Abstract
Kruppel like factor 6 (KLF6), a zinc finger transcription factor and tumor suppressor, is induced as an immediate-early gene during hepatic stellate cell (HSC) activation. The paradoxical induction of a tumor suppressor in HSCs during proliferation led us to explore the biology of wild type KLF6 (KLF6WT) and its antagonistic, alternatively spliced isoform KLF6SV1 in cultured HSCs and animal models.
Methods
The animal models generated include a global heterozygous KLF6 mouse (Klf6 +/−), and transgenic mice expressing either hKLF6WT or hKLF6SV1 under the control of the Collagen α2 (I) promoter to drive HSC-specific gene expression following injury.
Results
The rat Klf6 transcript has multiple splice forms that are homologous to those of the human KLF6 gene. Following a transient increase, all rat Klf6 isoforms decreased in response to acute CCl4 liver injury, and culture-induced activation. After acute CCl4, Klf6 +/− mice developed significantly increased fibrosis and enhanced fibrogenic mRNA and protein expression. In contrast, HSC-specific transgenic mice over-expressing KLF6WT or KLF6SV1 developed significantly diminished fibrosis with reduced expression of fibrogenic genes. Chromatin IP, and qRT-PCR in mouse HSCs over-expressing KLF6WT demonstrated KLF6WT binding to GC boxes in promoters of Colα1 (I), Colα2 (I), and β-Pdgfr with reduced gene expression, consistent with transcriptional repression by KLF6. Stellate cells over-expressing either KLF6WT or KLF6SV1 were more susceptible to apoptotic stress based on PARP cleavage.
Conclusion
KLF6 reduces fibrogenic activity of HSCs via two distinct mechanisms, direct transcriptional repression of target fibrogenic genes and increased apoptosis of activated HSCs. These results suggest that following its initial induction, sustained downregulation of KLF6 in liver injury may allow de-repression of fibrogenic genes and decreased stellate cell clearance by inhibiting apoptosis.
Introduction
Our previous efforts to understand the molecular basis of stellate cell activation utilized subtraction hybridization to clone a novel zinc finger transcription factor, Klf6 (previously called “Zf9”), which is induced as an immediate-early gene in hepatic stellate cells during liver injury in vivo (1). Subsequent studies have broadened KLF6's roles in injury to include growth responses of vascular endothelial cells and hepatocytes, among others (2, 3). KLF6 is a member of a large family of zinc finger transcription factors that have a conserved C-terminal C2H2 DNA binding domain recognizing “GC box” motifs in responsive promoters. Studies originally performed in liver ultimately led to the identification of KLF6 as a growth-inhibitory tumor suppressor gene that is inactivated in several human cancers (4, 5).
The discovery that Klf6, a growth suppressive gene, is rapidly induced when stellate cells undergo a proliferative burst, presented a paradox that the current study has sought to reconcile. In addition to full length KLF6, human KLF6 has several shorter antagonistic splice isoforms, KLF6SV1-3; among these, KLF6SV1 is the best studied (6, 7). Based on these information we hypothesized that the cloning of Klf6 in rat stellate cells (1) actually represented a growth-promoting splice isoform that contributes to stellate cell activation, yet rodent splice isoforms were not described.
In our studies exploring the role of KLF6 during stellate cell activation we employed Klf6 +/− mice, lacking one allele of Klf6 in all cells; these mice are viable, fertile and phenotypically normal whereas Klf6 null mice are embryonic lethal (8). We used these animals, in addition to novel stellate cell-specific transgenic mice to explore the role of KLF6 in hepatic stellate cell activation and fibrogenic gene induction.
Methods
Semi-quantitative PCR and cloning of rat Klf6 splice variants
PCR was performed using primers to rat Klf6 mRNA untranslated regions- 5' UTR: AACTTTCACCTGCGCTCCCG, 3'UTR: TGGCTGGTACAGGTATCCCTC with rat primary stellate cell complementary DNA (cDNA) as a template. The products of the reaction were separated using gel electrophoresis. Bands were excised and extracted using the QIAquick® Gel Extraction Kit (Qiagen, Chatsworth, CA) according to the manufacturer's protocol. The DNA fragments were then cloned into the pCR®8/GW/TOPO® TA vector (Invitrogen, Carlsbad, CA) for sequencing and subsequently into the ® pcDNA3.1/V5-His© TOPO© TA Expression Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Animal Studies
All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86–23 revised 1985).
Murine and rat stellate cell isolation
Mice were injected intra-peritoneally with CCl4 (5ul/g mouse of 10% CCl4 in corn oil) (Sigma) three times (alternating days), and mouse stellate cells were isolated 2 days after the last dose of CCl4. mHSCs were isolated from C57/Bl6 wild type, Klf6F/F, Cola2(I)-KLF6WT and Cola2(I)-KLF6SV1 mice by enzymatic digestion and Percoll density gradient centrifugation, (9) with modifications. For the transgenic mouse HSC studies, stellate cells were isolated and mRNA was harvested without culturing the cells. For the in vitro KLF6 deletion studies, Klf6F/F HSCs were cultured for 10 days and then infected with either Ad-lacZ or Ad-Cre at an MOI of 10. Viral transduction efficiency was 85% as assessed using Ad-EGFP and fluorescence microscopy. Protein lysates were collected for Western blot.
Rat Klf6 isoform quantitation
For liver injury experiments, Sprague-Dawley rats were injected with CCl4 (2μl/g of a 50% CCl4 solution in corn oil) or corn oil, followed by isolation of HSCs (see below) at 24 h or 48 h after injection (n=3 for each condition). Messenger RNA was isolated and quantitative RT-PCR (see next section) was performed using isoform-specific primers to identify any rat Klf6 isoforms (see Table 1 for list of primers). For culture activation experiments, HSCs were isolated from untreated rats and activated by growth in primary culture on non-coated plastic dishes for up to 1 week. Messenger RNA was harvested from the cells at days 0, 2, 4, & 7, in three separate cell isolates.
Table 1.
