Abstract
MicroRNAs have been globally profiled in cancers, usually by microarrays, but there is poor agreement between studies in even the same cancer, and very few targets of the microRNAs have been validated. To overcome the lack of reproducibility, we used two genomic strategies for microRNA profiling, next generation sequencing and Locked Nucleic Acid miRNA microarrays, and verified the concordant changes by quantitative RT-PCR. miR-125b and members of miR-99 family (miR-99a, -99b, -100) were down-regulated in all three assays in the more advanced prostate cancer cell line C4-2 relative to the parental LNCaP cells. Similar decrease was seen in the transformed prostate cancer cell line WPE1-NB26 compared to its untransformed parent, RWPE1. Interestingly, all four miRNAs were also down-regulated in human prostate tumor tissue compared to normal prostate. Transfection of miR-99a, -99b or -100 inhibited the growth of prostate cancer cells and decreased the expression of prostate-specific antigen (PSA) indicating their potential role as tumor suppressors. To identify validated targets, we combined the computational prediction of potential targets, microarrays to detect mRNAs down-regulated by miRNAs and an analysis of the loading of mRNA on polyribosomes. We validated three direct targets of the miR-99 family: chromatin remodeling factors SMARCA5 and SMARCD1 and a kinase involved in signal transduction, mTOR and demonstrated that the expression of PSA is post-transcriptionally regulated by miR-99 family at least partially through repression of SMARCA5.
Keywords: microRNA, prostate cancer, deep sequencing, locked nucleic acid microarray
Introduction
A major advance in biology in the last decade is the discovery of small regulatory non-coding RNAs including microRNA (miRNA), small interfering RNA (siRNA), tiny noncoding RNAs (tncRNAs) and Piwi-interacting RNA (piRNA) (1). miRNAs typically bind to the 3′-untranslated region (3′-UTR) of a target mRNA and posttranscriptionally regulate its expression by degrading the mRNA and repressing translation. Many studies have demonstrated that miRNAs play critical roles in cancer (2). Each type and/or stage of cancer exhibits a characteristic miRNA expression profile, which therefore has the potential to serve as a useful tool in cancer diagnosis and prognosis (3) (4).
Prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer-related deaths in the male population of the United States. Initially, prostate cancers depend upon androgens for their growth. Therefore, the primary treatment for metastatic prostate cancer is androgen deprivation therapy, achieved by orchiectomy or anti-androgens. However, prostate cancer often progresses into an androgen-refractory metastatic stage (5) (6). Therefore, understanding the mechanism of progression to androgen refractoriness may allow the treatment of advanced prostate cancer.
Several lines of evidence have noted the role of miRNAs in prostate cancer. Comparison of an aggressive PC3 line to LNCaP and 22Rv1 cell lines showed that up-regulation of miR-221 and miR-222 promoted cell proliferation through targeting a cell cycle regulator p27(7). Advanced cancer cell lines PC3 and C4-2B have a reduced level of miR-146a, whose ectopic overexpression suppresses their growth (8). Ectopic expression of miR-126* was also shown to suppress the proliferation and invasion of androgen-dependent LNCaP cells (9). miR-125b is differentially expressed between androgen-dependent cell line LNCaP and androgen-refractory cell lines Cds1 and Cds2, as well as in benign and malignant prostate tumor tissues (10). A recent review points out that there is extensive disagreement between the published profiles of microRNAs in prostate cancer (11). The differences could arise from limitations of the hybridization method for detecting the small RNAs and from the wide variability of malignant tissue content in clinical samples.
All published expression profiles of miRNAs in prostate cancer depended on microarray hybridization and none utilized the recently available profiling method of next generation sequencing of miRNAs. In this paper, we systematically examined the expression profile of miRNAs in an androgen-dependent human prostate cancer cell line LNCaP and its derivative androgen-refractory more advanced cell line C4-2 using both Roche 454 sequencing and miRCURY LNA microarray platform. We confirmed the changes that were concordant by the two methods by miRNA-specific quantitative RT-PCR to identify four miRNAs, miR-125b and members of miR-99 family (miR-99a, -99b, -100), that were down-regulated in C4-2 relative to LNCaP. These miRNAs were decreased in human prostate tumor tissue compared to normal prostate tissue as well, indicating their importance in the development of prostate cancer. One of the bottlenecks in the study of miRNAs is the identification of relevant miRNA targets. Computational methods make too many predictions with an unacceptable level of false positives. After it became apparent that miRNAs often lead to the degradation of target mRNAs, we and others used microarray analysis to find down-regulated mRNAs to improve target prediction. In this report, we have added a third approach, the transfer of mRNAs bound to microRNAs from polyribosomes to monoribosomes, to identify three bona fide targets of the miR-99 family. We propose that the miR-99 family regulates the growth and PSA production of prostate epithelial cells at least partially through repressing these three targets SMARCA5, SMARCD1 and mTOR.
Materials and Methods
Cells and Tissues
Human prostate cancer cells LNCaP and C4-2 were maintained in RPMI 1640 medium, supplemented with 10% fetal bovine serum. For experiments on androgen responsiveness, cells were cultured in phenol red-free RPMI 1640 medium supplemented with charcoal:dextran stripped fetal bovine serum (Hyclone) for 48 hours before the addition of the androgen analog R1881 (Perkin-Elmer). De-identified mid-Gleason grade prostate cancer and normal prostate were obtained from the University of Virginia mid Atlantic CHTN. A pathologist screened sections so that at least 70% of the cells in a cancer section were malignant.
mRNA microarray
mRNA microarray was performed with Affymatrix HG_U133 Plus 2.0 array.
