Abstract
Hepatitis C virus (HCV) is a major causative agent of severe liver disease including fibrosis, cirrhosis and liver cancer. Therapy has improved over the years, but continues to be associated with adverse side effects and variable success rates. Furthermore, a vaccine protecting against HCV infection remains elusive. Development of more effective intervention measures has been delayed by the lack of a suitable animal model. Naturally, HCV infects only humans and chimpanzees. The determinants of this limited host range are poorly understood in part due to difficulties of studying HCV in cell culture. Some progress has been made elucidating the barriers for the HCV lifecycle in non-permissive species which will help in the future to construct animal models for HCV infection, immunity and pathogenesis.
Keywords: Hepatitis C virus, Animal models, Humanized mice, Species tropism, Drug development, Vaccine development, Pathogenesis
Highlights
► Hepatitis C virus has a narrow host range limited to chimpanzees and humans. ► Constraints on chimpanzees research created a great need for alternative HCV models. ► HCV species tropism is restricted at multiple steps of the viral lifecycle. ► Species barriers can be overcome by viral and/or host adaptation.
Introduction
Hepatitis C virus (HCV) is a positive-sense, single stranded RNA virus classified within hepacivirus genus of the Flaviviridae family. HCV has a high propensity for establishing lifelong persistent infections, which are associated with a significant risk of progressive liver fibrosis and hepatocellular carcinoma. World-wide at least 130 million people are chronically infected resulting in an estimated 366,000 deaths due to cirrhosis and cancer annually (Perz et al., 2006). Epidemiological data is incomplete and in the United States alone the frequency of chronic carriers may be twice as high (Edlin, 2011). Thus, HCV poses a major public health threat. Treatment options have improved but are still limited. Furthermore, the current standard of care, consisting of a combination of pegylated interferon (IFN) alpha, the nucleoside analog, ribavirin (RBV), and one of two HCV NS3–4A protease inhibitors, boceprevir or telaprevir, is not well tolerated. Prior to the approval of these directly acting antivirals (DAAs) roughly 50% of genotype 1 infected patients – clinically, the hardest HCV sub-strain to treat – who underwent treatment, cleared the infection. Inclusion of a HCV protease inhibitor to the peg-IFN/RBV regiment has increased sustained virological response (SVR) rates in certain clinical trial cohorts to 60–70% (Jacobson et al., 2011, Kwo et al., 2010, McHutchison et al., 2009, Poordad et al., 2011). Currently, numerous viral and host proteins are being pursued as antiviral drug targets. Second-generation protease inhibitors and several compounds interfering with the functions of the NS5A phospho-protein and the RNA-dependent RNA polymerase, NS5B, are already being tested in clinical trials (Yang et al., 2011). A combination of orally administered DAAs with distinct mechanisms of action holds promise to boost SVR rates across all HCV patient cohorts. Despite these successes unexpected toxicity in late-stage clinical trials has led to the discontinuation or delays in development of some very potent compounds, including the protease inhibitor BILN2061 (Lamarre et al., 2003), the nucleotide polymerase inhibitor BMS-986094(Vernachio et al., 2011), and the cyclophilin A antagonist, alisporivir (Coelmont et al., 2009). These challenges highlight the need for thorough pre-clinical testing in predictive animal models. Likewise, the development of pan-genotypic prophylactic or therapeutic vaccines instrumental in containing the HCV epidemic in resource-poor communities would be greatly accelerated by a tractable animal model.
Chimpanzees are the only available immunocompetent in vivo experimental system, but their use is limited by ethical concerns, restricted availability and prohibitively high costs. Undoubtedly, more tangible animal models are needed not only to prioritize clinical development of vaccine and drug candidates, but also to gain deeper insights into virus–host biology. The study of HCV in conventional cell culture systems, i.e. human hepatoma cells, may not accurately reflect host responses to infection. With the advent of more sensitive detection methods for viral infection and improved technical ability to culture primary hepatocytes it is now possible to dissect HCV infection in a physiological-relevant environment (Jones et al., 2010a, Ploss et al., 2010). However, even in the most advanced tissue culture platforms, including micropatterned primary hepatocyte co-cultures (MPCCs; Jones et al., 2010a, Khetani and Bhatia, 2008, Ploss et al., 2010), cultures of human fetal hepatocytes (Andrus et al., 2011, Marukian et al., 2011), or hepatocyte-like cells derived from induced pluripotent-stem cells (iPSCs; Roelandt et al., 2012, Schwartz et al., 2012, Wu et al., 2012), important features of liver biology as yet cannot be adequately recapitulated. Within the liver, numerous cell types, including hepatocytes and various non-parenchymal cell subsets (i.e. liver sinusoidal endothelial cells, stellate cells, and oval cells), are arranged in an intricate three-dimensional architecture. Nutrient and oxygen gradients within the liver result in a compartmentalization of the liver, referred to as zonation, affecting metabolism, detoxification and response to injury. Whether those specific microenvironments impact HCV infection is not understood and may only be adequately modeled within the three-dimensional context of the liver. Furthermore, a tractable animal model may be suitable to decipher mechanisms of viral persistence and pathogenesis and assess disease states and co-morbidities, such as HIV, alcohol or nutritionally exacerbated viral hepatitis, or extrahepatic manifestations associated with HCV infection.
The chimpanzee model
The only hosts known to be naturally permissive to HCV infection are humans and chimpanzees ( Table 1). The basis for this limited species tropism is incompletely understood and the topic of this review. The chimpanzee model played an instrumental role in early characterization of non-A, non-B hepatitis (Houghton, 2009) which ultimately led to the discovery of HCV as the etiologic agent for the classically defined non-A non-B hepatitis by Michael Houghton and his team in 1989 (Choo et al., 1989). Subsequently, many important discoveries were facilitated by their use; for example it was first demonstrated in chimpanzees that in vitro transcribed RNA from a HCV cDNA clone was infectious (Kolykhalov et al., 1997). Furthermore, experimental infection of chimpanzees with HCV helped to define the nature of protective immunity (Farci et al., 1992, Prince et al., 1992) and to demonstrate the significance of cellular subsets, most stringently the role of CD4 and CD8 T cells in controlling chronic HCV infection (Grakoui et al., 2003, Shoukry et al., 2003). Chimpanzees remain the only fully immunocompetent animal model for HCV infection and consequently, have and continue to play an important role in evaluating the preclinical efficacy of vaccine candidates (reviewed in Houghton, 2011). Precedence for the efficacy of new treatment modalities, such as the IFN-free control of HCV infection with DAAs (Olsen et al., 2011) or interference with the liver specific microRNA (miR) 122 (Lanford et al., 2010) was first shown in chimpanzees. Despite their utility, the use of large apes in biomedical research has raised ethical concerns, which culminated in the ban of experiments conducted in chimpanzees in many countries. An NIH moratorium on ‘non-essential’ chimpanzee research (NIH, 2011) is likely to constrain HCV research in the future and has made the need for alternative animal models even more pressing.
Table 1.
HCV susceptibility of selected species.
Human 
|
Great ape(s) (chimpanzee) 
|
Tree shrew 
|
Monkeys (rhesus macaque) 
|
Mice 
|
|
|---|---|---|---|---|---|
| HCV susceptibility | yes | yes | yes | yes | no |
| Entry | yes | yes | yes | yes | no |
| Replication | yes | yes | yes | no | no |
| Assembly | yes | yes | yes | ? | yes |
HCV infection in other primate species
The natural host reservoir of HCV remains poorly defined. While chimpanzees can be experimentally infected with HCV, the prevalence in chimpanzees or other great apes, gorillas and orangutans in the wild is not known. In search of alternative, more readily accessible experimental models for HCV numerous species have been tested for their susceptibility to HCV. Woodchucks, old- and new-world monkeys, including Cynomolgus, Rhesus, Japanese, Green monkeys, Doguera (Abe et al., 1993), Chacma Baboons (Sithebe et al., 2002), Cottontop tamarins (Garson et al., 1997) and marmosets appear to be mostly resistant to HCV infection. Surprisingly, tree shrews (Tupaia belangeri) which are distantly related to primates (Schmitz et al., 2000, Xu et al., 2012) appear to support HCV infection to some level (Xie et al., 1998, Xu et al., 2007) (Table 1) and were recently shown to develop signs of severe liver disease, including steatosis, fibrosis and cirrhosis (Amako et al., 2010). While these data are encouraging additional studies with larger cohorts of this outbred and thus, genetically heterogeneous species, are needed to define more clearly the natural course of HCV infection in tree shrews.
