Abstract
Innate immune sensing of viral nucleic acids triggers type I interferon (IFN) production, which activates interferon-stimulated genes (ISGs) and directs a multifaceted antiviral response. ISGs can also be activated through IFN-independent pathways, although the precise mechanisms remain elusive. Here we found that the cytosolic exonuclease Trex1 regulates the activation of a subset of ISGs independently of IFN. Both Trex1−/− mouse and TREX1-mutant human cells express high levels of antiviral genes and are refractory to viral infections. The IFN-independent activation of antiviral genes in Trex1−/− cells requires STING, TBK1 and IRF3 and IRF7. We also found that Trex1-deficient cells display expanded lysosomal compartment, altered subcellular localization of the transcription factor EB (TFEB), and reduced mTORC1 activity. Together, our data identify Trex1 as a regulator of lysosomal biogenesis and IFN-independent activation of antiviral genes, and shows dysregulation of lysosomes can elicit innate immune responses.
Vertebrates are constantly facing challenges from pathogenic microbes that introduce a variety of microbial proteins and nucleic acids into the host cell. To counter this, eukaryotic cells express many different pattern-recognition receptors (PRRs) that detect microbial pathogen-associated molecular patterns (PAMP), which then activate antiviral interferon (IFN) and proinflammatory responses1. Mammalian PRRs include Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), and an emerging category of cytosolic DNA receptors2. In the case of viral infection, viral nucleic acids are the major PAMP detected by the host innate immune receptors, which include RLRs and DNA receptors in the cytosol and a subfamily of TLRs that localize to the endosomal membrane3. The central hub for cytosolic DNA sensing is the endoplasmic reticulum (ER) membrane protein STING (also known as MITA, MPYS or ERIS)2. A number of proteins have been proposed to detect double-stranded DNA in the cytosol and signal through STING, such as DAI, IFI16 and DDX414–6. STING can also directly recognize c-di-GMP, which is usually associated with bacterial infection and activates IFN expression7. Although IFN plays a major role in controlling viral infections, IFN-independent pathways also exist and are vital for antiviral defense. For example, STING can activate STAT6 independently of the IFN pathway; and activated STAT6 induces chemokine expression that primes adaptive immune responses8. Infection with enveloped viruses also triggers an IFN-independent pathway that involves the direct activation of a subset of IFN stimulatory genes (ISG) by IRF39. A recent study of MAVS-mediated innate immune responses to RNA viruses demonstrated that IFN-independent induction of antiviral genes occurs rapidly after infection and is functionally important for controlling viral replication before the onset of more robust and sustained IFN activation10.
Innate immune sensing pathways are carefully designed to distinguish self- versus non-self ligands, either by spatial separation (e.g. TLR7 and TLR9 reside in endosomes which are devoid of host nucleic acids), or by stringent ligand specificity (e.g. TLR9 recognizes CpG-containing DNA in bacteria; RIG-I recognizes 5′-ppp-containing RNA in viruses). However, how cytosolic DNA sensing pathways distinguish host and viral DNA remains unclear. We have previously identified Trex1, an exonuclease that resides on the ER, as a negative regulator of innate immune sensing of cytosolic HIV DNA. In Trex1−/− mouse embryonic fibroblasts or human CD4+ T cells and macrophages in which Trex1 was depleted by RNAi, cytosolic HIV DNA accumulated and triggers IFN through the STING-TBK1-IRF3 pathway11. These findings and other studies12 suggest that cells rely on negative regulatory mechanisms such as Trex1 to keep cytosolic DNA sensing pathways in check.
Trex1 deficiency has been implicated in the pathogenesis of autoimmunity. TREX1 mutations in humans are associated with a spectrum of autoimmune and inflammatory phenotypes including Aicardi-Goutières syndrome (AGS, an inflammatory brain disease that mimics the symptoms of congenital viral infection 13,14), systemic lupus erythematosus (SLE), familial chilbain lupus (FCL) and retinal vasculopathy with cerebral leukodystrophy (RVCL)15–17. TREX1 mutations were found in up to 2% of SLE patients with an extremely high odds ratio (OR=25)18, representing one of the highest disease risk recorded for a single susceptibility gene in complex polygenic SLE14. Studies using Trex1−/− mice revealed that Trex1−/− cells accumulate cytosolic ssDNA that might be derived from DNA repair in the nucleus or from endogenous retroelements19,20. Recent genetic evidence demonstrated that the STING-mediated DNA sensing pathway is essential for the pathogenesis of autoimmune disease in Trex1−/− mice12. Initiation of IFN expression is only detected in a subset of non-hematopotic cells in Trex1−/− mice, raising the question of what happens to the majority of other cells that also lack Trex1 function. We also wondered whether Trex1 inhibits IFN responses to other viruses besides HIV, and/or if the mere loss of Trex1 function in a cell would elicit innate immune responses and establish an antiviral state?
In this study, we found that Trex1-deficient or mutant cells display broad antiviral activity against many RNA viruses. The antiviral activity comes from elevated expression of ISGs in cells that lack Trex1 function, and is mediated through an IFN-independent signaling pathway that involves STING-TBK1-IRF3-IRF7. We also found that Trex1 regulates lysosomal biogenesis through TFEB and mTORC1 pathway, and provided evidence that dysregulation of lysosomes elicits innate immune response.
RESULTS
Impaired VSV replication in Trex1 deficient cells
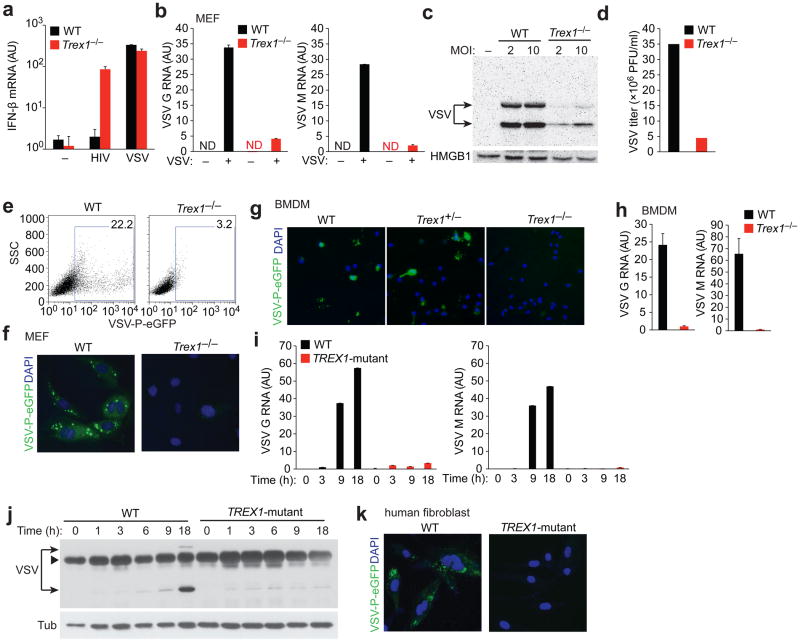
To investigate whether Trex1 is involved in the IFN response to RNA viruses, we infected wild-type (WT) and Trex1−/− MEFs with vesicular stomatitis virus (VSV, Indiana strain), a negative stranded RNA virus, with VSV-G pseudotyped HIV11, or with a mock infection, and measured levels of IFN-β mRNA 24 h post infection (hpi). As previously reported11, mock-infected WT and Trex1−/− cells did not express detectable levels of IFN-β mRNA, and HIV infection only stimulated IFN-β mRNA expression in Trex1−/− cells, but not in WT cells. In contrast, VSV infection stimulated strong IFN-β mRNA expression in both WT and Trex1−/− cells at similar levels (Fig. 1a), suggesting that Trex1 does not regulate the Type I IFN response to VSV. However, VSV replication was severely impaired in Trex1−/− cells compared to WT, even though IFN-β induction was indistinguishable between the two cell types (Fig. 1b–d). Specifically, mRNA levels of two major forms of VSV RNA, G and M, were reduced to 12% and 7% (of WT), respectively, in Trex1−/− as compared to in WT cells (Fig. 1b). We also detected markedly reduced amounts of VSV proteins in Trex1−/− as compared to in WT cells, using two different multiplicities of infection (MOI, 2 and 10) (Fig. 1c). VSV titers from infected Trex1−/− cells were also reduced compared to WT (Fig. 1d). To better quantify and visualize VSV replication, we infected WT and Trex1−/− cells with VSV-PeGFP, in which eGFP was fused in-frame to the VSV P protein that is usually associated with viral RNA replication foci in the cell21. We observed reduced VSV-PeGFP replication (14% of WT) in Trex1−/− cells compared to WT cells by fluorescence-activated cell sorting (FACS) analysis (Fig. 1e).
