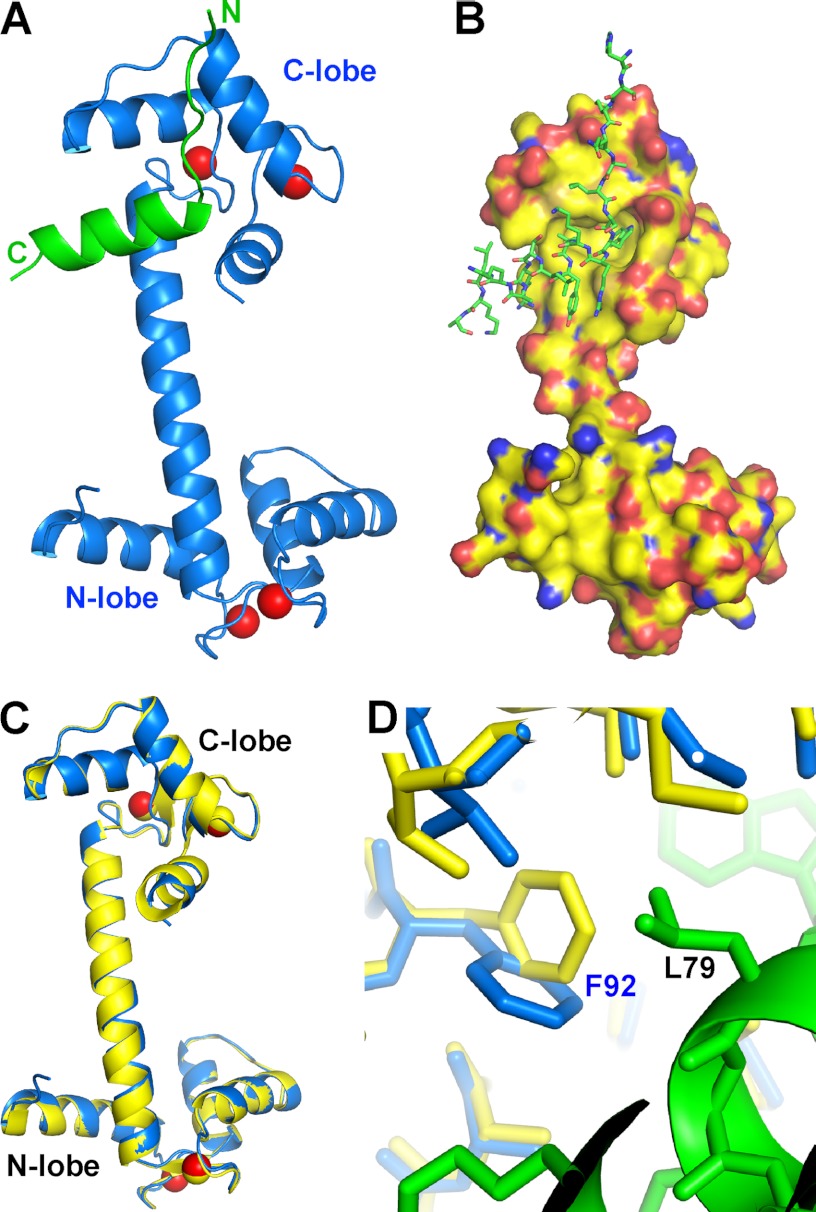
Surprisingly, CaM adopts an extended conformation in the complex, with its central linker forming an α helix (Fig. 1, A and B). The structure of CaM here is almost identical to Ca2+·CaM that is free of ligand (PDB code 3CLN) (28) (Fig. 1C). When our structure and that of ligand-free Ca2+·CaM were superimposed along their Cα, the superposition gave a root mean square deviation of 0.44 Å over 127 Cα. In fact, upon structural superposition, the surface of the C-lobe of Ca2+·CaM accommodates Orai-CMBD well, indicating that their side chains also adopt similar conformations. The major change in the hydrophobic pocket of CaM-C is the side chain of Phe92 that rotates about 90° along the Cβ-Cγ bond to accommodate the side chain of Leu79 of Orai1-CMBD (Fig. 1D). In one asymmetric unit, only CaM-C interacts with Orai1-CMBD; CaM-N has no interaction with either Orai1-CMBD or CaM-C. Orai1-CMBD, however, interacts with CaM-N of a symmetry-related CaM molecule (supplemental Fig. S2). This interaction consists of a hydrophobic interaction (between Leu81 of Orai1-CMBD and Leu4 and Leu70 of CaM) and two hydrogen bonds (CaM Lys13 NE-Orai1-CMBD Lys87 O, and CaM Asp64 OG1-Orai1-CMBD Lys78 NE) (supplemental Fig. S2A). The interface of CaM-C and Orai1-CMBD has a higher shape complementarity than that of Orai1-CMBD and symmetry-related CaM-N, resulting in a surface area of 780 Å2 for Orai1-CMBD/CaM-C interaction, and 420 Å2 for the Orai1-CMBD/symmetry-related CaM-N interaction (supplemental Fig. S2, B and C). Shape complementarity analysis using the PISA server from CCP4 indicates that Orai1-CMBD interaction with the N-lobe of symmetry-related CaM is unlikely to exist in solution. In addition, size exclusion chromatography of the CaM·Orai1-CMBD complex does not show the existence of a dimeric complex, which would have been formed by this interaction. Thus the interaction of Orai1-CMBD with CaM-N observed in the crystal structure is simply a consequence of crystal packing.
FIGURE 1.
Overall crystal structure of the CaM·Orai1-CMBD complex. A, ribbon representation. Orai1-CMBD is colored in green, CaM is colored in blue, and Ca2+ ions are represented as red spheres. The central linker of CaM adopts an extended α-helical conformation, and Orai1-CMBD interacts only with the C-terminal lobe of CaM. B, surface and stick representation. The colors of atoms are shown as: C, green; N, blue; O, red for Orai1-CMBD, and C, yellow; N, blue; O, red; and S, orange for CaM. C, structural superposition of CaM from CaM·Orai1-CMBD (blue) onto ligand-free Ca2+·CaM (yellow, PDB code 3CLN). D, zoomed-in view of conformational change of Phe92 of calmodulin. The aromatic ring of Phe92 in the CaM·Orai1-CMBD complex (blue) rotates about 90° along the Cβ-Cγ bond from the structure of the ligand-free calmodulin (yellow) to accommodate the side chain of Leu79 of Orai1-CMBD (green).
Orai1-CMBD Partially Adopts a Helical and Partially an Extended Conformation
The N-terminal part of the peptide (residues 69–74) adopts an extended conformation. Some residues (His69, Gln72, and Ala73) on this segment of the peptide interact with neighboring CaM and Orai1-CMBD molecules resulting from crystal packing. The C-terminal segment of Orai1-CMBD adopts α helical conformation. Most of the residues interacting with CaM are located in this helical segment (Fig. 1, A and B).
