Abstract
The identification of interactions between viral and host cellular proteins has provided major insights into papillomavirus research, and these interactions are especially relevant to the role of papillomaviruses in the cancers with which they are associated. Recent advances in mass spectrometry technology and data processing now allow the systematic identification of such interactions. This has led to an improved understanding of the different pathologies associated with the many papillomavirus types, and the diverse nature of these viruses is reflected in the spectrum of interactions with host proteins. Here we review a history of proteomic approaches, particularly as applied to the papillomaviruses, and summarize current techniques. Current proteomic studies on the papillomaviruses use yeast-two-hybrid or affinity purification-mass spectrometry approaches. We detail the advantages and disadvantages of each and describe current examples of papillomavirus proteomic studies, with a particular focus on the HPV E6 and E7 oncoproteins.
Keywords: papillomavirus, proteomics, interaction, HPV, mass spectrometry, yeast two hybrid
Papillomavirus Diversity and Disease Associations
The papillomaviruses (PVs) are a family of non-enveloped viruses containing double stranded, circular DNA genomes of approximately 8000 bp. PVs infect squamous epithelial cells and have a restricted host range. Although papillomaviruses have been identified that infect organisms from reptiles to humans, and PVs likely infect all amniotes, each virus can in general infect only a single species (Gottschling et al., 2011; Mengual-Chulia et al., 2012). The viral genomes share a conserved structure, and each PV identified to date encodes the core proteins E1, E2, L1, and L2 (Garcia-Vallve, Alonso, and Bravo, 2005). Many papillomaviruses also encode the accessory proteins E6 and E7; smaller numbers of papillomaviruses encode additional proteins and splice variants of the various viral genes. Most of the PV genomes contain 7–9 open reading frames. While the virus’ genome structure is conserved, the genome sequences differ from one another significantly. Two papillomaviruses are different ‘types’ if their L1 genes differ by 10% or more at the DNA sequence level (Bernard et al., 2010; de Villiers et al., 2004). L1 sequences were used to group the 189 PV types listed in the most recent classification into species and further into genera (Bernard et al., 2010). The number of PV types that have been identified and sequenced continues to increase rapidly.
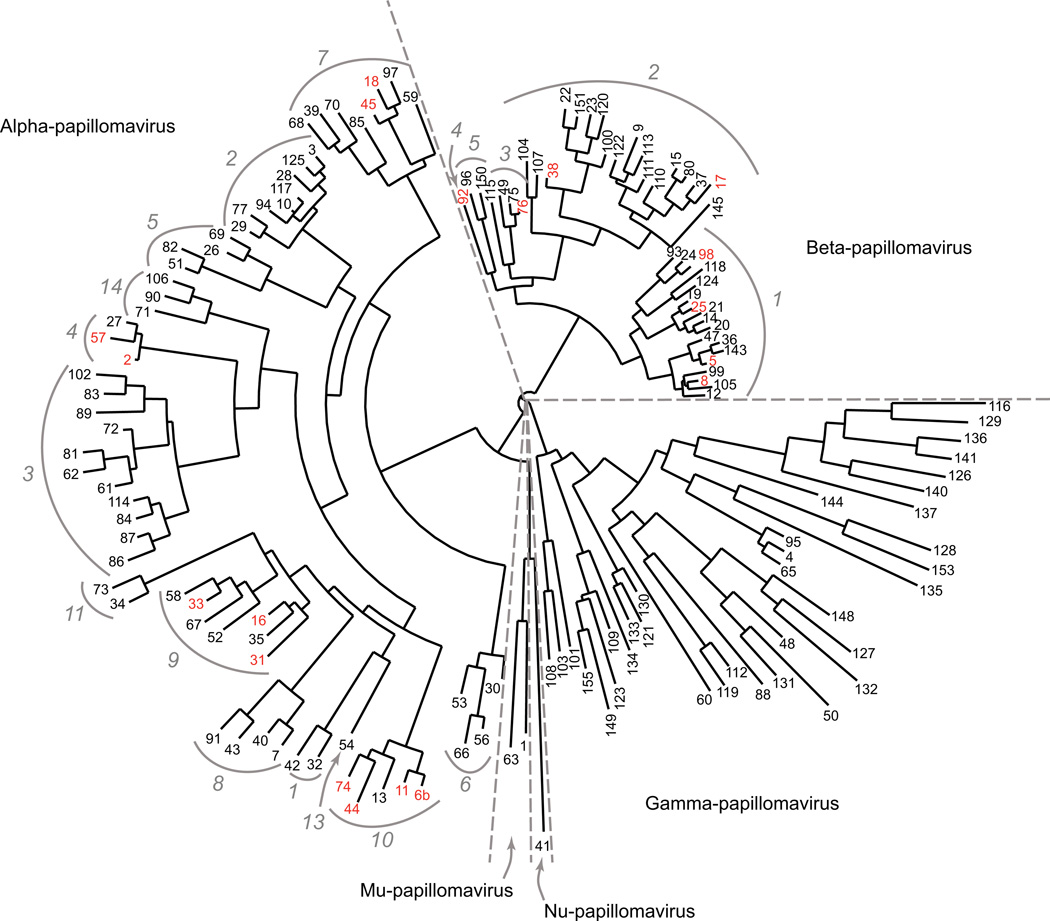
The majority of the human papillomaviruses (HPVs) fall into genus alpha and genus beta, although a few of the HPVs identified so far are members of the gamma, mu, and nu genera (Bernard et al., 2010). A phylogenetic tree representing HPV types from these five genera is shown in Figure 1. Most of the alpha genus HPVs infect mucosal tissues, and a subset of the alpha HPVs are the 15 ‘high-risk’ types that cause cervical cancer and other anogenital cancers (Lowy et al., 2008; Schiffman et al., 2007). Additionally HPV16 and HPV 18 (two of the alpha genus HPVs) have been now associated with a significant fraction of head and neck cancers, most notably oropharyngeal cancers. So far these have been associated with as many as 30% of these cancers worldwide and 60% in the United States (Gillison, Chaturvedi, and Lowy, 2008; Kreimer et al., 2005; Watson et al., 2008). Some of the other ‘low-risk’ HPVs from genus alpha cause genital warts and are infrequently associated with cancer. In contrast, the HPVs from genus beta infect the cutaneous epithelium [reviewed in (Howley and Lowy, 2006)]. Several of the beta HPVs are found in hyperproliferative lesions in patients with Epidermodysplasia Verruciformis (EV). These lesions in patients with EV may progress to squamous cell carcinoma (SCC), often in sun-exposed regions of patients’ skin. The beta HPVs have also been potentially implicated in non-melanoma skin cancers (NMSCs) in non-EV individuals (Akgul, Cooke, and Storey, 2006; Feltkamp et al., 2008). However, transcription of PV RNA was not detected in SCCs in one recent study (Arron et al., 2011). Infection by the beta HPVs is in general quite frequent but typically asymptomatic, either because of efficient clearance of infected cells or because of persistent infections that go undetected by the immune system and do not cause abnormal pathology (Doorbar, 2006).
Figure 1.
Sequence relatedness among HPV types. A phylogenetic tree based on the sequences of the L1 genes of the 143 human papillomavirus types currently listed in the Papillomavirus Episteme (PaVE) (van Doorslaer et al., 2012) was generated as described by (Dereeper et al., 2010; Dereeper et al., 2008). The HPVs are grouped into 5 genera (alpha, beta, gamma, mu, and nu), as shown. Genera are further subdivided into species, which are indicated (grey italics) for viruses in genus alpha and genus beta. Virus types discussed in the text are indicated in red.
Proteomic Approaches and Tools
The importance of protein-protein interactions to biological processes was not always appreciated. In the 1950s and 1960s, it was determined that proteins consisted of a polypeptide chain that could fold into a defined structure, and this led to the idea that these structures could form defined surfaces that might mediate protein-protein interactions (Braun and Gingras, 2012). In these early years, only a very few protein-protein interactions (PPIs) were understood and were studied individually. Many of the key early determinations of protein-protein interactions came from the study of oncogenic viruses. These include the discovery in the late 1970s of SV40 large T antigen bound to p53 (Lane and Crawford, 1979; Linzer and Levine, 1979), the finding that pRB1 bound to adenovirus E1A in the 1980s (Whyte et al., 1988), and others.
Early analysis of the composition of proteins in a mixture and of potential PPIs was possible using two-dimensional gel electrophoresis, but this technology was quite limited. Several technical advances allowed the study of proteins and their interactions to begin to progress (Patterson and Aebersold, 2003). Techniques including Edman degradation and mass spectrometry began to allow the determination of a protein or peptide sequence. When the peptide sequence could be matched to a known gene, this allowed the study of that specific protein. Researchers could advance the study of individual PPIs further using new tools including the generation of gene fusions, the development of protein and epitope tags either singly or in tandem, and the pairing of matched antibodies and epitope tags (Patterson and Aebersold, 2003). Gene fusions in particular paved the way for the development of yeast-two-hybrid technology, which was originally proposed by Fields and Song as a proof-of-principle experiment (Fields and Song, 1989) but went on to become a powerful and widely used technique. Two more recent key advances are the sequencing of the complete genomes of an ever-increasing number of organisms, and advances in mass spectrometric technology. In particular, identification of peptides from more and more complex mixtures became possible with the development and refinement of LC-MS/MS technology, in which a peptide mixture is injected by electrospray into a mass spectrometer without any previous separation (Coon et al., 2005; Wilm et al., 1996).