Surrogate virus systems
Genetically related viruses, which replicate more readily in non-human species have been considered as potential surrogates for the study of HCV in vivo. Most prominently, GB viruses (GBV), named after the surgeon George Baber (GB) who presented with acute viral hepatitis in the 1960s (Deinhardt et al., 1967), specifically GBV-B, can be transmitted to new world monkeys (Bukh et al., 2001, Karayiannis et al., 1989, Lanford et al., 2003, Schaluder et al., 1995, Simons et al., 1995). GBV-B and HCV both belong to the Flaviviridae family but share only approximately 28% amino acid identity within their polyprotein, which constrains the utility of this model for testing of drugs specific to HCV or vaccine candidates formulated with HCV specific antigens. Another caveat is the different course of GBV-B infection in tamarins and marmoset, which results in acute hepatitis but – in contrast to HCV – does not establish chronicity. More recently, other viruses that are genetically more related to HCV than GBV-B have been detected in other species. Non-primate hepaci-virus (NPHV), also known as canine hepacivirus (CHV) has been identified in dogs (Kapoor et al., 2011) and horses (Burbelo et al., 2012). Intriguingly, CHV/NPHV appears to have a broader tissue tropism in these species but has yet to be shown to cause hepatitis. Comparative analysis of CHV/NPHV and HCV may help to shed light on potentially common viral and host factors determining viral host and tissue tropism.
Murine xenotransplantation models
Since HCV does not readily replicate in non-human species conceptually the most straightforward strategy would be to introduce the relevant tissue compartment – a human liver – into mice. To this end various approaches have been taken including transplantation of human liver pieces under the kidney capsule (Ilan et al., 2002), engraftment of “human ectopic artificial livers” (HEALs) in the peritoneal cavity (Chen et al., 2011) or actual expansion of human hepatocytes within the liver parenchyma (reviewed in de Jong et al. (2010) and Meuleman and Leroux-Roels (2008b) of immunocompromised mice. For the latter approach, suitable xenorecipients combine two features; immunodeficiency to prevent graft rejection and liver injury to provide a growth stimulus for the normally quiescent hepatocytes as well as yield the transplanted human cells a competitive growth advantage over endogenous murine cells. The best-characterized liver injury mouse strains are urokinase-type plasminogen activator (uPA) transgenic mice (Heckel et al., 1990) and mice deficient in fumaryl acetoacetate hydrolase (FAH; Grompe et al., 1993). When crossed to an immunodeficient background the livers of either line can be highly repopulated with human hepatocytes, rendering the human liver chimeric mice susceptible to HCV infection (Bissig et al., 2010, Mercer et al., 2001, Meuleman et al., 2005). Such humanized mice have been invaluable to study basic aspects of HCV biology within the three-dimensional context of the liver and assessing preclinically the efficacy of antiviral compounds and biologics (reviewed in Meuleman and Leroux-Roels (2008a)). However, presumably due to their immunocompromised status HCV infected human liver chimeric mice generally do not exhibit signs of progressive liver disease, a process that is thought to be immune-mediated in humans. To overcome this caveat, engrafting components of a human immune system alongside human hepatocytes in a single recipient has been proposed (Legrand et al., 2009). Proof-of-concept for this strategy was provided in a recent study showing that mice co-injected with a mixture of human hematopoietic stem cells, fetal hepatoblasts and other non-parenchymal cells could be infected with HCV and develop signs of liver fibrosis (Washburn et al., 2011). While intriguing, human immune responses are generally weak in these reconstituted mice and additional modifications will be needed to improve both cellular complexity and functionality of the engrafted human immune system (reviewed in Shultz et al., 2007, Willinger et al., 2011). The utility of these xenotransplantation models is hampered by low through-put, intricate logistics, high costs and technical difficulties in their production but also the inherent donor-to-donor variability and differences in the human chimerism between animals.
Blocks in the HCV lifecycle in non-permissive species
An inbred mouse model for HCV would overcome the technical difficulties of the xenotransplantation model. Mice with inheritable susceptibility to HCV could be produced fairly easily in large quantities, would be amendable to genetic manipulations on defined genetic backgrounds and viral infection could be analyzed with a vast amount of existing tools and reagents. The challenge is to elucidate and overcome restrictions for HCV to complete its lifecycle in mouse cells and mice ( Fig. 1). In recent years progress has been made in explaining aspects of the restricted host range of HCV. In the following sections, we will highlight barriers in the HCV lifecycle in non-permissive species, focusing on mice, and discuss putative strategies to overcome them.
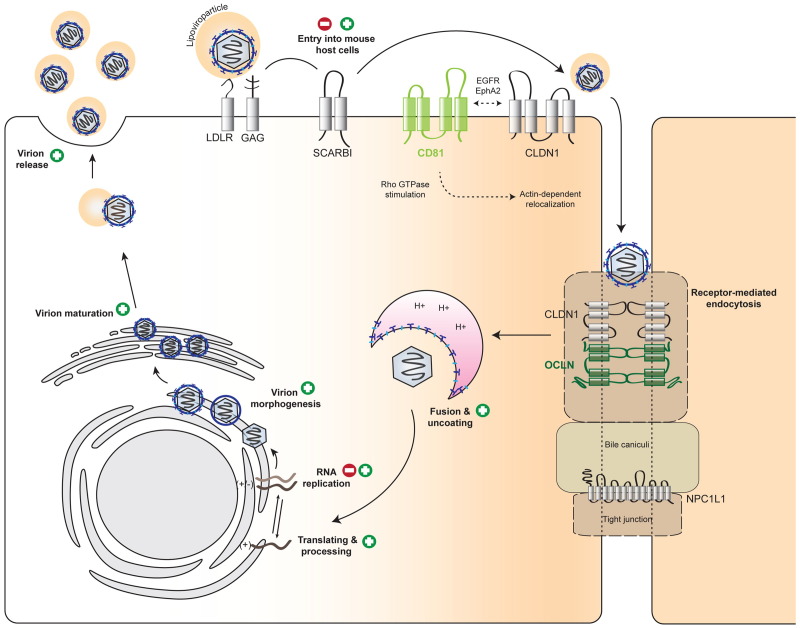
Fig. 1.
Blocks in HCV species tropism. Mouse hepatocytes support HCV replication cycle incompletely. CLDN1, claudin1; EGFR, epidermal growth factor receptor; EphA2, ephrin receptor A2; GAG, Glycosaminoglycans; LDLR, low-density lipoprotein receptor; NPC1L1, Niemann-Pick C1-Like 1; OCLN, occludin; SCARB1, scavenger receptor class B member 1. (+) supported, (−) blocked step in the HCV lifecycle in murine cells.
HCV entry
A large number of cellular factors has been implicated in facilitating HCV entry into human cells. In a coordinated multistep process ( Table 2, reviewed in Ploss and Evans (2012)) HCV, complexed with host lipoproteins in so called lipo-viroparticles, is thought to initially attach to glycosaminoglycans, then binds to low-density lipoprotein receptor (LDLR), the scavenger receptor class B type I (SCARB1; Scarselli et al., 2002) and the tetraspanin CD81 (Pileri et al., 1998) on the hepatocyte surface, before engaging the tight junction proteins, claudin-1 (CLDN1; Evans et al., 2007) and occludin (OCLN; Liu et al., 2009, Ploss et al., 2009), ultimately resulting in endocytotic uptake (Fig. 1). More recently, the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2; Lupberger et al., 2011) and the cholesterol uptake receptor Niemann-Pick C1 like 1 (NPC1L1; Sainz et al., 2012) have been implicated in the viral entry process by indirectly influencing the composition of membrane microdomains.
Table 2.