Orai1-CMBD Interacts with CaM-C Mainly through Hydrophobic Interactions
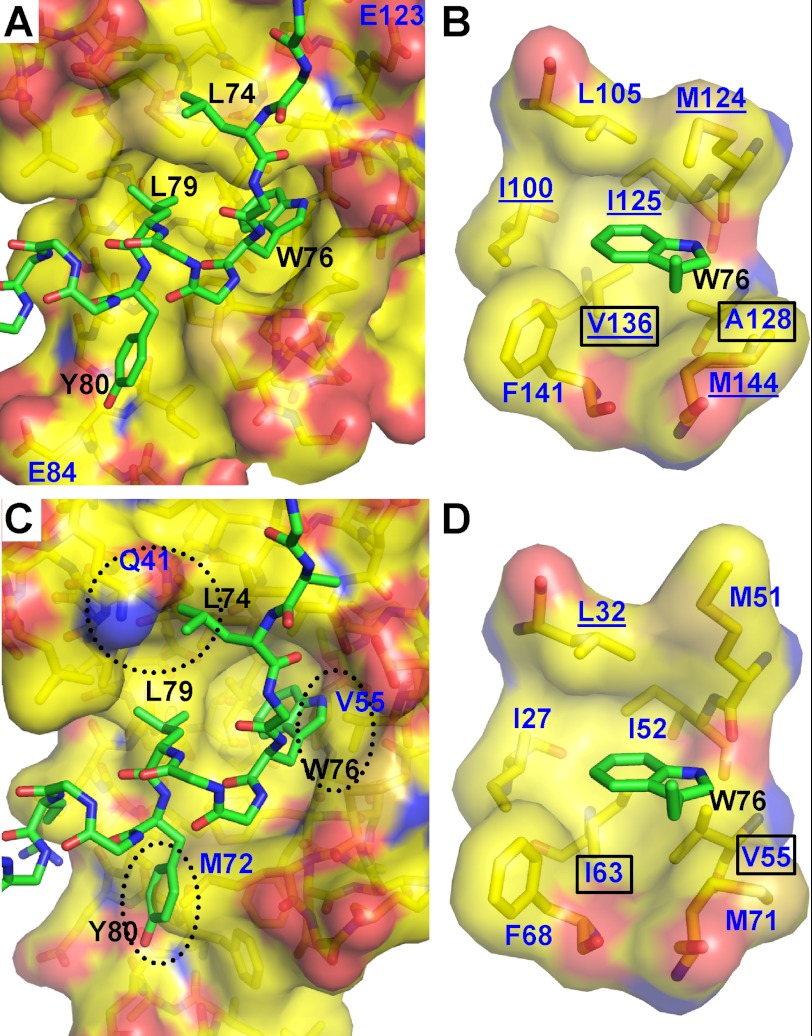
Residues Trp76 and Leu79 of Orai1-CMBD bind to the hydrophobic pocket of CaM-C (Fig. 2A). The side chain of Trp76 is deeply buried in this pocket (Fig. 2B), interacting with residues Ile100, Leu105, Met124, Ile125, Ala128, Val136, Phe141, and Met144 of CaM. NOE signals confirm the interaction of Trp76 with Ile100, Met124, Ile125, Val136, and Met144 (see below). Leu79 of Orai1-CMBD interacts with a hydrophobic surface that is composed of residues Phe92, Leu105, Val108, Met109, and Leu112 of CaM-C (supplemental Fig. S3B). Thus, Trp76 and Leu79 of Orai1-CMBD make the major contribution to its interaction with CaM. In addition, Leu74 of Orai1-CMBD makes hydrophobic interactions with residues Met109, Glu114, Leu116, and Met124 of CaM, and Tyr80 of Orai1-CMBD interacts with Ile85, Ala88, Phe141, and Met145 of CaM (supplemental Fig. S3, A and C).
FIGURE 2.
Interaction of Orai1-CMBD with CaM-C and CaM-N (hypothetical). A, interaction of Orai1-CMBD with CaM-C. Residues of Orai1-CMBD are labeled in black, and CaM in dark blue. Leu74, Trp76, and Leu79 of Orai1-CMBD make major contributions to the hydrophobic interaction between CaM and Orai1-CMBD. Labeled in dark blue are Glu123 and Glu84 of CaM that may have electrostatic repulsion for A73E and Y80E mutants of Orai1-CMBD, respectively. The color scheme of atoms is the same as described in the legend to Fig. 1B. B, zoomed-in view of the interaction between Trp76 of Orai1-CMBD (green) and the hydrophobic pocket of calmodulin C-terminal domain (yellow). The hydrophobic pocket is shown as a surface representation. The residues composing the hydrophobic pocket of calmodulin and the side chain of Trp76 are shown as sticks. Residues of Orai1-CMBD are labeled in black, and CaM in dark blue. The color scheme of atoms is the same as described in the legend to Fig. 1B. C, hypothetical interaction of Orai1-CMBD with CaM-N. Structure of CaM-N was superimposed onto the CaM-C in complex with Orai1-CMBD, to mimic the interaction of CaM-N with Orai1-CMBD. This simple superposition introduces three major clashes between CaM-N and Orai1-CMBD, as indicated by the dotted oval. D, zoomed-in view of the hypothetical interaction between Trp76 of Orai1-CMBD (green) and the hydrophobic pocket of the calmodulin N-terminal domain (yellow). The scheme of the representation is the same as B. Two residues of the hydrophobic pocket that are different between CaM-N and CaM-C are boxed. The CaM residues that show NOE signals with Trp76 of Orai1-CMBD are underlined in B and D.
CaM-N Binds Orai1-CMBD in Solution
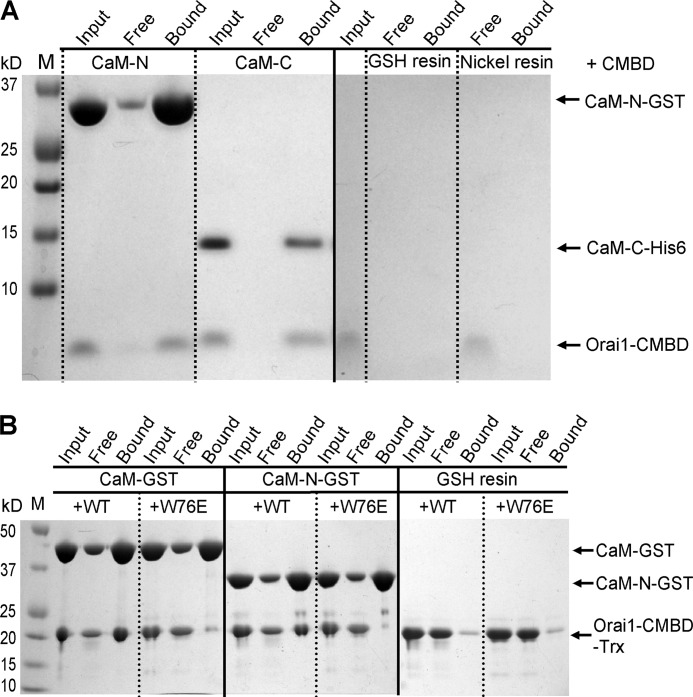
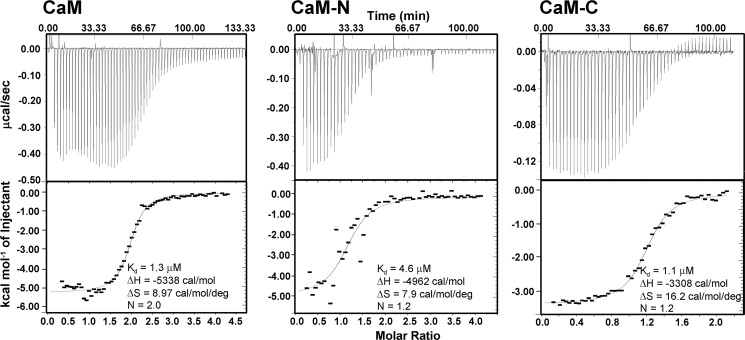
In the crystal structure, CaM-N interacts with Orai1-CMBD from a symmetry-related complex. To evaluate the interaction of Orai1-CMBD with CaM-N, we expressed and purified CaM-N (aa 1–75) as a GST fusion protein and CaM-C (aa 79–148) with a His tag. The individual lobes of CaM were used to pull down the synthetic Orai1-CMBD. Orai1-CMBD was pulled down not only by CaM-C, but also by CaM-N (Fig. 3A). It is noteworthy that the controls for the pulldown assay indicate a nonspecific binding of Orai1-CMBD to GSH resin. However, the nonspecifically bound Orai1-CMBD was removed from GSH resin during the wash. Thus, Orai1-CMBD was indeed pulled down by CaM-N GST fusion protein. ITC experiments also showed that both CaM-N and CaM-C alone bound Orai1-CMBD, with a Kd of 4.6 μm for CaM-N, and 1.1 μm for CaM-C (Fig. 4). This indicates that CaM-N alone can bind Orai1-CMBD, but with a slightly weaker affinity than CaM-C. The binding of CaM-N to Orai1-CMBD detected by pulldown assays and ITC suggested that the interaction between CaM-N and Orai1-CMBD observed in crystal packing might contribute to the interaction in solution. However, when we mutated Trp76 of Orai1-CMBD to Glu, a mutation reported to abolish the binding of Orai1-CMBD to CaM and thus disrupt CDI (13), the pulldown assay (Fig. 3B) and ITC results (data not shown) showed that the W76E mutant of Orai1-CMBD did not bind to CaM, CaM-N, or CaM-C, indicating that the interaction of CaM-N with Orai1-CMBD observed at the lattice contact, which does not involve Trp76 of Orai1-CMBD, differs from the interaction observed in solution. Based on these data, we concluded that CaM-N and CaM-C both bind Orai1-CMBD through Trp76, with CaM-C having a higher affinity than CaM-N. Because CaM-N and CaM-C share high sequence homology, we superimposed the CaM-N structure to CaM-C, and noticed that Trp76 of Orai1-CMBD also fits into the hydrophobic pocket of CaM-N (Fig. 2, C and D), making Trp76 a ligand for both CaM-N and CaM-C.