Transfection of siRNA and miRNA duplex
Transfection of siRNA, miRNA duplex or 2′-O-methyl antisense oligonucleotide was performed with Lipofectamine RNAiMax reagent (Invitrogen) as described (12).
Western blotting
The antibodies used were as follows: anti-SMARCD1 (BD Bioscience), anti-SMARCA5 (Santa Cruz), anti-mTOR (BD Bioscience), anti-PPFIA3 (ProteinTech Group), anti-AR (BD Bioscience) and anti-β-actin (Sigma). The western blot image was captured by G:Box iChemi XT gel documentation and analysis system. Signal intensity of western blots was quantified with GeneTools from SynGene.
RNA isolation and quantification of miRNA
Total RNA was extracted using TRIzol (Invitrogen). 1μg total RNA was reverse transcribed using NCode miRNA First-Strand cDNA Synthesis kit (Invitrogen). The expression level of miRNAs was measured by quantitative PCR using NCode SYBR GreenER miRNA qPCR kit (Invotrogen) in triplicate. U6 small nuclear RNA (snU6) was used to normalize the expression data of miRNAs. The primer sequence of snU6 is 5′-CTGCGCAAGGATGACACGCA-3′. miRNA microarray profiling was carried out using Exiqon miRCURY LNA array system (v.9.2).
Cloning of small RNAs and Roche 454 deep sequencing
Small RNA cloning was performed as described in Lau’s paper with minor modifications (13). Small RNA with a size of 17-26 nt was gel purified from 500 μg of total RNA. Purified small RNA was ligated with a modified 3′-adaptor, followed by a 5′-adaptor ligation, PCR amplification and concatamerization. Concatamerized DNAs with a size of 200-250nt were subjected to Roche 454 deep sequencing (VBI Core Lab at Virginia Bioinformatics Institute in Virginia Tech).
Luciferase reporter assay
The 3′-UTR fragments of SMARCD1, SMARCA5, mTOR and PPFIA3 containing miR-99 family binding sites were cloned into a modified vector pRL-CMV (12). The mutations were made to the miR-99 family binding sites in the 3′UTR-MUT clones. The primers used in 3′UTR or 3′UTR-MUT cloning are described in Table S4. The luciferase reporter assay was performed as previously described (12).
Polyribosome fractionation and qRT-PCR
48 hours after miRNA duplex or si-GL2 transfection in C4-2 cells, polysome fractionation assay was performed as described (14). The total RNAs from monoribosome and polyribosome fractionations were extracted separately, and subjected to qRT-PCR analysis for individual mRNAs.
BrdU Incorporation and PSA ELISA assay
BrdU incorporation was measured as previously described (15) and was normalized to cell density measured by MTT assay (Promega). PSA ELISA assay was performed using culture supernatant 72 hr after siRNA/miRNA duplex transfection using Human PSA ELISA Kit (Abazyme) according to the manufacturer’s instructions and normalized to MTT assay.
Results
Screening for differential expression of miRNAs in prostate cancer cells
Small RNAs of 17-26 nucleotides were cloned from two prostate cancer cell lines, LNCaP and C4-2, and subjected to 454 deep sequencing. A flowchart for the analysis of 454 deep sequencing data is outlined in Fig S1. The sequencing reads, ~190,000 from each sample, were compared with one another to yield unique sequences and their cloning frequencies. In further analysis, we only included unique sequences cloned five times or more, which add up to more than a thousand. Among them, a few hundred sequences were cloned more than 50 times (Table S1).
miRNAs were identified by BLASTing the unique sequences against the miRNA database (miRbase release 10.0, (16)) (Fig S1). As expected, miRNAs are the most abundant class of small RNAs in the cloned sequences; 37% of sequencing reads were mapped to miRNAs in the database (Table S1). A report on the analysis of the non-miRNA short RNAs has been published (17). Altogether, 293 miRNAs were cloned from the two cell lines. The most frequently cloned miRNAs were let-7 family members, miR-125b, -99a, -200c, -17, and -21, suggesting that these miRNAs are abundant in these cell lines (Table S2).
Significant change in the relative cloning frequency of several miRNAs between LNCaP and C4-2 (Table 1) suggested that these miRNAs are differentially expressed between the two cell lines. To confirm the changes in these miRNAs, we obtained miRNA expression profile from LNA (Locked Nucleic Acid)-microarray and compared the array data to the 454 sequencing data. Based on these two data sets, the expression of several miRNAs was measured by quantitative PCR (Fig 1A). The data are summarized in Table 1. In general, the qRT-PCR results are more consistent with the microarray data than 454 sequencing data, probably because both qRT-PCR and microarray are hybridization-based methods.
Table 1.