List of Primers
| Name | Sequence |
|---|---|
| rKlf6 Bot F | 5'-ACTCCTGATCGTTCACCTCCCTG-3' |
| rKlf6 Bot R | 5'-GTAAGGCTTTTCTCCTGTTGCCAAT-3' |
| rKlf6 Mid F | 5'-CGAAACGGGCTACTTCTCGGCT-3' |
| rKlf6 Mid R | 5'-TTCGGGAGAAGAAGGATTTTGGT-3' |
| rKlf6 Top F | 5'-CGGGAGAGGAAGGAGGAATCAGA-3' |
| rKlf6 Top R | 5'-AGAGGTAAACTTGGTCGTGGGC-3' |
| rGAPDH F | 5'-GGCATCGTGGAAGGGCTCAT-3' |
| rGAPDH R | 5'-AGGGATGATGTTCTGGGCTGC-3' |
| rCOLa2 (I) F | 5'-TGAGCCTGGTGAGCCCG-3' |
| rCOLa2 (I) R | 5'-TCTCGCCAGGTCTTCCAGG-3' |
| rCola1 (I) F | 5'-GTGGTAACGATGGTGCTGTC-3' |
| rCola1 (I) R | 5'-CTTCACCCTTAGCACCAGC-3' |
| rASMA F | 5'-GCTGTCTTCCCATCCATCGTG-3' |
| rASMA R | 5'-TGGGGTACTTCAGAGTCAGGATG-3' |
| rPDGFBR F | 5'-CATTGGGGACAGGGAAGTGGAC-3' |
| rPDGFBR R | 5'-CCTGATGGTGATGCTCTCGC-3' |
| rTGFBR1 F | 5'-CACAAACAGTGGCAGCGGGAC-3' |
| rTGFBR1 R | 5'-CAGAGGTGGCAGAAACACTGTAATG-3' |
| rMMP2 F | 5'-GACGGCAAATATGGCTTCTGTCC-3' |
| rMMP2 R | 5'-GCCCTCGGTGGTACAGC-3' |
| rTIMP1 F | 5'-GCCTACACCCCAGCCAT-3' |
| rTIMP1 R | 5'-ATGCCAGGGAACCAGGAAGC-3' |
| rTIMP2 F | 5'-GGCAACCCCATCAAGAGGATTCAAT-3' |
| rTIMP2 R | 5'-CACACTGCTGAGGAGGGG-3' |
| mGAPDH F | 5'-CAATGACCTTCATTGACC-3' |
| mGAPDH R | 5'-GATCTCGCTCCTGGAAGATG-3' |
| mCOLa1 (I) F | 5'-GTCCCTGAAGTCAGCTGCATA-3' |
| mCOLa1 (I) R | 5'-TGGGACAGTCCAGTTCTTCAT-3' |
| mASMA F | 5'-TCCTCCCTGGAGAAGAGCTAC-3' |
| mASMA R | 5'-TATGGTGGTTTCGTGGATGC-3' |
| mTGFB F | 5'-TGCGCTTGCAGAGATTAAAA-3' |
| mTGFB R | 5'-CTGCCGTACAACTCCAGTGA-3' |
| mMMP2 F | 5'-ACCCAGATGTGGCCAACTAC-3' |
| mMMP2 R | 5'-TACTTTTAAGGCCCGAGCAA-3' |
| mTIMP2 F | 5'-GCCAAAGCAGTGAGCGAGAAG-3' |
| mTIMP2 R | 5'-CACACTGCTGAAGAGGGGGC-3' |
| mTIMP1 F | 5'-ACGAGACCACCTTATACCAGCG-3' |
| mTIMP1 R | 5'-GCGGTTCTGGGACTTGTGGGC-3' |
| mCOLa2 (I) F | 5'-GTGTTCGTGGTTCTCAGGGT-3' |
| mCOLa2 (I) F | 5'-GTCTGAGTGAAGGCTGGGAG -3' |
| mTGFbR1 F | 5'-ACTGAAAGCTTCCCAGGGTT-3' |
| mTGFbR1 R | 5'-TAAGGGCTGGCAGTTGTCTT-3' |
| mPDGFRB F | 5'-CTTTGTGCCAGATCCCACCA-3' |
| mPDGFRB R | 5'-TCACTCGGCACGGAATTGTC-3' |
Reverse-Transcription and Real-Time Quantitative PCR
RNA was extracted from cells/liver tissue and reverse-transcribed into cDNA using an RNeasy® kit (Qiagen, Valencia, CA) and using Sprint™ RT Complete-Double PrePrimed tubes (Clontech, Mountain View, CA) respectively, and analyzed by quantitative PCR using SYBR green qPCR Master Mix (Roche) on the lightCycler®480 System (Roche). Data are represented as the relative expression of fibrogenic genes after normalizing to GAPDH. Please refer to Table 1 for a complete list of primers.
Western blot
Western blots of cell extracts were prepared by centrifuging the cells with lysis buffer complemented with protease inhibitor (Complete Lysis-M kit, Roche Diagnostics, Indianapolis, IN) Protein concentration was determined with a Bio-Rad DC kit (Bio-Rad). Antibodies used were as follows: Rabbit anti-Collagen I (1:1000)(Rockland), Rabbit anti-β-PDGFR (1:500)(Santa Cruz), Mouse anti-ASMA (1:500)(Milipore).
Rat KLF6 degradation analysis
293FT cells were plated in six-well plates and serum starved for four hours prior to transfection. Cells were transiently transfected using the Lipofectamine™ 2000 Reagent (Invitrogen, Chatsworth CA) according to the manufacturer's protocol with pcDNA3.1/rKlf6top-V5, pcDNA3.1/rKlf6mid-V5, pcDNA3.1/rKlf6bot-V5, or pcDNA3.1/V5. 24 hours later cells were either treated with proteasomal inhibitor MG132 (Sigma) at a final concentration of 10μM or an equal volume of DMSO. Protein was harvested 5 hours after addition of MG132.
Rat KLF6 cellular localization
293FT cells were plated in six-well plates and serum-starved for four hours prior to transfection. Cells were transiently transfected using the Lipofectamine™ 2000 Reagent (Invitrogen, Chatsworth CA) according to the manufacturer's protocol with pcDNA3.1/rKlf6top-V5, pcDNA3.1/rKlf6mid-V5, pcDNA3.1/rKlf6bot-V5, or pcDNA3.1/V5. Cells were fixed with ice-cold methanol, and stained first with monoclonal mouse anti-V5 antibody (1:50)(Abcam), followed by FITC-conjugated goat anti-mouse antibody (1:250) (Invitrogen). Cells were mounted using Vectashield containing DAPI (Invitrogen).