Figure 1.
VSV replication is impaired in Trex1 deficient cells. (a) Quantitative RT-PCR analysis of IFN-β mRNA in wild type (WT, black bars) and Trex1−/− MEFs (red bars) infected with VSV-G pseudotyped HIV-GFP11 or with VSV at an MOI of 2 for 24 h. AU, arbitrary units. ND, not detectable (b–c) Quantitative RT-PCR analysis of VSV G and M RNA (b), western blot analysis of VSV proteins (c) and virus titers in the supernatants (d) of WT and Trex1−/− MEFs mock-infected or infected with VSV for 18 h. (e, f) Fluorescence activated cell sorting (FACS) (e) and fluorescent microscopic (f) analysis of WT and Trex1−/− MEFs infected with VSV-PeGFP21 for 18 h. (g, h) fluorescent microscopic (g) and quantitative RT-PCR analysis of VSV G and M RNA (h) in WT, Trex1+/− and Trex1−/− MEFs infected with VSV-PeGFP (g) or VSV (h) for 18 h. (i, j) Quantitative RT-PCR analysis of VSV G and M RNA (i) and western blot analysis of VSV proteins (j) in WT and TREX1R114H/R114H primary human skin fibroblasts (TREX1-mutant, isolated from a healthy donor or from an AGS patient, respectively) at varying times post infection (0–18 hpi). Arrowhead, a non-specific band. (k) Fluorescent microscopic analysis of WT and TREX1R114H/R114H cells infected with VSV-PeGFP for 18 h. Data are representative of at least three independent experiments (error bars, s.d.).
Consistent with our FACS data, fluorescent microscopy analysis of infected WT cells revealed bright replication foci marked by PeGFP, whereas very little green fluorescent signal was detected in VSV-PeGFP infected Trex1−/− cells (Fig. 1f). We also infected bone marrow-derived macrophages (BMDMs) generated from WT, Trex1+/− and Trex1−/− mice and found that only Trex1−/− cells were resistant to VSV infection (Fig. 1g, h).
We next examined whether VSV entry is inhibited in Trex1−/− cells. This seemed unlikely as Trex1−/− cells did not inhibit entry of VSV-G pseudotyped HIV11, and VSV infection stimulated indistinguishable levels of IFN mRNA expression in WT and Trex1−/− cells (Fig. 1a). Nonetheless, to rule out the possibility of an entry defect, we labeled wild type VSV virions with a fluorescent dye DiL and followed the infection of VSV-DiL in WT and Trex1−/− cells by live cell fluorescence microscopy (Supplementary Fig. 1a). We observed no differences in intracellular VSV-DiL comparing WT and Trex1−/− cells at 1 hpi. We also observed similar levels of VSV G and M RNA at 1 hpi in both cell types (Supplementary Fig. 1b). These data suggest that VSV replication was blocked at an early stage post entry, such as uncoating or RNA replication, in Trex1−/− cells. We also found that in contrast to infected WT cells, VSV infected Trex1−/− cells did not show detectable cytopathic effects (Supplementary Fig. 2), consistent with the notion that Trex1−/− cells were protected against viral infection.
To investigate whether Trex1 is also required for VSV replication in human cells, we infected WT and TREX1R114H/R114H (TREX1-mutant) skin fibroblasts from an AGS patient with VSV or VSV-PeGFP and measured levels of viral RNA. Arginine 114 is a critical residue at the interface of the Trex1 dimer, and the R114H mutation severely disrupts Trex1 function in vitro22. R114H represents the most common Trex1 mutation in patients with AGS, and has also been associated with SLE14. Both VSV and VSV-PeGFP infection was decreased in TREX1R114H/R114H cells compared to WT cells, as reflected by reduced levels of viral RNA, reduced amounts of viral proteins, and reduced numbers of viral replication foci (Fig. 1i–k). Taken together, we conclude that VSV replication is impaired at an early post entry step in both mouse and human cells lacking Trex1 function.
Trex1 deficient cells display broad antiviral resistance
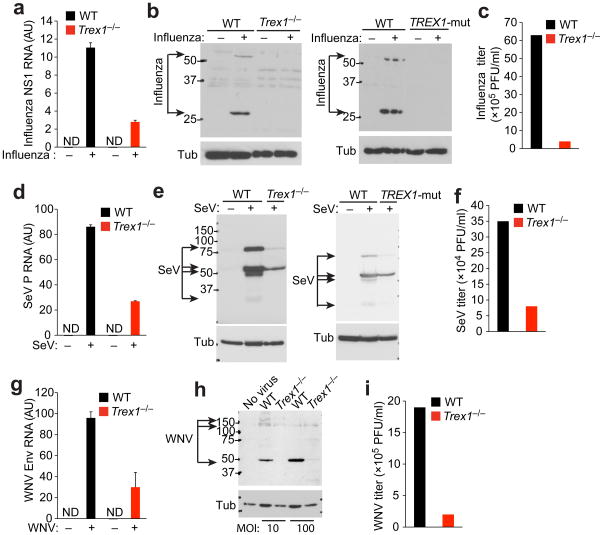
To determine whether the replication block in Trex1−/− and TREX1R114H/R114H cells was unique for VSV, we infected WT and Trex1−/− MEFs or WT and TREX1R114H/R114H human fibroblasts with three additional RNA viruses that contain either positive- or negative-stranded genomes, and measured levels of viral RNA, amounts of viral proteins, and viral titers in the supernatant. All three viruses, namely influenza virus (A/WSN/1933 strain), Sendai virus (SeV) and West Nile virus (WNV/TX02 strain), failed to replicate efficiently in Trex1−/− or TREX1R114H/R114H cells compared to WT cells (Fig. 2). These results demonstrate that cells lacking Trex1 function are resistant to infection with several different types of RNA viruses.
Figure 2.
Trex1 deficient cells display broad antiviral resistance. (a–c) Quantitative RT-PCR analysis of influenza NS1 RNA (a), western blot analysis of influenza proteins (b) and viral titers in the supernatants (c) of WT and Trex1−/− MEFs or WT and TREX1R114H/R114H (TREX1-mut) human fibroblasts infected with influenza virus (A/WSN/1933 strain) at an MOI of 1. AU, arbitrary units. ND, not detectable. (d–f) Quantitative RT-PCR analysis of Sendai virus P RNA (d), western blot analysis of Sendai virus proteins (e) and viral titers in the supernatants (f) of WT and Trex1−/− MEFs or WT and TREX1R114H/R114H human fibroblasts infected with Sendai virus at MOI of 10. (g–i) Quantitative RT-PCR analysis of West Nile virus Env RNA (g), western blot analysis of West Nile virus proteins (h) and viral titers in the supernatants (i) of WT and Trex1−/− MEFs infected with West Nile virus (WNV/TX02 strain) at an MOI of 10 or 100, as indicated. Data are representative of at least two independent experiments (error bars, s.d.).