Expression of HCV entry factor candidates in human and mice.
|
Gene |
Most abundant tissue expression |
Liver expression |
Sequence identity (%) | Reference | |||
|---|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Human | Mouse | ||
| CD81 | cd81 | Ubiquitous | Ubiquitous | + | + | 96.6 | Pileri et al. (1998) |
| OCLN | ocln | All tight junctions | All tight junctions | + | + | 94.5 | Ploss et al. (2009) |
| CLDN1 | cldn1 | Skin, liver | Skin, liver | + | + | 97.2 | Evans et al. (2007) |
| SCARB1 | scarb1 | Liver, adrenal tissue,placenta | Liver, adrenal tissue, ovary | + | + | 88.6 | Scarselli et al. (2002) |
| LDLR | ldlr | Liver, intestine | Liver, intestine | + | + | 86.7 | Molina et al. (2007) and Agnello et al. (1999) |
| NPC1L1 | npc1l1 | Small intestine, liver, stomach | Small intestine | + | − | 84.7 | Sainz et al. (2012) |
| EGFR | egfr | Ubiquitous, except central nervous tissue | Ubiquitous, except central nervous tissue | + | + | 95.1 | Lupberger et al. (2011) |
| EphA2 | epha2 | Epithelial endothelial cells | Epithelial endothelial cells | + | + | 95.8 | Lupberger et al. (2011) |
| PCDHB5 | pcdhb5 | Connective tissue | Connective tissue | − | − | 82.0 | Wong-Staal et al. (2008) |
| TfR1 | tfr1 | Immature erythroid cells, placenta, liver | Immature erythroid cells, placenta, liver | + | + | 87.0 | Danyelle et al. (2008) |
| CLDN6 | cldn6 | Embryonic tissue, pituitary gland, ovary | Embryonic tissue, pituitary gland, ovary | − | − | 95.0 | Zheng et al. (2007) and Meertens et al. (2008) |
| CLDN9 | cldn9 | Pituitary gland, lung,intestine | Pituitary gland, lung,intestine | − | − | 99.1 | Zheng et al. (2007) and Meertens et al. (2008) |
| CD209 | cd209 | Dendritic cells | Dendritic cells | − | − | 52.3 | Gardner et al. (2003) |
| CD209L/DC-SIGNR | signr1(signr2–4) | Lymph node, liver,placenta | Lymph node, spleen, pancreas | + | + | 54.5 | Gardner et al. (2003) |
The C-type lectins liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin (L-SIGN, CD209L; Cormier et al., 2004), which is expressed on liver sinusoidal endothelial cells (LSECs) and the dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN, CD209), expressed by dendritic and Kupffer cells have been shown to bind to HCV E2 (Gardner et al., 2003, Lozach et al., 2003, Pohlmann et al., 2003). C-type lectins on sinusoidal cells may be able to trap HCV in the liver, but it is not known whether this can lead to productive infection of hepatocytes in trans or whether sequestered HCV is subjected to lysosomal degradation. Other molecules such as CLDN6, CLDN9 (Meertens et al., 2008, Zheng et al., 2007), transferrin receptor (TfR; Martin and Uprichard, 2010) and protocadherin β 5 (PCDHB5; Wong-Staal et al., 2008) have been proposed to be involved in influencing HCV entry. However, CLDN6, CLDN9 nor PCDHB5 are expressed at appreciable levels in the liver (Table 2). Thus, strong experimental evidence supporting their suggested relevance for viral entry and independent confirmation of these results still remain pending.
In contrast to human hepatocytes, murine cells do not support HCV entry thereby creating a first and important barrier for a broader host tropism of the virus. In an attempt to define the determinants restricting HCV entry into non-human cells a cDNA complementation screen was conducted in mouse cells ectopically expressing human CD81, SCARB1 and CLDN1 (Ploss et al., 2009). Human OCLN was identified as the missing factor needed to facilitate viral entry into human and mouse cells. While CD81, SCARB1, CLDN1 and OCLN are all required, only CD81 and OCLN need to be of human origin to promote viral entry into mouse cells (Ploss et al., 2009). These observations are in line with previous reports demonstrating that in contrast to CD81 of various non-human primate species, mouse CD81 does not support HCV pseudoparticle entry into human HepG2 hepatoma cells, which lack endogenous expression of CD81 (Flint et al., 2006). Similarly, in human cells lacking endogenous OCLN expression, primate sequences function equivalently to human OCLN, whereas canine, hamster, mouse and rat OCLN had intermediate to low activities, and guinea pig OCLN was completely nonfunctional (Michta et al., 2010). For both CD81 and OCLN, differences in the second extracellular loops are in part responsible for this apparent functional activity. In contrast, the residues (I32 and E48) within the first extracellular loop of CLDN1 required for HCV entry (Evans et al., 2007) are conserved between mice and man. Thus, it is not surprising that mouse CLDN1 supports HCV uptake. Originally, it was shown that human but not mouse SCARB1 can bind to soluble HCV E2 (Scarselli et al., 2002). Nonetheless, in both in vitro (Ploss et al., 2009) and in vivo (Dorner et al., 2011) infection assays mouse and human SCARB1 were equally functional. These apparently conflicting data are likely attributable to the read-out system and highlight that HCV E2 binding may not be the most reliable proxy for functional entry assays. To date any differences in the susceptibility of cells of non-human origin to HCV entry are largely accounted for by differences in critical positions within respective orthologues of CD81, SCARB1, CLDN1 and OCLN or their limited or absent expression.
Genetically humanized mice for HCV entry
The observation that CD81 and OCLN comprise the minimal set of human factors required to render mouse cells permissive to HCV entry in vitro provided the blue print for engineering a mouse that at least supports viral uptake in vivo. Genetically humanized mice have been successfully constructed by expression of receptors and/or coreceptors in mice for other human pathogens, such as poliovirus (hCD155; Racaniello and Ren, 1994), measles virus (hCD46/CD150; Sellin and Horvat, 2009), human coronavirus (hCD13; Lassnig et al., 2005), human immunodeficiency virus (hCD4/CCR5/CXCR4; Klotman and Notkins, 1996) and Listeria monocytogenes (Lecuit et al., 2001). Earlier attempts to render mice susceptible to HCV infection by transgenic expression of CD81 were not successful (Masciopinto et al., 2002). Expression of human CD81 in a wide variety of tissues increased the binding of recombinant E2 to liver, thymocytes and splenic lymphocytes in comparison to non-transgenic controls, but due to the lack of human OCLN human CD81-transgenic mice were resistant to HCV infection.
Recently, it was demonstrated that adenoviral delivery of human CD81 and OCLN is sufficient to allow HCV infection of fully-immunocompetent inbred mice (Dorner et al., 2011). Using intergenotypic virus chimeras expressing CRE recombinase to activate a cellular reporter, in vivo uptake of diverse HCV genotypes could be conveniently quantified by bioluminescent imaging. Utilizing blocking antibodies specific to CD81 or the viral envelope protein E2, expression of entry factor mutants and mice with a targeted disruption of the SCARB1 gene validated uptake of HCV into murine hepatocytes in an HCV glycoprotein-mediated fashion. Furthermore, it established a precedent for applying mouse genetics to dissect the viral entry process, in addition to validating the role of SCARB1 for HCV uptake in vivo for the first time. Beyond studying HCV entry, this model can be employed to evaluated passive and active immunization strategies (Dorner et al., 2011, Dorner et al., in press, Giang et al., 2012).
The transient adenoviral delivery approach is high-throughput and allows rapid evaluation of mutant genes. However, in order to accurately reproduce the complex process of HCV entry in vivo, it will be important to achieve native expression patterns of the human HCV entry factor orthologues by using transgenic and/or knock-in approaches. Recently, transgenic mice expressing human CD81, SCARB1, CLDN1 and OCLN were generated, but did not appear to be permissive to HCV infection in vivo (Hikosaka et al., 2011). This apparent discrepancy between transient adenoviral and stable transgenic expression approaches can be in part explained by the lower level of entry factor expression in the transgenic mice and the requirement for a very sensitive reporter system to quantify viral entry (Dorner et al., 2011).
Adaptation of HCV to infection of mouse cells
As an alternative to this host adaptation approach to overcome species barriers it may be possible to adapt HCV to usage of entry factor orthologs from other species. Using an unbiased selection approach, a laboratory strain of HCV was adapted to utilize mouse CD81 (Bitzegeio et al., 2010). Taking advantage of the high mutational plasticity of HCV, three adaptive mutations in the viral glycoproteins E1 and E2 were identified that allowed the virus to enter cells expressing human SCARB1, CLDN1, OCLN and mouse CD81. Interestingly, the resulting murine-tropic (mt) HCV was also capable of efficiently utilizing mouse OCLN and had a lower dependency on SCARB1. Whether mtHCV is capable of infecting mouse hepatocytes in vitro or in vivo has yet to be shown. Clearly, this study provides an important proof-of-concept for the validity of the viral adaptation approach but also raises a few potential caveats. The adaptive mutations within the viral envelope appear to have resulted in conformational changes. Consequently, it remains to be tested whether mtHCV faithfully recapitulates viral entry and raises concerns of whether the neutralizing capacity of antibodies or human specific therapeutics to block HCV entry can be adequately tested.