Generation of HSC-specific KLF6 transgenic mice
Colα2(I)-KLF6WT and Colα2(I)-KLF6SV1 constructs were generated by cloning KLF6WT, and KLF6SV1 downstream of the Colα2(I) enhancer promoter. These constructs were then used to generate transgenic mice on a C57/Bl6 background. (Mouse Genetics Shared Resource Facility at Mt. Sinai) The same transgene specific primers were used to identify both KLF6WT, and KLF6SV1 transgene positive founder mice. Geno Fwd: GGAACGGTCCACGATTGCCAAGTCT, Geno Rev: TAACGTTCCAGCTCTAGGCAGGTCTG. Transgenic lines were selected based on good transgene induction following a single dose of CCl4.
Chronic CCl4 injury
For chronic CCl4 experiments, mice were injected intra-peritoneally twice per week for 8 weeks with a dose of (5ul/g mouse of 10% CCl4 in corn oil) Mice were sacrificed 48h after the last dose of CCl4, and liver tissue was collected for mRNA/protein analysis, H&E staining, Sirius Red Staining, and ASMA immunohistochemistry. For the Klf6 +/− mice (n=7 per group), and for the HSC specific transgenic animals (n=6 per group).
Quantitative fibrosis morphometry in liver sections
Formalin fixed, paraffin embedded liver tissue was stained with Sirius Red. For each animal, 40 images were taken at low magnification (10× objective lens), and were subsequently used for quantifying percentage of fibrotic tissue using BioQuant Software.
Chromatin immunoprecipitation assays (ChIP)
Chromatin immunoprecipitation assays (ChIP) were performed using a commercial kit according to the manufacturer's instructions (Active Motif-ChIP-IT™ Express). Proteins cross-linked to DNA were immunoprecipitated with 10 μg of anti-KLF6 antibody (0.2 μg/μl) (R-173) (Santa Cruz Biotechnology), 10 μg anti-histone H3 rabbit antiserum (0.2 μg/μl) (07-690) (Upstate Biotechnology, Inc), 10 μg anti-KLF6SV1 antibody (0.5 μg/μl) (Invitrogen) or 10 μg control IgG (0.4 μg/μl) (sc-2027) (Santa Cruz Biotechnology) and protein-G-magnetic beads. Genomic sequence primers encompassing “GC-boxes” in promoter regions (within 2 kbp) upstream of transcriptional start site were used to amplify immunoprecipitated DNA. Colα1(I) Fwd: 5'-CCCTTCCTTTCCCTCCTCC-3', Colα1(I) Rev: 5'- TGGCCTGGGCCCCTTTTAT-3'; Colα2(I) Fwd: 5'-CCAGGTCCCTCAACACCTG-3', Colα2(I) Rev: 5'-AGTGCTGGGAGGAGACTGC-3'; β-PDGFR Fwd: 5'-GCGAAAGTGGAAGAGAACCAG-3', β-PDGFR Rev: 5'-GGACTAAGTTGTCTGGAACCAC-3'.
PARP cleavage studies
We employed human HSC line (LX2) stably transfected with Adenovirus containing pBabe, pBabe-KLF6WT, or pBabe-KLF6SV1. Briefly LX2 cells were infected with adenovirus containing the above sequences and cell selection was performed using puromycin at a concentration of 0.45 μg/ml. Stable colonies were selected at one week, amplified and used in further experiments.
Results
Rat Klf6 is alternatively spliced, with all splice isoforms decreased during stellate cell activation
Klf6 was first cloned from primary rat stellate cells (1) using subtractive hybridization (10), which yielded a cDNA fragment that was used to screen a full-length, sized cDNA library from stellate cells; this approach led to the isolation of the Klf6 full length rat cDNA. Using this cloning strategy, we were not able to recognize that the rat KLF6 transcript might be alternatively spliced. Subsequently, three alternative splice isoforms of human KLF6 (hKLF6) were discovered (KLF6SV1-3) in the context of carcinogenesis (11). Among these, KLF6SV1, is the best studied, and has been further identified as a growth promoting isoform that antagonizes the function of full length KLF6 (“KLF6WT”) (6, 7, 12). Based on this finding, we then surmised that KLF6 might also be alternatively spliced during hepatic stellate cell activation in response to tissue injury, leading to increased generation of an antagonistic splice form that promotes stellate cell activation.
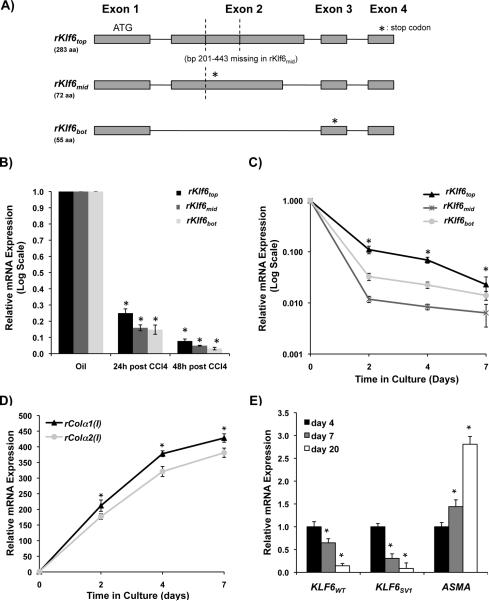
To test this hypothesis, we explored whether Klf6 was alternatively spliced in rat stellate cells by using RT-PCR based on flanking primer sequences in the rat 5' and 3' KLF6 untranslated regions (UTRs). Using cDNA primary rat stellate cells from untreated animals we consistently detected 3 distinct bands, which were cloned, sequenced, and aligned against the rat genome. This approach yielded 3 novel rKlf6 splice forms. The rKlf6 splice forms were labeled `Top', `Middle', and `Bottom' based on their size, shown in Fig. 1A.
Figure 1. KLF6 isoforms in human and rat stellate cells decrease with activation of these cells.