Trex1 regulates IFN-independent activation of ISGs
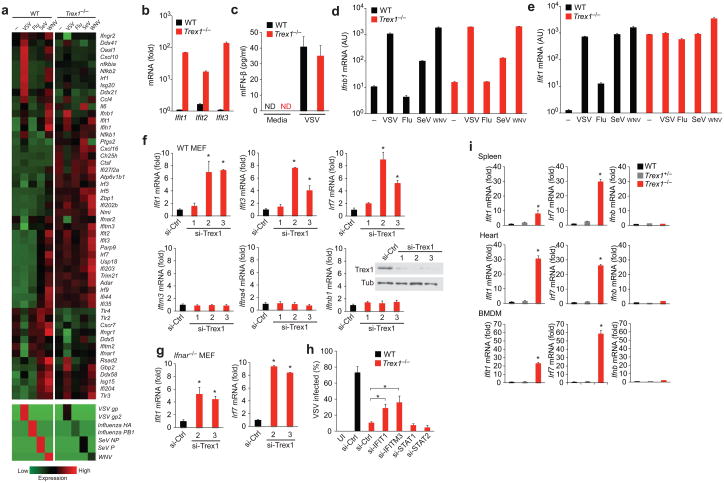
Next, we investigated the mechanism of antiviral resistance in Trex1−/− cells. We first examined gene expression profiles by isolating total RNA from WT or Trex1−/− MEFs that were mock-infected or infected with VSV, influenza virus, Sendai virus or West Nile virus, and performed RNA-SEQ analysis, which offers quantitative measurement of both host and viral RNAs simultaneously (Fig. 3a). Gene expression fold-change values validated by qPCR were remarkably similar to those from RNA-SEQ (Fig. 3b, Supplementary Fig. 3, data not shown), which underscores the quantitative power of our RNA-SEQ analysis. We first analyzed gene expression profiles of uninfected WT and Trex1−/− samples by Ingenuity Pathway Analysis (IPA) and found that the most enriched gene network in Trex1−/− cells compared to WT cells is ‘antimicrobial response, inflammatory response, infectious diseases’ consisting mostly of ISGs (Supplementary Fig. 5a,b). ‘Interferon signaling’ and ‘cytosolic pattern recognition receptors’ are also among top ranked canonical pathways with high ‘hit ratio’ (determined by the percentage of genes in a pathway that are represented in a dataset, Supplementary Fig. 5c). We then constructed a heatmap of genes involved in ‘antimicrobioal response’ network based on expression values and standard deviations of each gene across all samples (Fig. 3a). All four RNA viruses replicated less efficiently in Trex1−/− cells (Fig. 3a lower panel, Supplementary Fig. 4), with values ranging from 4–22% of those observed in WT cells. We also found that many ISGs, such as Ifit1, Ifit3 Isg15, Zbp1, and Usp18, were highly induced in uninfected Trex1−/− cells (Fig. 3b, Supplementary Fig. 3). Remarkably, uninfected Trex1−/− cells display an ISG activation signature that resembled infected WT cells (Supplementary Fig. 6), suggesting that the lack of Trex1 function alone is enough to initiate an antiviral state. The establishment of this antiviral state appeared to be independent of IFN, because we did not detect any activation of IFN genes or IFN proteins (Fig. 3c, Supplementary Fig. 3). Moreover, Ifnb mRNA induction patterns by different viruses were indistinguishable between WT and Trex1−/− cells (Fig. 3d; influenza virus is known to inhibit IFN activation23). In contrast, Ifit1 mRNA was low in WT cells and increased after viral infection, whereas Trex1−/− cells started with very high Ifit1 (that appeared to be at a level that is equivalent to that observed in infected WT cells), and it remained high after viral infection (Fig. 3e). Trex1−/− cells treated with increasing dose of recombinant IFN-β showed further increase of Ifit1 expression, suggesting that Trex1−/− cells are capable of responding to IFN signaling (Supplementary Fig. 7). ISGs that were highly induced by Trex1 deficiency such as IFIT family members have intrinsic antiviral activity against RNA viruses24,25. Of note, not all known ISGs are activated in Trex1−/− cells; IFITM family members were expressed at similar amounts in WT and Trex1−/− cells (Supplementary Fig. 3). Together, our data suggested that a subset of ISGs are activated at very high levels in Trex1−/− cells independently of the IFN response.
Figure 3.
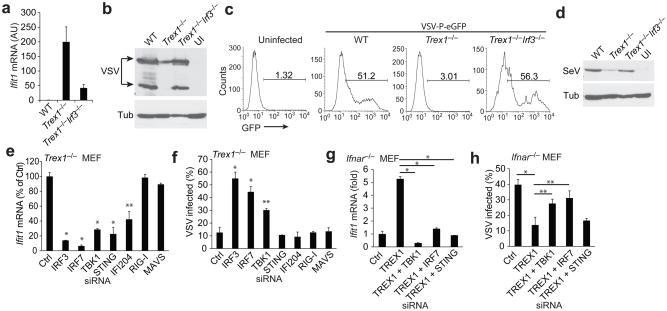
IFN-independent activation of a subset of ISGs in Trex1 deficient cells. (a) A heatmap of selected RNA expression values measured by RNA-SEQ analysis from WT and Trex1−/− MEFs uninfected or infected with viruses indicated on top for 18 h. (b) Quantitative RT-PCR validation of IFIT family gene expression in uninfected WT and Trex1−/− MEFs. (c) Mouse IFN-β protein measured by ELISA in supernatants from WT and Trex1−/− MEFs uninfected (‘Media’) or infected with VSV. ND, not detectable. (d, e) Quantitative RT-PCR validation of Ifnb1 (d) and Ifit1 (e) mRNA levels in WT and Trex1−/− MEFs mock-infected or infected with indicated virus. AU, arbitrary units. (f) Quantitative RT-PCR analysis of selected ISGs and IFN genes in WT MEFs 72 h after transfection with a control siRNA or Trex1-specific siRNAs. si-Ctrl was normalized to 1 in all panels. *P < 0.05 (Student’s t-test). Data are representative of three independent experiments (error bars, s.d.). Insert shows western blot analysis of Trex1 knockdown. (g) Quantitative RT-PCR analysis of Ifit1 in Ifnar−/− MEFs 72 h after transfection with a control siRNA or Trex1-specific siRNAs. si-Ctrl was normalized to 1. *P < 0.05 (Student’s t-test). Data are representative of three independent experiments (error bars, s.d.). (h) FACS analysis of VSV-PeGFP replication in WT and Trex1−/− MEFs transfected with indicated siRNA. Cells were transfected with siRNA for 48 h and mock-infected or infected with VSV-PeGFP for 18 h before FACS analysis. Percentages of GFP positive cells are shown. *P < 0.05 (Student’s t-test). Data are representative of two independent experiments (error bars, s.d.). (i) Quantitative RT-PCR analysis of Ifit1, Irf7 and Ifnb1 mRNA in spleen, heart and BMDM isolated from WT, Trex1+/− and Trex1−/− mice. *P < 0.05 (Student’s t-test). Data are representative of two independent experiments (error bars, s.d.).
To further confirm that the ISG activation is specific to the loss of Trex1 function, we knocked down Trex1 expression in WT MEFs using three different siRNAs and observed that the expression of Ifit1, Ifit3 and Irf7 (also an ISG) were increased significantly while the expression of Ifitm3, Ifna4 and Ifnb1 were not increased (Fig. 3f). We also knocked down Trex1 expression in Ifnar−/− MEFs and observed a similar increase in Ifit1 and Irf7 expression (Fig. 3g), further suggesting that the ISG activation regulated by Trex1 is IFN-independent. To determine whether the ISG activation or the IFN pathway contributed to viral infection control in Trex1−/− cells, we transfected WT and Trex1−/− cells with a control siRNA or a specific siRNA against two ISGs (IFIT1 and IFITM3) or two key components of the IFN signaling pathway (STAT1 and STAT2). We then infected cells with VSV-PeGFP and measured infectivity by FACS (Fig. 3h). The knockdown of IFIT1 and IFITM3 in Trex1−/− cells partially alleviated the block in VSV replication, consistent with their known antiviral functions24–26. In contrast, STAT1 or STAT2 knockdown had no effect on VSV replication, further demonstrating that the IFN response is not required for control of viral infection in Trex1−/− cells. To determine whether this ISG-induction signature occurs in primary immune cells and tissues from Trex1−/− mice, we isolated total RNA from whole spleen, heart and BMDMs from WT, Trex1+/− and Trex1−/− mice and measured Ifit1, Irf7 and Ifnb mRNA levels. We observed up to 30-fold induction of ISGs in whole tissues and up to 60-fold induction of ISGs in primary immune cells only in Trex1−/− mice compared to WT mice (Fig. 3i). We also observed very low levels of Ifnb expression in all samples from Trex1−/− mice (Fig. 3i), consistent with a previous report where IFN expression was restricted to a subset of heart muscle cells12.