These developments now leave the proteomics community at a key crossroad. With the ability to generate increasing amounts of data on more and more protein-protein interactions, there is a need to implement a range of different computational solutions for the analysis of these data. Current and ongoing challenges include refining the ability to remove background or false-positive interactions, managing and sharing proteomic data, integrating proteomic data with gene expression data, and comparing data across proteomic platforms.
Today, yeast-two-hybrid and affinity purification-mass spectrometry are the two basic strategies typically used to define PPIs on a large scale (Sardiu and Washburn, 2011). Each has advantages and disadvantages, and below we will compare the types of data generated by the two approaches with the PVs. In both methods the goal is to determine interactors of the ‘bait’ protein, and these interactors are often referred to as the ‘prey’.
The first high-throughput method for the detection of protein-protein interactions was the yeast-two-hybrid (Y2H) system (Fields and Song, 1989). It is based on the separable nature of the yeast GAL4 transcription factor and the ability to make protein fusions. The bait protein of interest is fused to the GAL4 DNA-binding domain (DBD) and potential prey proteins are fused to GAL4’s transactivation domain. Interactions in a yeast cell between bait and prey thus reconstitute GAL4 function and lead to the activation of a reporter gene whose promoter is decorated with GAL4 binding sites. Genome sequencing efforts and the availability of large open reading frame (ORF) collections have made Y2H approaches more powerful. It became possible using these tools to rapidly screen large numbers of potential bait and/or prey proteins, and the first large-scale yeast-two hybrid studies were published in 2000–2001 (Ito et al., 2001; Ito et al., 2000; Uetz et al., 2000; Walhout et al., 2000).
One advantage of Y2H screening is that it is potentially a high-throughput, low cost experiment. Researchers can screen a cloned genome library generated from any cell type of interest, helping to increase the number of potential prey interactions that might be of physiologic significance. The yeast-two-hybrid system does however have significant limitations. In particular, there are many opportunities to detect false-positive or false-negative interactions, either because yeast proteins promote or inhibit the interaction between a bait and prey or because a bait or prey may itself contribute to transcriptional activation or repression of the reporter gene. In addition, the binding of two proteins may require post-translational modifications that are not present in yeast. Nonetheless, yeast-two-hybrid analysis is widely used today, and data analysis approaches will need to handle comparisons that include Y2H data. To circumvent some of these limitations, the yeast-two-hybrid approach has been adapted to a so-called mammalian two-hybrid system. Using mammalian cells allows researchers to consider protein modifications that may impact binding, to use a relevant cell type, and to manipulate the cells with drugs or other treatments during an experiment. The reconstituted transcription factor typically is comprised of the herpes simplex virus VP16 activation domain and the GAL4-DBD. The mammalian system is less scalable than the yeast system, and thus far has been used mostly for smaller scale studies or validation of other experiments. Mammalian-two-hybrid systems have been comprehensively reviewed in (Lievens, Lemmens, and Tavernier, 2009).
Affinity purification coupled with mass spectrometry (AP-MS) is the second major approach to detecting protein-protein interactions, and for a more detailed review of AP-MS we direct the reader to (Dunham, Mullin, and Gingras, 2012). A bait protein of interest is purified from cells using an epitope tag coupled with antibody or other affinity purification, or less often using an antibody to the endogenous protein. Mass spectrometry is then used to identify bound prey proteins. Early implementations of this approach relied on separating proteins on a gel, then identifying proteins present in individual bands. More recently, high-throughput sequencing of complex protein mixtures has become possible using LC-MS/MS technology. Now hundreds of potential interacting proteins can be identified from a single sample.
The advantages of AP-MS are that it takes place in mammalian cells where native protein modifications are present and cells can be treated or manipulated as desired. It is unbiased in the sense that any protein present in a cell that can be detected by mass spectrometry is a potential prey. Disadvantages include the potentially laborious task of tagging and expressing each potential bait protein of interest, either transiently or in a stable cell line, and the relatively low throughput compared to a Y2H experiment. An interaction needs to be preserved through many purification steps to be detected, so transient interactions can be difficult to detect, and when multiple protein isoforms exist for a prey protein it can be difficult to distinguish them by MS.
Post-collection analysis is required for both Y2H and AP-MS data sets. These analyses generate a large amount of data to be compiled and visualized, and both types of data sets may contain a large number of false-positive or background/nonspecific interactions. Early global yeast-two-hybrid studies included little computational analysis. More recent, larger scale yeast-two-hybrid analyses have begun to detect thousands of interactions, leading to the idea that these must be filtered or assigned a confidence score in some way. For example, Giot and colleagues (Giot et al., 2003) mapped Drosophila binary interactions, generated a statistical model based on a training set, and used this to refine the 20,439 interactions they detected to a high-confidence list of 4780 interactions. Most of the computational analysis of Y2H data sets so far has been in the form of post-publication comparison and analysis of several data sets to one another or to literature curated or other reference data sets (Braun et al., 2009; Yu et al., 2008). The analysis is generally not integrated into the initial Y2H data collection and processing protocol.
More methods have been developed for the analysis of AP-MS data, and these are discussed in greater detail in (Nesvizhskii, 2012). This was less important in early low-throughput experiments and in some tandem affinity purification (TAP) strategies still used today, as these experiments relied on dual purifications, often followed by separation on an acrylamide gel and analysis of individual protein bands. Researchers have moved to higher-throughput techniques, often using purification with a single tag and analysis of the resulting complex sample, and so separating real data from background interactions has become paramount. One estimate is that 10% of all proteins identified after AP-MS are bona fide interactors of a given bait (Trinkle-Mulcahy et al., 2008).
The methods that attempt to eliminate nonspecific interactions range from the very simple to the very complex. The first and easiest analysis method was to subtract all negative-control interactors from the list of interactors identified for the bait of interest. Early genome-wide AP-MS studies in yeast advanced this analytical approach by integrating varying amounts of data processing into their workflows, but in general considered only the presence or absence of an interaction, not the quantity of the protein detected. These approaches ranged from performing replicate experiments to some statistical analysis and machine learning methods (Gavin et al., 2006; Ho et al., 2002; Krogan et al., 2006). Collins and colleagues (Collins et al., 2007) next combined these two approaches and refined the yeast genomic interaction data set.
Current analytical methods use the quantitative or semi-quantitative information present in a mass spectrometry data set to generate confidence scores for a given PPI, and some of the key methods are highlighted here. These use the spectral count that is generated for each protein identified by MS as the basis for quantification (Breitkreutz et al., 2010; Sardiu et al., 2008; Sardiu et al., 2009; Sardiu and Washburn, 2011; Sowa et al., 2009). The AP-MS studies covered in this review have used several of these methods, although some of the studies have adapted an approach that was originally developed to analyze a different large data set. CompPASS was first used to define the interactomes of the human deubiquitinating enzymes (Sowa et al., 2009) and has subsequently been used by our group to analyze HPV protein-human host cell protein interactions. MiST was developed and has been used for the analysis of interactions between HIV-1 proteins and cellular proteins (Jager et al., 2012a). A survey of immune regulatory proteins from a number of viruses used an adaptation of an earlier normalized spectral abundance factor (NSAF) method (Pichlmair et al., 2012; Zybailov et al., 2006). In addition to our studies with the HPV proteins described below, the other recent HPV protein/host cell protein interaction study used tandem affinity purification and considered interactions to be significant when they were detected in duplicate experiments (Rozenblatt-Rosen et al., 2012; Zhou et al., 2010). CompPASS, MiST, and a third computational platform, SAINT (Choi et al., 2011), determine confidence scores for each interaction differently, but each has been shown to be a powerful tool in separating bona fide interactors from background.
Papillomaviruses as Candidates for Proteomic Analysis
The PV replication is invariably linked to and dependent upon the differentiation of squamous epithelial cells that the virus infects (Doorbar, 2006). HPV particles infect cells in the basal layer of a stratified epithelium, either mucosal or cutaneous, and progress through the subsequent stages of their life cycle in the differentiating cells. Virus particle production and release occurs at the top layer of the desquamating epithelium. This connection between differentiation and viral replication has limited the ability to recapitulate the viral life cycle in tissue culture. Several systems have been implemented to model this life cycle in differentiating cells, but on a much smaller scale than would be required for proteomic experiments. Nonetheless, papillomaviruses are excellent candidates for proteomic analysis. Many PV genomes have been cloned and completely sequenced, and their small open reading frames are easily subcloned and epitope tagged. More importantly, it appears that nearly all of the HPV-mediated changes in the host cellular environment must occur through virus-host PPIs, since only one PV protein is known to encode an enzymatic activity: the helicase function of the E1 protein.