A. Primary rat HSC cDNA from a normal rat was used as template in PCR reaction with primers designed to the 5' and 3' UTRs of rat Klf6 transcript. Three DNA products were present, which were cloned, sequenced and aligned with rat genome, confirming 3 Klf6 splice isoforms shown; B. Rats were injected once with either oil, or CCl4 followed by HSC isolation 24h and 48h thereafter (n=3 for each treatment and time point). Rat Klf6 isoform levels were assessed by qRT-PCR with isoform-specific primers, demonstrating a significant decrease for all 3 isoforms (p-value<0.001) (data was normalized to GAPDH, fold change was calculated by comparing CCl4 and oil samples); C. Primary rat stellate cells (3 separate isolates) from untreated rats were plated in primary culture for 7 days, and all Klf6 isoforms decreased similar to stellate cells analyzed following liver injury in vivo (p-value<0.01) (data was normalized to GAPDH, fold change was calculated by comparing days 2, 4, and 7 to day 0); D. Collagen (I) mRNA increases during primary culture confirming that cells responded as expected to culture-induced activation; E. Human KLF6 isoforms (KLF6WT and KLF6SV1) are both down-regulated following primary culture of human stellate cells isolated from normal liver (3 separate isolations, p-value<0.05).
Even though the splicing pattern for rKlf6 is different from hKLF6, there are several key similarities. The rKlf6top (283 aa) corresponds to hKLF6WT, with a homology of 93% at the amino acid level. A 243 bp stretch of nucleotides of exon 2 (bps 201–443) is spliced out in rKlf6mid (72aa) and this splicing pattern leads to a frame-shift with a premature stop codon (depicted by * in Fig. 1A). For rKlf6bot (55aa), there is a complete skipping of exon 2, also leading to a frame-shift and premature stop codon. Interestingly, rKlf6bot is very similar to hKLF6SV1 (the shortest variant for human KLF6). The hKLF6SV1 isoform has a unique 21 amino acid stretch at its C-terminus that is thought to be critical for its function; rKlf6bot shares 19 of these 21 amino acids.
To determine whether the shorter rKlf6mid and rKlf6bot isoforms are translated into proteins we transfected 293FT cells with V5-tagged expression constructs encoding each of the three rKlf6 cDNA isoforms. Cells were grown in the presence or absence of proteasomal inhibitor MG132 to account for potential rapid degradation by the proteasome, since this pathway is important in the degradation of hKLF6 (13). All three rat Klf6 isoforms were translated into protein, and were stabilized in the presence of the proteasomal inhibitor, indicating their degradation by the proteasome (Supplemental Fig. 1A). Moreover, the putative nuclear localization signal for hKLF6 lies in the C-terminus region, which is absent in rKlf6mid and rKlf6bot, indicating that the isoforms should be cytoplasmic. To verify the localization of these rat KLF6 isoforms, confocal microscopy was performed on 293FT cells transfected with V5-tagged rKlf6 expression constructs that were immunostained for the V5 epitope tag. As predicted, rKLFtop was present in both the nucleus and cytoplasm, while the two lower molecular weight isoforms, rKLF6mid and rKLF6bot, were unable to enter the nucleus where they displayed a cytoplasmic punctate pattern (Supplemental Fig. 1B).
By exploiting the unique splice junctions within the rKlf6 isoforms, real-time PCR primers were designed to perform isoform-specific real time PCR (see Table 1 for list of primers). With these tools, we next assessed rKlf6 isoform expression during stellate cell activation following liver injury, and during activation induced by primary culture on plastic (14). Rat stellate cells were isolated 24 h and 48 h after a single dose of oil (control) or CCl4. As shown in Fig. 1B, there was a dramatic decrease in levels of all rKlf6 isoforms at 24h and 48h post injection of CCl4, and following activation in primary culture (Fig. 1C). To confirm that the cells were activating in response to culture, we assessed collagen (I) expression levels, which increased progressively during the culture activation as expected (Fig. 1D). To establish the relevance of these findings in rats to human stellate cells, expression of hKLF6WT and hKLF6SV1 mRNAs was assessed in primary human stellate cells during culture activation. As for rKlf6, both human KLF6 isoforms decreased significantly with progressive activation in culture (Fig. 1E).
Loss of Klf6 in hepatic stellate cells leads to more fibrogenic activity and increased liver fibrosis following liver injury
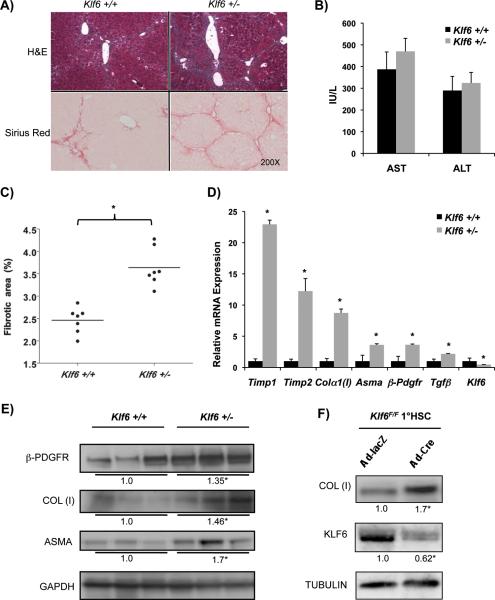
To further define the contribution of KLF6 to stellate cell activation in vivo, we next assessed the response of Klf6 +/− mice to chronic CCl4, reasoning that loss of a single allele of Klf6 in stellate cells might modify their fibrogenic behavior in this model. Remarkably, there was a marked increase in inflammation along the fibrotic tracts in the Klf6 +/− animals (Fig. 2A). Likewise, Klf6 +/− mice displayed a marked increase in fibrotic area compared to wild type mice, as assessed by Sirius Red staining and morphometry (3.6% vs. 2.5%, p-value<0.01) with comparable levels of AST and ALT between the two groups (Fig. 2B &C). Consistent with the increased fibrosis accumulation, stellate cell-derived fibrogenic mRNAs were significantly increased in the Klf6 +/− mice, including Asma, Colα1 (I), Timp1, Timp2, β-Pdgfr, and Tgf-β mRNA transcripts, along with a decrease in levels of Klf6 mRNA, as expected (Fig. 2D). Western blot analysis of whole liver protein lysates confirmed an increased expression of the fibrogenic proteins β-PDGFR, COL(I), and alpha- smooth muscle actin (ASMA) in Klf6 +/− animals (Fig. 2E).