The contribution of other host factors implicated in viral entry to the species tropism of HCV is not well understood. Most human and murine orthologues share a high degree of amino acid sequence similarity (Table 2) but, as discussed above, differences in individual residues can drastically affect viral uptake efficiency. The exact roles of EGF receptor and ephrin receptor A2, which are both expressed in mouse and human hepatocytes, in this process are incompletely defined. Both receptor tyrosine kinases (RTKs) appear to mediate entry by regulating CD81–CLDN1 co-receptor associations and viral glycoprotein-dependent membrane fusion (Lupberger et al., 2011). Recently, it was shown that EGFR activation is dependent on interactions between HCV and CD81 but not CLDN1 (Diao et al., 2012). Although, it is conceivable that expression of human EGFR and/or ephrin receptor A2 could increase HCV entry efficiency in vivo their murine orthologues appear to exert sufficient functionality in genetically humanized mice.
Interestingly, although NPC1L1 has been shown to play a role in the viral entry in human cells in vitro and in vivo (Sainz et al., 2012) it appears to be dispensable for entry into mouse cells. In contrast to humans, the murine ortholog of NPC1L1 has approximately 85% sequence similarity to the human protein and is abundantly expressed in the gut but not in the liver (Altmann et al., 2004). Nonetheless, HCV is still capable of entering into hepatocytes of mice modified to express a combination of human CD81 and OCLN (Dorner et al., 2011) suggesting some level of redundancy for NPC1L1 in this process. It is conceivable that HCV entry would be affected, perhaps becoming more efficient, if NPC1L1 along with human CD81 and OCLN was ectopically co-expressed in the liver, which remains to be experimentally tested. It was recently demonstrated that hepatic overexpression of human NPC1L1 in transgenic mice (Temel et al., 2007) or via adenoviral delivery modulates cellular and plasma lipid metabolism (Kurano et al., 2012). Altered cellular lipid profiles can arguably affect the composition of membrane microdomains and consequently, the trafficking of membrane proteins including HCV entry factors.
Fusion, uncoating and initial HCV RNA translation
Following receptor mediated endocytosis the viral membrane has to fuse with the endosome to eventually release the HCV RNA genome from the nucleocapsid into the cytoplasm. It has been known that viral RNA translation occurs in vivo, when the RNA is directly introduced into mouse hepatocytes, e.g. by hydrodynamic delivery (McCaffrey et al., 2002). This suggests that the HCV internal ribosome entry site (IRES) within the 5′ untranslated region is functional in mouse cells. Recently, a genetically humanized mouse model was constructed utilizing cell culture produced recombinant hepatitis C virus to activate a cellular encoded reporter (Dorner et al., 2011, Dorner et al., in press). In this model HCV is taken up into murine hepatocytes in a viral glycoprotein-dependent fashion providing further evidence that fusion and uncoating work efficiently in this background.
HCV RNA replication
The advent of HCV replicons, i.e. subgenomic or full-length genomes containing a dominant selectable marker such as neomycin amino transferase (Blight et al., 2000, Lohmann et al., 1999), allowed for assessing whether HCV is capable of replicating in non-human cells. Indeed, when these replicons were introduced into mouse hepatoma cell lines and embryonic fibroblasts, cell clones harboring replicating HCV RNA could be identified at very low frequency (Uprichard et al., 2006, Zhu et al., 2003). These data suggest that all necessary host factors are present in mouse cells and sufficiently similar to support HCV RNA replication. However, it is conceivable that the viral RNA replication machinery cooperates more robustly with essential human host factors than their murine counterparts. Targeted and genome wide loss of function screens led to the discovery of numerous cellular factors putatively promoting or restricting HCV replication in human cells (Berger et al., 2009, Borawski et al., 2009, Coller et al., 2009, Jones et al., 2010b, Li et al., 2009, Ng et al., 2007, Randall et al., 2007, Reiss et al., 2011, Supekova et al., 2008, Tai et al., 2009, Trotard et al., 2009, Vaillancourt et al., 2009). Unfortunately, independent studies are often minimally overlapping and the relevance of many of these interactions to HCV biology remains to be demonstrated. Strong experimental evidence has been provided for the critical role of cyclophilin A (CypA) in RNA replication and virion assembly (Kaul et al., 2009, Yang et al., 2008). Another essential co-factor for RNA replication independently identified by several groups (Berger et al., 2009, Borawski et al., 2009, Reiss et al., 2011, Tai et al., 2009, Trotard et al., 2009), is phosphatidylinositol 4 kinase IIIα (PI4KIIIα). PI4KIIIα is recruited to the membranous web via interaction with NS5A. The human and murine orthologues of CypA and PI4KIIIα share a high degree of sequence similarity at 98.2% and 98.6%, respectively. It is yet to be shown whether ectopic expression of human CypA and PI4KIIIα augments HCV RNA replication in mouse cells. Likewise, there is little evidence for the accumulation of adaptive mutations boosting viral RNA replication in mouse cells by fostering more efficient interactions with mouse orthologues of host factors.
Cellular microRNAs can drastically influence virus replication and pathogenesis (Gottwein and Cullen, 2008). A liver specific microRNA, miR-122, was shown to be critical for efficient HCV RNA replication (Jopling et al., 2005). miR-122 is derived from a liver-specific noncoding polyadenylated RNA transcribed from the gene hcr and interacts with two sites in the viral 5′ non-translated region (NTR) (Jopling et al., 2008, Jopling et al., 2005). Those interactions presumably protect the 5′ terminal viral sequences from nucleolytic degradation or from inducing innate immune responses to the RNA terminus (Machlin et al., 2011), resulting in greater viral RNA abundance in both infected cultured cells and in the liver of infected chimpanzees (Lanford et al., 2010). The exact sequence of miR-122 is highly conserved in vertebrate species; comparatively, mouse miR-122 is sequence-wise identical and with 50,000 copies per hepatocytes similarly expressed as the human counterpart (Chang et al., 2004). Consequently, miR-122 is not likely to contribute to the restricted host tropism in non-permissive species.
It is possible that cell-intrinsic, species-specific restriction factors, similar to those that control retroviral infection, such as Fv1, TRIM5α or APOBEC3 cytidine deaminases (reviewed in Bieniasz (2004)), interfere with HCV RNA replication in murine cells. However, heterokaryons of mouse and human cells capable of supporting the HCV lifecycle suggest that dominant negative inhibitors of HCV replication do not exist (Frentzen et al., 2011).
Antiviral cellular defenses limiting HCV RNA replication
Differences in the magnitude, nature and kinetics of an antiviral response between hosts are known to restrict tropism of certain viruses, such as myxoma virus, which is only permissive in mouse cells that have impaired IFN responses (Wang et al., 2004). The interferon system also plays a pivotal role in limiting HCV both under physiological conditions and when triggered during therapeutic intervention. Hundreds of IFN stimulated genes (ISGs) are induced in the liver during HCV infection (reviewed in Schoggins and Rice (2011)) and ISG products can potently inhibit HCV infection (Schoggins et al., 2011; Fig. 2). Pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs)-3 and 7 in endosomes as well as a family of RIG-I like receptors (RLRs), including the retinoic-acid-inducible gene I (RIG-I) and the melanoma-differentiation-associated gene 5 (MDA5) sense the presence of viral RNAs. Subsequently, host antiviral responses are activated resulting in the production of cytokines, such as type I and III interferons (Yoneyama et al., 2004). When IFN signaling is blunted, HCV replicates more efficiently in both human hepatoma cells (Blight et al., 2002) and primary human hepatocytes (Andrus et al., 2011, Marukian et al., 2011). Similarly, in mouse embryonic fibroblasts with targeted disruptions in protein kinase R (PKR; Chang et al., 2006) or interferon regulatory factor 3 (IRF3; Lin et al., 2010) RNA replication is enhanced. Whether disruption of these or other innate signaling pathways are sufficient to render mice expressing human HCV entry factors permissive to HCV infections has yet to be shown.