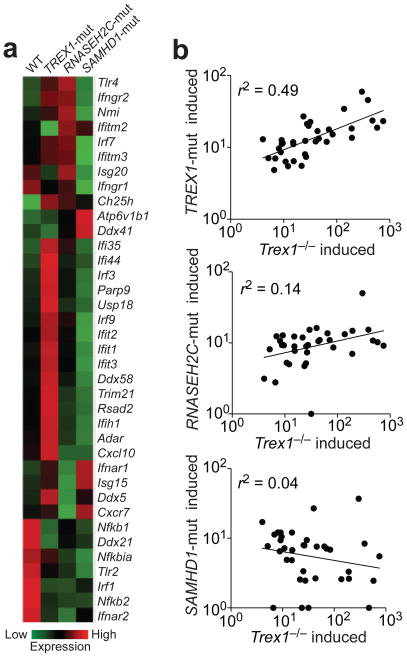
We also performed RNA-SEQ to analyze total RNA from uninfected WT or AGS patient fibroblasts carrying mutations in TREX1 (R114H/R114H), or AGS causing genes including RNASEH2C (D39Y/D115fs) or SAMHD1 (R290H/Q548X)27. We again found strong up-regulation of a subset of ISGs, but not IFN genes, in TREX1R114H/R114H cells (Fig. 4a). Interestingly, the ISG activation signature was weak in RNASEH2C mutant cells and not present in SAMHD1 mutant cells (Fig. 4a). To determine whether the same group of ISGs are activated in Trex1−/− and TREX1R114H/R114H cells, we selected 35 ISGs that are expressed in both mouse and human cells, and compared their induction in Trex1−/− versus TREX1R114H/R114H cells. ISGs that were induced in Trex1−/− MEFs were also induced in TREX1R114H/R114H fibroblasts, with a correlation r-squared value of 0.49 (Fig. 4b). We observed a weak correlation between gene induction in Trex1−/− versus RNASEH2CD39Y/D115fscells (r2=0.14) and no correlation in Trex1−/− versus SAMHD1R290H/Q548X cells (r2=0.04). Our data demonstrated that Trex1 also regulates ISG activation in human fibroblasts.
Figure 4.
Selective ISG activation in TREX1 mutant fibroblasts. (a) A heatmap of selected RNA expression values measured by RNA-SEQ analysis from skin fibroblasts isolated from a healthy donor (WT) or from AGS patients carrying mutations in TREX1 (R114H/R114H), RNASEH2C (D39Y/D115fs) or SAMHD1 (R290H/Q548X). (b) Correlation analysis of ISGs induced by Trex1 deficiency in MEFs versus AGS mutations in humans. Thirty-five ISGs that were expressed in both human and mouse cells were selected for the dot plot. Each dot represents a gene: the x-axis value is fold-increase in Trex1−/−MEFs compared to WT, and the y-axis value is fold-increase in AGS mutant cells compared to WT. r-squared values represent the quality of correlation observed by fitting a power trend line through all data points.
IFN-independent ISG activation requires STING, TBK1, IRF3 and IRF7
We next wanted to identify innate immune factors that are required for IFN-independent ISG activation in Trex1− deficient cells. We chose to measure Ifit1 mRNA as an example of Trex1-regulated ISGs because it is the most highly up-regulated mRNA by Trex1 deficiency. We first examined IRF3, which activates ISG directly9,28. We measured Ifit1 mRNA in WT, Trex1−/− and Trex1−/−Irf3−/− MEFs, and found that Ifit1 is induced in Trex1−/− single knockout cells and the induction was inhibited by Trex1−/−Irf3−/− double knockout, suggesting that IRF3 is required for Ifit1 activation (Fig. 5a). To determine whether IRF3 is also required for antiviral activity in the setting of Trex1 deficiency, we infected WT, Trex1−/− and Trex1−/−Irf3−/− MEFs with VSV or SeV, and measured viral proteins by western blot or FACS analysis (Fig. 5b, c, d). Both VSV and SeV infections were inhibited in Trex1−/− cells, and both were restored to close to wild-type levels in Trex1−/−Irf3−/− cells. Therefore, IRF3 is a key component of antiviral resistance in Trex1−/− cells. We next wanted to explore which innate immune pathway upstream of IRF3 is involved. IRF3 is activated mostly by cytosolic DNA or RNA sensing pathways that are mediated by STING-TBK1 or RIG-I-MAVS, respectively. Therefore, we knocked down key components of each pathway by siRNAs in Trex1−/− cells, and measured Ifit1 expression. The knockdown of IRF3, IRF7, TBK1, STING, IFI204 significantly reduced Ifit1 expression, whereas RIG-I and MAVS knockdown had no effect (Fig. 5e). We also did not observe any effect on Ifit1 expression in Trex1−/− cells by knocking down TLR7 or TLR9 (data not shown). IRF3, IRF7 and TBK1 knockdown also decreased the VSV replication block in Trex1−/− cells (Fig. 5f). These results suggest that the cytosolic DNA, but not RNA, sensing machinery, is required for IFN-independent ISG activation in Trex1-deficient cells. STING knockdown did not appear to increase VSV replication in Trex1−/− cells, likely because the VSV replication assay measures the entire life cycle of VSV and many host factors may contribute to it, or because STING also regulates many other genes8 that could be required for VSV replication. The same cytosolic DNA sensing pathway is also involved in the activation of IFN genes during viral infection2,11, which can then activate ISGs. Therefore, we performed double knockdowns of Trex1 plus components of the cytosolic DNA sensing pathway in Ifnar−/− MEFs. Ifit1 expression was increased by Trex1 knockdown, and this increase was reduced by further knockdown of TBK1, IRF7 OR STING (Fig. 5g). Trex1 knockdown in Ifnar−/− MEFs also inhibited VSV replication, and further knockdown of TBK1 or IRF7 alleviated that inhibition (Fig. 5h). Taken together, our data suggest that the core cytosolic DNA sensing machinery, STING-TBK1-IRF3-IRF7, is involved in activating ISGs directly in cells with reduced or no Trex1 activity.
Figure 5.
IFN-independent ISG activation in Trex1 deficient cells requires STING, TBK1, IRF3 and IRF7. (a) Quantitative RT-PCR analysis of Ifit1 in uninfected WT, Trex1−/− and Trex1−/−Irf3−/− MEFs. AU, arbitrary units. (b) Western blot analysis of VSV proteins in WT, Trex1−/− and Trex1−/−Irf3−/− MEFs infected with VSV for 18 h. UI, uninfected. (c) FACS analysis of VSV replication in WT, Trex1−/− and Trex1−/−Irf3−/− MEFs infected with VSV-PeGFP for 18 h. (d) Western blot analysis of Sendai virus proteins in WT, Trex1−/− and Trex1−/−Irf3−/− MEFs infected with Sendai virus for 18 h. (e,f) Quantitative RT-PCR analysis of Ifit1 in uninfected (e), or FACS analysis of VSV-PeGFP infected (f) Trex1−/− MEFs transfected with indicated siRNA. Trex1−/− MEFs were transfected with indicated siRNAs for 72 h and RNA was extracted for qRT-PCR analysis (e) or infected with VSV-PeGFP for 18 h and analyzed by FACS (f). *P< 0.01, **P< 0.05 (Student’s t-test). Data are representative of two independent experiments (error bars, s.d.). (g,h) Quantitative RT-PCR analysis of Ifit1 in uninfected (g), or FACS analysis of VSV-PeGFP infected (h) Ifnar−/− MEFs transfected with one or two siRNAs as indicated. *P< 0.01, **P< 0.05 (Student’s t-test). Data are representative of two independent experiments (error bars, s.d.).
Trex1 regulates lysosomal biogenesis through TFEB and mTORC1
We next wanted to identify the underlying basis for ISG activation in Trex1−/− or TREX1R114H/R114H cells. We first considered the possibility that Trex1 directly inhibits the cytosolic DNA sensing machinery. To test this, we used 293T cells, in which the overexpression of STING induced a 6-fold increase in levels of Ifit1 mRNA (Supplementary Fig. 8). We then co-expressed STING and increasing amounts of Trex1 to examine whether Trex1 overexpression inhibits STING-mediated IFIT1 activation. We did not observe any effect on Ifit1 induction by overexpressing Trex1 (Supplementary Fig. 8). The same level of Trex1 overexpression inhibited HIV-mediated activation of IFN genes11. These results suggest that Trex1 does not directly inhibit the cytosolic DNA sensing machinery.