FIGURE 3.
Interaction of Orai1-CMBD with CaM-N revealed by pulldown assays. A, pulldown of synthetic Orai1-CMBD by CaM-N-GST with GSH resin or by CaM-C-His6 with nickel resin. Orai1-CMBD can be pulled down by either CaM-N or CaM-C, suggesting that in solution CaM-N also interacts with Orai1-CMBD. As a control, the peptide was added to GSH resin or nickel resin in the absence of CaM-N or CaM-C. The control experiments showed a nonspecific binding of Orai1-CMBD to GSH resin. However, this nonspecifically bound peptide was washed off the resin during the wash. B, pulldown of recombinantly expressed Orai1-CMBD or Orai1-CMBD(W76E) mutant by CaM-GST or CaM-N-GST with GSH resin. Wild type (WT) Orai1-CMBD can be pulled down by CaM or CaM-N, consistent with the pulldown results of the synthetic peptide. The W76E mutation disrupts the interaction of Orai1-CMBD with both CaM and CaM-N, suggesting that both CaM-N and CaM-C bind to Trp76 of Orai1-CMBD.
FIGURE 4.
The affinity of Orai1-CMBD for CaM, CaM-N, and CaM-C determined by ITC. ITC data show that CaM binds Orai1-CMBD as a 1:2 complex, whereas CaM-N or CaM-C binds Orai1-CMBD as a 1:1 complex. CaM and CaM-C have similar affinity for Orai1-CMBD, whereas CaM-N has slightly lower affinity.
CaM Binds Orai1-CMBD at a 1:2 Ratio
The binding of CaM-N and CaM-C to Orai1-CMBD described above suggests that full-length CaM binds Orai1-CMBD at a 1:2 ratio in solution, and that the crystal structure reflects an intermediate toward this 1:2 complex. Indeed, ITC data show that one CaM molecule binds two Orai1-CMBDs with a Kd of 1.3 μm (Fig. 4).
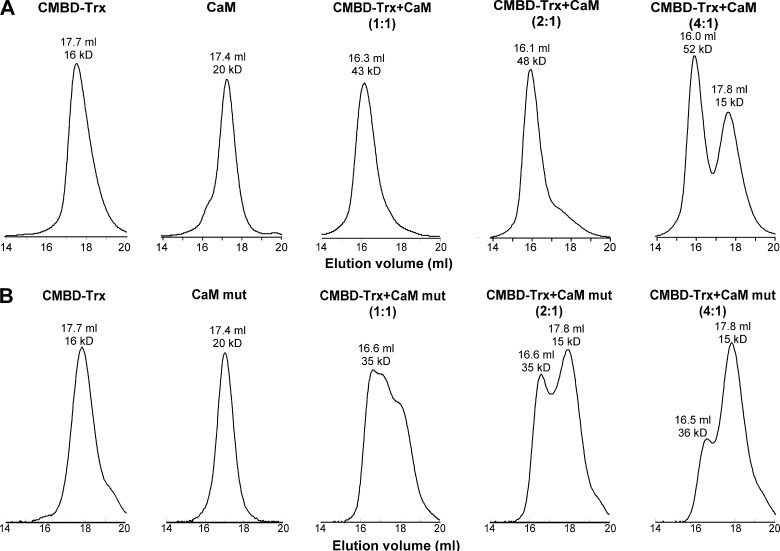
To further confirm the 1:2 binding ratio of CaM to Orai1-CMBD, we checked complex formation of CaM with the Orai1-CMBD-Trx fusion protein (Orai1-CMBD fused to the C terminus of thioredoxin encoded in the pET32 vector) by size exclusion chromatography (Fig. 5). Purified Orai1-CMBD-Trx, CaM, and a mixture of these two proteins were individually run on Superdex 200 column, and their molecular weights were calculated from the elution volumes using a standard curve. CaM has a theoretical molecular mass of 16.7 kDa, and Orai1-CMBD-Trx of 19.1 kDa. Their molecular weights calculated from size exclusion chromatograms are consistent with their theoretical molecular weights (Fig. 5). The calculated molecular mass of CaM is 20 kDa, slightly higher than the theoretical value. This could be due to the elliptical rather than the globular shape of CaM in the absence of target peptide. The calculated molecular weight of Orai1-CMBD-Trx is 16 kDa, slightly smaller than the theoretical value. At 1:1 molar ratio, the calculated molecular mass of the CaM·Orai1-CMBD-Trx mixture is 43 kDa, a number between the molecular masses of 1:1 and 1:2 CaM·Orai1-CMBD complexes. Because the affinity of Orai1-CMBD to CaM-C is only 4-fold stronger than that to CaM-N, at 1:1 molar ratio, a major fraction of Orai1-CMBD-Trx will bind to CaM-C, and the other fraction of Orai1-CMBD-Trx will bind to CaM-N, resulting in a mixture of 1:1 and 1:2 complexes. We suggest that there is equilibrium between the 1:1 and 1:2 complexes. This suggestion is supported by the NMR chemical shift perturbation data in Fig. 6, which indicates weak binding of CaM-N to Orai1-CMBD with an intermediate exchange rate. This model is consistent with our failure to observe two separate peaks for 1:1 and 1:2 complexes on the size exclusion chromatogram of the 1:1 mixture. The calculated molecular mass of the CaM·Orai1-CMBD-Trx mixture at a 1:2 molar ratio is 48 kDa, indicating a 1:2 CaM·Orai1-CMBD-Trx complex. When CaM and Orai1-CMBD-Trx were mixed at a 1:4 molar ratio, an additional elution peak containing Orai1-CMBD-Trx appeared, representing the excess of Orai1-CMBD-Trx. These results indicate that CaM and Orai1-CMBD-Trx form a 1:2 complex, rather than a 1:1 complex. Because Orai1-CMBD does not form dimers or oligomers, as indicated by its size exclusion chromatogram, we propose that upon binding to Orai1-CMBD, CaM remains in its extended conformation.