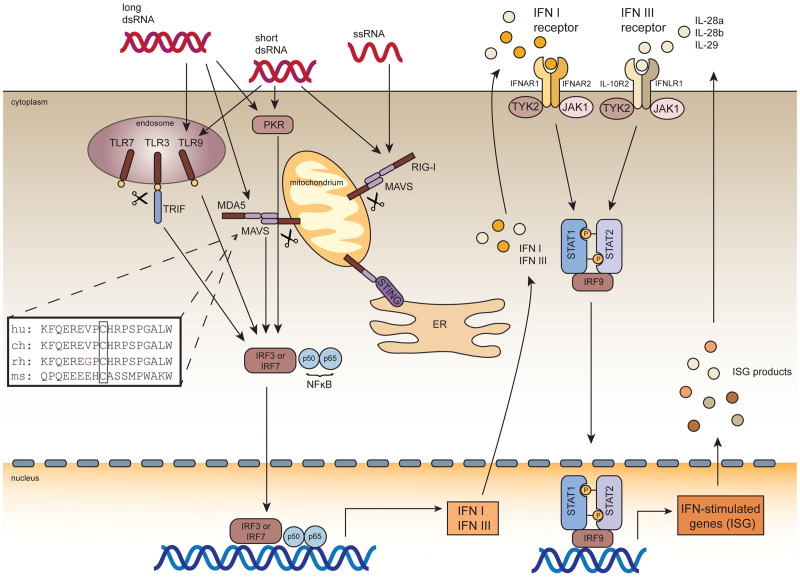
Fig. 2.
Innate immune response to HCV infection. Ch; chimpanzee, dsRNA, double-stranded RNA; ER, endoplasmatic reticulum; hu, human; IFN, interferon; IFNAR, interferon-alpha receptor; IL, interleukin; IRF, interferon regulatory factor; JAK, Janus kinase; ms, mouse; MAVS, mitochondrial associated antiviral signaling protein; MDA5, melanoma-differentiation-associated gene 5; PKR, protein kinase R; rh, rhesus macaque; RIG-I, retinoic-acid-inducible gene I; ssRNA, single-stranded RNA; STAT1, signal transducer and activator of transcription protein-1; STAT2, signal transducer and activator of transcription protein-2; STING, stimulator of interferon genes; TLR, toll-like receptor; TRIF, TIR-domain-containing adapter-inducing interferonβ; TYK, tyrosine kinase. Inlet: sequence alignment of the NS3–4a cleavage motif within MAVS from different permissive and non-permissive species.
HCV has devised mechanisms to counteract innate defenses but those may not work equally efficient in cells from different species. For example, NS3–4A serine protease is not only essential for processing of the HCV polyprotein but has been shown to cleave the mitochondrial associated antiviral signaling protein (MAVS; Meylan et al., 2005), the T-cell protein tyrosine phosphatase (TC-PTP; Brenndorfer et al., 2009) and TIR domain-containing adapter inducing IFNβ (TRIF; Li et al., 2005) thereby modulating antiviral signaling. The cleavage motifs of MAVS and TRIF are partially conserved between mice and man/chimpanzees (Fig. 2) and it remains to be demonstrated whether these antiviral evasion strategies are functional in murine cells. Contrarily, MAVS from certain new world monkey species, including rhesus macaques and baboons, both of which are resistant to HCV infection, is functional for interferon signaling even in the presence of heterologously expressed NS3/4A protease in human epithelial cells. Thus, escape from HCV antagonism of innate immune sensing may explain at least part of HCV species restriction (Patel et al., 2012). Furthermore, other aspects of HCVs antiviral evasion strategies may work less efficiently in mouse cells. For example, NS4B was shown to inhibit RLR-mediated IFN signaling through association with the stimulator of interferon genes (STING; Nitta et al., in press). Similarly, overexpression data suggests that the HCV core protein interferes with IFN signaling downstream of the IFNαβ receptor, presumably through direct interaction with STAT1, leading to reduced phospho-STAT1. The protein sequences of mouse and human STAT1 are highly conserved (>90% sequence identity), mouse and human STING are only ca. 60% identical (ca. 80% similar) which may result in a weaker interaction of NS4B with the murine orthologue and consequently, more exacerbated RLR signaling.
HCV assembly, egress and release from mouse cells
The development of the infectious cell culture system, which recapitulates all parts of the viral lifecycle paved the way to study HCV assembly, egress and release. Increasing evidence points to a connection between the viral lifecycle and the biogenesis of host very low-density lipoproteins (VLDL) (Huang et al., 2007, Merz et al., 2011, Ye, 2007). Very low-density viral particles can be detected in sera from HCV+ patients using antibodies targeting lipoprotein-associated proteins, notably apolipoproteins (apo) B and E (Nielsen et al., 2006). The role of specific apolipoproteins in virion assembly is incompletely understood. Loss of function experiments have pointed towards apoA1 (Mancone et al., 2011), apoB (Huang et al., 2007), and apoE (reviewed in Bartenschlager et al. (2011)) as potential players in this process.
While human cells with a functional VLDL pathway are capable of producing infectious HCV it was unclear whether other species would also support late stages of the HCV lifecycle. To investigate this process mouse Hep56.1D cells harboring a subgenomic HCV replicon were transcomplemented with HCV core, E1, E2, p7 and NS2 proteins but only marginal amounts of infectious HCV were released (Long et al., 2011). A comparative transcriptome analysis between naïve mouse cells and those containing HCV replicons hinted that low levels of apoE in the replicon-containing cells might limit HCV assembly. Indeed, complementation with human or mouse apoE was sufficient to rescue infectious HCV production from these cells, yielding infectious titers similar to those observed in the widely used human hepatoma cell line, Huh-7.5. These data confirm the critical importance of apoE in HCV assembly and provide evidence that mouse cells can support the late stages of HCV lifecycle, if critical components of the VLDL pathway are present. However, it remains unclear whether HCV can assemble in more physiologically relevant systems and importantly if mice are capable of producing infectious particles in vivo.
Conclusions and future perspectives
Substantial progress has been made in our understanding of the HCV lifecycle which is being translated into improved therapies. Experimental systems devised to study HCV in cell culture have enabled comparative studies to elucidate the barriers that restrict HCV infection to humans and chimpanzees. While murine cells can be engineered to express the human orthologs of the entry molecules CD81 and occludin to allow entry, viral RNA replication is still low in these cells. This is at least in part due to varying antiviral defenses. Murine cells with an intact VLDL pathway can produce infectious virions. Taken together these observations raise the hope that an inbred mouse model for HCV infection can be achieved. Recapitulating the entire viral lifecycle in a mouse would certainly be of great importance but it remains to be seen whether such a model would also mimic clinical relevant disease aspects. In humans pathogenesis in the context of chronic HCV infection is thought to be driven by persistent liver inflammation ultimately resulting in fibrosis, cirrhosis and liver cancer. Thus, establishing HCV persistence in the presence of a functional immune system is likely necessary to model adequately HCV pathogenesis. Studies characterizing chronicity in humanized mice engrafted with both human hepatocytes and components of a human immune system (Washburn et al., 2011) as well as long-term follow up studies in tree shrews (Amako et al., 2010) hint that it may become possible to dissect liver disease progression in small animal models. The course of HCV infection has been studied in great detail in chimpanzees and by and large mirrors adequately acute and chronic HCV infection in humans. Whether HCV infection in other species such as inbred mice will follow a similar clinical course is unclear. However, even if HCV infections were to cause e.g. mostly acute hepatitis in mice inheritable traits rendering animals permissive to HCV could be combined with genetic backgrounds that are more prone for chronicity and liver disease progression. Different approaches, including both viral and host adaptations are likely needed to construct a selection of HCV permissive animal models, which in combination may be suitable to replace the chimpanzee. A cost-effective animal model that is amendable to genetic manipulations and can be reproducibly produced at higher throughput would undoubtedly lend itself to address currently intractable problems in HCV research.
Acknowledgments
The authors thank Dr. Cynthia de la Fuente for critical discussions and editing. L.S. is a fellow of the German National Merit Foundation and is supported by a stipend from the Organization of Supporters of the Hannover Medical School (Gesellschaft der Freunde der Medizinischen Hochschule Hannover e.V.). A.P. is a recipient of the Astellas Young Investigator Award of the Infectious Disease Society of America and a Liver Scholar Award from the American Liver Foundation. Work in the laboratory is supported in part by the Starr Foundation, the Greenberg Medical Institute, the National Institutes of Health and the Bill and Melinda Gates Foundation. The funding sources were not involved in the writing of the report.