Figure 2. KLF6 deficiency in Klf6 +/− mice leads to increased fibrogenesis following CCl4 liver injury with similar results obtained using knockdown of Klf6 in cultured Klf6F/F stellate cells.
A&C. Increased fibrosis Klf6 +/− mice following 8 weeks of chronic CCl4, compared to Klf6 +/+ mice (n=7/group) (3.6% vs. 2.5%, p-value<0.01)(For each animal, 40 images were taken at low magnification (10X objective lens), and were subsequently used for quantifying percentage of fibrotic tissue using BioQuant Software) B. Both groups showed comparable liver injury levels as demonstrated by equally elevated AST and ALT; D. Whole liver sample qRT-PCR from the same mice, shows an increase in HSC-derived fibrogenic mRNA in Klf6 deficient livers (p-value<0.01); E. Whole liver protein Western blot of 3 representative samples from Klf6 +/− and Klf6 +/+ mice after chronic CCl4 documenting increased fibrogenic proteins COL (I), β-PDGFR, and ASMA; F. Klf6 F/F stellate cells were isolated from mice and allowed to activate for 1 week in culture, followed by infection with Ad-lacZ or Ad-Cre (MOI=10) and analyzed 48h later. Cells with a Klf6 knockdown show an increase in COL (I).
Because Klf6 depletion in Klf6 +/− mice is not restricted to stellate cells, we sought to exclude the possibility that allelic loss of Klf6 in hepatocytes might contribute to the observed phenotype, even though the extent of injury based on serum AST and ALT levels was not affected by Klf6 heterozygosity. To do so, we examined the impact of chronic CCl4 on mice with hepatocyte-specific deletion of Klf6 generated by crossing mice with a floxed allele of (Klf6F/+) (15) to animals expressing Cre-recombinase driven by the albumin promoter. In these mice, chronic CCl4 did not alter fibrogenic gene expression or the extent of fibrosis as observed in the Klf6 +/− animals (data not shown). These findings implicate loss of KLF6 in a non-parenchymal cell(s) of the liver as the reason for the observed increase in fibrosis.
To more specifically characterize the impact of KLF6 depletion on stellate cells, mouse stellate cells were isolated from Klf6F/F mice and activated by primary culture on plastic for 7 days. The stellate cells were then infected with adenoviruses expressing either LacZ or Cre-recombinase, and the expression of COL(I) protein was assessed by Western. As predicted, Cre-mediated deletion of Klf6 in culture-activated stellate cells led to increased collagen expression (Fig. 2F).
Transgenic mice with over-expression of KLF6WT or KLF6SV1 in stellate cells have reduced fibrosis after injury
Despite numerous attempts to clone and characterize Klf6 splice forms in the mouse, we repeatedly detected only one Klf6 isoform by 5' / 3' flanking PCR. Therefore, we instead established mice expressing human isoforms as a relevant model by first establishing that the biologic activity of human splice variants in mouse stellate cells is identical to their known activity in human stellate cells. In doing so, we thereby validated the use of transgenes in which the human isoforms were expressed. In fact, the absence of multiple mouse Klf6 isoforms facilitated our ability to attribute biology directly to human isoforms rather than through their potential interactions with mouse isoforms.
Based on our initial hypothesis that injury might be stimulating a change in the splicing pattern of KLF6, we examined the impact of over-expressing of KLF6WT or KLF6SV1 on the fibrogenic activity of stellate cells. We employed the Colα2(I) enhancer promoter, which is active as early as 12h after injury (16). The KLF6WT or KLF6SV1 isoforms were cloned downstream of this promoter. After validating expression of the expression constructs in a mouse stellate cell line (JS1) (17) (Supplemental Fig. 2A), stellate cell specific transgenic mice were generated. At least two transgenic mouse lines were selected for efficient expression of the transgene by qRT-PCR (Supplemental Fig. 2B), and all experiments were validated in these two lines for each genotype. Because experimental findings were consistent between both mouse lines only results from a single line of Colα2(I)- KLF6WT and Colα2(I)- KLF6SV1 mice are included.
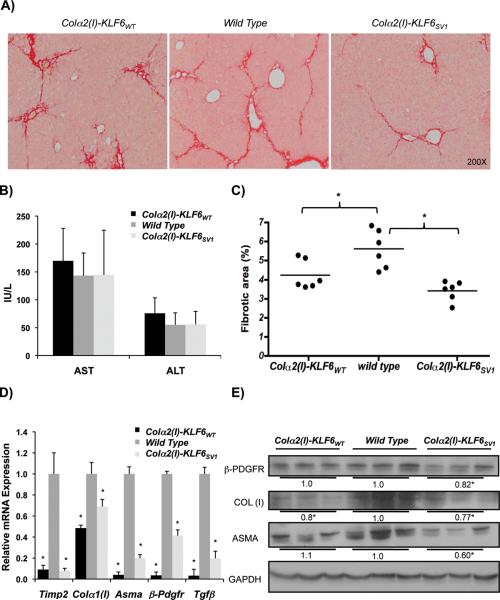
The loss-of-function studies in Klf6 +/− mice suggested that Klf6 is anti-fibrotic in stellate cells, and its depletion augments their fibrogenic activity. Therefore, we next examined whether over-expression of KLF6WT in stellate cells would yield the opposite result, namely decreased fibrosis. Based on our characterization of the endogenous rat isoforms, as well as results from the chronic CCl4 experiments in the Klf6 +/− mice, we anticipated decreased fibrosis in the Colα2(I)-KLF6WT mice. As predicted, increased KLF6WT expression in stellate cells reduced the amount of fibrosis in mouse livers following chronic CCl4 injury (Fig. 3A&C). Moreover, we anticipated the opposite phenotype in Colα2(I)-KLF6SV1 transgenic mice, since in human cancer, KLF6SV1 antagonizes the activity of KLF6WT. Surprisingly, these Colα2(I)-KLF6SV1 mice also had dramatically decreased fibrosis, similar to Colα2(I)-KLF6WT transgenic mice (Fig. 3A&C). Injury levels assessed by AST and ALT were similar between the three groups (Fig. 3B). The reduced fibrosis based on morphometry was correlated with decreased expression of fibrogenic mRNAs and proteins in whole liver by real-time PCR (Asma, Colα1 (I), Timp2, β-Pdgfr, and Tgf-β) (Fig. 3D) and Western blot (Fig. 3E).