Protein-protein interactions have revealed the functions of the papillomavirus proteins since the earliest days that these viruses were studied. The E6 and E7 oncoproteins engage in key PPIs. E7 proteins bind to and promote the degradation of pRB, releasing E2F transcription factors thus promoting S phase entry and DNA replication (Dyson et al., 1992; Dyson et al., 1989; Munger et al., 1989). High-risk E6 proteins bind the cellular ubiquitin ligase E6AP to form a complex that targets p53 for degradation, thereby blocking signaling through the apoptotic pathways that would otherwise be triggered by E7 (Huibregtse, Scheffner, and Howley, 1991; Scheffner et al., 1990; Werness, Levine, and Howley, 1990). When the normal regulation of E6 and E7 expression is lost, these protein-protein interactions can then trigger cellular immortalization and genomic instability that may ultimately result in cellular transformation and cancer. Some of these functions, such as E7 binding to Rb, are conserved across all HPV types examined to date (White et al., 2012). Others, like the targeted degradation of p53 by E6, are restricted to a subset of the HPVs. Bovine papillomavirus type 1 (BPV1) is often used as a model virus, but its oncoproteins function slightly differently. BPV1 E7 does not bind to pRB1 or the other RB related pocket proteins (DeMasi et al., 2005), and BPV1 E6 neither binds p53 nor targets it for degradation.
Proteomic studies on a limited number of PV proteins in the early 2000s revealed several important interactions. An AP-MS experiment using BPV1 E2 as the bait revealed that BPV1 E2 binds to and uses Brd4 to tether its genomes to host mitotic chromosomes in dividing cells (You et al., 2004), and the E2/Brd4 interaction has since been characterized for all PVs that have been studied (McPhillips et al., 2006). Other early AP-MS studies on a few HPV and BPV E6 and E7 proteins detected other PPIs (DeMasi et al., 2005; Huh et al., 2007; Huh et al., 2005; McLaughlin-Drubin, Huh, and Munger, 2008; Vos et al., 2009), but relied on an abundant cellular binding partner and a stable interaction, since the bound proteins were detected after one-dimensional SDS-PAGE and excision and sequencing of an individual protein band. Perhaps most importantly, these studies reinforced the idea that many PV functions are mediated through protein-protein interactions and indicated that larger-scale proteomic experiments would reveal new information about PV biology, virus-host cell interactions, and possibly links to disease.
Designing Systems for Proteomic Analysis of Papillomaviruses
The first choice in designing a proteomic study is whether to use a Y2H system, an AP-MS approach, or both. In the case of an AP-MS experiment, several parameters are more flexible than in the two-hybrid case. An AP-MS experiment begins with the choice of analysis platform, from mass spectrometry instrumentation to data analysis and processing. Next, the cell line should be chosen early in the AP-MS design. A cell line might be chosen for ease of culture, for biological relevance, for ready comparison of newly generated data to large existing data sets, for cost considerations, or for other study-specific reasons. The investigators must decide whether the tagged bait proteins of interest will be expressed in cells stably or transiently, as some cell lines may be compatible with one method but not the other. The type of vector and cloning system required will be based on the cell line and expression method chosen. Epitope tag(s) can then be chosen based on the available vectors and perhaps the need to compare to existing data sets. Finally, individual bait proteins may require other adaptations. For example, some protein functionality may only be compatible with N-terminal, C-terminal, or even internal epitope tags. Other proteins may be toxic when maintained in cells, indicating that they may require transient or inducible expression prior to an experiment. Finally, different analysis platforms require different numbers and types of controls, including negative controls, replicates, unrelated protein baits, or others. A proteomic experiment will have the maximum chance of success when as many of these factors as possible are considered before the experiment even begins.
Discovery of Novel Protein-Protein Interactions through Papillomavirus Proteomics
Several recent publications have reported proteomic analyses of the papillomaviruses, and these have used different approaches to generate data sets with some overlap and some unique features. We will concentrate on two large-scale AP-MS studies, with references to related work and with mention of yeast-two-hybrid approaches ongoing in other groups. These two are the work of a large multi-lab consortium (Gulbahce et al., 2012; Rozenblatt-Rosen et al., 2012) and our own studies (Martinez-Noel et al., 2012; Tan et al., 2012; White et al.; White et al., 2012). Similarities and differences both in implementation and in results are highlighted below and are summarized in Table 1.
Table 1.
Recent virus-host interaction studies discussed in the review
| Virus | System | Analytical approach | References | Summary |
|---|---|---|---|---|
| HPV | AP-MS | CompPASS (Sowa et al., 2009) | (Tan et al., 2012; White et al., in press; White et al., 2012) | Seventeen different HPV E7 baits and sixteen different HPV E6 baits were analyzed by AP-MS and CompPASS with the goal of understanding HPV diversity though PPIs. The E7 study was conducted in keratinocytes and identified largely conserved interactions, with some species- and type-specific PPIs. The E6 study identified genus-specific interactions and a higher proportion of species- and type-specific PPIs than for the E7 baits. |
| HPV | TAP-MS and Y2H | TAP-MS: tandem purification and comparison of duplicate runs Y2H: experimental and analytical platform as in (Dreze et al., 2010) | (Rozenblatt-Rosen et al., 2012) | E6, E7, and several other ORFs from six (TAP-MS) or seven (Y2H) HPV types were analyzed for binding to host cellular proteins. The TAP-MS study was performed in IMR-90 fibroblasts stably expressing tagged HPV ORFs. This work is part of a larger study aimed at identifying pathways that are perturbed by four different tumor viruses and as such, are likely to drive cancer development when pathway function is lost. |
| HIV-1 | AP-MS | MiST (Jager et al., 2012a) | (Jager et al., 2012a; Jager et al., 2011; Jager et al., 2012b) | All 18 HIV-1 proteins were analyzed by AP-MS in Jurkat and 293 cells and data was processed using MiST. This study provides a global survey of HIV-host interactions. |
The Rozenblatt-Rosen study of PV interacting proteins was part of a larger effort to study proteins encoded by four different families of tumor viruses (the papillomaviruses, the polyomaviruses, the herpesviruses, and the adenoviruses), with the overall goal of identifying genes that drive cancer (Rozenblatt-Rosen et al., 2012). The authors analyzed interactions for proteins from the four virus families by both Y2H and AP-MS approaches, but in this review we will discuss only their HPV interaction results. The AP-MS component of their study included 31 ORFs from 6 different PVs (two high-risk: HPV16 and 18, two low-risk: HPV6b and 11, and two genus beta, species 1: HPV5 and 8). The E6 and E7 oncoproteins and E6 mutants account for about half of these ORFs, and this is the PV interaction data that the authors highlight in their analysis. E6s were tagged with Flag and HA at their N-termini, and the high-risk HPV E6s were additionally analyzed with a C-terminal Flag-HA tag to block the association of PDZ proteins that bind to the C-terminus of high-risk HPV E6s. HPV E7 proteins were tagged at their C-termini with Flag and HA. Each of these choices is appropriate, as N-terminal tags on E6 proteins allow interactions with PDZ domain proteins, and C-terminal tags on E7 have been show to allow the interaction of E7 with cellular proteins including the large cellular protein UBR4 (originally named p600). Adding amino acids (aa) at the N-terminus of E7 blocks the E7/UBR4 interaction (DeMasi et al., 2005).
These tagged ORFs were incorporated into retroviral vectors and used to create stable cell lines in IMR-90 human diploid fibroblasts. These cells have the advantage of being ‘normal’, i.e. they have a limited lifespan and are not transformed, but they are fetal lung fibroblasts, which are mesenchymal in origin. Thus they are relatively un-related to the squamous epithelial cells that are the natural host of PV infection, and epithelial-specific interactors of the HPV proteins will not be detected in this assay. Presumably, the authors chose to use IMR-90 cells for ease of culture and to use one cell type for proteins from all four tumor virus families they examined. Viral proteins were purified by a two-step affinity purification using both Flag and HA tags and only proteins that were detected in both of two independent experiments were considered in subsequent analyses.
Our studies took a similar approach but with several key differences. The goal of our studies is to better understand the biological differences among various HPV types through their PPIs. To this end we cloned 160 ORFs from 20 different viral genomes, about 80% coverage. We planned to perform our analysis using CompPASS software developed by Sowa and colleagues (Sowa et al., 2009). As in the Rozenblatt-Rosen study we began with the analysis of HPV E6 and E7 interactors, hypothesizing that virus-specific interactions would be most apparent for the oncoproteins. Our HPV E7 study included seventeen different HPV types and our HPV E6 study included sixteen different E6 baits, both listed below. We made similar tagging decisions as above. E7 proteins were tagged at the C-terminus with Flag and HA, and E6 proteins were tagged at the N-terminus with HA. Our protocol used a single anti-HA AP step, so the Flag tag was extraneous and was removed from the next-generation E6 expression vector.
In our studies, we have used N/Tert-1 human keratinocytes, a cell line derived from normal human foreskin keratinocytes that was immortalized by the addition of hTert, the catalytic subunit of telomerase (Dickson et al., 2000). N/Tert-1 cells do not express p16INK4a, a common feature of immortalized epithelial cells (Kiyono et al., 1998). Thus, our analysis was conducted in squamous epithelial cells, the normal host cell of a PV infection. One requirement of the CompPASS system is that each IP-MS/MS experiment is compared to a library of interaction data generated from a large number of comparable IP-MS/MS experiments, as introduced above. The first step in establishing this system was to generate approximately 40 different N/Tert-1 cell lines each expressing an HA-tagged cellular protein. Each cell line was processed for an IP-MS/MS experiment and used to generate the CompPASS ‘stats table’ as the basis for comparison for the individual viral baits.