References
- Abe K., Kurata T., Teramoto Y., Shiga J., Shikata T. Lack of susceptibility of various primates and woodchucks to hepatitis C virus. J. Med. Primatol. 1993;22:433–434. [PubMed] [Google Scholar]
- Altmann S.W., Davis H.R., Jr., Zhu L.J., Yao X., Hoos L.M., Tetzloff G., Iyer S.P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M.P. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- Amako Y., Tsukiyama-Kohara K., Katsume A., Hirata Y., Sekiguchi S., Tobita Y., Hayashi Y., Hishima T., Funata N., Yonekawa H., Kohara M. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J. Virol. 2010;84:303–311. doi: 10.1128/JVI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrus L., Marukian S., Jones C.T., Catanese M.T., Sheahan T.P., Schoggins J.W., Barry W.T., Dustin L.B., Trehan K., Ploss A., Bhatia S.N., Rice C.M. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Penin F., Lohmann V., Andre P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Berger K.L., Cooper J.D., Heaton N.S., Yoon R., Oakland T.E., Jordan T.X., Mateu G., Grakoui A., Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P.D. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Bissig K.D., Wieland S.F., Tran P., Isogawa M., Le T.T., Chisari F.V., Verma I.M. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J. Clin. Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzegeio J., Bankwitz D., Hueging K., Haid S., Brohm C., Zeisel M.B., Herrmann E., Iken M., Ott M., Baumert T.F., Pietschmann T. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K.J., Kolykhalov A.A., Rice C.M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Blight K.J., McKeating J.A., Rice C.M. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C.J., Myer V., Cornellataracido I., Baryza J., Tallarico J., Joberty G., Bantscheff M., Schirle M., Bouwmeester T., Mathy J.E., Lin K., Compton T., Labow M., Wiedmann B., Gaither L.A. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenndorfer E.D., Karthe J., Frelin L., Cebula P., Erhardt A., Schulte am Esch J., Hengel H., Bartenschlager R., Sallberg M., Haussinger D., Bode J.G. Nonstructural 3/4A protease of hepatitis C virus activates epithelial growth factor-induced signal transduction by cleavage of the T-cell protein tyrosine phosphatase. Hepatology. 2009;49:1810–1820. doi: 10.1002/hep.22857. [DOI] [PubMed] [Google Scholar]
- Bukh J., Apgar C.L., Govindarajan S., Purcell R.H. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Dubovi E.J., Simmonds P., Medina J.L., Henriquez J.A., Mishra N., Wagner J., Tokarz R., Cullen J.M., Iadarola M.J., Rice C.M., Lipkin W.I., Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R., Zaret K.S., Taylor J.M. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chang K.S., Cai Z., Zhang C., Sen G.C., Williams B.R., Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J. Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.A., Thomas D.K., Ong L.L., Schwartz R.E., Golub T.R., Bhatia S.N. Humanized mice with ectopic artificial liver tissues. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11842–11847. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Coelmont L., Kaptein S., Paeshuyse J., Vliegen I., Dumont J.M., Vuagniaux G., Neyts J. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob. Agents Chemother. 2009;53:967–976. doi: 10.1128/AAC.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K.E., Berger K.L., Heaton N.S., Cooper J.D., Yoon R., Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. doi: 10.1371/journal.ppat.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier E.G., Durso R.J., Tsamis F., Boussemart L., Manix C., Olson W.C., Gardner J.P., Dragic T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14067–14072. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong Y.P., Rice C.M., Ploss A. New horizons for studying human hepatotropic infections. J. Clin. Invest. 2010;120:650–653. doi: 10.1172/JCI42338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinhardt F., Holmes A.W., Capps R.B., Popper H. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J. Exp. Med. 1967;125:673–688. doi: 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Pantua H., Ngu H., Komuves L., Diehl L., Schaefer G., Kapadia S.B. Hepatitis C Virus (HCV) induces Epidermal Growth Factor Receptor (EGFR) activation via CD81 binding for viral internalization and entry. J. Virol. 2012 doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M., Horwitz J.A., Robbins J.B., Barry W.T., Feng Q., Mu K., Jones C.T., Schoggins J.W., Catanese M.T., Burton D.R., Law M., Rice C.M., Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner, M., Rice, C.M., Ploss, A. Study of hepatitis C virusentry in genetically humanized mice. Methods 10.1016/j.ymeth.2012.05.010, in press. [DOI] [PMC free article] [PubMed]
- Edlin B.R. Perspective: test and treat this silent killer. Nature. 2011;474:S18–S19. doi: 10.1038/474S18a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., von Hahn T., Tscherne D.M., Syder A.J., Panis M., Wolk B., Hatziioannou T., McKeating J.A., Bieniasz P.D., Rice C.M. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Farci P., Alter H.J., Govindarajan S., Wong D.C., Engle R., Lesniewski R.R., Mushahwar I.K., Desai S.M., Miller R.H., Ogata N., Purcell R.H. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- Flint M., von Hahn T., Zhang J., Farquhar M., Jones C.T., Balfe P., Rice C.M., McKeating J.A. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 2006;80:11331–11342. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen A., Huging K., Bitzegeio J., Friesland M., Haid S., Gentzsch J., Hoffmann M., Lindemann D., Zimmer G., Zielecki F., Weber F., Steinmann E., Pietschmann T. Completion of Hepatitis C virus replication cycle in heterokaryons excludes dominant restrictions in human non-liver and mouse liver cell lines. PLoS Pathog. 2011;7:e1002029. doi: 10.1371/journal.ppat.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.P., Durso R.J., Arrigale R.R., Donovan G.P., Maddon P.J., Dragic T., Olson W.C. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4498–4503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garson J.A., Whitby K., Watkins P., Morgan A.J. Lack of susceptibility of the cottontop tamarin to hepatitis C infection. J. Med. Virol. 1997;52:286–288. [PubMed] [Google Scholar]
- Giang E., Dorner M., Prentoe J.C., Dreux M., Evans M.J., Bukh J., Rice C.M., Ploss A., Burton D.R., Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E., Cullen B.R. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microb. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Shoukry N.H., Woollard D.J., Han J.H., Hanson H.L., Ghrayeb J., Murthy K.K., Rice C.M., Walker C.M. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- Grompe M., al-Dhalimy M., Finegold M., Ou C.N., Burlingame T., Kennaway N.G., Soriano P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- Heckel J.L., Sandgren E.P., Degen J.L., Palmiter R.D., Brinster R.L. Neonatal bleeding in transgenic mice expressing urokinase-type plasminogen activator. Cell. 1990;62:447–456. doi: 10.1016/0092-8674(90)90010-c. [DOI] [PubMed] [Google Scholar]
- Hikosaka K., Noritake H., Kimura W., Sultana N., Sharkar M.T., Tagawa Y., Uezato T., Kobayashi Y., Wakita T., Miura N. Expression of human factors CD81, claudin-1, scavenger receptor, and occludin in mouse hepatocytes does not confer susceptibility to HCV entry. Biomed. Res. 2011;32:143–150. doi: 10.2220/biomedres.32.143. [DOI] [PubMed] [Google Scholar]
- Houghton M. The long and winding road leading to the identification of the hepatitis C virus. J. Hepatol. 2009;51:939–948. doi: 10.1016/j.jhep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Houghton M. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol. Rev. 2011;239:99–108. doi: 10.1111/j.1600-065X.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- Huang H., Sun F., Owen D.M., Li W., Chen Y., Gale M., Jr., Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan E., Arazi J., Nussbaum O., Zauberman A., Eren R., Lubin I., Neville L., Ben Moshe O., Kischitzky A., Litchi A., Margalit I., Gopher J., Mounir S., Cai W., Daudi N., Eid A., Jurim O., Czerniak A., Galun E., Dagan S. The hepatitis C virus (HCV)-trimera mouse: a model for evaluation of agents against HCV. J. Infect. Dis. 2002;185:153–161. doi: 10.1086/338266. [DOI] [PubMed] [Google Scholar]