Figure 3. Stellate cell specific KLF6WT and KLF6SV1 transgenic mice have decreased fibrosis following CCl4 liver injury.
A&C. Decreased fibrosis based on Sirius Red morphometry in Colα2(I)-KLF6WT and Colα2(I)-KLF6SV1 mice following 8 weeks of chronic CCl4 compared to wild type littermates (n=6/group) (4.2% and 3.4% respectively compared to 5.6%, p-value<0.01)(For each animal, 40 images were taken at low magnification (10X objective lens), and were subsequently used for quantifying percentage of fibrotic tissue using BioQuant Software); B. Levels of AST and ALT were comparable between WT and transgenic mice after chronic CCl4 injury; D. Decreased HSC-derived fibrogenic mRNAs in whole liver from Colα2(I)-KLF6WT and Colα2(I)-KLF6SV1 mice compared to wild type littermates (p-value<0.01); E. Parallel decrease in fibrogenic proteins COL (I), β-PDGFR, and ASMA for Colα2(I)-KLF6SV1 mice and a decrease in COL (I) for Colα2(I)-KLF6WT mice based on western blot of whole liver lysates from 3 representative mice/group after chronic CCl4.
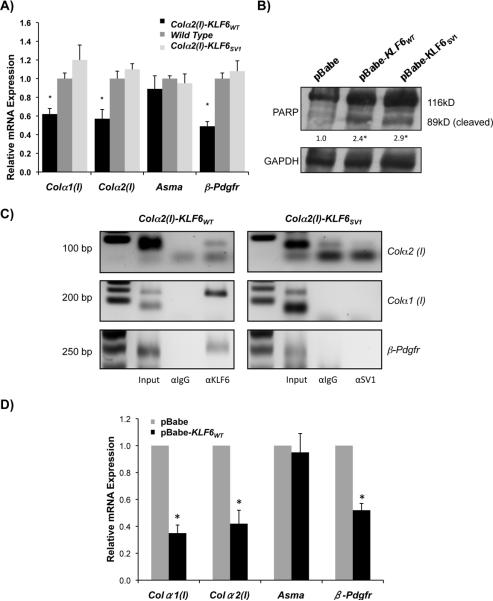
The reduced fibrosis in mice over-expressing KLF6WT or KLF6SV1 in stellate cells could result from a decrease in total number of stellate cells following injury, a decrease in fibrogenic potential of each cell, or a combination of the two. In order to address these possibilities, we examined the fibrogenic activity of stellate cells isolated from both Colα2(I)-KLF6WT and Colα2(I)-KLF6SV1 mice following acute CCl4 administration. Whereas stellate cells isolated from the Colα2(I)-KLF6WT mice expressed significantly less Colα1 (I), Colα2 (I), and β-Pdgfr, fibrogenic gene expression was not altered in cells from the Colα2(I)-KLF6SV1 animals (Fig. 4A). In order to further clarify this issue, we performed ASMA immunohistochemistry on liver sections of transgenic and wild type mice following chronic CCl4 administration. Interestingly, there was a decrease in the number of ASMA positive cells in both the Colα2(I)-KLF6WT and Colα2(I)-KLF6SV1 livers (Supplemental Fig. 3A & B) consistent with the possibility that the decreased fibrosis might reflect decreased numbers of activated cells in the presence of either KLF6WT or KLF6SV1.
Figure 4. KLF6WT transcriptionally inhibits Colα1(I),Colα2(I), and β-Pdgfr; both KLF6 isoforms lead to increased apoptosis of stellate cells in vitro.
A. Mouse stellate cells were isolated from Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1, and wild type mice (5 mice each) following 3 doses of CCl4. A decrease in fibrogenic gene expression in Colα2(I)-KLF6WT, but not Colα2(I)-KLF6SV1 transgenic stellate cells is evident in fresh HSC isolates; B. Increased apoptosis in human stellate cell lines (LX2) following stable transfection of pBabe-KLF6WT, or pBabe-KLF6SV1, based on the presence of cleaved form of PARP; C. Immortalized mouse stellate cells (JS1) transfected with Colα2(I)-KLF6WT , or Colα2(I)-KLF6SV1 constructs, and samples were prepared for chromatin IP 48h after transfection. Primer sets specific for GC boxes in promoter regions (2kb region upstream of start site) of Colα1(I), Colα2(I), and β-Pdgfr demonstrate binding of KLF6WT, but not KLF6SV1 to all three promoters; D. mouse stellate cells (JS1) over-expressing pBabe-KLF6WT show decreased transcript levels of Colα1(I), Colα2(I), and β-Pdgfr (p-value<0.05).
These findings suggested that transgenic stellate cells expressing KLF6WT or KLF6SV1 might be more prone to apoptosis. We therefore assessed the impact of KLF6WT or KLF6SV1 on apoptosis in a human HSC line (LX2) that was stably transfected with pBabe, pBabe-KLF6WT, or pBabe-KLF6SV1. Indeed, apoptosis was increased in stellate cells expressing pBabe-KLF6WT and pBabe-KLF6SV1 as indicated by increased PARP cleavage (Fig. 4B).
In order to confirm that KLF6WT was acting as a transcription factor that binds to GC boxes in the promoter regions of Colα1 (I), Colα2 (I), and β-PDGFR, we performed chromatin immunoprecipitation (CHIP). For this experiment, the JS1 mouse stellate cell line (JS1) was transfected with either Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1, or Colα2(I)-GFP constructs. Based on this analysis, KLF6WT but not KLF6SV1 bound to the promoters of Colα1 (I), Colα2 (I), and β-Pdgfr (Fig. 4C). The lack of chromatin binding by KLF6SV1 is consistent with its lack of a DNA-binding domain. To reinforce the conclusion that KLF6WT can repress endogenous promoters associated with stellate cell activation, JS1 cells were transfected with pBabe-KLF6WT or the pBabe control plasmid. Similar repression was seen in the levels of Colα1 (I), Colα2 (I), and β-Pdgfr but not Asma transcripts (Fig. 4D).