Identification of PV E7 interacting proteins by large scale proteomics
We have published the results of the proteomic analysis of HPV E7 proteins, conducted in N/Tert-1 cells (White et al., 2012). We tested seventeen different E7 baits, ten HPV E7s from genus alpha (HPV18 and 45 from species 7; HPV16, 31, and 33 from species 9; HPV6b, 55 (a subtype of HPV44), and 74 from species 10; and HPV2a and 57 from species 4) and seven HPV E7s from genus beta (HPV8, 25, and 98 from species 1; HPV17a and 38 from species 2; HPV76 from species 3; and HPV92 from species 4). Rozenblatt-Rosen and colleagues tested six E7s in their TAP-MS study, as described above. We highlight several of our findings and compare them to those of Rozenblatt-Rosen and colleagues in the sections below. Some of the interactions detected in one or both studies are summarized in Figure 2.
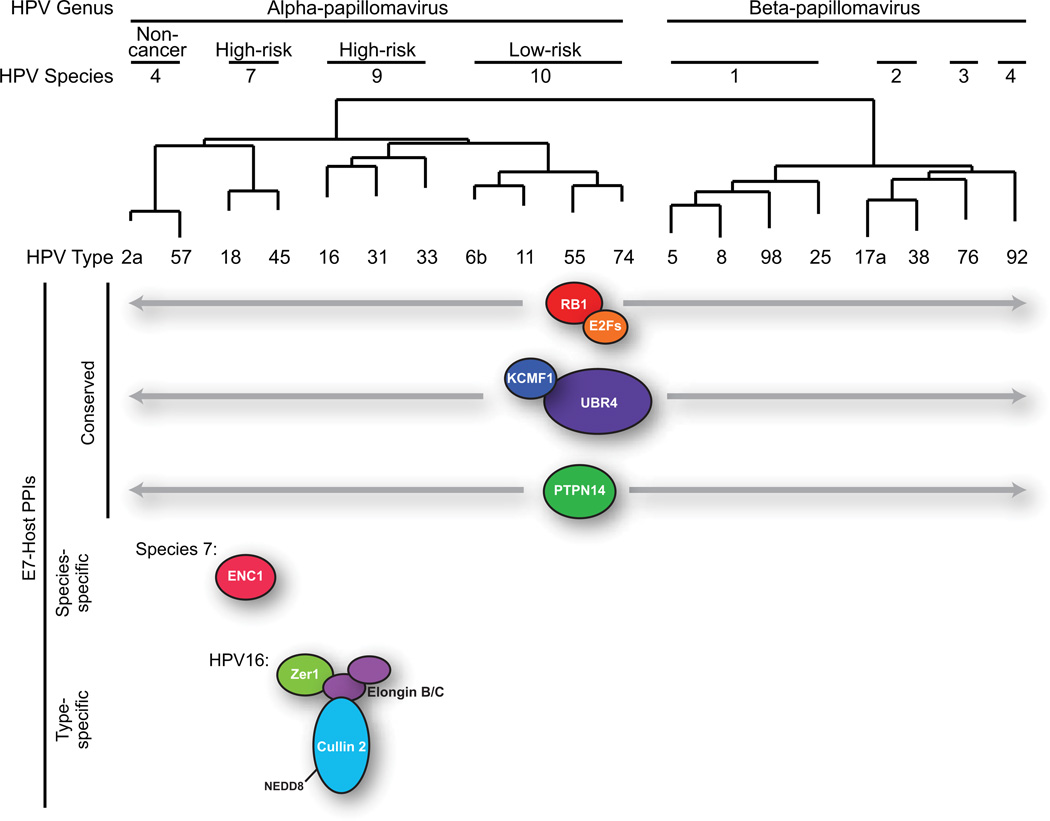
Figure 2.
Selected binding partners of HPV E7 proteins. HPV types used in the studies of Rozenblatt-Rosen et al and White et al on E7 interacting proteins (Rozenblatt-Rosen et al., 2012; White et al., 2012) were arranged into a phylogenetic tree based on the sequences of their L1 genes. Selected conserved, species-specific, or type-specific PPIs involving E7 proteins are indicated.
pRB1 and UBR4
Our experiments identified several known interactors of HPV E7s, including pRB1 and UBR4. E7 proteins share an LXCXE motif responsible for binding to pRB1 and related ‘pocket proteins’ including pRBL1 (p107) and pRBL2 (p130). This motif is conserved among the seventeen different HPV E7s in our study, and binding to pRB1 therefore served as a positive control in these experiments. Each E7 tested bound to pRB1. pRBL1 and pRBL2 each bound to 10 of the 17 E7 baits, although not the same 10 E7s in each case. A more revealing result was that UBR4 bound to each HPV E7 tested. Prior to this report, UBR4 was known to interact with HPV16, 6b, and 11 E7s and with bovine papillomavirus (BPV) E7 (DeMasi et al., 2005; Huh et al., 2005). UBR4 contains a UBR box, a motif that is common to proteins involved in the N-end rule-mediated degradation of proteins (Tasaki et al., 2005; Tasaki et al., 2009), but UBR4 itself has not yet been definitively shown to be a functional E3 ubiquitin ligase or to act in the N-end rule pathway. Based on the conservation of the UBR4-E7 interaction across several PV genera, it seems that this critical interaction has PV-related functions that are yet to be revealed. Other newly identified cellular proteins also interact with all E7s tested. KCMF1, another putative E3 ubiquitin ligase, bound to all seventeen E7s. Preliminary IP-MS/MS and CompPASS experiments in our laboratory suggest that KCMF1 interacts with UBR4 in the absence of E7; this binding may or may not be related to the function of the UBR4-E7 interaction.
PTPN14
The non-receptor tyrosine phosphatase PTPN14 also bound to many of the E7 baits. It was detected with 12 of 17 E7s as a HCIP, and the PPI confidence scores were higher for alpha than beta E7s. PTPN14 has recently been implicated in density-dependent cell growth (Huang et al., 2012; Liu et al., 2012; Wang et al., 2012). It binds to YES1, a regulator of Hippo signaling, and negatively regulates YES1 when cells are at high density. The interaction of PTPN14 with E7 therefore raises the question of how E7 might control cell density, possibly promoting proliferation, through the Hippo pathway.
The six E7s in the Rozenblatt-Rosen TAP-MS study also bound to pRB1, p600/UBR4, KCMF1, and PTPN14. Five E7s bound to RBL1, three bound to RBL2, four bound to KCMF1, and four bound to PTPN14, indicating similar results as our own study. From their data, this group concludes that HPV E7s do not target groups of interacting proteins in a ‘class-specific’ manner, that is, there are not significant differences among E7 interactors between the high-risk, low-risk, and cutaneous E7 proteins. Our own results are consistent with this idea insofar as the predominant and most easily detected cellular binding partners of the E7s are those proteins whose interactions are conserved across virus types.
ZER1 and ENC1
Nonetheless, there are specific interactions exhibited by the E7 proteins from viruses of different types. In both our study, which tested 17 E7s, and that of Rozenblatt-Rosen and colleagues, which tested six E7s, Zer1 bound only to HPV16 E7. We analyzed the function of Zer1 (also known as Zyg11BL), the substrate specificity component of a CUL2-based cullin-RING ligase (CRL). Zer1 contains a BC-box and a CUL2-box and interacts with Elongin B, Elongin C, and CUL2 (Mahrour et al., 2008). In cells that express HPV16 E7, we demonstrated that Zer1 contributes to the E7-mediated degradation of pRB1. siRNA-mediated knockdown of Zer1 resulted in an increased level of pRB1, particularly the hypophosphorylated form that is bound by E7. We thus refined the understanding of the mechanism by which HPV16 E7 targets pRB1 for degradation (White et al., 2012). Curiously, although degradation of hypophosphorylated pRB1 is observed in cells expressing other high-risk E7s, the Cul2-Zer1 ligase is specific for HPV16, and our data does not suggest other CRL complexes in association with other E7s. All E7s bind to CUL3, but our preliminary evidence has not suggested that this interaction contributes to pRB1 degradation. In our studies HPV genus alpha, species 7 E7s bind to the BTB protein ENC1. BTB proteins bind to CUL3, but since the E7/ENC1 interaction is not universal, we do not believe that all E7s bind to CUL3 using ENC1. Further experiments will be required to understand how other E7s target pRB1 for degradation and why E7s bind to CUL3.
Proteomic identification of interactions between HPV E6 and host cellular proteins and characterization of genus-, species-, and type-specific binding patterns
The publication by Rozenblatt-Rosen and colleagues also reported TAP-MS data for six different E6 proteins, from the same HPV types as used in their analysis of E7s. Our group has also recently reported the AP-MS based identification of host cellular proteins that bind to E6 proteins from sixteen different HPV types (White et al.). The sixteen E6 baits in our study are nine E6s from genus alpha: HPV16, 31, 33, and 52 (high-risk, species 9); HPV18 and 45 (high-risk, species 7); HPV6b (low-risk, species 10); HPV2a, 57 (species 4) and eight E6s from genus beta: HPV8, 20, 25, and 98 (species 1); HPV17a and 38 (species 2); HPV76 (species 3); and HPV92 (species 4). Thus, we can again compare the two studies and the E6 interactions they identified. The Rozenblatt-Rosen study was conducted in fibroblasts whereas our experiments were performed using keratinocytes. In our study, in addition to the analysis of E6 interactions from untreated E6-expressing cells, we also analyzed each E6 bait after the cell lines were treated with the proteasome inhibitor MG132. We hypothesized that it might be easier to detect E6 interactors that are targeted for degradation by E6, such as p53 or PDZ domain proteins, when the proteasome was not active. Selected interactions detected in one or both of the studies are summarized in Figure 3.