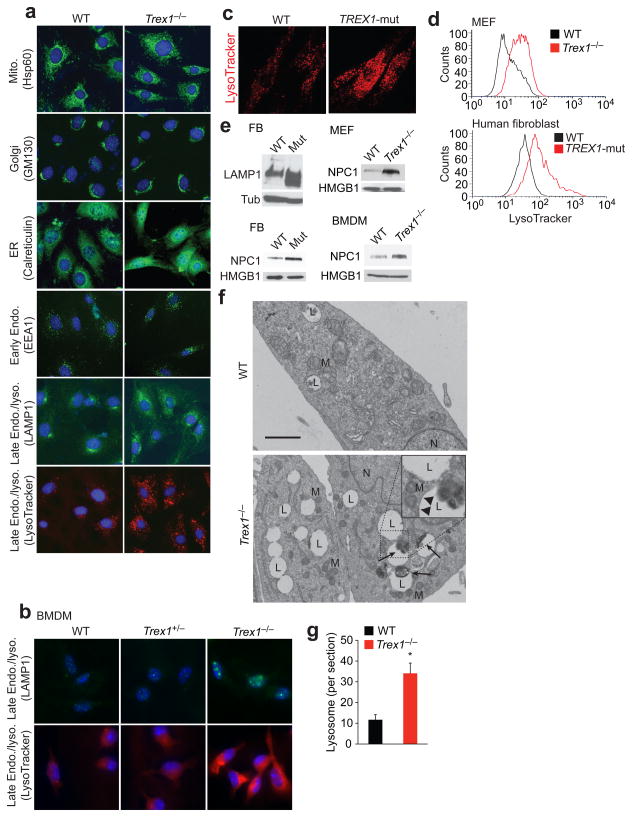
We then hypothesized that perhaps accumulation of self-ligands or any cellular abnormality in Trex1−/− or TREX1R114H/R114H cells could be detected by the STING-TBK1-IRF3-IRF7 pathway. We first looked at the abundance and morphology of cellular organelles, comparing WT and Trex1−/− cells, by immunofluorescence staining using well-defined organelle markers (Fig. 6a). We did not observe great differences in mitochondria, Golgi, ER, and early endosomes. However, late endosomes (identified by anti-LAMP1 staining) and lysosomes (identified by LysoTracker staining) appeared to be more abundant in Trex1−/− cells compared to WT cells (Fig. 6a). Similar increase in the late endosome/lysosome compartment was also observed in Trex1−/− BMDMs, but not WT or Trex1+/− BMDMs (Fig. 6b). To examine if this is also observed in human cells, we stained WT and TREX1R114H/R114H human fibroblasts, or control and Trex1 knockdown (by siRNAs) HeLa cells with LysoTracker; in both cases, we observed a marked increase in LysoTracker staining (Fig. 1c, Supplementary Fig. 9), indicating that the late endosome/lysosome compartment is expanded in cells that lack Trex1 function. We quantified lysosome expansion by LysoTracker FACS using live cells; Trex1−/− and TREX1R114H/R114H cells contained 3–5 fold more lysosomes compared to WT cells (Fig. 6d). We also detected increased LAMP1 and NPC1 (lysosomal membrane proteins) protein level in TREX1R114H/R114H and Trex1−/− cells compared to WT cells by western blot (Fig. 6e), suggesting enhanced lysosomal biogenesis in Trex1 deficient cells. To further confirm the increase in the lysosome compartment, we analyzed WT and Trex1−/− cells by electron microscopy (EM). Trex1−/− cells contained significantly more lysosome vacuolar structures (Fig. 6f,g). These structures are surrounded by single-layer membranes, some of which contain electron dense cellular materials that are commonly found in lysosomes (Fig. 6f inset). Lysosomes are important organelles for the breakdown and turnover of other cellular organelles (e.g. mitochondria), proteins and nucleic acids29. Of note, we did not observe excessive accumulation of undigested cellular materials in these lysosomes, which were often found in cells associated with lysosomal storage diseases30. We also did not detect an increase in autolysosomes in Trex1−/− cells compared to WT as measured by EM, GFP-LC3 dot formation and by western blots analyzing p62 and LC3 protein levels (Supplementary Fig. 10, data not shown).
Figure 6.
Trex1 negatively regulates lysosomal biogenesis. (a-c) Fluorescent microscopic images of WT and Trex1−/− MEFs (a) and BMDMs (b) stained with indicated organelle markers, WT and TREX1R114H/R114H (TREX1-mut) human fibroblasts stained with LysoTracker Red (c). (d) FACS analysis of live WT and Trex1−/− MEFs or WT and TREX1R114H/R114H human fibroblasts stained with LysoTracker Red. (e) Western blot analysis of lysosomal membrane proteins, LAMP1 and NPC1, in WT and Trex1−/−MEFs and BMDMs and WT and TREX1R114H/R114H (Mut) human fibroblasts (FB). (f) Electron microscopic images of WT and Trex1−/− MEFs. The lysosome vacuoles were surrounded by single membrane (arrowheads in insert). Undigested cellular materials (electron dense) were found in some lysosome vacuoles (arrows). N, nucleus. M, mitochondrion. L, lysosome. Scale bar, 1 μm. (g) Number of lysosome vacuoles in thin sections per cell. Averages of 20 cells are shown (error bars, s.d.). *P< 0.05 (Student’s t-test).
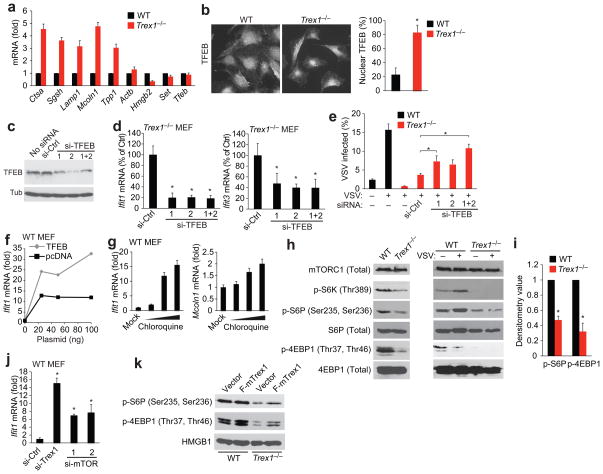
To determine whether the lysosome expansion phenotype in Trex1−/− cells was caused by the induction of lysosome genes, we measured Ctsa, Sgsh, Lamp1, Mcoln1 and Tpp1 expression which encode enzymes or structural proteins of the lysosome. All five genes were up-regulated 3–5 fold in Trex1−/− cells compared to WT cells, whereas other non-lysosomal genes did not (Fig. 7a). Many other genes involved in lysosomal biogenesis were also up-regulated in Trex1−/− cells compared to WT cells (Supplementary Fig. 11). Lysosome genes are regulated by the transcription factor EB (TFEB) through the recognition of conserved binding sites in their promoters. TFEB is a master regulator of the Coordinated Lysosomal Expression and Regulation (CLEAR) gene network31; and its overexpression increases lysosome gene expression and promotes lysosome expansion32. TFEB resides mostly in the cytoplasm and translocates into the nucleus upon complex post-translational modifications33,34. We did not observe any difference in TFEB mRNA or protein amounts in WT and Trex1−/− cells (Fig. 7a, data not shown). To examine the subcellular localization of TFEB, we stained WT and Trex1−/− cells with anti-TFEB and found that endogenous TFEB became predominately nuclear in Trex1−/− cells (Fig. 7b). This result strongly suggests that the increase in lysosomal gene expression and lysosomal compartment expansion were connected to altered TFEB localization in Trex1−/− cells. We did not detect any interaction between Trex1 and TFEB by immunoprecipitation from WT MEFs (data not shown), suggesting Trex1 is unlikely to regulate TFEB translocation through direct binding and retention in the cytosol.
Figure 7.
Trex1 regulates lysosomal biogenesis through TFEB and mTORC1. (a) Quantitative RT-PCR analysis of lysosomal and non-lysosomal genes in WT and Trex1−/− MEFs. (b) Fluorescent microscopic analysis of endogenous TFEB localization in WT and Trex1−/− MEFs. Right panel shows percentage of nuclear TFEB in the cell. Averages of 13 cells are shown (error bars, s.d.) *P < 0.05 (Student’s t-test). (c,d) Western blot analysis of TFEB knockdown (c) and qRT-PCR analysis of Ifit1 and Ifit3 mRNA (d) in Trex1−/− MEFs transfected with control or TFEB specific siRNAs. (e) FACS analysis of VSV-PeGFP replication in WT and Trex1−/− MEFs transfected with control or TFEB specific siRNAs for 72 h and mock-infected or infected with VSV-PeGFP for 18 h. *P< 0.05 (Student’s t-test). Data are representative of three independent experiments (error bars, s.d.). (f) Quantitative RT-PCR analysis of Ifit1 expression in WT MEFs transfected with myc-TFEB or pcDNA3 vector plasmid at indicated amount for 24 h. (g) Quantitative RT-PCR analysis of Ifit1 and Mcoln1 in WT MEFs treated with chloroquine at 10, 50 and 100 uM for 16 h. (h, i) Western blot (h) and densitometry (i) analysis of proteins involved in the mTORC1 pathway. WT and Trex1−/− MEFs were uninfected or infected with VSV for 16 h. Densitometry analysis was performed using Image J on 6 independent western blots. WT normalized to 1. *P< 0.05 (Student’s t-test). Data are representative of 6 independent experiments (error bars, s.d.). (j) Quantitative RT-PCR analysis of Ifit1 expression in WT MEFs transfected with indicated siRNAs for 72 h. *P< 0.05 (Student’s t-test). Data are representative of two independent experiments (error bars, s.d.). (k) Western blot analysis of proteins involved in the mTORC1 pathway. WT and Trex1−/− MEFs were transfected with vector or Flag-Trex1 plasmid for 24 h. A representative gel image of 4 independent experiments is shown.