FIGURE 5.
Formation of 1:2 CaM·Orai1-CMBD complex revealed by size exclusion chromatography. A, chromatograms of wild type CaM/Orai1-CMBD mixtures. B, chromatograms of CaM mutant/Orai1-CMBD mixtures. The two calcium binding sites of CaM-N were abolished in the CaM mutant so that only its C-lobe can bind Orai1-CMBD. The Orai1-CMBD-Trx fusion protein, CaM (or CaM mutant), and Orai1-CMBD-Trx/CaM mixture at 1:1, 2:1, or 4:1 molar ratios were individually subjected to a Superdex 200 column. The molecular weights of proteins in each elution peak were calculated from the elution volumes as described under “Experimental Procedures.” The results show that two Orai1-CMBDs bind to one CaM.
FIGURE 6.
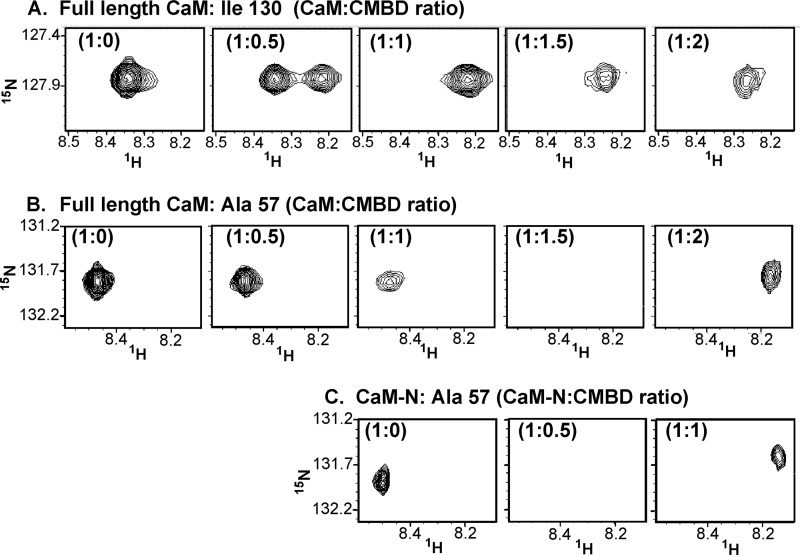
Selected NMR 15N-1H chemical shift perturbations of CaM and CaM-N due to Orai1-CMBD. A, titration behavior for the amide resonance of Ile130 (located in the C-lobe) of the full-length CaM is shown upon addition of Orai1-CMBD at protein to peptide ratios of 1:0, 1:0.5, 1:1, 1:1.5, and 1:2.0. B, titration behavior for the amide resonance of Ala57 (located in the N-lobe) in the full-length CaM titrated as in A. C, behavior of the amide resonance of Ala57 in isolated CaM-N is shown upon titration of Orai-CMBD at protein to peptide ratios of 1:0, 1:0.5, and 1:1. The panels are aligned for comparison with Ala57 in full-length CaM. Note that the N-terminal Ala57 amide resonance is not significantly perturbed until the peptide:protein ratio exceeds 1.
The property of proteins can make the elution volume on size exclusion chromatography deviate from the regular elution volume, resulting in a wrong calculated molecular weight. To rule out this possibility, we mutated the two calcium binding sites in CaM-N. The mutant, CaM(Ca2+-Nmut), will have only CaM-C capable of binding to Orai1-CMBD, and therefore can only form a 1:1 complex with Orai1-CMBD. When we subjected the 1:1 molar ratio mixture of CaM(Ca2+-Nmut):Orai1-CMBD-Trx to size exclusion chromatography, the calculated molecular weight of the complex was 35 kDa, corresponding to the 1:1 complex (Fig. 5B). At 1:2 molar ratio of CaM(Ca2+-Nmut):Orai1-CMBD-Trx, an additional peak corresponding to free Orai1-CMBD-Trx appeared, and no complex appeared larger than the 1:1 complex formed (Fig. 5B). At the 1:4 molar ratio of CaM(Ca2+-Nmut):Orai1-CMBD-Trx, the dominant peak on the size exclusion chromatogram was the free Orai1-CMBD-Trx (Fig. 5B). These results confirmed that complexes in the 1:2 and 1:4 CaM·Orai1-CMBD-Trx mixtures observed on size exclusion chromatograms were 1:2 CaM·Orai1-CMBD-Trx complexes.
NMR data provided confirmation of the binding of two peptides to one CaM, and confirmed that CaM-N has a weaker affinity than CaM-C for Orai1-CMBD. Fig. 6 tracks a representative resonance from the 15N-1H heteronuclear single quantum coherence of CaM-C (Ile130) and CaM-N (Aal57) upon titration of Orai1-CMBD using full-length CaM. The full 15N-1H heteronuclear single quantum coherence is provided in supplemental Fig. S4. Fig. 6A shows two peaks for Ile130 at a 1:0.5 molar ratio of the CaM·Orai1-CMBD mixture indicative of slow exchange between a bound and unbound conformation for Ile130; this is common for tight binding. At a 1:1 ratio, the C-terminal domain is fully saturated and the peak changes very little upon further addition of peptide to 1:1.5 and 1:2 molar ratios. In comparison, Fig. 6B shows that the Ala57 peak from the N-terminal domain broadens slightly at 1:0.5 and 1:1 but broadens significantly at 1:1.5, but does recover full intensity when a ratio of 1:2 is reached. This result indicates that binding to the C-terminal domain is tighter and occurs first in the titration. Broadening of the amide resonances of the N-terminal domain upon addition of peptide can also be observed with the CaM-N construct (Fig. 6C). At a 1:0.5 ratio, the Ala57 peak of CaM-N is broadened due to intermediate exchange, consistent with a weaker binding to this domain. The difference is most striking between the Ile130 of full-length CaM and Ala57 of CaM-N at the same molar ratio (Fig. 6, A and C). Based on these comparisons, it appears that Orai1-CMBD interacts independently with the N- and C-terminal domains.
To model interactions of the N-terminal domain with the peptide, the N-terminal domain was superimposed on the C-terminal domain with the peptide bound to it. Inspection of this model suggests that several methyl groups of the N-terminal domain would potentially interact with Trp76 of Orai1-CMBD, particularly Ile27, Leu32, Ile52, Val55, and Ile63 (Fig. 2D). Indeed, an examination of the methyl region of the 13C-1H HMQC spectra in Fig. 7A showed that the resonances of Ile52 and Ile63 shift considerably upon addition of Orai1-CMBD to CaM-N. To confirm this interaction in full-length CaM, Fig. 7B shows similar shifts for resonances of the N-terminal domain, at a ratio of 1:2 CaM:Orai1-CMBD.
FIGURE 7.
NMR 13C-1H chemical shift perturbations of CaM due to Orai1-CMBD. A, the methyl resonances of CaM-N apo (black) and CaM-N/Orai1-CMBD at a 1:1 molar ratio (red) are compared in this 13C-1H HMQC. B, similar methyl shifts of the N-terminal residues are observed in CaM with a 2-fold excess of Orai1-CMBD (red). The apo spectra of CaM are in black.