- Jacobson I.M., McHutchison J.G., Dusheiko G., Di Bisceglie A.M., Reddy K.R., Bzowej N.H., Marcellin P., Muir A.J., Ferenci P., Flisiak R., George J., Rizzetto M., Shouval D., Sola R., Terg R.A., Yoshida E.M., Adda N., Bengtsson L., Sankoh A.J., Kieffer T.L., George S., Kauffman R.S., Zeuzem S. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- Jones C.T., Catanese M.T., Law L.M., Khetani S.R., Syder A.J., Ploss A., Oh T.S., Schoggins J.W., MacDonald M.R., Bhatia S.N., Rice C.M. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.M., Domingues P., Targett-Adams P., McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J. Gen. Virol. 2010;91:2238–2248. doi: 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
- Jopling C.L., Schutz S., Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microb. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J.A., Firth C., Hirschberg D.L., Rice C.M., Shields S., Lipkin W.I. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiannis P., Petrovic L.M., Fry M., Moore D., Enticott M., McGarvey M.J., Scheuer P.J., Thomas H.C. Studies of GB hepatitis agent in tamarins. Hepatology. 1989;9:186–192. doi: 10.1002/hep.1840090204. [DOI] [PubMed] [Google Scholar]
- Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Klotman P.E., Notkins A.L. Transgenic models of human immunodeficiency virus type-1. Curr. Top. Microbiol. Immunol. 1996;206:197–222. doi: 10.1007/978-3-642-85208-4_11. [DOI] [PubMed] [Google Scholar]
- Kolykhalov A.A., Agapov E.V., Blight K.J., Mihalik K., Feinstone S.M., Rice C.M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- Kurano M., Hara M., Tsuneyama K., Okamoto K., Iso O.N., Matsushima T., Koike K., Tsukamoto K. Modulation of lipid metabolism with the over-expression of NPC1L1 in mice liver. J. Lipid Res. 2012;53:2275–2285. doi: 10.1194/jlr.M026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwo P.Y., Lawitz E.J., McCone J., Schiff E.R., Vierling J.M., Pound D., Davis M.N., Galati J.S., Gordon S.C., Ravendhran N., Rossaro L., Anderson F.H., Jacobson I.M., Rubin R., Koury K., Pedicone L.D., Brass C.A., Chaudhri E., Albrecht J.K. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376:705–716. doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- Lamarre D., Anderson P.C., Bailey M., Beaulieu P., Bolger G., Bonneau P., Bös M., Cameron D.R., Cartier M., Cordingley M.G., Faucher A.M., Goudreau N., Kawai S.H., Kukolj G., Lagacé L., LaPlante S.R., Narjes H., Poupart M.A., Rancourt J., Sentjens R.E., St.George R., Simoneau B., Steinmann G., Thibeault D., Tsantrizos Y.S., Weldon S.M., Yong C.L., Llinàs-Brunet M. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- Lanford R.E., Chavez D., Notvall L., Brasky K.M. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology. 2003;311:72–80. doi: 10.1016/s0042-6822(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassnig C., Kolb A., Strobl B., Enjuanes L., Muller M. Studying human pathogens in animal models: fine tuning the humanized mouse. Transgenic Res. 2005;14:803–806. doi: 10.1007/s11248-005-1676-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M., Vandormael-Pournin S., Lefort J., Huerre M., Gounon P., Dupuy C., Babinet C., Cossart P. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- Legrand N., Ploss A., Balling R., Becker P.D., Borsotti C., Brezillon N., Debarry J., de Jong Y., Deng H., Di Santo J.P., Eisenbarth S., Eynon E., Flavell R.A., Guzman C.A., Huntington N.D., Kremsdorf D., Manns M.P., Manz M.G., Mention J.J., Ott M., Rathinam C., Rice C.M., Rongvaux A., Stevens S., Spits H., Strick-Marchand H., Takizawa H., van Lent A.U., Wang C., Weijer K., Willinger T., Ziegler P. Humanized mice for modeling human infectious disease: challenges, progress, and outlook. Cell Host Microb. 2009;6:5–9. doi: 10.1016/j.chom.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Foy E., Ferreon J.C., Nakamura M., Ferreon A.C., Ikeda M., Ray S.C., Gale M., Jr., Lemon S.M. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Brass A.L., Ng A., Hu Z., Xavier R.J., Liang T.J., Elledge S.J. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.T., Noyce R.S., Pham T.N., Wilson J.A., Sisson G.R., Michalak T.I., Mossman K.L., Richardson C.D. Replication of subgenomic hepatitis C virus replicons in mouse fibroblasts is facilitated by deletion of interferon regulatory factor 3 and expression of liver-specific microRNA 122. J. Virol. 2010;84:9170–9180. doi: 10.1128/JVI.00559-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yang W., Shen L., Turner J.R., Coyne C.B., Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V., Korner F., Koch J.O., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Long G., Hiet M.S., Windisch M.P., Lee J.Y., Lohmann V., Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141(3):1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Lozach P.Y., Lortat-Jacob H., de Lacroix de Lavalette A., Staropoli I., Foung S., Amara A., Houles C., Fieschi F., Schwartz O., Virelizier J.L., Arenzana-Seisdedos F., Altmeyer R. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 2003;278:20358–20366. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- Lupberger J., Zeisel M.B., Xiao F., Thumann C., Fofana I., Zona L., Davis C., Mee C.J., Turek M., Gorke S., Royer C., Fischer B., Zahid M.N., Lavillette D., Fresquet J., Cosset F.L., Rothenberg S.M., Pietschmann T., Patel A.H., Pessaux P., Doffoel M., Raffelsberger W., Poch O., McKeating J.A., Brino L., Baumert T.F. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin E.S., Sarnow P., Sagan S.M. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA–target RNA complex. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancone C., Steindler C., Santangelo L., Simonte G., Vlassi C., Longo M.A., D'Offizi G., Di Giacomo C., Pucillo L.P., Amicone L., Tripodi M., Alonzi T. Hepatitis C virus production requires apolipoprotein A-I and affects its association with nascent low-density lipoproteins. Gut. 2011;60:378–386. doi: 10.1136/gut.2010.211292. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Uprichard, S., 2010. The role of transferrin receptor in Hepatitis C virus entry. In: Proceedings of the 18th International Symposium on Hepatitis C Virus & Related Viruses, Yokohama, Japan.
- Marukian S., Andrus L., Sheahan T.P., Jones C.T., Charles E.D., Ploss A., Rice C.M., Dustin L.B. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54(6):1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciopinto F., Freer G., Burgio V.L., Levy S., Galli-Stampino L., Bendinelli M., Houghton M., Abrignani S., Uematsu Y. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology. 2002;304:187–196. doi: 10.1006/viro.2002.1631. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Ohashi K., Meuse L., Shen S., Lancaster A.M., Lukavsky P.J., Sarnow P., Kay M.A. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- McHutchison J.G., Everson G.T., Gordon S.C., Jacobson I.M., Sulkowski M., Kauffman R., McNair L., Alam J., Muir A.J. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- Meertens L., Bertaux C., Cukierman L., Cormier E., Lavillette D., Cosset F.L., Dragic T. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J. Virol. 2008;82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer D.F., Schiller D.E., Elliott J.F., Douglas D.N., Hao C., Rinfret A., Addison W.R., Fischer K.P., Churchill T.A., Lakey J.R., Tyrrell D.L., Kneteman N.M. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- Merz A., Long G., Hiet M.S., Brugger B., Chlanda P., Andre P., Wieland F., Krijnse-Locker J., Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman P., Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antivir. Res. 2008;80(3):231–238. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Meuleman P., Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antivir. Res. 2008;80:231–238. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Meuleman P., Libbrecht L., De Vos R., de Hemptinne B., Gevaert K., Vandekerckhove J., Roskams T., Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Michta M.L., Hopcraft S.E., Narbus C.M., Kratovac Z., Israelow B., Sourisseau M., Evans M.J. Species-specific regions of occludin required by hepatitis C virus for cell entry. J. Virol. 2010;84:11696–11708. doi: 10.1128/JVI.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.I., Mo H., Pilot-Matias T., He Y., Koev G., Krishnan P., Mondal R., Pithawalla R., He W., Dekhtyar T., Packer J., Schurdak M., Molla A. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- Nielsen S.U., Bassendine M.F., Burt A.D., Martin C., Pumeechockchai W., Toms G.L. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH, 2011. 〈http://grants.nih.gov/grants/guide/notice-files/NOT-OD-12-025.html〉.