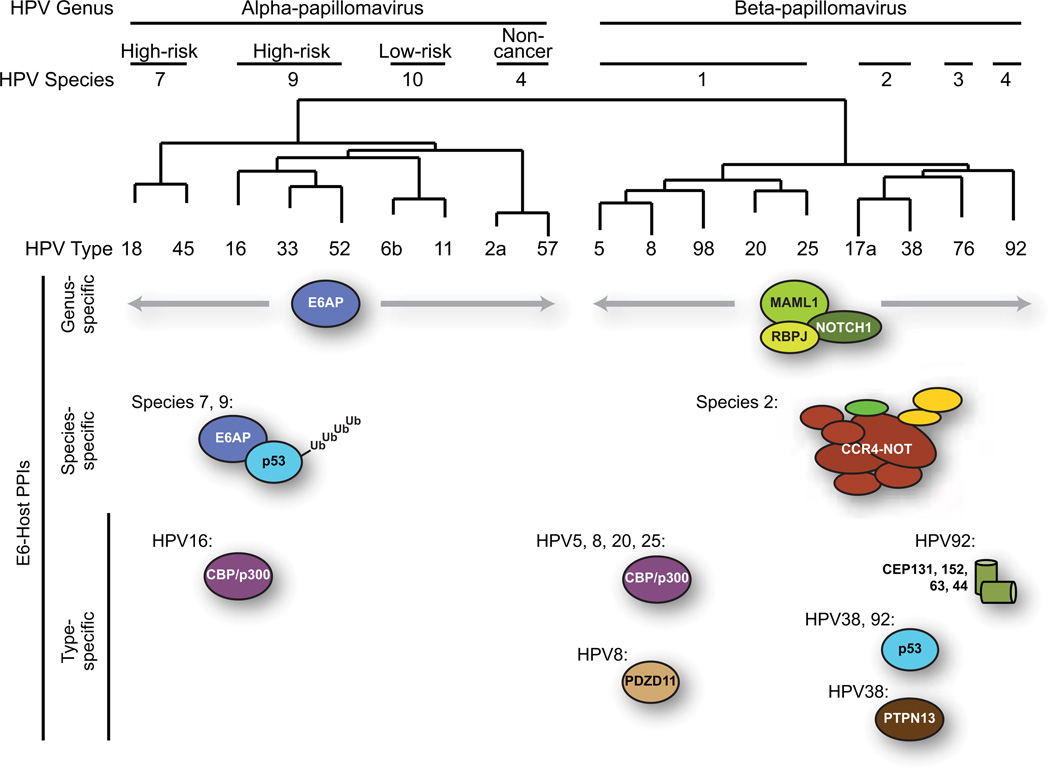
Figure 3.
Selected binding partners of HPV E6 proteins. HPV types used in the studies of Rozenblatt-Rosen et al and White et al on E6 interacting proteins (Rozenblatt-Rosen et al., 2012; White et al., in press) were arranged into a phylogenetic tree based on the sequences of their L1 genes. Selected conserved, species-specific, or type-specific PPIs involving E6 proteins are indicated.
E6AP, p53, and the proteasome
Both studies detected known E6 binding partners. All genus alpha E6s bound to E6AP, although in our study the interactions with the non-cancer types (species 4) were not scored as HCIPs by the CompPASS software. This reinforces the idea that the interaction between E6 and E6AP has different consequences for different genus alpha viruses, since only for the viruses from high-risk species does the complex also contain p53. Rozenblatt-Rosen and colleagues detect p53 in complex only with HPV18 E6, highlighting the difficulty of detecting this interaction when the proteasome is active. We found p53 with all five of our high-risk genus alpha E6s, but only in one case could we detect this interaction in cells with an active proteasome. Other known interacting partners of E6AP including HERC2 (Kuhnle et al., 2011) were identified in these studies. HERC2 bound to HPV18 E6 in the Rozenblatt-Rosen paper and bound to all five of the high-risk alpha E6s in our study. We also detected HERC2 in complex with the low-risk HPV6b E6, although not as an HCIP.
E6AP is known to interact with the proteasome (Besche et al., 2009; Kleijnen et al., 2000; Martinez-Noel et al., 2012; Scanlon et al., 2009; Tai et al., 2010; Wang et al., 2007), and consistent with that data we observed that E6 immunoprecipitated some proteasome subunits. We concluded that E6AP mediates the interaction of E6 with the proteasome, that this interaction is better detected after MG132 treatment, and that an E6AP binding-deficient mutant of E6 (HPV16 E6 I128T) no longer binds to the proteasome. These ideas are consistent with the Rozenblatt-Rosen publication, which detected proteasome subunits in complex with HPV16, 8, and 6b E6s.
MAML1
Recently, our group and that of Scott Vande Pol have shown that beta genus E6 proteins as well as the BPV E6 bind to the cellular protein MAML1, a transcriptional regulator that is involved in several cell signaling pathways (Brimer et al., 2012; Tan et al., 2012). This interaction is also noted in both proteomic studies, and the downstream effect of the beta E6-MAML interaction is inhibition of the Notch signaling pathway (Brimer, Lyons, and Vande Pol, 2007; Rozenblatt-Rosen et al., 2012; Tan et al., 2012). Notch acts as an oncogene in some settings such as T-cell leukemia but as a tumor suppressor elsewhere, including in epithelial cells (McElhinny, Li, and Wu, 2008; Wu and Griffin, 2004). Two recent studies have shown that Notch is frequently mutated in head and neck cancers, highlighting the importance of this pathway in the epithelial cells infected by beta HPVs (Agrawal et al., 2011; Stransky et al., 2011). Notch is a determinant of keratinocyte differentiation, and Notch activation leads to induction of the cell cycle inhibitor p21 and the expression of differentiation markers (Devgan et al., 2006; Nguyen et al., 2006; Rangarajan et al., 2001; Restivo et al., 2011). Now that the two E6 proteomic studies have extended the binding observations to a larger number of E6s, we can generalize to say that nearly all of the genus beta E6s that were tested in either study bound to MAML1. Our study was also able to detect MAML1 binding partners including Notch1 and RBPJ for most of the beta E6s tested.
These two genus-specific interactions involve the same binding motif. Genus alpha E6s bind to E6AP, and this interaction has been demonstrated to occur through the LXXLL motif in E6AP (Chen et al., 1995). The Tan and Brimer studies demonstrated that MAML1 contains an LXXLL motif that is responsible for the interaction with beta E6s (Brimer et al., 2012; Tan et al., 2012). The LXXLL motif in either case is thought to be bound by the flexible linker region between the two globular domains in the N- and C-terminal halves of E6s (Nomine et al., 2006). It is interesting to speculate, then, that the ability of E6 to bind to either an LXXLL motif like the one in E6AP or like the one in MAML1 arose early in the evolution of genus alpha versus genus beta HPVs, and that this relates to their respective tropisms for mucosal or cutaneous epithelium.
CBP/p300
Both our data and that of Rozenblatt-Rosen indicate that HPV5 and HPV8 E6 proteins (both from genus beta, species 1) bind to the acetyltransferases CBP and p300. The Rozenblatt-Rosen study proposes that this interaction is related to the impact of beta E6 proteins on Notch signaling, and this is consistent with their data since CBP/p300 contribute to the MAML1-dependent transactivation of Notch responsive genes (Oswald et al., 2001). In contrast, our results show that nearly all beta HPV E6 proteins bind to MAML1, but only three of four beta E6 proteins from species 1 and no other E6s from genus beta bound to CBP/p300. Furthermore, our proteomic data and previous studies indicate that HPV16 E6 binds to CBP/p300 (Patel et al., 1999; Zimmermann et al., 1999). Thus, if a subset of beta E6s bind to CBP and p300 as an additional way to affect Notch signaling, other beta E6s must not require CBP/p300 to impact the Notch pathway and HPV16 must use CBP/p300 for other purposes. Studies on HPV16 E6 binding to p300/CBP suggest that it blocks p53 and NFkB transcription and inhibits cellular differentiation (Patel et al., 1999) or that it downregulates p53 activity and p53-dependent transcription (Thomas and Chiang, 2005; Zimmermann et al., 1999). p300 binding to HPV5 and 8 E6s has been proposed to result in the degradation of p300 and the loss of Akt binding to p300 (Howie et al., 2011).
PDZ proteins and p53
Through examining additional HPV types, our study of E6 interactions identified a number of new binding partners of genus beta HPV E6s. Some of these are proteins that were previously thought to bind only to alpha type E6s; others are proteins not previously shown to bind to any E6. For example, p53 binds to HPV38 and HPV92 E6; cells that express these E6s appear to stabilize p53 rather than target it for degradation. We also observed that several genus beta E6s bound to proteins that contain PDZ domains. This is a surprise in light of the fact that it is the C-terminal PDZ binding motif present in high-risk HPV E6 proteins that mediates their interaction with PDZ proteins. Our study validates one of these interactions, that of the beta genus HPV38 E6 with the PDZ protein PTPN13. The HPV8 E6/PDZD11 interaction was detected in both our proteomic study and that of Rozenblatt-Rosen and colleagues. Beta papillomavirus E6 proteins do not contain the classical C-terminal PDZ binding motif, so this interaction may be a result of binding through a protein intermediate or because the PDZ protein is binding to a different sequence in E6. Even non-oncogenic viruses encode factors that bind to PDZ proteins, and these interactions can contribute to virus replication in ways unrelated to transformation (Javier and Rice, 2011). Beta HPVs have been shown to alter PDZ functions, with HPV8 E6 able to reduce transcription of the gene encoding the PDZ protein Syntenin 2 (Lazic et al., 2012).