Ratios of miRNAs in C4-2 relative to LNCaP.
| miRNA | microarray | sequencing | qPCR | |
|---|---|---|---|---|
| miRNAs signficantly decreased in C4-2 relative to LNCaP |
hsa-miR-100 | 0.38 | NA | 0.17 |
| hsa-miR-125b | 0.34 | 0.31 | 0.21 | |
| hsa-miR-19b | 0.48 | 0.43 | 0.23 | |
| hsa-miR-99a | 0.39 | 0.21 | 0.26 | |
| miRNAs modestly decreased in C4-2 relative to LNCaP |
hsa-miR-99b | 0.66 | 0.62 | 0.43 |
| hsa-miR-106a | 0.49 | 0.97 | 0.41 | |
| hsa-miR-21 | 0.71 | 0.7 | 0.5 | |
| hsa-miR-16 | 0.86 | 0.61 | 0.59 | |
| miRNAs with no signficant change | hsa-miR-222 | 1.01 | 2.19 | 0.92 |
| hsa-miR-29a | 1.35 | 2.3 | 1.12 | |
| hsa-miR-15b | 1.07 | 0.84 | ||
| hsa-let-7b | 1.02 | 1.51 | ||
| hsa-miR-200b | 1.09 | 1.07 | 0.79 | |
| hsa-miR-22 | 1.12 | 2.17 | 0.81 | |
| miRNAs signficantly increased in C4- 2 relative to LNCaP |
hsa-miR-196b | 1.39 | 5.32 | 2.48 |
| hsa-miR-557 | 1.97 | NA | 3.28 | |
| hsa-miR-9 | 2.76 | NA | 5.86 |
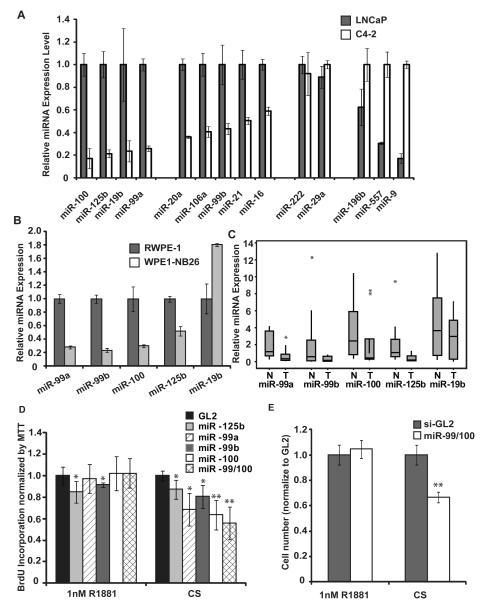
Figure 1. Identifying miR-99a family as potential tumor suppressors.
A-B. qPCR was used to measure the expression level of miRNAs. The value was normalized to that of snU6. The mean and standard deviation from triplicate samples are indicated. C. The expression of five miRNAs was measured by qPCR in 10 human normal prostate tissue samples (N) and 10 human prostate tumor samples (T). The value was normalized to that of snU6. The data is presented as a box-plot showing quartiles and the median (Minitab). The vertical line indicates the range. Asterisk (*) indicates the outliers, which are beyond the outerquartile by > 3 times the interquartile range (Minitab). The p-values between normal and tumor for miR-99a, -99b, -100, -125b and -19b are 0.0086, 0.11, 0.012, 0.015 and 0.13 respectively (excluding outliers). D-E. Upon transfection of indicated miRNAs or si-GL2, C4-2 cells were cultured in charcoal-stripped serum with or without 1nM R1881. D. After 72 hrs, BrdU incorporation was measured and normalized to cell density from MTT assay (y-axis). The mean and standard deviation from triplicate samples are shown. The value of si-GL2 is set as 1. * indicates p-value of difference from si-GL2 < 0.05; ** indicates p-value < 0.01. E. After 72 hrs, cell number was counted in a hemacytometer. The mean and standard deviation from triplicate samples are shown. ** indicates p-value of difference from si-GL2 < 0.01.
miR-100, -125b, -19b, and miR-99a were the most down-regulated miRNAs in C4-2 relative to LNCaP. miR-20a, -106a, -99b, -21 and miR-16 were modestly decreased in C4-2. In contrast, miR-9, -557 and -196b were the most up-regulated in C4-2 relative to LNCaP (with more than two fold changes confirmed by at least two methods). The differential expression of these miRNAs between LNCaP and the more advanced C4-2 lead us to hypothesize that the change of these miRNAs may be important in the progression of prostate cancer.
Confirmation of miRNA changes in other prostate cell line model and cancer tissue
To further test whether the changes in the expression level of the miRNAs we have seen in C4-2 and LNCaP are correlated with the progression of prostate cancer, we measured miRNA expression by qRT-PCR in the immortalized prostate epithelial cell RWPE-1 and the invasive cancer cell line WPE1-NB26 derived from RWPE-1. miR-125b and members of miR-99 family (miR-99a, miR-99b, and miR-100) also exhibited a significant decrease in WPE1-NB26 compared to RWPE1 cells (Fig 1B).
To evaluate if the differential expression of miRNAs in the cell lines was also seen in human tumor specimens, we performed miRNA qRT-PCR from 10 human prostate tumor samples and 10 normal prostate tissue samples. miR-125b and miR-99 family were significantly decreased in the human prostate tumor samples compared to normal tissue (Fig 1C). Our data are consistent with another study where miR-125b and the miR-99 family were decreased in prostate cancer compared to normal tissue (Table S3) (18) (19). Therefore, miR-125b and miR-99 family may play an important role during the genesis and progression of prostate cancer.
miR-99 family as potential tumor suppressors
Having observed a decrease of miR-99 family and miR-125b in human prostate cancer cells relative to normal prostate tissue, we tested whether these miRNAs affect the proliferation of prostate cancer cells. We transfected these miRNAs in C4-2, where their initial expression was low, and measured the growth of cells by BrdU incorporation assay and counting cell numbers. Unlike miR-125b, transfection of miR-99a, -99b or -100 inhibits the growth of C4-2 cells more markedly in the absence of androgen (CS) than in the presence of 1nM synthetic androgen R1881 (Fig 1D, E). This inhibition of androgen-independent growth by the miR-99 family requires the presence of AR, as the miR-99 family does not affect the growth of PC3 and Du145 cells (Fig S2). Thus, the reduction of miR-99 family, seen during the progression from LNCaP to C4-2, could provide a growth advantage under androgen-depleted condition. This result encouraged us to identify relevant target genes that are regulated by the miR-99 family.