To examine whether TFEB function is required for ISG activation and antiviral activity in Trex1−/− cells, we knocked down TFEB expression in Trex1−/− cells and measured Ifit1 and Ifit3 mRNA in uninfected cells, and VSV replication in the same knockdown cells. TFEB knockdown in Trex1−/− cells reduced both Ifit1 and Ifit3 expression and increased VSV replication (Fig. 7c, d, e). Knockdown of TFEB in WT MEFs did not affect Ifit1 or other innate immune genes that were predicted to be TFEB targets31, suggesting that TFEB is unlikely to regulate ISGs directly (Supplementary Fig. 12). Moreover, We found that Trex1−/−Irf3−/− cells express elevated levels of lysosome genes and LAMP1 protein similar to that in Trex1−/− cells, while ISG expression is drastically reduced in Trex1−/−Irf3−/− cells compared to Trex1−/− cells, suggesting that lysosomal biogenesis (regulated by TFEB) acts upstream of ISG expression (regulated by IRF3/7, Supplementary Fig. 11).
TFEB overexpression promotes lysosomal biogenesis32. To examine whether manipulating TFEB expression or nuclear translocation in WT cells also induces ISG expression, we overexpressed TFEB in WT MEFs and found that Ifit1 expression was increased in a dose dependent manner (Fig. 7f). We also treated WT MEFs with chloroquine, which induces TFEB nuclear translocation35, and observed dose-dependent increase of Mcoln1 (a lysosomal gene) and Ifit1 expression (Fig. 7g). These data further support the link between TFEB function in lysosomal biogenesis and ISG induction. Of note, chloroquine treatment of Trex1−/− cells did not rescue VSV replication (Supplementary Fig. 13), likely due to its known antiviral effect36–38.
One of the upstream regulators of TFEB nuclear translocation is mTORC1, and inhibition of mTORC1 activity under many conditions promotes TFEB nuclear transport35,39. We thus examined mTORC1 activity in infected and uninfected WT and Trex1−/− cells. We found that VSV infection induces mTORC1 activity in WT MEFs, consistent with mTORC1 being a pro-viral factor40 (Fig. 7h). mTORC1 activity is greatly reduced in both uninfected and infected Trex1−/− cells compared to uninfected and infected WT cells (measured by reduced p-S6K, p-S6P and p-4EBP1 levels. Fig. 7h, 7i). We also knocked down mTOR with two independent siRNAs and found that mTOR knockdown in WT MEFs increased Ifit1 expression (Fig. 7j). Moreover, expression of Flag-Trex1 enhanced mTORC1 activity in WT cells and restored mTORC1 activity in Trex1−/− cells compared to vector plasmid controls (Fig. 7k). Our data suggest that Trex1 is important for maintaining mTORC1 activity, and that reduced mTOR leads to ISG induction. Consistent with our data, reduced mTORC1 activity has been associated with antiviral effect40. Collectively, our data suggested that Trex1 regulates lysosomal biogenesis through TFEB and mTORC1, and lysosomal biogenesis plays a critical role in innate immunity and antiviral defense (Supplementary Fig. 14).
DISCUSSION
It is well established that IFN plays an important role in antiviral immunity. Our cells are equipped with an extensive network of innate immune sensing mechanisms for detecting invading pathogens through recognition by PRRs. When a PRR is engaged, it triggers a signaling pathway that often leads to the activation of IFN expression3. Infection with enveloped viruses also trigger an IFN-independent pathway that involves the direct activation by IRF3 of a subset of ISGs28. In fact, IRF3 can bind promoters of many ISGs in addition to IFN genes41. Promoters of IFN genes are complex (e.g. Ifnb1), containing both positive and negative regulatory elements for IRFs, NF-κB and AP-1, and a concerted effort of multiple transcription factors is often required for their stimulation. In contrast, promoters of many ISGs are simpler (e.g. Ifit1), and can be easily turned on by IRFs independently of IFN9,41. Direct activation of antiviral genes is important for nonprofessional IFN-producing cells such as fibroblasts to effectively defend themselves against viral infection, or for cells to defend themselves against viruses that have evolved mechanisms to disrupt the IFN response. It is also advantageous for cells to rapidly induce some ISGs upon viral infection before a stronger and more sustained response can be established by IFN signaling pathways. A recent study of the cytosolic RNA sensing pathway provided strong evidence that IFN-independent activation of ISGs mediated by peroxisomal MAVS is functionally important for defense against RNA virus infections10.
Very little is known about whether IFN-independent activation of ISGs occurs in the absence of infection, and how it is regulated. Here, we identified Trex1, a cytosolic protein associated with the ER, as a key negative regulator of IFN-independent activation of Ifit1 and other ISGs in uninfected cells. When the function of Trex1 is disrupted, either by genetic knockout in mice, or by a homozygous mutation in humans, or by siRNA knockdown in a variety of cell types, a subset of ISGs were activated independently of IFN, leading to an antiviral state. Remarkably, the ISG induction in Trex1-deficient cells is sustained at very high amounts and achieves an antiviral state that is comparable to that caused by the IFN-dependent pathway. This is in contrast to the viral infection induced IFN-independent response in WT cells that appears to be temporary and less robust10. We have also challenged WT and Trex1−/− mouse cells or TREX1R114H/R114H human cells with a variety of RNA viruses including VSV, influenza, Sendai and West Nile virus; and they all failed to replicate in cells that have lost Trex1 function.
We have also identified an innate immune pathway, involving STING-TBK1-IRF3-IRF7 that is important for the IFN-independent ISG activation in Trex1-deficient cells. STING is a critical adaptor protein for sensing pathogen-associated DNA or cyclic di-GMP in the cytosol and subsequent induction of IFN expression2,7. Our data expands the function of the STING-TBK1-IRF3-IRF7 pathway to include both IFN-dependent and independent branches as downstream pathways. A recent study also showed that STING activates STAT6 phosphorylation upon viral infection, which then induces chemokines such as CCL2, CCL20 and CCL26 and immune cell homing8. We did not observe induction of these chemokines in Trex1−/− or TREX1R114H/R114H cells compared to WT (data not shown). Together, STING, and associated innate immune factors are becoming a versatile machinery that can activate multiple distinct downstream pathways.
Our data also shed some light on the potential endogenous trigger of IFN-independent ISG activation. We found that Trex1-deficient or mutant cells contain excessive amount of lysosomal vacuoles and expanded lysosomal compartments as determined by immunofluroscence and immunoblot analysis of lysosomal markers, quantitative RT-PCR analysis of lysosomal genes and by electron microscopy. Consistent with elevated lysosome biogenesis, the master regulator of lysosome genes, TFEB translocates to become predominantly nuclear in Trex1−/− cells. We also found that mTORC1 activity is reduced in Trex1−/− cells and is restored after Flag-Trex1 expression in Trex1−/− cells, suggesting that Trex1 plays an important role in maintaining mTORC1 activity, which regulates TFEB nuclear translocation35,39. We also provided several lines of evidence to demonstrate that TFEB-regulated lysosomal biogenesis is functionally linked to ISG activation: TFEB knockdown in Trex1 deficient cells tempered ISG activation and antiviral immunity; TFEB overexpression in WT cells, which promotes lysosomal biogenesis32, increased Ifit1 expression; chloroquine treatment of WT cells, which induces nuclear translocation of TFEB35 and exhibits antiviral activity36–38, increased Ifit1 expression up to 15-fold; and mTOR knockdown by siRNA in WT cells also increased Ifit1 expression. Furthermore, based on our observation of elevated lysosomal genes and protein expression and lack of excess accumulation of undigested contents, Trex1−/− cells are likely to have enhanced lysosomal function. One could imagine that the release of abnormally high amounts of processed peptide or nucleic acids into the cytosol, or into the extracellular space (via exocytosis42), might break cellular homeostasis or immune tolerance or exceed the threshold for cytosolic DNA sensing. The exact identity of these cytosolic DNA remains unclear, and previous studies have indicated DNA replication debris20 and endogenous retroelements19. Aberrant functions of lysosomes have been indicated in lupus nephritis where lysosomal contents mimic viral particles and activate innate immunity43. It is also possible that increased lysosome vacuoles could cause membrane perturbation that would elicit IFN-dependent or -independent antiviral response9,44. Further studies are required to distinguish these possibilities. Together, our work demonstrate an important link between lysosomal biogenesis and innate immune activation of ISGs, as well as a novel role for TREX1 in regulating lysosomal biogenesis through TFEB and mTORC1.