Ccr4-Not
Finally, in our broad survey of beta E6 proteins we identified cellular proteins that bound only to E6s of a single HPV species. Our study includes two genus beta, species 2 HPV E6s, and these bind to a unique subset of cellular proteins including all 10 subunits of the 1 MDa form of the Ccr4-Not complex. Ccr4-Not is conserved from yeast to humans and has various functions, the most well-characterized being its function as a deadenylase. This deadenylase activity is a focus of study for many labs, and recently Ccr4-Not has been established as a major factor in the removal of polyA tails from messenger RNAs that are being targeted by a cognate microRNA (Behm-Ansmant et al., 2006; Braun et al., 2011; Chekulaeva et al., 2011; Fabian et al., 2011; Fabian et al., 2009). Other functions, including a ubiquitin ligase activity associated with the CNOT4 subunit (Albert et al., 2002), are less well understood. We are not aware of any other reports of a viral protein that binds to or affects the function or expression of Ccr4-Not, and we are continuing to study the downstream effects of this E6 interaction.
Centrosome and keratinocyte-specific proteins
Other species-specific interactions exist; for example HPV 92 is the only virus type in genus beta, species 4 that has yet been identified, and correspondingly it was the only HPV E6 in our study that bound to another distinct subset of cellular proteins. These interactors include HIF1α and ARNT (HIF1β), which together comprise the HIF1 heterodimer (Wang et al., 1995). Protein components of the centrosome including AZI1 (CEP131), CEP152, CEP63, and KIAA1712 (CEP44) bound to HPV92 E6, and perhaps this is related to the microtubule binding protein KIAA1543 (CAMSAP3) that bound as well. Cell adhesion proteins such as JUB, AAMP, and the bovine papillomavirus type 1 (BPV1) E6-interacting protein Paxillin bound to HPV92 E6, as did a cation transporter that regulates keratinocyte proliferation (SLC12A8), and DUSP3, a negative regulator of MAPK. Our group has recently shown that MAPK6 and HIF1AN bind to E6AP (Martinez-Noel et al., 2012), so the presence of DUSP3 and HIF1 here raise the question of whether HPV92 E6 uses other binding events to target pathways impacted by a high-risk HPV E6.
Comparison of HPV affinity purification-mass spectrometry and yeast-two-hybrid studies
As described above there is significant overlap between the two large HPV AP-MS studies completed to date. In contrast, there is less overlap between the AP-MS data and the yeast-two-hybrid data included in the Rozenblatt-Rosen et al. manuscript. Their Y2H study detected 454 binary interactions involving 123 viral baits. 28 of the 123 baits were HPV proteins from 7 virus types (the six types used in the TAP-MS study plus HPV33), and at least one cellular interactor was detected for 16 of these 28 HPV proteins. This resulted in a total of 267 interactions between HPV and cellular proteins detected by Y2H. Although interactions were detected for all seven E7 proteins and five of the E6 proteins tested, there was no overlap between the Y2H and TAP-MS datasets, at least for the HPV component of the study. In the study overall, only 6 interactions were in common between the 454 Y2H and 3787 TAP-MS interactions that were identified. This means that for the HPV baits, none of the known control interactors detected in the TAP-MS experiments (e.g. pRB1, p600, KCMF1, Zer1, or PTPN14 for E7; or p53, p300, CBP, or MAML1 for E6) were detected in the yeast two hybrid studies.
Some groups continue to conduct Y2H studies only, and a recent report described binding partners of the E2 proteins from 12 HPV types (eight from genus alpha: types 16 and 33 from high-risk species 9, types 18 and 39 from high-risk species 7, low-risk types 6 and 11 from species 10, type 32 from species 1 (associated with the oral mucosa), and type 3 from species 2; three from genus beta: types 5 and 8 from species 1 and type 9 from species 2; and one virus, HPV1, from genus mu) (Muller et al., 2012). Although their experiments began with an unbiased yeast-two-hybrid study in which they screened a cDNA library generated from HaCaT keratinocytes, the authors quickly note that only five of 53 known E2 binding partners were detected in their screen. In light of that potentially low discovery rate, they continued by using a complementary assay in mammalian cells. They chose about 25% (48) of their 202 potential interactors identified from the Y2H on the basis of having interacted with multiple E2s, then rescued another ~25% on the basis of having a potential functional relationship to E2 or its binding partners. These plus 19 ‘Gold Standard’ or known E2 interactors not identified in the screen resulted in a total of 121 proteins to be re-screened in a High-Throughput Gaussia princeps luciferase-based Complementation Assay (HT-GPCA). This assay is similar to a mammalian two-hybrid. The E2 bait was fused to one half of a luciferase reporter, while the prey was fused to the second half. A bait-prey interaction thus reconstitutes measurable luciferase activity. A cutoff value derived from a separate study was used to determine significant vs. non-significant interactions.
The HT-GPCA detected 72% of the gold standard interactions, and based on that result the authors continue to test their larger set of potential interactors in this assay. Here 42% of the potential interactions, representing 98 of the 121 potential cellular proteins, bound to one or more E2s. The authors compare this to a set of 10 cellular proteins chosen randomly and tested for interaction with E2, in which they detect ~6% of the possible interactions. They therefore establish this as their false negative rate, but it is important to note that the two sets of cellular proteins to be tested were chosen in very different ways: the potential E2 interactors as proteins involved in transcription, DNA replication, or as relatives of an initial E2 interactor; and the negative controls randomly. Thus the representation by cellular compartment, association with chromatin, and other factors are potentially quite different between the two sets, meaning that they are biased in different ways for association with E2. Since many potential interactors are associated with chromatin, proteomic experiments on the E2s will require special considerations to address the relative insolubility of chromatin-associated proteins and the transient nature of their interactions (Lambert, Pawson, and Gingras, 2012), and these experiments will be more powerful when they are conducted in a truly unbiased way.
Mutant HPV proteins are valuable proteomic tools
Additional experimental design steps can help to eliminate issues related to background or contaminant proteins. In one study from our laboratory, the use of mutant ORFs allowed Powell and colleagues to demonstrate that the HPV E8^E2C protein uses the NCoR/HDAC3 complex to repress the HPV long control region (LCR) E6/E7 promoter (Powell et al., 2010). E8^E2C is a protein that is encoded by at least a few of the alpha genus HPV types and by BPV1 (Choe et al., 1989; Doorbar et al., 1990; Rotenberg, Chow, and Broker, 1989; Stubenrauch et al., 2000). A 12 aa E8 ORF is joined through splicing to the E2 hinge and C-terminal DNA binding domains of E2; E8 replaces the full-length E2 transactivation domain. Before this study began several groups including that of Stubenrauch and colleagues had observed that E8^E2C repressed transcription from the LCR and had characterized two repression-incompetent mutant forms of E8^E2C (Choe et al., 1989; Doorbar et al., 1990; Lace et al., 2008; Rotenberg, Chow, and Broker, 1989; Stubenrauch et al., 2000; Stubenrauch et al., 2007; Stubenrauch, Zobel, and Iftner, 2001; Zobel, Iftner, and Stubenrauch, 2003). One is a 3 amino acid substitution or ‘KWK’ mutant (aa 5–7 changed from KWK to AEA); the other is a deletion of E8 aa 3–12. The Powell study used a proteomic approach in which C33A cells stably expressed wild type (wt), KWK, or deletion mutant E8^E2C proteins. These proteins were tagged with an internal HA epitope tag located in the E2 flexible hinge region. This tag had previously been shown not to interfere with E8^E2C repression functions and is a reminder that tag choice and location is a critical feature of a proteomic experiment design. Here, C-terminal tags are known to interfere with the function of the E2 DNA binding domain; an N-terminal tag on E8 interfered with the repression function.
The C33A cells were processed for anti-HA immunoprecipitation, mass spectrometry, and CompPASS analysis using an existing stats table consisting of data from 293T cells. The wt and KWK mutant E8^E2C baits recovered 12 HCIPs each, but the deletion mutant immunoprecipitated 144 HCIPs. Comparison of the wt to mutant forms established that six of the wt interacting proteins (NCOR1, TBLR1, ARG1, BLMH, TGM3, and CASP14) were not found in either of the repression-deficient mutants. Further validation experiments showed that NCOR1 and HDAC3 are critical mediators of E8^E2C repression. They bind to wild type, but not mutant E8^E2C proteins in IP-western blot experiments. When NCOR1 and/or HDAC3 were depleted from E8^E2C-expressing cells by siRNA knockdown, repression of the LCR was relieved. NCOR1 and HDAC3 did not affect LCR repression in cells expressing full-length E2. Even though the experiments were conducted in a different cell type than the 293T cells in which the comparison data was generated, this experimental design based on known biological functions allowed the authors to determine the molecular basis of E8^E2C-mediated repression of the HPV LCR.