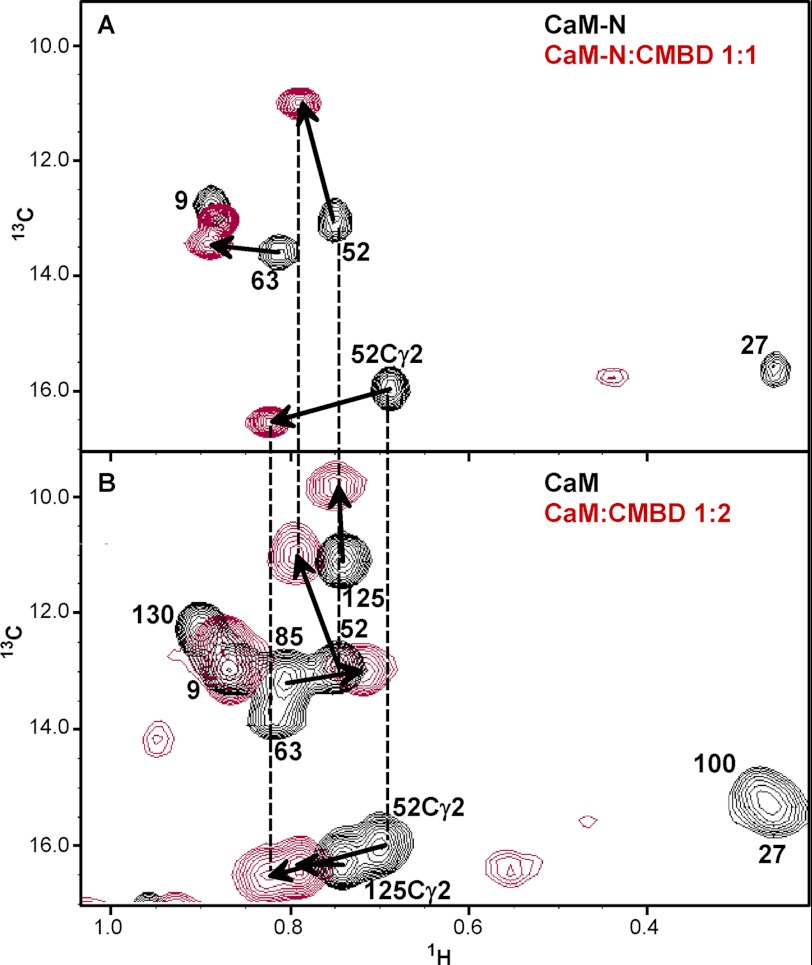
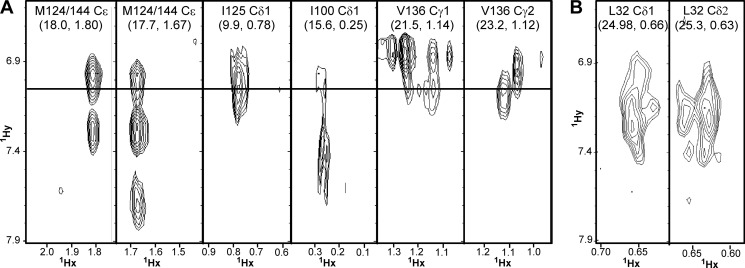
To more precisely understand the sites of interaction, an isotope-filtered NOESY study was performed, with the 13C,15N doubly labeled protein and the unlabeled Orai1 peptide. This allows for detection of intermolecular signals that connect the resonances in the (labeled) calmodulin with resonances in the (unlabeled) peptide. There were clear signals from the C-terminal lobe Met124, Met144, Ile100, Ile125, and Val136 to aromatic proton shifts typical of a tryptophan side chain; Trp76 is the only tryptophan in Orai1-CMBD (Fig. 8A). This is consistent with what we observed in the crystal structure (Fig. 2B). Isotope-filtered NOEs were also detected between CaM-N alone and Orai1-CMBD. Fig. 8B shows the interactions of Trp76 of Orai1-CMBD with the N-terminal Leu32 of CaM. This is consistent with the homology model described above (Fig. 2D). However, we did not observe NOE signals between Trp76 of Orai1-CMBD and other residues in the hydrophobic pocket of CaM-N (see “Discussion” below). In total, these results confirm that CaM interacts with two Orai1-CMBD peptides via the N- and C-terminal domains, and that CaM-N has a weaker affinity for the peptide than CaM-C.
FIGURE 8.
The CN-filtered NOESY spectra. A, CaM with Orai1-CMBD at a 1:1 molar ratio (including the methyl resonance of Met, Ile, and Val in the C-lobe of CaM). The methionines were not assigned. Therefore, Met124 and Met144 cannot be distinguished. B, the isolated CaM-N with Orai1-CMBD at a 1:1 molar ratio.
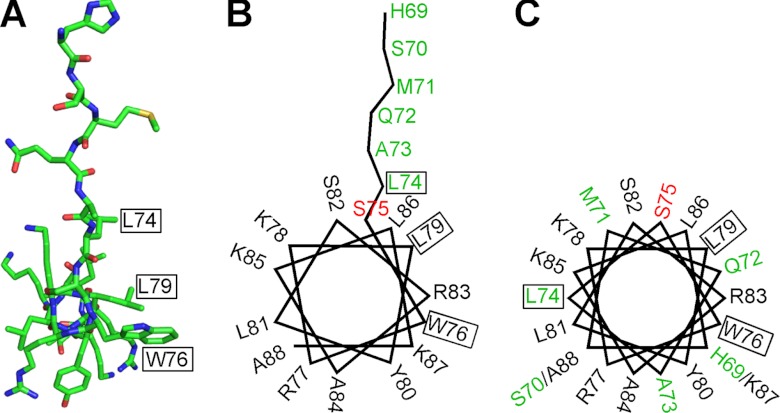
Structure of Orai1-CMBD Reveals a Modified Helical Wheel for Interaction with Camodulin
Most of calmodulin binding domains adopt a helical structure. Mullins et al. (13) predicted a helical wheel model of Orai1-CMBD. Based on the crystal structure (Fig. 9A), we made a helical wheel presentation of Orai1-CMBD (Fig. 9B), and compared it with the helical wheel model predicted by Mullins et al. (13) (Fig. 9C). The helical part of the peptide in the structure is in agreement with the prediction. However, the residues N-terminal to Ser75 of Orai1-CMBD adopt an extended conformation in the crystal structure (Fig. 9B), rather than the predicted helical conformation (Fig. 9C).
FIGURE 9.
Comparison between the structure of Orai1-CMBD and the reported hypothetical helical wheel model. A, structure of Orai1-CMBD in stick presentation viewed along the axis of α-helix. The colors of atoms are shown as: C, green; N, blue; O, red; S, yellow. B, a helical wheel model of Orai1-CMBD derived from the crystal structure. C, the hypothetical helical wheel model of Orai1-CMBD proposed by Mullins et al. (13). In both B and C, the residues are labeled with their amino acid numbers according to human Orai1 sequence. Ser75 is colored in red. Note that, N-terminal to Ser75, the crystal structure differs from the hypothetical model. The residues that reflect the discrepancy between the structure and the hypothetical model are colored in green. In all three panels, residues Leu74, Trp76, and Leu79 involved in hydrophobic interaction with CaM are boxed.