Discussion
Our data implicate KLF6WT as a transcription factor that preserves the quiescent phenotype of HSCs by inhibiting expression of fibrogenic genes, including Colα1 (I), Colα2 (I), and β-Pdgfr, as well as by inducing apoptosis of stellate cells. KLF6SV1 normally acts as a splice form that antagonizes KLF6WT. Interestingly, however, unlike transformed cancer cells where KLF6SV1 promotes survival (6, 18), in stellate cells KLF6SV1 promotes apoptosis. Moreover, because endogenous levels of KLF6 decrease with activation, there is diminishing KLF6WT for KLF6SV1 to antagonize.
Our initial hypothesis that KLF6 alternative splicing yields increased antagonistic splice forms that promote stellate cell activation is refuted by this data. In fact, there are parallel decreases in full-length KLF6 (rKlf6top), and the shorter KLF6 isoforms (rKlf6mid and rKlf6bot) with progressive activation. The parallel decreases in expression of all isoforms suggests that the pre-mRNA for Klf6 is reduced, resulting in similar changes for all splice isoforms, rather than due to a change in the splicing pattern.
Reduced Klf6 mRNA levels during activation at first seem to contradict our earlier finding of transient induction during early stellate cell activation (i.e., 6–12 hours after CCl4)(1). KLF6 may have a unique role and distinct transcriptional targets early in stellate cell activation, which are different from its activities later during the activation. However, the transgenic Colα2(I)-promoter is only expressed at least 12 h after injury, so that the contribution of Klf6 to very early stellate cell activation cannot be assessed. Indeed, the findings suggest that Klf6 may be solely anti-fibrogenic. The early, transient peak of Klf6 mRNA immediately following injury could serve to preserve cellular quiescence in order to prevent unwanted, premature activation of stellate cells. Thereafter, decreasing expression of Klf6 mRNA could enable cells to activate. This role of KLF6 as a potential `quiescence-preserving' gene is consistent with findings for another transcription factor, Lhx2, whose absence in development leads to spontaneous neonatal liver fibrosis (19).
The findings of reduced fibrosis in Klf6 +/− mice suggest that allelic loss of Klf6 in stellate cells might contribute to increased fibrosis. However, because heterozygosity is present in all cell types, the observed phenotype cannot be definitively ascribed directly to KLF6 loss in stellate cells. Moreover, Klf6 +/− mice displayed an increased inflammatory infiltrate in liver after CCl4, compared to wild type animals, raising the prospect that loss of KLF6 in the inflammatory cells might alter their response to injury even though AST and ALT levels after CCl4 were unaffected by Klf6 heterozygosity. A more specific way to establish that stellate cell specific loss of KLF6 leads to increased fibrosis would be to cross Klf6F/F mice with GFAP-Cre mice. However, this attempt resulted in a profound neurologic phenotype in mice, which precluded analysis of liver injury (data not shown). Studies using isolated stellate cells from Klf6F/F mice support the idea that loss of KLF6 in stellate cells is an important contributor to enhanced fibrosis.
Data from mice with transgenic expression of KLF6WT or KLF6SV1 is complementary and supports an antifibrotic role of this gene. Reduced fibrosis in mice over-expressing KLF6WT or KLF6SV1 in stellate cells reflects either a decrease in stellate cell numbers following injury, a decrease in fibrogenic potential of each cell, or the combination of the two. The experiments utilizing stable over-expression of KLF6WT or KLF6SV1 in hepatic stellate cells demonstrate an increase in apoptosis in both cases. Furthermore, although stellate cells isolated from the Colα2(I)-KLF6WT mice expressed significantly less Colα1 (I), Colα2 (I), and β-Pdgfr, fibrogenic gene expression was not altered in cells from the Colα2(I)-KLF6SV1 animals. KLF6WT but not KLF6SV1 interacts directly with `GC' boxes in the promoter regions of Colα1 (I), Colα2 (I), and β-Pdgfr. These data, combined with findings following the over-expression of KLF6WT in JS1 cells demonstrating transcriptional inhibition, suggest that KLF6WT directly represses these promoters. In short, the phenotype in the KLF6WT transgenic mice is due to both a decrease in fibrogenic potential, as well as decreasing stellate cell number, which is likely as a result of increased apoptosis.
KLF6 is a well studied transcription factor that can repress or activate target genes by interacting with co-regulator proteins such as HDACs (20). A recent paper implicates ligand-dependent corepressor (LCoR), in the regulation of KLF6 target genes (21), notably p21, and E-cadherin. Promoter bound KLF6 inhibits downstream genes by tethering a transcriptional co-repressor complex containing LCoR, with specific contributions by C-terminal binding protein (CtBP1), and HDACs. It would be informative to determine which co-regulator proteins interact with KLF6 during the inhibition of fibrogenic genes in quiescent stellate cells.
In summary, our findings clarify the role of KLF6 and its isoforms during hepatic stellate cell activation. Moreover, the parallel activities of both KLF6WT and its splice isoforms contrasts entirely with their behavior in human cancer (6, 7, 12, 18), reinforcing the conclusion that activities of splice isoforms may be highly context-dependent not only for KLF6, but possibly other genes that are alternatively spliced.
Supplementary Material
A. 293FT cells were transfected with pcDNA3.1/rKlf6top-V5, pcDNA3.1/rKlf6mid-V5, pcDNA3.1/rKlf6bot-V5, or pcDNA3.1/V5. 24 hours later cells were either treated with proteasomal inhibitor MG132 (10μM) or an equal volume of DMSO. Protein was harvested 5 hours after addition of MG132, and SDS-PAGE was performed followed by a western blot for V5. All 3 isoforms show degradation by the proteasomal pathway, as indicated by increasing protein in presence of MG132; B. 293FT cells were transfected as before with each of the three rat Klf6 expression vectors, and V5 immunofluorescence was performed 24h later. Confocal microscopy failed to show nuclear staining of rKLF6mid and rKLF6bot.