The two E6 studies introduced above also take advantage of some existing E6 mutants. E6s consist of two globular domains bridged by a flexible linker, and this limits the availability of exposed sites that can be mutated without affecting the overall structure of E6 (Nomine et al., 2006). A few useful mutants have been characterized and are widely used, including mutants that alter the predominant splice donor site in E6 (forcing the production of only full-length E6 and not spliced E6*) (Sedman et al., 1991), an HPV16 E6 mutant that is impaired in its binding to E6AP (I128T) (Liu et al., 1999), and a mutant lacking the flexible C-terminal PDZ binding domain (in HPV16 E6, this is Δ146–151) (Kiyono et al., 1997). Both our study and that of Rozenblatt-Rosen use the I128T mutation; in the Rozenblatt-Rosen study the I128T mutant is studied in the context of the splice donor mutation, while in our study it is compared to the wild-type E6 ORF. The data from both studies suggests that introduction of the I128T mutation both eliminates E6AP binding and the binding of nearly all other E6 interactors. In the Rozenblatt-Rosen study, 16E6 nonsplice bound to 23 cellular proteins and 16E6 nonsplice with I128T bound to seven; of the three proteins in common to the two E6s, two are E6AP and PSMD3, a subunit of the proteasome. The number of peptides detected for E6AP and PSMD3 in the presence of the I128T mutation is just 10–15% of the peptides detected for the nonsplice mutant alone. Similarly, introducing the I128T mutation into the E6 in our study resulted in loss of binding to E6AP and no proteins detected as HCIPs in the absence of MG132.
These results indicate that I128 is a critical residue for HVP16 E6 interactions. Nearly all other protein interactions are lost when E6 does not bind to E6AP. While we do not believe that every interaction with E6 is mediated through E6AP (e.g., PDZ binding proteins), it is possible that high-risk HPV E6s binding to E6AP could either mediate interactions with other proteins or could stabilize E6 in a conformation that allows these interactions, and this is consistent with recent data from the Vande Pol laboratory (Ansari, Brimer, and Vande Pol, 2012). It will be interesting to characterize a similar mutant for a beta E6 that interacts with the LXXLL motif in MAML1, and to see if binding to MAML1 or another LXXLL protein is also critical for interactions with beta E6s.
Proteomic approaches to the study of other viruses
HPV researchers are certainly not alone in applying powerful proteomic techniques to better understand virus-host PPIs. Although beyond the scope of this review, we direct readers to several other studies that have used this technology successfully. Nevan Krogan’s group (Jager et al., 2012a; Jager et al., 2011) conducted a comprehensive AP-MS study using all ORFs of HIV-1 expressed in 293 and Jurkat cells. In a companion paper, they reported that HPV-1 Vif allows the recruitment of the transcription factor CBP-β to a ubiquitin ligase complex that is specific for APOBEC3G but not APOBEC3A (Jager et al., 2012b). Pichlmair and coworkers recently reported a proteomic study using 70 different viral ORFs from 30 viruses (Pichlmair et al., 2012). The ORFs were chosen for their ability to modulate the host innate immune response, and the analysis revealed a large number of host cellular pathways, some previously unreported, that are involved in the innate immune response to viral infection. Other more targeted studies have answered specific questions about herpesvirus biology (Kramer et al., 2011; Loret, Guay, and Lippe, 2008; Salsman et al., 2012) or measles biology (Komarova et al., 2011). Still other groups have applied yeast two hybrid techniques to the large-scale study of influenza and EBV (Calderwood et al., 2007; Shapira et al., 2009). However, the diversity of the HPVs and the direct comparisons between HPV types that is allowed by AP-MS does make them exceptionally well-suited to comparative proteomic studies.
Coupling systematic proteomic analysis to global gene expression analysis
As high-throughput mass spectrometry approaches become more common and as the cost and availability of high-throughput sequencing improves, we anticipate that more and more studies will combine proteomics and gene expression studies into a single systematic investigation of cellular changes mediated by virus infection and viral proteins. Rozenblatt-Rosen and colleagues demonstrate the potential of this approach by coupling gene expression data to their proteomic results. They use the same viral ORF-expressing fibroblast cell lines to generate RNA which is analyzed on microarrays. In keeping with the discovery that beta HPV E6s modulate signaling through the Notch pathway, they show that Notch transcriptional targets are decreased both in beta-E6 expressing fibroblasts and in cells depleted of MAML1. The study of influenza interactors by Shapira and coworkers also used gene expression profiling data and was able to identify several pathways (including NFkB signaling, MAPK signaling, and p53-dependent apoptosis) that are regulated both through PPIs and altered gene expression (Shapira et al., 2009).
Conclusions and future directions
The advent of large-scale proteomic techniques has broadened and deepened progress in the study of papillomavirus host cell interactions. The field has progressed from the initial discoveries describing single protein-protein interactions, to later studies identifying several host proteins in complex with a single HPV bait, and now to the studies examining multiple baits from many HPV types. Mass spectrometry advances have allowed the detection of more and more interactors with higher confidence than ever before. We are confident that the continued development of such powerful technologies and the in-depth study of the interactions they define will propel research on the papillomaviruses and papillomavirus-associated diseases.
Research Highlights.
-
-
Papillomavirus proteins engage in key interactions with host cell proteins
-
-
Affinity purification-mass spectrometry or yeast-two-hybrid are current strategies
-
-
Published proteomics studies have focused on the HPV E6 and E7 oncoproteins
-
-
HPV diversity and disease association is reflected by protein-protein interactions
Acknowledgement
Studies related to the papillomaviruses conducted in the Howley laboratory referred to in this review were supported by grants from the NIH to PMH: 1RC1 CA145188 and P01 CA50661.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Elizabeth A. White, Email: elizabeth_white@hms.harvard.edu.
Peter M. Howley, Email: peter_howley@hms.harvard.edu.
REFERENCES
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgul B, Cooke JC, Storey A. HPV-associated skin disease. J Pathol. 2006;208(2):165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21(3):355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari T, Brimer N, Vande Pol SB. Peptide Interactions Stabilize and Restructure Human Papillomavirus Type 16 E6 To Interact with p53. J Virol. 2012;86(20):11386–11391. doi: 10.1128/JVI.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arron ST, Ruby JG, Dybbro E, Ganem D, Derisi JL. Transcriptome sequencing demonstrates that human papillomavirus is not active in cutaneous squamous cell carcinoma. J Invest Dermatol. 2011;131(8):1745–1753. doi: 10.1038/jid.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20(14):1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401(1):70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48(11):2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44(1):120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Braun P, Gingras AC. History of protein-protein interactions: from egg-white to complex networks. Proteomics. 2012;12(10):1478–1498. doi: 10.1002/pmic.201100563. [DOI] [PubMed] [Google Scholar]
- Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, Venkatesan K, Rual JF, Vandenhaute J, Cusick ME, Pawson T, Hill DE, Tavernier J, Wrana JL, Roth FP, Vidal M. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6(1):91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimer N, Lyons C, Vande Pol SB. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology. 2007;358(2):303–310. doi: 10.1016/j.virol.2006.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimer N, Lyons C, Wallberg AE, Vande Pol SB. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene. 2012;31 doi: 10.1038/onc.2011.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A. 2007;104(18):7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18(11):1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Reid CE, Band V, Androphy EJ. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269(5223):529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- Choe J, Vaillancourt P, Stenlund A, Botchan M. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J Virol. 1989;63(4):1743–1755. doi: 10.1128/jvi.63.4.1743-1755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6(3):439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Coon JJ, Syka JE, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. Biotechniques. 2005;38(4):519, 521, 523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- DeMasi J, Huh KW, Nakatani Y, Munger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci U S A. 2005;102(32):11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan V, Nguyen BC, Oh H, Dotto GP. p21WAF1/Cip1 suppresses keratinocyte differentiation independently of the cell cycle through transcriptional up-regulation of the IGF-I gene. J Biol Chem. 2006;281(41):30463–30470. doi: 10.1074/jbc.M604684200. [DOI] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20(4):1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110(5):525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- Doorbar J, Parton A, Hartley K, Banks L, Crook T, Stanley M, Crawford L. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology. 1990;178(1):254–262. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. High-quality binary interactome mapping. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12(10):1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66(12):6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243(4893):934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18(11):1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, 3rd, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35(6):868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltkamp MC, de Koning MN, Bavinck JN, Ter Schegget J. Betapapillomaviruses: innocent bystanders or causes of skin cancer. J Clin Virol. 2008;43(4):353–360. doi: 10.1016/j.jcv.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallve S, Alonso A, Bravo IG. Papillomaviruses: different genes have different histories. Trends Microbiol. 2005;13(11):514–521. doi: 10.1016/j.tim.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Chaturvedi AK, Lowy DR. HPV prophylactic vaccines and the potential prevention of noncervical cancers in both men and women. Cancer. 2008;113(10 Suppl):3036–3046. doi: 10.1002/cncr.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Bravo IG, Schulz E, Bracho MA, Deaville R, Jepson PD, Van Bressem MF, Stockfleth E, Nindl I. Modular organizations of novel cetacean papillomaviruses. Mol Phylogenet Evol. 2011;59(1):34–42. doi: 10.1016/j.ympev.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Gulbahce N, Yan H, Dricot A, Padi M, Byrdsong D, Franchi R, Lee DS, Rozenblatt-Rosen O, Mar JC, Calderwood MA, Baldwin A, Zhao B, Santhanam B, Braun P, Simonis N, Huh KW, Hellner K, Grace M, Chen A, Rubio R, Marto JA, Christakis NA, Kieff E, Roth FP, Roecklein-Canfield J, Decaprio JA, Cusick ME, Quackenbush J, Hill DE, Munger K, Vidal M, Barabasi AL. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol. 2012;8(6):e1002531. doi: 10.1371/journal.pcbi.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Howie HL, Koop JI, Weese J, Robinson K, Wipf G, Kim L, Galloway DA. Beta-HPV 5 and 8 E6 promote p300 degradation by blocking AKT/p300 association. PLoS Pathog. 2011;7(8):e1002211. doi: 10.1371/journal.ppat.1002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM, Lowy DR. Papillomaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5th edition 2006. [Google Scholar]
- Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, Zhang H, Karlan B, Greene MI, Wang Q. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2012 doi: 10.1038/onc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81(18):9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci U S A. 2005;102(32):11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10(13):4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98(8):4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97(3):1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D'Orso I, Fernandes J, Fahey M, Mahon C, O'Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Global landscape of HIV-human protein complexes. Nature. 2012a;481(7381):365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Gulbahce N, Cimermancic P, Kane J, He N, Chou S, D'Orso I, Fernandes J, Jang G, Frankel AD, Alber T, Zhou Q, Krogan NJ. Purification and characterization of HIV-human protein complexes. Methods. 2011;53(1):13–19. doi: 10.1016/j.ymeth.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012b;481(7381):371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol. 2011;85(22):11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396(6706):84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci U S A. 1997;94(21):11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6(2):409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Komarova AV, Combredet C, Meyniel-Schicklin L, Chapelle M, Caignard G, Camadro JM, Lotteau V, Vidalain PO, Tangy F. Proteomic analysis of virus-host interactions in an infectious context using recombinant viruses. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M110.007443. M110 007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer T, Greco TM, Enquist LW, Cristea IM. Proteomic characterization of pseudorabies virus extracellular virions. J Virol. 2011;85(13):6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(2):467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuhnle S, Kogel U, Glockzin S, Marquardt A, Ciechanover A, Matentzoglu K, Scheffner M. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J Biol Chem. 2011;286(22):19410–19416. doi: 10.1074/jbc.M110.205211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lace MJ, Anson JR, Thomas GS, Turek LP, Haugen TH. The E8--E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J Virol. 2008;82(21):10841–10853. doi: 10.1128/JVI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Pawson T, Gingras AC. Mapping physical interactions within chromatin by proteomic approaches. Proteomics. 2012;12(10):1609–1622. doi: 10.1002/pmic.201100547. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Lazic D, Hufbauer M, Zigrino P, Buchholz S, Kazem S, Feltkamp MC, Mauch C, Steger G, Pfister H, Akgul B. Human papillomavirus type 8 e6 oncoprotein inhibits transcription of the PDZ protein syntenin-2. J Virol. 2012;86(15):7943–7952. doi: 10.1128/JVI.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Lemmens I, Tavernier J. Mammalian two-hybrids come of age. Trends Biochem Sci. 2009;34(11):579–588. doi: 10.1016/j.tibs.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17(1):43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2012 doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen JJ, Gao Q, Dalal S, Hong Y, Mansur CP, Band V, Androphy EJ. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73(9):7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret S, Guay G, Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82(17):8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy DR, Solomon D, Hildesheim A, Schiller JT, Schiffman M. Human papillomavirus infection and the primary and secondary prevention of cervical cancer. Cancer. 2008;113(7 Suppl):1980–1993. doi: 10.1002/cncr.23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283(12):8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32(15):3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27(38):5138–5147. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- McLaughlin-Drubin ME, Huh KW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82(17):8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol. 2006;80(19):9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengual-Chulia B, Garcia-Perez R, Gottschling M, Nindl I, Bravo IG. Novel animal papillomavirus sequences and accurate phylogenetic placement. Mol Phylogenet Evol. 2012 doi: 10.1016/j.ympev.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Muller M, Jacob Y, Jones L, Weiss A, Brino L, Chantier T, Lotteau V, Favre M, Demeret C. Large scale genotype comparison of human papillomavirus e2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog. 2012;8(6):e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8(13):4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12(10):1639–1655. doi: 10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20(8):1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, Orfanoudakis G, Kieffer B, Trave G. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21(5):665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21(22):7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18(18):5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SD, Aebersold RH. Proteomics: the first decade and beyond. Nat Genet. 2003;33(Suppl):311–323. doi: 10.1038/ng1106. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Burckstummer T, Muller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487(7408):486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- Powell ML, Smith JA, Sowa ME, Harper JW, Iftner T, Stubenrauch F, Howley PM. NCoR1 mediates papillomavirus E8;E2C transcriptional repression. J Virol. 2010;84(9):4451–4460. doi: 10.1128/JVI.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, Miele L, Aguet M, Radtke F, Dotto GP. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo G, Nguyen BC, Dziunycz P, Ristorcelli E, Ryan RJ, Ozuysal OY, Di Piazza M, Radtke F, Dixon MJ, Hofbauer GF, Lefort K, Dotto GP. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011;30(22):4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg MO, Chow LT, Broker TR. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology. 1989;172(2):489–497. doi: 10.1016/0042-6822(89)90191-8. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487(7408):491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, Frappier L. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. J Virol. 2012;86(2):806–820. doi: 10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Gilmore JM, Carrozza MJ, Li B, Workman JL, Florens L, Washburn MP. Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics. PLoS One. 2009;4(10):e7310. doi: 10.1371/journal.pone.0007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Washburn MP. Building protein-protein interaction networks with proteomics and informatics tools. J Biol Chem. 2011;286(27):23645–23651. doi: 10.1074/jbc.R110.174052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon TC, Gottlieb B, Durcan TM, Fon EA, Beitel LK, Trifiro MA. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp Cell Res. 2009;315(2):176–189. doi: 10.1016/j.yexcr.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- Sedman SA, Barbosa MS, Vass WC, Hubbert NL, Haas JA, Lowy DR, Schiller JT. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J Virol. 1991;65(9):4860–4866. doi: 10.1128/jvi.65.9.4860-4866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenrauch F, Hummel M, Iftner T, Laimins LA. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J Virol. 2000;74(3):1178–1186. doi: 10.1128/jvi.74.3.1178-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenrauch F, Straub E, Fertey J, Iftner T. The E8 repression domain can replace the E2 transactivation domain for growth inhibition of HeLa cells by papillomavirus E2 proteins. Int J Cancer. 2007;121(10):2284–2292. doi: 10.1002/ijc.22907. [DOI] [PubMed] [Google Scholar]
- Stubenrauch F, Zobel T, Iftner T. The E8 domain confers a novel long-distance transcriptional repression activity on the E8E2C protein of high-risk human papillomavirus type 31. J Virol. 2001;75(9):4139–4149. doi: 10.1128/JVI.75.9.4139-4149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Front Mol Neurosci. 2010;3:12. doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MJ, White EA, Sowa ME, Harper JW, Aster JC, Howley PM. Cutaneous beta-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc Natl Acad Sci U S A. 2012;109(23):E1473–E1480. doi: 10.1073/pnas.1205991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25(16):7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Zakrzewska A, Dudgeon DD, Jiang Y, Lazo JS, Kwon YT. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284(3):1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17(2):251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, Lamond A. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183(2):223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Doorslaer K, Gopalan V, Bandaru S, Mahmoud Y, Tan Q, Xirasaga S, McBride AA. Papillomavirus Episteme (PaVE), vol. 2012. 2012 doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol. 2009;83(17):8885–8892. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287(5450):116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26(17):1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- Watson M, Saraiya M, Ahmed F, Cardinez CJ, Reichman ME, Weir HK, Richards TB. Using population-based cancer registry data to assess the burden of human papillomavirus-associated cancers in the United States: overview of methods. Cancer. 2008;113(10 Suppl):2841–2854. doi: 10.1002/cncr.23758. [DOI] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- White EA, Kramer RE, Tan MJ, Hayes SD, Harper JW, Howley PM. Comprehensive Analysis of Host Cellular Interactions with Human Papillomavirus E6 Proteins Identifies New E6 Binding Partners and Reflects Viral Diversity. J. Virol. 86 doi: 10.1128/JVI.02172-12. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Sowa ME, Tan MJ, Jeudy S, Hayes SD, Santha S, Munger K, Harper JW, Howley PM. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A. 2012;109(5):E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334(6178):124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wu L, Griffin JD. Modulation of Notch signaling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumorigenesis. Semin Cancer Biol. 2004;14(5):348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117(3):349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Cardoza JD, Ficarro SB, Adelmant GO, Lazaro JB, Marto JA. Online nanoflow RP-RP-MS reveals dynamics of multicomponent Ku complex in response to DNA damage. J Proteome Res. 2010;9(12):6242–6255. doi: 10.1021/pr1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Degenkolbe R, Bernard HU, O'Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73(8):6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel T, Iftner T, Stubenrauch F. The papillomavirus E8-E2C protein represses DNA replication from extrachromosomal origins. Mol Cell Biol. 2003;23(22):8352–8362. doi: 10.1128/MCB.23.22.8352-8362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5(9):2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]