DISCUSSION
Unique Binding Mode of Orai1-CMBD to CaM
We show a unique binding mode of CaM to the target peptide Orai1-CMBD: binding of one peptide to each lobe of CaM (Fig. 10A). The binding mode observed here is unique in two aspects: each lobe of CaM binds one target peptide to form a 1:2 complex, and CaM adopts an extended conformation in the complex rather than a collapsed conformation. It is well known that CaM has plasticity to bind different target peptides. Based on the number of residues of CMBDs that are critical for interaction with CaM-N and CaM-C, CaM target peptides can be divided into different groups: 1–7, 1–10, 1–14, 1–16, and 1–18 (Fig. 10B)(29, 30). Because each Orai1-CMBD interacts with only one lobe of CaM, we categorize it as group 1–0. To date, there has been only one structure of 1 CaM:2 target peptides reported that falls into this group, CaM/glutamate decarboxylase (GAD)-CMBD (Fig. 10B). The structure of this CaM·GAD complex shows that CaM-N and CaM-C each binds one GAD-CMBD. The two GAD-CMBDs interact with each other to induce CaM to adopt a collapsed conformation (Fig. 10C) (18). CaM-N and CaM-C interact with GAD-CMBD in similar ways. The tryptophan residue on GAD-CMBD, however, is not deeply buried in the hydrophobic pocket of CaM-N or CaM-C (Fig. 10D). CaM·Orai1-CMBD may resemble the structure of CaM·GAD-CMBD in that Orai1-CMBD interacts with CaM-N and CaM-C in similar ways. The extended conformation in the CaM·Orai1-CMBD complex instead of a collapsed conformation (Fig. 10A) may result from the fact that Orai1-CMBD does not form a dimer in solution (Fig. 5) as GAD-CMBD does. Therefore, CaM·Orai1-CMBD displays a unique binding mode of CaM to its target peptides, and again shows the flexibility of CaM to bind a large variety of target proteins. In addition to Orai1-CMBD and GAD-CMBD, IQ calmodulin binding peptides from NaV1.5 (31), NaV1.2 (32), and PEP-19 (33–35) bind selectively to the C-lobe of CaM, leaving the N-lobe unoccupied, and thus fall in this group 1–0.
FIGURE 10.
Conformational flexibility of CaM to interact with different target peptides. A, proposed model of the 1:2 CaM·Orai1-CMBD complex. CaM-C·Orai1-CMBD was superimposed to CaM-N to obtain the binding model of CaM-N with Orai1-CMBD. CaM is shown as a blue ribbon, the peptide is shown as a green ribbon, and calcium ions are shown as red spheres. Because Orai1-CMBD does not dimerize in solution, we propose that CaM adopts an extended conformation in this 1:2 complex with Orai1-CMBD. B, sequence alignment of different groups of CMBD. The sequences are aligned according to the residue interacting with the hydrophobic pocket of CaM-C, which is shown in red letters. Residues that interact with the hydrophobic pocket of CaM-N are highlighted in yellow. The red line in Orai1 and PMCA (in Ref. 31) sequences indicates where CMBDs used for structural studies ended. The numbers above the sequences indicate the space plus 1 between the two residues that interact with the hydrophobic pockets of CaM-C and CaM-N. The sequences shown here are representatives for each group: 1–7, 1–10, 1–14, 1–16, and 1–18. The CMBDs of Orai1 and GAD interact with only one lobe of CaM, and are therefore considered as group 1–0. C, NMR structure of CaM·GAD-CMBD. CaM-N and CaM-C each bind one GAD-CMBD. The two GAD-CMBDs interact with each other, inducing CaM into a collapsed conformation, in contrast to the extended conformation in the proposed 1:2 CaM·Orai1-CMBD complex in A. The color scheme is the same as in A. D, zoomed-in view of the interaction of GAD-CMBD with CaM-C. CaM-N and CaM-C interact with GAD-CMBD similarly. CaM is displayed in surface representation, and GAD-CMBD is in sticks. The side chains of other peptide residues not involved in hydrophobic interaction with CaM are omitted for clarity. The colors of atoms are: C, green for GAD-CMBD and yellow for calmodulin; N, blue; O, red; and S, orange.
The NMR studies indicate that the interactions of each lobe of the calmodulin with the Orai1-CMBD are homologous, so that the two binding lobes each compete for the same set of residues. This type of interaction requires that binding be sequential, with the first Orai1-CMBD interacting with the stronger binding C-terminal lobe, and the second binding event involving the N-terminal lobe. This sequential binding effect is particularly apparent from the titration study shown in Fig. 6.
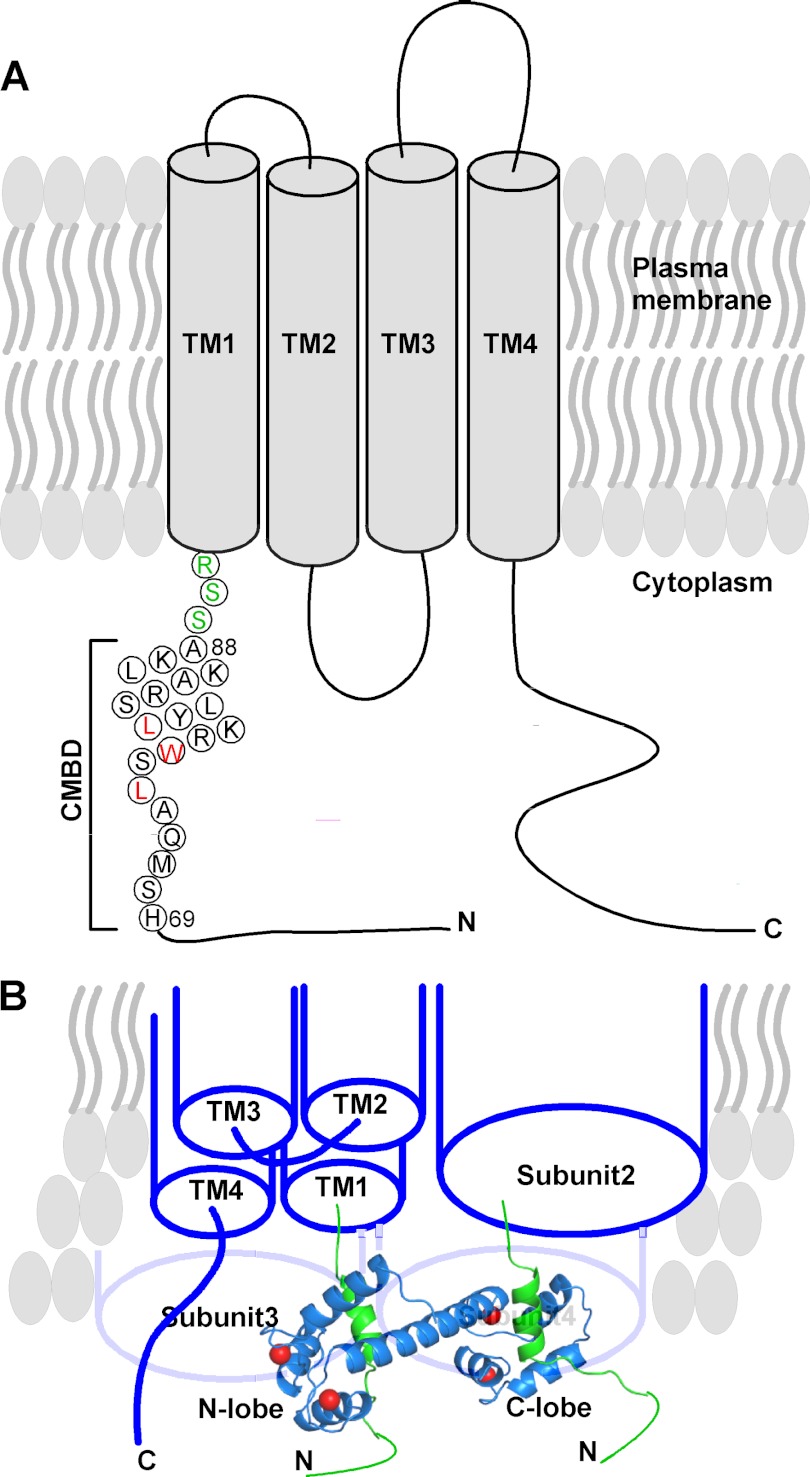
The first structure reported for CaM·PMCA-CMBD showed that CaM interacted with PMCA-CMBD through CaM-C only (36). Because the PMCA-CMBD used for their studies was a splice variant of PMCA lacking additional C-terminal residues (Fig. 10B), it was suggested that the structure of the complex was an intermediate toward the collapsed structure (36). As shown in Fig. 10B, when additional residues were added, the Val residue could have served as a binding site for CaM-N, resulting in a collapsed complex, as observed in the structure of CaM·smMLCK complex (37) with CaM wrapped around the peptide. In reality, the anchor residue for CaM-N binding was a Phe rather than Val, 17 residues away from the anchoring Trp residue at CaM-C (Fig. 10B), creating a new 1-18 binding mode (38). However, in our case, extension of residues at the C terminus of Orai1-CMBD is not feasible as Orai1-CMBD is immediately N-terminal to the 1st transmembrane helix (Fig. 11A). Therefore, no additional residues can be added to the C terminus of Orai1-CMBD to provide a binding site for CaM-N to allow CaM to adopt a collapsed structure without incurring a clash between CaM-N and the plasma membrane.