Identification of targets by bioinformatics and microarray
miR-99a, miR-99b and miR-100 belong to the same family with a shared seed sequence (nucleotides 2-7 of the miRNA), which is known to be the critical determinant in recognition of target mRNAs (Fig S3A). Therefore, miR-99 family members are predicted to target a common list of genes according to the computational target prediction program Targetscan. We employed two filtering methods to obtain a shorter list of potential targets of miR-99 family. Intersection of mRNAs down-regulated by the miRNA with in silico predicted targets was previously shown to yield a significantly shorter list containing bona fide targets (12). We performed a microarray analysis to detect mRNAs decreased after transfection of miR-99a compared to control siRNA (si-GL2) in C4-2 cells. Among the hundreds of targets predicted by TargetScan, 19 were down-regulated by at least a third by miR-99a (Table 2).
Table 2.
Genes downregulated at mRNA level and/or blocked at translational initiation by miR-99a.
| Target gene | Gene name | miR-99a/GL2 |
miR-99a monosome/polysome si-GL2 monosome/polysome |
|---|---|---|---|
| TRIB1 | tribbles homolog 1 (Drosophila) | 0.17 | 1.41 |
| HOXA1 | homeobox A1 | 0.28 | 2.74 |
| INSM1 | insulinoma-associated 1 | 0.29 | |
| ADCY1 | adenylate cyclase 1 (brain) | 0.42 | |
| CTDSPL | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
0.49 | |
| PPFIA3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
0.5 | 5.29 |
| SMARCA5 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
0.51 | 6.83 |
| SMARCD1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
0.51 | 6.5 |
| KBTBD8 | kelch repeat and BTB (POZ) domain containing 8 | 0.52 | |
| C4orf16 | chromosome 4 open reading frame 16 | 0.52 | |
| FRAP1 | FK506 binding protein 12-rapamycin associated protein 1/mTOR |
0.54 | 5.31 |
| SLC44A1 | solute carrier family 44, member 1 | 0.57 | |
| BMPR2 | bone morphogenetic protein receptor, type II (serine/threonine kinase) |
0.61 | |
| BAZ2A | bromodomain adjacent to zinc finger domain, 2A | 0.65 | |
| ICMT | isoprenylcysteine carboxyl methyltransferase | 0.66 | 3.64 |
| MBNL1 | muscleblind-like (Drosophila) | 0.66 | |
| FZD8 | frizzled homolog 8 (Drosophila) | 0.66 | 3.34 |
| C1orf34 | chromosome 1 open reading frame 34 | 0.67 | |
| ZZEF1 | zinc finger, ZZ-type with EF-hand domain1 | 0.67 |
Filtering targets by polyribosome/monoribosome loading
miRNAs regulate gene expression at post-transcriptional stage. The mechanisms include blocking translational initiation, ribosome loading, or translational elongation (20) (21) (22) (23) (24). Therefore, we reasoned that targets of a microRNA will shift from polyribosome to monoribosome fractions if the microRNA blocks translation initiation. To test this, we first measured the ribosome profile of three validated targets of miR-206 (DNA Polα, MMD and CX43) (Fig S3B) (12). Compared to the control transfection, miR-206 induced significant accumulation of all the three mRNAs in the monoribosome fraction (Fig S3C) encouraging us to add this assay to our filters.
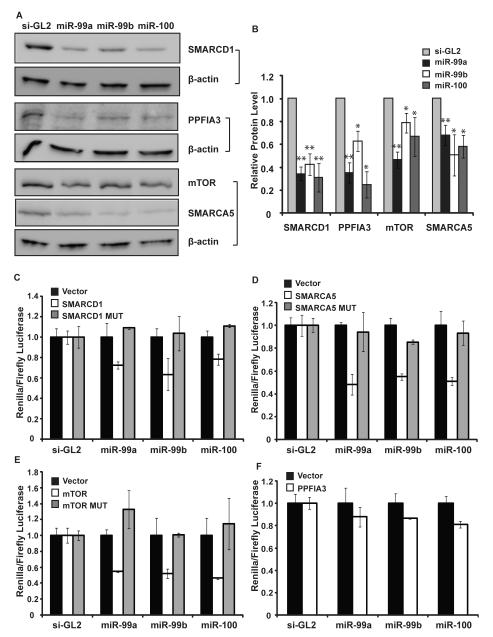
We tested 8 out of 19 genes in Table 2 by the ribosome fractionation assay before and after miR-99a transfection. These genes were selected based on previous literature implicating their involvement in prostate cancer. Upon the transfection of miR-99a, all eight genes were accumulated in the monosome fraction (Table 2). Among these genes, SMARCA5, SMARCD1, PPFIA3, and FRAP1/mTOR exhibited more than 5 fold accumulations in the monoribosome fraction. For comparison, their mRNA levels were reduced by about 2 fold after introduction of miR-99a (Table 2). Consistent with the decreased loading of ribosomes onto these mRNAs, the protein levels of all four genes were all decreased by one or more members of the miR-99 family (Fig 2A and B). Thus, these four genes are likely to be direct targets of miR-99 family and were further tested in the following experiments.