We have previously shown that Trex1 inhibits HIV-mediated IFN activation11. Here, we validated this finding, and further uncovered a novel function of Trex1 in the regulation of IFN-independent innate immune activation through lysosomal biogenesis in uninfected cells, which results in a broad-spectrum antiviral state in which the replication of several different RNA viruses is inhibited. Both functions of Trex1 share a similar innate immune signaling pathway that involves STING-TBK1-IRF3, which can activate multiple downstream pathways. The upstream stimuli for HIV-mediated IFN activation is HIV DNA from nonproductive reverse transcription11, whereas the upstream stimuli for the IFN-independent pathway likely involves lysosome function.
Our work also provides further insight into pathogenetic mechanisms underlying systemic autoimmunity associated with TREX1 mutation such as SLE, a prototypic autoimmune disease. Central to SLE pathogenesis is that ineffective waste disposal due to impaired apoptosis or defective clearance of cellular debris leads to excessive release of autoantigens which activate innate immune sensors and trigger immune responses leading to formation of autoantibodies45. Our findings unravel a novel mechanism for a cell-intrinsic mechanism of initiation of autoimmunity due to enhanced lysosome function. Moreover, the constitutive type I IFN-independent ‘ISG-signature’ which is detectable in a variety of cell types and tissues may potentially represent a valuable biomarker that could be applied as clinical outcome measure.
In summary, our study uncovered a signaling cascade that involves the biogenesis of a cellular organelle (e.g. lysosome) and cytosolic innate immune detection. Both segments of the cascade function together to establish an antiviral state in Trex1-deficient cells independently of IFN activation or viral infection. We identified many components of this cascade, some of which (e.g. TFEB and mTORC1) have not been directly implicated in intrinsic antiviral immunity. We have also uncovered novel functions of known innate immune regulators such as Trex1 and STING. Further understanding of the mechanism by which this signaling cascade is regulated will have important implications not only for understanding antiviral defense but also pathogenic mechanisms underlying autoimmune diseases.
METHODS
Cells and viruses
Wild-type, Trex1−/− and Trex1−/−Irf3−/− MEFs and Trex1+/− mice were provided by D. Stetson (U. Washington) under an agreement with D. Barnes and T. Lindahl. Ifnar−/− MEFs were provided by Z. Chen (UT Southwestern). HeLa and 293T cells have been described11. All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) heat-inactivated fetal calf serum (FCS), 2 mM L-glutamine, 10 mM HEPES and 1 mM sodium pyruvate (complete DMEM) with the addition of 100 U/ml penicillin, 100 mg/ml streptomycin and cultured at 37°C with 5% CO2. WT and AGS human fibroblasts were derived by surgical removal of a piece of skin tissue from healthy donors or AGS patients with indicated mutations, and subsequently cultured in DMEM with 10% (v/v) FCS. For generation of bone marrow-derived macrophages, femurs and tibias were harvested from 8 to 10 weeks old mice. Bone marrow was flushed from the bones with cold DMEM supplemented with 20% L-929 cell-conditioned medium, 10% (v/v) heat-inactivated FCS, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 mg/ml streptomycin. Bone marrow cells were cultured in 10-cm petri dishes (10 ml vol) at 37°C, 5% CO2 for 7 d. At days 3 and 6, fresh medium was added to the cultured cells. Experiments involving human and mouse materials were approved by the Institutional Review Boards of UT Southwestern Medical Center, Dallas, Texas, USA and the Children’s Hospital, Technical University Dresden, Dresden, Germany. WT VSV21, VSV-PeGFP21, influenza virus40, Sendai virus46, West Nile virus47 were generated as described. Cells were infected with the indicated virus, MOI and time points and washed three times with 1X PBS before subsequent analysis. Experiments carried out in BSL2 and BSL3 conditions were approved by the Environmental Health & Safety Committee at UT Southwestern Medical Center.
Reagents and antibodies
Reagents: TRI Reagent, MG132, 3-MA, NH4Cl, Chloroquine, Wortmannin, Rapamycin (Sigma), Lipofectamine 2000, Vybrant Dil Cell labeling solution, Lysotracker (Invitrogen), mouse interferon ELISA kit (PBL interferon source), recombinant mouse interferon β (Millipore). Antibodies: mouse anti-Trex1 (mouse; 1:1,000 dilution; 29; 611987; BD Biosciences), anti-HMGB1 (rabbit; 1:2,000 dilution; ab18256; Abcam), anti-LAMP1 (rabbit; 1:500 dilution; ab24170; Abcam), anti-SQSTM1/p62 (mouse; 1:1,000 dilution; ab56416; Abcam), anti-LC3 (rabbit; 1:500 dilution; NB100-2220; Novus Biologicals), anti-NPC1 (rabbit; 1:1,000 dilution; # 3878-1, Epitomics), anti-Tubulin (mouse; 1:2,000 dilution; B-5-1-2; Sigma), α-VSV (rabbit; 1:4,000 dilution; R4006-F, kind gift from M. Whitt, University of Tennessee Health Science Center, Memphis, TN), anti-TFEB (rabbit; 1:2,000 dilution; generated in house), anti-Influenza A (goat; 1:250 dilution; B65141G, Meridian Life sciences), anti-Sendai (rabbit; 1:2,000 dilution; PD029, MBL), anti-WNV (rabbit; 20ug total; C19367, Lifespan Biosciences), anti-mTOR (7C10) (rabbit; 1:1,000 dilution; #2983; Cell Signaling), anti-S6 ribosomal protein (5G10) (rabbit; 1:1,000 dilution; #2217; Cell Signaling), anti-phospho-S6 ribosomal protein (Ser235/236) (rabbit; 1:1,000 dilution; #2211; Cell Signaling), anti-phospho-p70 S6 kinase (Thr389) (rabbit; 1:1,000 dilution; #9205; Cell Signaling), anti-phospho-4E-BP1 (Thr37/46) (rabbit; 1:1,000 dilution; #2855; Cell Signaling), anti-4E-BP1 (rabbit; 1:1,000 dilution; #9644; Cell Signaling) and secondary antibodies (1:4,000 dilution; GE Healthcare) were used for immunoblot analysis according to standard protocols.
RNA isolation, qRT-PCR analysis and Cytokine detection assay
Total RNA from different mouse tissues was extracted with the RNeasy Mini kit (Qiagen, # 74104) and total RNA from cells was isolated using TRI Reagent (Sigma) according to the manufacturer’s instructions. RNA quantity and quality were confirmed using a NanoDrop® ND-1000 Spectrophotometer. cDNAs were synthesized from 1 μg total RNA using SuperScript® III First-Strand Synthesis (Invitrogen). Real-time PCR was performed using the Power SYBR Green PCR master mix in an ABI-7500 fast real-time PCR machine (Applied Biosystems) with specific gene primers (sequences, Supplementary Table 1, Sigma) and values are presented relative to GAPDH mRNA. Mouse IFN-β protein in culture supernatants was measured by ELISA (42400-1; PBL interferon source) according to the manufacturer’s recommendations.
RNA-SEQ and data analysis
Total RNA was isolated from cells using Trizol. The quality of RNA was determined to be RIN=8 or higher by Bio-Analyzer. One microgram of total RNA was used for producing RNA-SEQ cDNA library using standard protocols that include cDNA synthesize, fragmentation, adding adaptors, size selection, amplification and QC (Illumina). SE50 pair-ended sequencing was done using HI-SEQ 2000 (Illumina) with > 18,000,000 reads/sample. Basic data analysis was done using CLC-Biosystems Genomic Workbench analysis programs to generate quantitative data for all genes, including RPKM values, unique and total gene reads, annotated transcripts and detected transcripts, median coverage, chromosomal location, and putative exons. Uninfected WT and Trex1−/− dataset were analyzed by Ingenuity Pathway Analysis software package (Ingenuity Systems, Inc). Heat maps were produced by first normalizing RPKM values of each gene by average and standard deviation across all treatment conditions, and then hierarchy-clustered heat maps were generated using Spotfire software.