FIGURE 11.
Proposed mechanism for the interaction of CaM with Orai1 to carry out CDI. A, schematic presentation of the proposed Orai1 topology. The sequence of Orai1-CMBD from the structure is shown in circles. Leu74, Trp76, and Leu79 that are involved in hydrophobic interaction with CaM are colored in red. The three residues between the Orai1-CMBD and the first transmembrane helix (TM1) are colored in green. B, proposed binding of one CaM to two Orai1. Orai1 is shown as a tetramer. Subunit 1 is displayed with four transmembrane helices. The remaining three subunits are displayed as cylinders. Here, one CaM (blue ribbon) binds to two CMBDs (green ribbons) of the two adjacent Orai1 subunits, displacing STIM1 from binding to Orai1 to inactivate the channel. The other CaM for binding to subunits 3 and 4 is not displayed for clarity.
Possible Mechanism for Ca2+·CaM-dependent Inhibition (CDI) of Orai1
Based on this unique feature of the CaM/Orai1 interaction, we propose a mechanism for the CDI of Orai1. Orai1 exists as a dimer in the resting state, and upon activation by STIM1, forms a tetramer (39). STIM1 was reported to interact with the cytosolic N and C termini of Orai1 to activate ion flux (11, 12). We therefore propose that CaM-C interacts with one subunit of Orai1 followed by CaM-N interacting with another adjacent subunit in the Orai1 oligomer (Fig. 11B). In so doing CaM displaces STIM1 from interacting with Orai1, and thus inactivates the channel.
Interpretation of Different Binding Affinities of Orai1-CMBD for CaM-N and CaM-C
ITC data showed that the binding affinity of Orai1-CMBD for CaM-C is four times stronger than that for CaM-N. To interpret this difference, we superimposed CaM-N with CaM-C to obtain the model of CaM-N·Orai1-CMBD complex (Fig. 2). By doing so, we noticed three significant clashes between CaM-N and the peptide (Fig. 2C). Two of these clashes (between Leu74 of Orai1-CMBD and Gln41 of CaM, and between Tyr80 of Orai1-CMBD and Met72 of CaM) can be resolved by rotating the side chains of CaM residues to adopt different rotamer conformations. The third clash is between Trp76 of Orai1-CMBD with Val55 in the hydrophobic pocket of CaM-N. When we compared the hydrophobic pocket of CaM-N with that of CaM-C, we found that the amino acid compositions of the two pockets are identical except for two amino acids, Val55(CaM-N)/Ala128(CaM-C) and Ile63(CaM-N)/Val136(CaM-C). Val55 has a more bulky side chain than Ala128, creating a clash with the side chain of Trp76 of Orai1-CMBD. This clash, unlike the two mentioned above, cannot be resolved by rotating the side chain of Val55. Thus, the side chain of Val55 may prevent Trp76 of Orai1-CMBD from going deeply into the hydrophobic pocket of CaM-N, resulting in a weaker affinity. This proposal may also explain why we did not observe the NOE signals between Trp76 of Orai1-CMBD and Met51, Ile52, Ile63, and Met71 of CaM, whose equivalent residues in CaM-C show NOE signals with Trp76 of Orai1-CMBD (Figs. 2, B and D, and 8).
Interpretation of Reported Biochemical Data of Orai1 Inactivation Using the Structural Information
The structure of CaM·Orai1-CMBD is consistent with the biochemical and electrophysiological data of Mullins et al. (13). First, in the structure, Trp76 of Orai1-CMBD interacts with a hydrophobic pocket of CaM-C. This hydrophobic pocket is made of side chains of residues Ile100, Leu105, Met124, Ile125, Ala128, Val136, Phe141, and Met144 (Fig. 2B). This shows that Trp76 is critical for the interaction of Orai1-CMBD with CaM. In fact, Mullins et al. (13) showed that W76A, W76E, and W76S mutants of Orai1 all lost their capability of binding to CaM, and CDI of Orai1 was abolished by these mutations.
Tyr80 of Orai1 is another residue that was predicted to be important for interacting with CaM. When Tyr80 of Orai1-CMBD was mutated to Ser or Ala (13), Orai1 mutants retained their binding affinity for CaM, whereas the Y80E mutation disrupted the Orai1 interaction with CaM (13). In the structure, the aromatic ring of Tyr80 of Orai1-CMBD interacts with Ile85, Ala88, and Phe141 and Met145 of CaM-C through hydrophobic interactions (supplemental Fig. S3C). The hydroxyl group of Tyr80 also forms a hydrogen bond with the backbone oxygen atom of Glu84 of CaM. Based on the structure, Y80A or Y80S mutations will not disturb this hydrophobic interaction significantly enough to disrupt the interaction of Orai1 with CaM. The electron density of OE1 and OE2 of Glu84 of CaM is not well defined, indicating the side chain of Glu84 adopts different rotamers in solution. When Tyr80 is mutated to Glu, there will be electrostatic repulsion between Y80E and Glu84 (Fig. 2A), which may be significant enough to disrupt the interaction between Orai1 and CaM. Alternatively, because Tyr80 is located in a hydrophobic environment formed by CaM residues (supplemental Fig. S3C), the Y80E mutation will place the hydrophilic and charged side chain of glutamic acid in this hydrophobic environment, which will be disfavored, and thus disrupt interaction between Orai1 and CaM.
Ala73 of Orai1 was the third amino acid that was predicted to interact with CaM. Mutation A73E also disrupts the interaction of Orai1 with CaM (13). In the crystal structure, Ala73 does not interact with CaM. However, the CB atom of Ala73 is 4.8 Å away from the OE2 atom of Glu123 of CaM (Fig. 2A). The A73E mutation will create electrostatic repulsion between Glu73 of the Orai1 mutant and Glu123 of CaM, disrupting the interaction of Orai1 with CaM. We predict that mutation of Ala73 to amino acids with short side chains, such as Ser, Val, or Thr, should not disrupt the interaction of Orai1 with CaM.