Figure 2. Confirming three direct targets of miR-99 family.
A. Western blot was used to detect changes of four target proteins after transfecting miR-99a, -99b or -100 in C4-2 cells. β-actin was used as a loading control. Full western blot see supplementary Fig S7. B. Western blots were quantified: the level of the indicated protein normalized to β-actin. The mean and SEM (error bar) of western blots are presented. * indicates p-value < 0.05; ** indicates p-value < 0.01. C-F. Luciferase assay was performed with control luciferase vector, vector with 3′UTR of four targets (indicated by gene names), or 3′UTR with mutation in the predicted target sites (indicated by MUT). The ratio of the renilla luciferase to firefly luciferase (transfection control) was normalized to that in the si-GL2 transfection.
Confirming targets as directly repressed by miR-99 family in luciferase reporter assay
Targetscan predicted one recognition site of miR-99 family in the 3′-UTR region of SMARCD1, SMARCA5, mTOR and PPFIA3 (Fig S4A). We inserted the 3′UTR fragments downstream of luciferase ORF in a reporter plasmid in order to test whether they are directly repressed by the miR-99 family. For FRAP1/mTOR, SMARCA5, and SMARCD1, the 3′-UTRs conferred repression of the heterologous luciferase ORF after transfection of miR-99a, miR-99b or miR-100 (Fig 2C-E). In all three cases, the repression by miR-99 family was abolished when we mutated the predicted target sites (Fig S4B, Fig 2C-E). In case of PPFIA, we did not observe any significant reduction of luciferase expression by miR-99 family (Fig 2F). Thus, the reduction of PPFIA3 mRNA and protein by miR-99 family (Table 2 and Fig 2A and B) was either due to an indirect effect or due to a target site in the open reading frame.
Together with the data from mRNA expression micorarray, ribosome profiling and protein measurements, the luciferase results clearly demonstrate that FRAP1/mTOR, SMARCA5, and SMARCD1 are direct targets of miR-99 family.
The miR-99 family decrease expression of PSA
miR-99a, -100 and -125b were down-regulated in C4-2 relative to LNCaP. Interestingly, the expression of these three miRNAs was repressed by an androgen analog R1881 in LNCaP cells, in a dose dependent manner (Fig 3A). As a positive control for androgen activity, we checked that R1881 stimulated the level of PSA mRNA, an androgen-responsive gene (Fig S6A). All three miRNAs are repressed by androgen and during the progression of prostate cancer, which suggests that prostate cancer progression is accompanied by the cells spontaneously phenocopying the effect of androgen. It is also possible that the reduction of these miRNAs in C4-2 relative to LNCaP may be due to hyper- and/or constitutive activation of the androgen receptor (AR) in C4-2, and conversely, these miRNAs may play an active role in androgen refractoriness in C4-2.
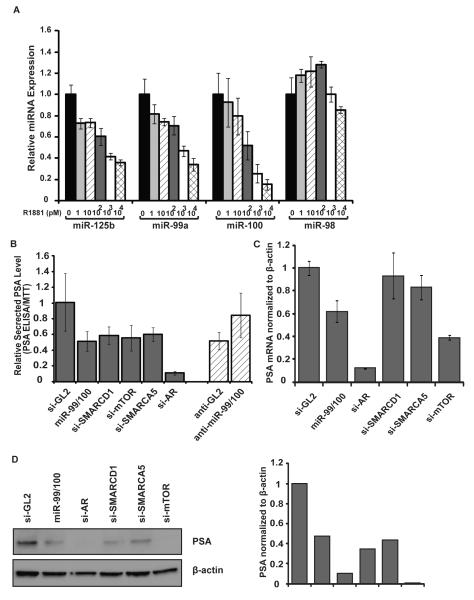
Figure 3. miR-99 family decreases PSA level.
A. qPCR assays of selected miRNAs after treating LNCaP with androgen analog R1881 at indicated concentrations. The miRNA level at no R1881 is set as 1. B. 72 hr after transfection of indicated siRNAs, miR-99 family or si-GL2 in C4-2 cells (first six bars) or transfection of 2′-O-methyl anti-sense oligonucleotide in LNCaP cells (the last two bars), PSA ELISA was measured using the culture supernatant and normalized to cell density from MTT assay. The average and standard deviation from triplicate samples are shown. C. RT-qPCR was used to determine the mRNA of PSA after transfecting indicated miRNA or siRNA in C4-2 cells. Results were normalized to β-actin. D. Western blot of PSA was performed in C4-2 cells after transfection of indicated miRNA or siRNA. β-actin was used as a loading control. Quantification of western blots. The value is normalized to β-actin.