A. Mouse stellate cell line (JS1) was transfected with Colα2(I)-GFP, Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1 construct and protein was harvested 48h later for SDS-PAGE followed by KLF6 immunoblotting. The constructs expressed the appropriate KLF6 isoforms; B. Efficient induction (˜40 fold) of transgene 24h after a single injection of CCl4 in 2 transgenic lines from each of the Colα2(I)-KLF6WT mice Colα2(I)-KLF6SV1 mice (2 mice per line per treatment, data was normalized to wild type littermates receiving oil injection) (p-value<0.01).
A. Livers from Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1 and wild type mice following 8 weeks of chronic CCl4 were immunostained with ASMA as described. Transgenic mice over-expressing either isoform show reduced number of ASMA positive cells. B. Quantification of ASMA positive cells is shown. For each animal 30 images taken at high magnification (40× objective lens) were employed (p-value<0.01).
Acknowledgments
Grant support: MSTP Grant to ZGN, DK56621, DK47340 and P20AA017067 to SLF, Summer Student support from MSSM
Abbreviations
- KLF6
Kruppel like factor 6
- KLF6WT
KLF6 wild type
- KLF6SV1
KLF6 splice variant 1
- HSC
Hepatic stellate cell
REFERENCES
- 1.Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, et al. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botella LM, Sanchez-Elsner T, Sanz-Rodriguez F, Kojima S, Shimada J, Guerrero-Esteo M, Cooreman MP, et al. Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood. 2002;100:4001–4010. doi: 10.1182/blood.V100.12.4001. [DOI] [PubMed] [Google Scholar]
- 3.Kremer-Tal S, Narla G, Chen Y, Hod E, Difeo A, Yea S, Lee JS, et al. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645–654. doi: 10.1016/j.jhep.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 5.Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 6.Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, Hod E, et al. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, Kalir T, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006;12:3730–3739. doi: 10.1158/1078-0432.CCR-06-0054. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blomhoff R, Berg T. Isolation and cultivation of rat liver stellate cells. Methods Enzymol. 1990;190:58–71. doi: 10.1016/0076-6879(90)90009-p. [DOI] [PubMed] [Google Scholar]
- 10.Lalazar A, Wong L, Yamasaki G, Friedman SL. Early genes induced in hepatic stellate cells during wound healing. Gene. 1997;195:235–243. doi: 10.1016/s0378-1119(97)00159-5. [DOI] [PubMed] [Google Scholar]
- 11.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 12.Narla G, DiFeo A, Yao S, Banno A, Hod E, Reeves HL, Qiao RF, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 13.Banck MS, Beaven SW, Narla G, Walsh MJ, Friedman SL, Beutler AS. KLF6 degradation after apoptotic DNA damage. FEBS Lett. 2006;580:6981–6986. doi: 10.1016/j.febslet.2006.10.077. [DOI] [PubMed] [Google Scholar]
- 14.De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, Schwabe RF. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 15.Leow CC, Wang BE, Ross J, Chan SM, Zha J, Carano RA, Frantz G, et al. Prostate-specific Klf6 inactivation impairs anterior prostate branching morphogenesis through increased activation of the Shh pathway. J Biol Chem. 2009;284:21057–21065. doi: 10.1074/jbc.M109.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inagaki Y, Truter S, Bou-Gharios G, Garrett LA, de Crombrugghe B, Nemoto T, Greenwel P. Activation of Proalpha2(I) collagen promoter during hepatic fibrogenesis in transgenic mice. Biochem Biophys Res Commun. 1998;250:606–611. doi: 10.1006/bbrc.1998.9345. [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Loke J, Zheng F, Hong F, Yea S, Fukata M, Tarocchi M, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, Narla G. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68:965–970. doi: 10.1158/0008-5472.CAN-07-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wandzioch E, Kolterud A, Jacobsson M, Friedman SL, Carlsson L. Lhx2−/− mice develop liver fibrosis. Proc Natl Acad Sci U S A. 2004;101:16549–16554. doi: 10.1073/pnas.0404678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, et al. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- 21.Calderon MR, Verway M, An BS, DiFeo A, Bismar TA, Ann DK, Martignetti JA, et al. Ligand-dependent corepressor (LCoR) recruitment by Kruppel-like factor 6 (KLF6) regulates expression of the cyclin-dependent kinase inhibitor CDKN1A gene. J Biol Chem. 287:8662–8674. doi: 10.1074/jbc.M111.311605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. 293FT cells were transfected with pcDNA3.1/rKlf6top-V5, pcDNA3.1/rKlf6mid-V5, pcDNA3.1/rKlf6bot-V5, or pcDNA3.1/V5. 24 hours later cells were either treated with proteasomal inhibitor MG132 (10μM) or an equal volume of DMSO. Protein was harvested 5 hours after addition of MG132, and SDS-PAGE was performed followed by a western blot for V5. All 3 isoforms show degradation by the proteasomal pathway, as indicated by increasing protein in presence of MG132; B. 293FT cells were transfected as before with each of the three rat Klf6 expression vectors, and V5 immunofluorescence was performed 24h later. Confocal microscopy failed to show nuclear staining of rKLF6mid and rKLF6bot.
A. Mouse stellate cell line (JS1) was transfected with Colα2(I)-GFP, Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1 construct and protein was harvested 48h later for SDS-PAGE followed by KLF6 immunoblotting. The constructs expressed the appropriate KLF6 isoforms; B. Efficient induction (˜40 fold) of transgene 24h after a single injection of CCl4 in 2 transgenic lines from each of the Colα2(I)-KLF6WT mice Colα2(I)-KLF6SV1 mice (2 mice per line per treatment, data was normalized to wild type littermates receiving oil injection) (p-value<0.01).
A. Livers from Colα2(I)-KLF6WT, Colα2(I)-KLF6SV1 and wild type mice following 8 weeks of chronic CCl4 were immunostained with ASMA as described. Transgenic mice over-expressing either isoform show reduced number of ASMA positive cells. B. Quantification of ASMA positive cells is shown. For each animal 30 images taken at high magnification (40× objective lens) were employed (p-value<0.01).