In summary, we show here that Orai1-CMBD binds to CaM through a unique binding mode: it interacts only with CaM-C in the crystal structure, but interacts with both CaM-N and CaM-C in solution, forming a 1:2 CaM·Orai1-CMBD complex. Based on the structure and biochemical data, we propose a mechanism for CaM to displace STIM1 and thus inactivate the Orai1 tetramer: in a first step the C-lobe of CaM would interact with the CMBD of one Orai1, which would be followed by interaction of the N-lobe of the same CaM with the CMBD of a neighboring Orai1 (Fig. 11B). Our data cannot address whether one or two Ca2+·CaM complexes are required for channel Orai1 inactivation. The structural information is useful for further studies on understanding how Orai1, STIM1, and CaM interact with each other to carry out CDI.
Supplementary Material
Acknowledgments
We thank Drs. Lars Pedersen, Chen Qiu, Joseph Krahn, and Andrea Moon for the help with x-ray diffraction data collection, processing, and structure determination; Drs. Traci Hall, Huancheng Wang, and Vijayakanth Pagadala for useful discussion; Dr. Jenny Yang, Georgia State University, for useful information and discussion; Drs. Traci Hall and Robert S. Williams for critical reading of the manuscript; Dr. Robert Petrovich, Protein Expression Core Facility, for help with the circular dichroism instrument; and Dr. Jason Williams and Katina Johnson, Protein Microcharacterization Core Facility, for protein identification using mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program Grant Z01-ES101684 and Collaborative Crystallography Group NIEHS Grant Z01 ES102645.

This article contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (code 4EHQ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CDI
- calcium-dependent fast inactivation
- CaM
- calmodulin
- CaM-N
- N-terminal lobe of calmodulin
- CaM-C
- C-terminal lobe of calmodulin
- CMBD
- calmodulin binding domain
- GAD
- glutamate decarboxylase
- HMQC
- heteronuclear multiple quantum coherence
- ITC
- isothermal titration calorimetry
- PMCA
- plasma membrane calcium ATPase
- Trx
- thioredoxin
- aa
- amino acid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Takemura H., Putney J. W., Jr. (1989) Capacitative calcium entry in parotid acinar cells. Biochem. J. 258, 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 3. Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yeromin A. V., Zhang S. L., Jiang W., Yu Y., Safrina O., Cahalan M. D. (2006) Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature 443, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang S. L., Yeromin A. V., Zhang X. H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K. A., Cahalan M. D. (2006) Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. U.S.A. 103, 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437, 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liou J., Fivaz M., Inoue T., Meyer T. (2007) Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. U.S.A. 104, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luik R. M., Wang B., Prakriya M., Wu M. M., Lewis R. S. (2008) Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature 454, 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu M. M., Luik R. M., Lewis R. S. (2007) Some assembly required: constructing the elementary units of store-operated Ca2+ entry. Cell Calcium 42, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mullins F. M., Park C. Y., Dolmetsch R. E., Lewis R. S. (2009) STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. U.S.A. 106, 15495–15500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krebs J., Buerkler J., Guerini D., Brunner J., Carafoli E. (1984) 3-(Trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine, a hydrophobic, photoreactive probe, labels calmodulin and calmodulin fragments in a Ca2+-dependent way. Biochemistry 23, 400–403 [DOI] [PubMed] [Google Scholar]
- 15. Kuboniwa H., Tjandra N., Grzesiek S., Ren H., Klee C. B., Bax A. (1995) Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 2, 768–776 [DOI] [PubMed] [Google Scholar]
- 16. Zhang M., Tanaka T., Ikura M. (1995) Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2, 758–767 [DOI] [PubMed] [Google Scholar]
- 17. Schumacher M. A., Rivard A. F., Bächinger H. P., Adelman J. P. (2001) Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 410, 1120–1124 [DOI] [PubMed] [Google Scholar]
- 18. Yap K. L., Yuan T., Mal T. K., Vogel H. J., Ikura M. (2003) Structural basis for simultaneous binding of two carboxyl-terminal peptides of plant glutamate decarboxylase to calmodulin. J. Mol. Biol. 328, 193–204 [DOI] [PubMed] [Google Scholar]
- 19. Hayashi N., Matsubara M., Takasaki A., Titani K., Taniguchi H. (1998) An expression system of rat calmodulin using T7 phage promoter in Escherichia coli. Protein Expr. Purif. 12, 25–28 [DOI] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 24. Press W. H., Flannery B. P., Teukolsky S. A., Vetterling W. T. (1989) Numerical recipes in FORTRAN. The art of scientific computing. Cambridge University Press, Cambridge, UK [Google Scholar]
- 25. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 26. Johnson B. A., Blevins R. A. (1994) NMRVIEW. A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 27. Lee W., Revington M. J., Arrowsmith C., Kay L. E. (1994) A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett. 350, 87–90 [DOI] [PubMed] [Google Scholar]
- 28. Babu Y. S., Bugg C. E., Cook W. J. (1988) Structure of calmodulin refined at 2.2-Å resolution. J. Mol. Biol. 204, 191–204 [DOI] [PubMed] [Google Scholar]
- 29. Ataman Z. A., Gakhar L., Sorensen B. R., Hell J. W., Shea M. A. (2007) The NMDA receptor NR1 C1 region bound to calmodulin. Structural insights into functional differences between homologous domains. Structure 15, 1603–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoeflich K. P., Ikura M. (2002) Calmodulin in action. Diversity in target recognition and activation mechanisms. Cell 108, 739–742 [DOI] [PubMed] [Google Scholar]
- 31. Chagot B., Chazin W. J. (2011) Solution NMR structure of apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 406, 106–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feldkamp M. D., Yu L., Shea M. A. (2011) Structural and energetic determinants of apo calmodulin binding to the IQ motif of the NaV1.2 voltage-dependent sodium channel. Structure 19, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Putkey J. A., Kleerekoper Q., Gaertner T. R., Waxham M. N. (2003) A new role for IQ motif proteins in regulating calmodulin function. J. Biol. Chem. 278, 49667–49670 [DOI] [PubMed] [Google Scholar]
- 34. Putkey J. A., Waxham M. N., Gaertner T. R., Brewer K. J., Goldsmith M., Kubota Y., Kleerekoper Q. K. (2008) Acidic/IQ motif regulator of calmodulin. J. Biol. Chem. 283, 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X., Kleerekoper Q. K., Xiong L. W., Putkey J. A. (2010) Intrinsically disordered PEP-19 confers unique dynamic properties to apo and calcium calmodulin. Biochemistry 49, 10287–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elshorst B., Hennig M., Försterling H., Diener A., Maurer M., Schulte P., Schwalbe H., Griesinger C., Krebs J., Schmid H., Vorherr T., Carafoli E. (1999) NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38, 12320–12332 [DOI] [PubMed] [Google Scholar]
- 37. Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. (1992) Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 38. Juranic N., Atanasova E., Filoteo A. G., Macura S., Prendergast F. G., Penniston J. T., Strehler E. E. (2010) Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif. J. Biol. Chem. 285, 4015–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Penna A., Demuro A., Yeromin A. V., Zhang S. L., Safrina O., Parker I., Cahalan M. D. (2008) The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 456, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.