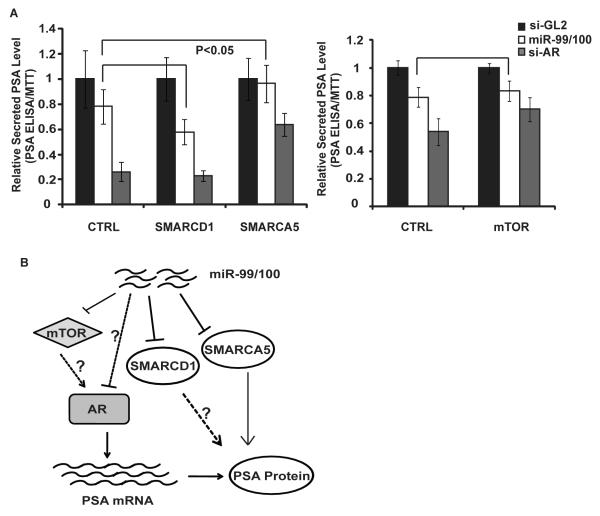
To measure the androgen-response upon modulation of the miR-99 family, miR-99a, -99b and -100 duplex were transfected to C4-2 cells in the presence of 1nM R1881. Prostate specific antigen (PSA) is an androgen-responsive secreted protein and important marker for prostate cancer detection. Its secretion was measured by ELISA and normalized to cell numbers assessed by MTT assay. When miR-99 family was ectopically expressed in C4-2, the PSA level was significantly repressed to the level of LNCaP (Fig 3B). Conversely, the PSA level was up-regulated in LNCaP cells upon inhibition of miR-99 family by treating with 2′-O-methyl antisense oligonucleotides against them (Fig 3B). To test whether the change in the secreted PSA level was due to the impaired AR activity, we tested the mRNA expression of two AR regulated genes PSA and SARG. We observed a similar decrease in mRNA level of both PSA and SARG after transfection of miR-99 family miRNAs (Fig 3C and Fig S6B). The protein level of PSA also showed a decrease after overexpression of miR-99/100 (Fig 3D). We next tested whether repression of the targets of these miRNAs phenocopied the effects of the miRNAs. The siRNA-mediated knockdown of SMARCD1 or SMARCA5 in C4-2 cells specifically decreased the PSA protein without affecting the mRNA level, suggesting a post-transcriptional regulation on PSA expression (Fig 3B-D). Knockdown of mTOR by siRNA decreases both the PSA mRNA and protein level (Fig 3B-D). Thus, repression of these targets could contribute to PSA repression by the miR-99 family, though the chromatin remodeling factors SMARCD1 and SMARCA5 appear to be required for expression of PSA protein at a post-transcriptional step. To test which of the three targets was rate-limiting after miR-99/100 expression, we ectopically expressed the open reading frame (ORF) of each of the three targets and then transfected the miRNAs of the miR-99 family. The absence of the 3′UTRs makes these exogenous genes resistant to the repression by the miR-99 family. Ectopic expression of the ORF region of SMARCA5 alone rescued the repression of miR-99/100 on the PSA protein level (Fig 4A). miRNA-resistant SMARCD1 or mTOR expression did not rescue the effect of miR-99 family on PSA expression (Fig 4A, Fig S6C-D). Additional targets of miR-99 family may contribute to the selective repression of AR activity by miR-99 family, as none of the three identified targets rescued the repression of PSA mRNA (Fig S6C-D). Our results suggest that loss of miR-99 family affects AR-driven gene expression, particularly the expression of PSA at both mRNA and protein level (Fig 4B). The de-repression of SMARCA5 by the decrease of miR-99 family in C4-2 clearly contributes to the elevated expression of PSA in this more advanced prostate cancer cell line.
Figure 4. SMARCA5 rescues the miR-99 family-induced reduction of PSA.
A. PSA ELISA assay was performed using culture supernatant of C4-2 cells stably expressing miRNA-resistant form of SMARCD1, SMARCA5 or mTOR 72hr after transfection of indicated miRNA or siRNA. Square brackets indicate p-values of the differences: <0.05 or >0.05 (unlabeled). B. Schemetic to show the regulation of miR-99 family on AR and PSA.
Discussion
In the last few years, the miRNA expression profiles have been studied by several groups in prostate cancer cell lines and clinical samples mainly using miRNA microarrays and quantitative PCR analysis (4, 11, 19, 25-28). In this study, we applied cloning and deep sequencing method besides the Locked Nucleic Acid based miRNA microarray to profile the microRNAs. We also avoided heterogeneity between prostate cancer samples by first doing the comparison between two cell lines, LNCaP and C4-2 and then following the validated changes in other cell lines and human tumors. Based on this conservative strategy with multiple iterative loops, we determined that the miR-99 family members (miR-99a, -99b and -100) were decreased in most advanced prostate cancer relative to normal prostate epithelium. Our results were supported by other profiling studies where miR-99a and miR-100 were shown to be reduced in prostate carcinomas and more aggressive and metastatic prostate tumors (27). The consistent changes of these miRNAs in prostate cancer in several independent studies suggest that decrease of miR-99 family is a signature for the genesis and progression of prostate cancer. Moreover, a frequent down-regulation of miR-99a and miR-100 was also seen in other types of cancer such as ovarian cancer, lung cancer and squamous cell carcinoma of tongue (29-31), suggesting the potential involvement of miR-99 family in the genesis and progression of cancers.
Identifying bona fide miRNA targets has been a difficult step in studying miRNAs. Bioinformatics methods utilize the sequence complement to identify targets and often produce hundreds of predicted targets. Upon experimental validation, the majority of the predicted targets appear not to be suppressed significantly by miRNAs, yielding a high false positive rate. The Bartel group reported that genes whose proteins were repressed more than 50% by miRNAs also exhibit mRNA degradation (32). Therefore, mRNA microarray is widely used to determine the actual targets of the miRNA. In many cases, however, the modest change in mRNA level is not due to a direct degradation by the miRNA and not accompanied by a decrease in protein. In this study, we thus added a polyribosome fractionation method to filter targets that were repressed at translational initiation stage. Binding of the miRNA to the 3′UTR of the target mRNA shifted the majority of target mRNA from polyribosome to monoribosome fraction without affecting the non-target mRNAs (23, 24). By using this method, we focused on four genes SMARCD1, SMARCA5, mTOR and PPFIA3. Out of the four genes we tested further by luciferase reporter assay, three proved to be direct targets of the miR-99 family. The exception, PPFIA3 was decreased at the protein level by the miR-99 family but not in the luciferase reporter with the 3′UTR of PPFIA3. A potential explanation is that the target site of the microRNA is in the open reading frame (ORF) of the gene. Indeed, the ORF region has a site with perfect match with the seed sequence of miR-99a and miR-100, which could be the primary binding site of miR-99 family members responsible for reducing PPFIA3 mRNA and protein. By adding the polyribosome fractionation screen, we were able to increase the true positive rate of candidate genes to >75%. A large scale screening of candidate genes by subjecting the RNAs from polyribosome and monoribosome fractions to sequencing or microarray would be very useful to identify target genes of miRNAs in a future study.