- Nitta, S., Sakamoto, N., Nakagawa, M., Kakinuma, S., Mishima, K., Kusano-Kitazume, A., Kiyohashi, K., Murakawa, M., Nishimura-Sakurai, Y., Azuma, S., Tasaka-Fujita, M., Asahina, Y., Yoneyama, M., Fujita, T., Watanabe, M. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type-I interferon-dependent innate immunity. Hepatology 10.1002/hep.26017, in press. [DOI] [PubMed]
- Olsen D.B., Davies M.E., Handt L., Koeplinger K., Zhang N.R., Ludmerer S.W., Graham D., Liverton N., MacCoss M., Hazuda D., Carroll S.S. Sustained viral response in a hepatitis C virus-infected chimpanzee via a combination of direct-acting antiviral agents. Antimicrob. Agents Chemother. 2011;55:937–939. doi: 10.1128/AAC.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.R., Loo Y.M., Horner S.M., Gale M., Jr., Malik H.S. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., Bell B.P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A.J., Houghton M., Rosa D., Grandi G., Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A., Evans M.J. Hepatitis C virus host cell entry. Curr. Opin. Virol. 2012;2:14–19. doi: 10.1016/j.coviro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A., Evans M.J., Gaysinskaya V.A., Panis M., You H., de Jong Y.P., Rice C.M. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploss A., Khetani S.R., Jones C.T., Syder A.J., Trehan K., Gaysinkaya V.A., Mu K., Ritola K., Rice C.M., Bhatia S.N. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl. Acad. Sci. USA. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann S., Zhang J., Baribaud F., Chen Z., Leslie G.J., Lin G., Granelli-Piperno A., Doms R.W., Rice C.M., McKeating J.A. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poordad F., McCone J., Jr., Bacon B.R., Bruno S., Manns M.P., Sulkowski M.S., Jacobson I.M., Reddy K.R., Goodman Z.D., Boparai N., DiNubile M.J., Sniukiene V., Brass C.A., Albrecht J.K., Bronowicki J.P. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince A., Brotman B., Huima T., Pascual D., Jaffery M., Inchauspé G. Immunity in hepatitis C infection. J. Infect. Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- Racaniello V.R., Ren R. Transgenic mice and the pathogenesis of poliomyelitis. Arch. Virol. Suppl. 1994;9:79–86. doi: 10.1007/978-3-7091-9326-6_9. [DOI] [PubMed] [Google Scholar]
- Randall G., Panis M., Cooper J.D., Tellinghuisen T.L., Sukhodolets K.E., Pfeffer S., Landthaler M., Landgraf P., Kan S., Lindenbach B.D., Chien M., Weir D.B., Russo J.J., Ju J., Brownstein M.J., Sheridan R., Sander C., Zavolan M., Tuschl T., Rice C.M. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., Kaul A., Buhler S., Pepperkok R., Lengauer T., Albrecht M., Eils R., Schirmacher P., Lohmann V., Bartenschlager R. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microb. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P., Obeid S., Paeshuyse J., Vanhove J., Van Lommel A., Nahmias Y., Nevens F., Neyts J., Verfaillie C.M. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Sainz B., Jr., Barretto N., Martin D.N., Hiraga N., Imamura M., Hussain S., Marsh K.A., Yu X., Chayama K., Alrefai W.A., Uprichard S.L. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E., Ansuini H., Cerino R., Roccasecca R.M., Acali S., Filocamo G., Traboni C., Nicosia A., Cortese R., Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaluder G.G., Dawson G.J., Simons J.N., Pilot-Matias T.J., Gutierrez R.A., Heynen C.A., Knigge M.F., Kurpiewski G.S., Buijk S.L., Leary T.P. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J. Med. Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Ohme M., Zischler H. The complete mitochondrial genome of Tupaia belangeri and the phylogenetic affiliation of scandentia to other eutherian orders. Mol. Biol. Evol. 2000;17:1334–1343. doi: 10.1093/oxfordjournals.molbev.a026417. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R.E., Trehan K., Andrus L., Sheahan T.P., Ploss A., Duncan S.A., Rice C.M., Bhatia S.N. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin C.I., Horvat B. Current animal models: transgenic animal models for the study of measles pathogenesis. Curr. Top. Microbiol. Immunol. 2009;330:111–127. doi: 10.1007/978-3-540-70617-5_6. [DOI] [PubMed] [Google Scholar]
- Shoukry N.H., Grakoui A., Houghton M., Chien D.Y., Ghrayeb J., Reimann K.A., Walker C.M. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz L.D., Ishikawa F., Greiner D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- Simons J.N., Leary T.P., Dawson G.J., Pilot-Matias T.J., Muerhoff A.S., Schlauder G.G., Desai S.M., Mushahwar I.K. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- Sithebe N.P., Kew M.C., Mphahlele M.J., Paterson A.C., Lecatsas G., Kramvis A., de Klerk W. Lack of susceptibility of Chacma baboons (Papio ursinus orientalis) to hepatitis C virus infection. J. Med. Virol. 2002;66:468–471. doi: 10.1002/jmv.2167. [DOI] [PubMed] [Google Scholar]
- Supekova L., Supek F., Lee J., Chen S., Gray N., Pezacki J.P., Schlapbach A., Schultz P.G. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J. Biol. Chem. 2008;283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- Tai A.W., Benita Y., Peng L.F., Kim S.S., Sakamoto N., Xavier R.J., Chung R.T. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microb. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temel R.E., Tang W., Ma Y., Rudel L.L., Willingham M.C., Ioannou Y.A., Davies J.P., Nilsson L.M., Yu L. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J. Clin. Invest. 2007;117:1968–1978. doi: 10.1172/JCI30060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotard M., Lepere-Douard C., Regeard M., Piquet-Pellorce C., Lavillette D., Cosset F.L., Gripon P., Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780–3789. doi: 10.1096/fj.09-131920. [DOI] [PubMed] [Google Scholar]
- Uprichard S.L., Chung J., Chisari F.V., Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol. J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt F.H., Pilote L., Cartier M., Lippens J., Liuzzi M., Bethell R.C., Cordingley M.G., Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Vernachio J.H., Bleiman B., Bryant K.D., Chamberlain S., Hunley D., Hutchins J., Ames B., Gorovits E., Ganguly B., Hall A., Kolykhalov A., Liu Y., Muhammad J., Raja N., Walters C.R., Wang J., Williams K., Patti J.M., Henson G., Madela K., Aljarah M., Gilles A., McGuigan C. INX-08189, a phosphoramidate prodrug of 6-O-methyl-2′-C-methyl guanosine, is a potent inhibitor of hepatitis C virus replication with excellent pharmacokinetic and pharmacodynamic properties. Antimicrob. Agents Chemother. 2011;55:1843–1851. doi: 10.1128/AAC.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ma Y., Barrett J.W., Gao X., Loh J., Barton E., Virgin H.W., McFadden G. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 2004;5:1266–1274. doi: 10.1038/ni1132. [DOI] [PubMed] [Google Scholar]
- Washburn, M.L., Bility, M.T., Zhang, L., Kovalev, G.I., Buntzman, A., Frelinger, J.A., Barry, W., Ploss, A., Rice, C.M., Su, L., 2011. A humanized mouse model to study Hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 140 (4), 1334–1344 [DOI] [PMC free article] [PubMed]
- Willinger T., Rongvaux A., Strowig T., Manz M.G., Flavell R.A. Improving human hemato-lymphoid-system mice by cytokine knock-in gene replacement. Trends Immunol. 2011;32:321–327. doi: 10.1016/j.it.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Wong-Staal, F., Chang, J., Rijnbrand, R., Shin, C., Lee, H., Syder, A.J., Yang, J.P., 2008. Characterization of the role of the protocadherin B famility in HCV entry. In: Proceedings of the 16th International Symposium on Hepatitis C Virus & Related Viruses, San Antonio, TX, USA.
- Wu X., Robotham J.M., Lee E., Dalton S., Kneteman N.M., Gilbert D.M., Tang H. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.C., Riezu-Boj J.I., Lasarte J.J., Guillen J., Su J.H., Civeira M.P., Prieto J. Transmission of hepatitis C virus infection to tree shrews. Virology. 1998;244:513–520. doi: 10.1006/viro.1998.9127. [DOI] [PubMed] [Google Scholar]
- Xu L., Chen S.Y., Nie W.H., Jiang X.L., Yao Y.G. Evaluating the phylogenetic position of Chinese tree shrew (Tupaia belangeri chinensis) based on complete mitochondrial genome: implication for using tree shrew as an alternative experimental animal to primates in biomedical research. J. Genet. Genomics=Yi chuan xue bao. 2012;39:131–137. doi: 10.1016/j.jgg.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Xu X., Chen H., Cao X., Ben K. Efficient infection of tree shrew (Tupaia belangeri) with hepatitis C virus grown in cell culture or from patient plasma. J. Gen. Virol. 2007;88:2504–2512. doi: 10.1099/vir.0.82878-0. [DOI] [PubMed] [Google Scholar]
- Yang F., Robotham J.M., Nelson H.B., Irsigler A., Kenworthy R., Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.L., Gao M., Lin K., Liu Q., Villareal V.A. Anti-HCV drugs in the pipeline. Curr. Opin. Virol. 2011;1:607–616. doi: 10.1016/j.coviro.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 2007;3:e108. doi: 10.1371/journal.ppat.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zheng A., Yuan F., Li Y., Zhu F., Hou P., Li J., Song X., Ding M., Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Guo J.T., Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]