Immunostaining, fluorescence microscopy and FACS
Cells grown on coverslips were fixed in 4% (wt/vol) paraformaldehyde and were permeabilized and stained by standard protocols. Samples mounted in Vectashield mounting medium containing DAPI (4,6-diamidino-2-phenylindole; Vector Laboratories) were imaged with a Zeiss Imager.M2 fluorescence microscope equipped with AxioVision software. The following antibodies were used for immunostaining; Anti-HSP60 (Santacruz, 13966), anti-GM130 (BD, 610822), anti-calreticulin (Abcam, Ab4-100), anti-EEA1 (Abcam, ab), anti-LAMP1 (Abcam, ab24170), anti-TFEB (generated in house) with Alexa Fluor 488 and 546 tagged secondary antibodies (Invitrogen, A21202, A21206, A10036 and A10040). Live cell fluorescence microscopy was done by growing cells in 35 mm glass bottom dish and imaging with a Zeiss LSM510 confocal microscope. For visualizing VSV infection in live cells, VSV was incubated with 2 mM Vybrant Dil Cell labeling solution (Invitrogen) in PBS for 10 min, followed by Quick Spin Sephadex G-50 column (Roche) purification to remove residual dye. Labeled VSV-DiL virions were subsequently added to target cells, incubated for 1 h before imaging. In some experiments, LysoTracker Green was used to visualize lysosomes in cells with red-labeled virus. For FACS analysis of lysosomes or VSV-PeGFP-infected cells, cells were incubated with LysoTracker Red (40 nmol/ml for 1 hour) or VSV-PeGFP for indicated time point. Cells were then washed 2 times with PBS and fixed with 1% paraformaldehyde. Cell acquisition was performed in a FACS Calibur (BD Biosciences). For all samples, 20,000 events were computed and analyzed by FlowJo software.
Transfections and western blot analysis
Cells were grown on 24-well plates and transfected with 50 nM siRNA (sequences, Supplementary Table 2. Sigma) using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction. Cells were harvested after 48–72 h and used for infection, or processed for qRT-PCR or western blot analysis. Plasmid transfections were done with Lipofectmine 2000 (Invitrogen) or with Lonza Amaxa nuclearfector according to manufacture’s instructions. For western blot analysis, cells were washed with ice-cold PBS and lysed with 100 μl of 1X SDS-PAGE reduced sample buffer. Lysates were incubated at 95°C for 5 min prior to resolving by 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and immunoblotted with the indicated antibodies. Bands were visualized using either the ECL detection reagent (Pierce) or Supersignal West Pico Chemiluminescence western blotting detection system (Thermo Scientific, Rockford, IL) and exposed to X-ray film. Films were scanned and images were assembled in Photoshop.
Electron Microscopy
Wild-type, Trex1−/− MEFs were washed with PBS and fixed in 2% glutaraldehyde (in 0.1 M phosphate buffered saline) for a minimum of 4 hours and post-fixed in 1% osmium tetroxide (in 0.1 M phosphate buffered saline) for 1 hour. After rinsing and dehydrating with graded ethanol solutions (50%, 70%, 95%, 100%), the specimens were infiltrated sequentially with propylene oxide, 1:1 mixture of catalyzed Eponate 12:propylene oxide, and 100% catalyzed Eponate 12. The specimens were then embedded in embedding molds and polymerized in a 60 °C oven overnight. Thick sections (1.0–1.5 micron) were cut on Leica Ultramicrotome with a glass knife, mounted on glass slides, and stained with toluidine blue stain. Thin sections (60–90 nm) were cut using a Leica Ultramicrotome with a diamond knife, mounted on copper grids, and stained with uranyl acetate and lead citrate. Ultrastructural examination was performed on a Hitachi H-7500 Transmission Electron Microscope.
Statistical methods
Data are presented as the mean ± s.d. Statistical significance was determined by Student’s t-test. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank D. Stetson (University of Washington) for wild-type, Trex1−/− and Trex1−/−Irf3−/− primary MEFs and Trex1+/− mice, under agreement with D. Barnes and T. Lindahl (Cancer Research UK); We thank R. Orchard and N. Alto (UT Southwestern) for assistance with live cell confocal microcopy; R. Sumpter (UT Southwestern) for assistance with fluorescence microscopy; Z.J. Chen (UT Southwestern) for Sendai virus, Ifnar−/− MEFs and valuable discussions; M. Gale (University of Washington) for West Nile virus; B. Fontoura (UT Southwestern) for influenza virus; Z. Zou (UT Southwestern) for technical assistance; M. Whitt for anti-VSV antibody; Electron Microscopy Laboratory at Children’s Medical Center for EM studies; J. Lieberman for critical reading of the manuscript and members of the Yan lab for discussions. Supported by the Rita C. and William P. Clements, Jr. Endowed Scholar Award from UT Southwestern to N.Y., the US National Institute of Health (AI093795 and AI098569 to N.Y., CA129387 to J.B., AI057156 to B.L.), The Alliance for lupus research foundation (N.Y.) and Deutsche Forschungsgemeinschaft (LE 1074/4-1 to M. L.-K.).
Footnotes
AUTHOR CONTRIBUTIONS
M.H. and N.Y. designed and performed most experiments; J.K. helped do the experiments; M.L-K. provided human cells and scientific advice; D.R. performed EM analysis. A.K.P. and B.L. contributed reagents and scientific advice; E.K.W. and I.D. helped analyze the data; M.H. and N.Y. wrote the paper.
References
- 1.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 3.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 5.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011 doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Noyce RS, Collins SE, Mossman KL. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. Journal of virology. 2006;80:226–235. doi: 10.1128/JVI.80.1.226-235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gall A, et al. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow Y, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 14.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18:R130–136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 16.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 17.Richards A, et al. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 18.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes and immunity. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Lindahl T, Barnes D. Trex1 Exonuclease Degrades ssDNA to Prevent Chronic Checkpoint Activation and Autoimmune Disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Das SC, Nayak D, Zhou Y, Pattnaik AK. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J Virol. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orebaugh CD, Fye JM, Harvey S, Hollis T, Perrino FW. The TREX1 Exonuclease R114H Mutation in Aicardi-Goutieres Syndrome and Lupus Reveals Dimeric Structure Requirements for DNA Degradation Activity. J Biol Chem. 2011;286:40246–54. doi: 10.1074/jbc.M111.297903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreijtz JHCM, Fouchier RAM, Rimmelzwaan GF. Immune responses to influenza virus infection. Virus research. 2011;162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 25.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramantani G, et al. Expanding the phenotypic spectrum of lupus erythematosus in aicardi-goutières syndrome. Arthritis and rheumatism. 2010;62:1469–77. doi: 10.1002/art.27367. [DOI] [PubMed] [Google Scholar]
- 28.Paladino P, Cummings DT, Noyce RS, Mossman KL. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J Immunol. 2006;177:8008–8016. doi: 10.4049/jimmunol.177.11.8008. [DOI] [PubMed] [Google Scholar]
- 29.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 30.Cox TM, Cachón-González MB. The cellular pathology of lysosomal diseases. The Journal of pathology. 2012;226:241–254. doi: 10.1002/path.3021. [DOI] [PubMed] [Google Scholar]
- 31.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Human molecular genetics. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 32.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 33.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peña-Llopis S, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roczniak-Ferguson A, et al. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Science signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent MJ, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi EE, Chew JS, Loh JP, Chua RC. In vitro inhibition of human influenza A virus replication by chloroquine. Virol J. 2006;3:39. doi: 10.1186/1743-422X-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mata MA, et al. Chemical inhibition of RNA viruses reveals REDD1 as a host defense factor. Nat Chem Biol. 2011;7:712–719. doi: 10.1038/nchembio.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grandvaux N, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina DL, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Migliorini A, Anders HJ. A novel pathogenetic concept-antiviral immunity in lupus nephritis. Nat Rev Nephrol. 2012;8:183–189. doi: 10.1038/nrneph.2011.197. [DOI] [PubMed] [Google Scholar]
- 44.Holm CK, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller BC, et al. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol. 2006;80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.