Hyperactivity of AR is one of the reasons that prostate cancer cells become androgen independent (5, 6, 33). C4-2 cells possess a higher activity of AR and respond to much lower concentration of androgen compared to LNCaP cells (34, 35). Restoring the level of miR-99 family members significantly reduced the AR activity in C4-2 cells as measured on the PSA and SARG promoter. Thus, repression of the miR-99 family may promote the hyperactivity of AR, which may in turn lead to the androgen independence of advanced prostate cancer. The growth effect of the miR-99 family in prostate cancer cells was tested by ectopically expressing them in C4-2 cells, where their initial expression level was low. Restoring the expression miR-99 family members reduced the cell growth in androgen depleted media, suggesting a potential tumor suppressive role of miR-99 family in prostate cancer cells. Taken together, the miR-99 family represses both AR responsiveness and cell growth in the absence of androgen. This raises the possibility of treating prostate cancer with anti-androgen along with gene-therapy vectors over-expressing the miR-99 family of miRNAs.
PSA is a serine protease belonging to the kallikrein family. As an androgen regulated gene, it has been used as a biomarker for prostate cancer diagnosis. Previous studies showed that PSA may assist the invasion of prostate cancer cells by degrading the extracellular matrix components fibronectin and laminin (36). PSA is also known to release IGF-1 and TGF-β from their binding partners and thus involved in osteoblastic lesions (37). C4-2 has higher metastatic capacity compared to LNCaP in the mouse model (38). Intrafemoral injection of C4-2 forms PSA-producing osteoblastic tumors (38). In this study, we found that miRNAs of miR-99 family decrease PSA expression at both mRNA and protein level. A decrease of miR-99 family may thus contribute to the elevation of PSA production in C4-2 compared to LNCaP, suggesting the potential involvement of miR-99 family in the bone metastasis of prostate cancer. This, of course, will need experimental testing in the future. Although siRNA against mTOR also decreased the PSA mRNA level, the miRNA-resistant forms of non of the three targets rescued PSA mRNA level (Fig S6C, D). Thus, at least one other unknown target of miR-99/100 must be important for decreasing AR activity at the PSA promoter.
SMARCA5 (hSNF2H) is a member of SWI/SNF family, containing helicase and ATPase activities. As part of a chromatin remodeling complex, it facilitates ATP-dependent nucleosome remodeling and transcription initiation (39). It is overexpressed in ovarian cancer and promotes tumor growth in ovarian cancer through interacting with remodeling and spacing factor 1 (Rsf1) (40). In this paper, we showed that as a direct target of miR-99 family, SMARCA5 regulates the PSA protein level, which contributes to the elevated expression of PSA in C4-2 cells. No direct interaction between SMARCA5 with AR or effect of SMARCA5 on translation or protein stability has been shown. Our data reveals a novel mechanism where SMARCA5 appears to regulate the expression of PSA post-transcriptionally in prostate cancer cells. Most likely this is through the regulation of expression of genes whose products are involved in the translation and/or stability of PSA protein.
SMARCD1 (BAF60a) is a member of SWI/SNF family of proteins and known to interact with the Ligand Binding Domain (LBD) of AR through its FxxFF motif in an androgen dependent manner (41). It is also known to interact with glucocorticoid receptor (GR) and provides the docking site for chromatin remodeling BRG1 complex (42). SMARCD1 is repressed by hepatocyte-specific miRNA miR-122 (43). In this paper, we showed that the miR-99 family also represses the expression of SMARCD1 at both mRNA and protein level. SMARCD1 is required for PSA protein expression but is not sufficient by itself to restore PSA expression in miR-99/100 overexpression cells.
FRAP1 (mTOR) is a phosphatidylinositol kinase-related kinase known to mediate cellular responses to growth factors and regulate cell proliferation, metabolism and angiogenesis. AKT/mTOR signaling was shown to be important during the development of androgen independence of prostate cancer. Inhibition of mTOR along with anti-androgen additively represses the prostate tumor growth in PTEN-null mouse, suggesting there might be crosstalk between mTOR and AR pathways (44). mTOR promotes translation through phosphorylation of S6K1 and 4EBP1, which may also enhance the expression of AR-responsive genes. Several studies suggest that the inhibition of mTOR combined with anti-androgen could be useful in prostate cancer treatment (45, 46). The repression of mTOR signaling by miR-100 was previously reported in clear cell ovarian cancer model (47). In this study, we showed that mTOR is repressed not only by miR-100, but also its family members miR-99a and miR-99b. mTOR is required for expression of PSA mRNA (Fig 3B), but not for maintaining AR levels (Fig S5A). It will be interesting to elucidate how mTOR impact on AR activity at the PSA promoter.
In summary, we implicated that the miRNAs of miR-99 family are repressed and three validated targets are de-repressed during the genesis and progression of prostate cancer. We also showed that miR-99 family represses AR activity and independently repress PSA protein level through SMARCA5 (Fig 4B). The consistent decrease of miR-99 family in the human prostate tumors increases the possibility of using them as a signature of prostate cancer progression. Our study also underlines the possible treatment of prostate cancer by restoring the level of miR-99 family members. Finally, in pursuing the targets of these tumor suppressive miRNAs, we make the exciting discovery that the SMARCA5 chromatin remodeling factor is important for post-transcriptional regulation of a metastatic factor, PSA.
Supplementary Material
Acknowledgements
We thank Dr. Yeou Cherng Bor for helping us in polyribosome fractionation assay. We also want to thank Drs. Jiandie Lin, Isa Hussaini for providing SMARCD1and mTOR antibodies and Dr. Bryce Paschal for many helpful discussions and experimental suggestions. This study was supported by grants PC050114 from the US Army DOD and R01 GM84465 to AD and PC094499 to DS from the US Army DOD.
This work was supported by PC050114 from the US Army DOD and R01 GM84465 to AD and DOD PC094499 from the US Army DOD to DS.
Reference
- 1.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 2.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–77. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 4.Tong AW, Fulgham P, Jay C, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–16. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Ueda T, Ichikawa T, Ito H. Androgen receptor involvement in the progression of prostate cancer. Endocr Relat Cancer. 2003;10:209–16. doi: 10.1677/erc.0.0100209. [DOI] [PubMed] [Google Scholar]
- 6.Agoulnik IU, Weigel NL. Androgen receptor action in hormone-dependent and recurrent prostate cancer. J Cell Biochem. 2006;99:362–72. doi: 10.1002/jcb.20811. [DOI] [PubMed] [Google Scholar]
- 7.Mercatelli N, Coppola V, Bonci D, et al. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One. 2008;3:e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–22. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi XB, Xue L, Yang J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:19983–8. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol Med. 2009;15:381–90. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjold ML. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–7. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 15.Machida YJ, Chen Y, Machida Y, Malhotra A, Sarkar S, Dutta A. Targeted comparative RNA interference analysis reveals differential requirement of genes essential for cell proliferation. Mol Biol Cell. 2006;17:4837–45. doi: 10.1091/mbc.E06-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. <http://microrna.sanger.ac.uk/sequences/>. [cited; Available from.
- 17.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 21.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 22.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–14. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 23.Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 24.Kong YW, Cannell IG, de Moor CH, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A. 2008;105:8866–71. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 27.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 28.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 30.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–92. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 31.Yamada H, Yanagisawa K, Tokumaru S, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47:810–8. doi: 10.1002/gcc.20582. [DOI] [PubMed] [Google Scholar]
- 32.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory CW, Johnson RT, Jr., Mohler JL, French FS, Wilson EM. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 2001;61:2892–8. [PubMed] [Google Scholar]
- 34.Gotoh A, Ko SC, Shirakawa T, et al. Development of prostate-specific antigen promoter-based gene therapy for androgen-independent human prostate cancer. J Urol. 1998;160:220–9. [PubMed] [Google Scholar]
- 35.Periyasamy S, Hinds T, Jr., Shemshedini L, Shou W, Sanchez ER. FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene. 2009 doi: 10.1038/onc.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webber MM, Waghray A, Bello D. Prostate-specific antigen, a serine protease, facilitates human prostate cancer cell invasion. Clin Cancer Res. 1995;1:1089–94. [PubMed] [Google Scholar]
- 37.Williams SA, Singh P, Isaacs JT, Denmeade SR. Does PSA play a role as a promoting agent during the initiation and/or progression of prostate cancer? Prostate. 2007;67:312–29. doi: 10.1002/pros.20531. [DOI] [PubMed] [Google Scholar]
- 38.Wu TT, Sikes RA, Cui Q, et al. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int J Cancer. 1998;77:887–94. doi: 10.1002/(sici)1097-0215(19980911)77:6<887::aid-ijc15>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 39.LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–90. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 40.Sheu JJ, Choi JH, Yildiz I, et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68:4050–7. doi: 10.1158/0008-5472.CAN-07-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Wijngaart DJ, Dubbink HJ, Molier M, de Vos C, Trapman J, Jenster G. Functional screening of FxxLF-like peptide motifs identifies SMARCD1/BAF60a as an androgen receptor cofactor that modulates TMPRSS2 expression. Mol Endocrinol. 2009;23:1776–86. doi: 10.1210/me.2008-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–20. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatfield D, Le Martelot G, Vejnar CE, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–26. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Zhu J, Efferson CL, et al. Inhibition of tumor growth progression by antiandrogens and mTOR inhibitor in a Pten-deficient mouse model of prostate cancer. Cancer Res. 2009;69:7466–72. doi: 10.1158/0008-5472.CAN-08-4385. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Mikhailova M, Bose S, Pan CX, deVere White RW, Ghosh PM. Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene. 2008;27:7106–17. doi: 10.1038/onc.2008.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, Birle DC, Tannock IF. Effects of the mammalian target of rapamycin inhibitor CCI-779 used alone or with chemotherapy on human prostate cancer cells and xenografts. Cancer Res. 2005;65:2825–31. doi: 10.1158/0008-5472.CAN-04-3137. [DOI] [PubMed] [Google Scholar]
- 47.Nagaraja AK, Creighton CJ, Yu Z, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 24:447–63. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.