The ability to fabricate protein micro and nano arrays in a low-cost and high throughput manner is important for a wide variety of applications, including drug screening, materials assembly, medical diagnostics, biosensors, and fundamental biological studies.[1-3] Traditional approaches to making protein microarrays include photolithography and inkjet printing. Recently, studies also have focused on the miniaturization of protein patterns into the nanometer regime because high density protein arrays can provide increased detection sensitivity and, in principle, allow one to screen millions of biomarkers with one chip.[4] Protein nanopatterns also can provide insight into important fundamental biological processes,[5] such as cell adhesion and differentiation.[6-9] Indeed, the ability to place an array of proteins or even multiple protein structures underneath a single cell opens up the opportunity to study multivalent interactions between a cell and a surface, and points to a major capability of nanoarray technology not afforded by analogous microscale structures. Herein, we report a novel and rapid strategy for inking nanoscale probes with different proteins, which can be transferred to a surface via the technique known as Polymer Pen Lithography (PPL).[10] Using this approach, we have generated sub-100 nm structures at a rate of 150,000 features per second.
Many new techniques have been explored for miniaturizing single component protein features and micropatterning of multiple proteins, including microcontact printing,[11-14] nanoimprint lithography,[15] e-beam lithography,[16] and a variety of scanning probe lithographies.[4, 17-22] To date, only a few examples of nanopatterning multiple proteins have been reported, and the majority among these examples uses destructive strategies, which require multiple cycles to deposit multiple proteins. As such, the throughput is relatively low and cross-contamination between different proteins is of great concern. Dip-pen nanolithography (DPN)[22] and PPL are particularly versatile “direct write” methods which allow one to generate protein structures over large areas with sub-micrometer resolution using as many as 11 million pens in parallel.[23-25] Lee et al.[26] showed that one can use DPN to generate nanoarrays of two different proteins in two sequential steps on a surface. This approach was extended to PPL in the context of single ink structures. Importantly, the “direct write” nature of DPN and PPL minimizes ink cross contamination. By combining the advantages of inkwell inking and inkjet printing with DPN, we have demonstrated multiplexed patterning of small molecules.[27]
Patterning multiple proteins by DPN over large areas remains a significant challenge for several reasons. (1) The opacity of Si and Si3N4 cantilevers makes it difficult, if not impossible, to align a 2D cantilever array for inking multiple proteins using inkwells. (2) The 2D Si3N4 cantilever array required for large scale parallel DPN experiments is relatively costly and fragile.
In principle, PPL is well-suited for patterning protein structures in a multiplexed manner. Instead of relying on hard Si3N4 cantilevers, PPL utilizes a soft polymer pen array to deliver inks onto a surface by controlling the movement of the pen array with a scanning probe microscope. Unlike DPN and conventional contact printing, the feature size in a PPL experiment not only depends upon probe-substrate contact time, but also contact force (which results in the reversible flattening of the tip). In addition, the same mould used to make the array can be used as an inkwell that can be addressed and filled via inkjet printing. In this way, one can achieve perfect registry between the pens in the array and the inkwells. Herein, we demonstrate that one can use PPL to pattern multiplexed protein arrays in one writing step with control over feature size (spanning the sub-100 nm to the μm length scale).
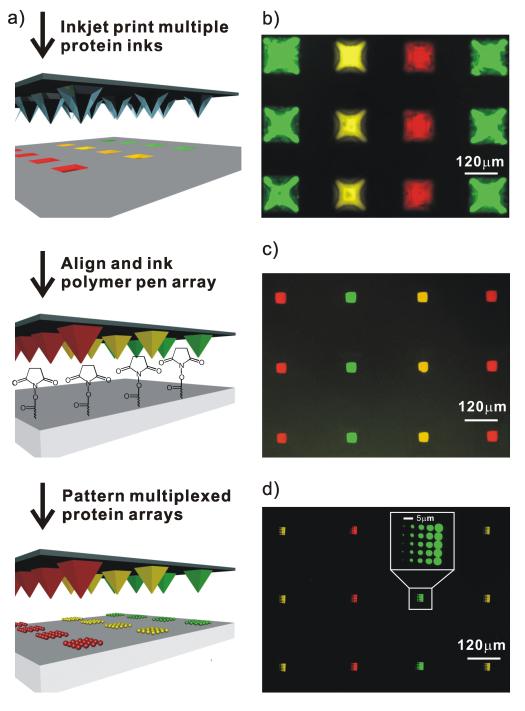
In a typical experiment, the pyramid-shaped wells in a Si mould used to make a PPL array were first filled with protein inks by inkjet printing (Figure 1a). The ink solution was composed of 0.1 to 0.5 mg/mL of protein molecules and 5 wt% of glycerol in phosphate buffered saline (PBS, pH 8.0). Note that the glycerol molecules serve as a carrier to increase the mobility of the ink on the polymer pens. A Piezorray (PerkinElmer, Waltham, MA) inkjet printer was used to selectively address and ink each well without contaminating neighbouring wells. Subsequently, the pen array was treated with oxygen plasma for 30 s to render the surface hydrophilic, and minimize nonspecific adhesion of proteins. The pen array was placed in an NSCRIPTOR™ (NanoInk, Skokie, IL) nanolithography instrument and dipped in the wells. We then used the inked polymer pen array to write directly on a Codelink™ slide, which has a surface terminated with N-hydroxysuccinimide (NHS) ester functional groups. The patterned slide was incubated at 4°C for 8 hours, according to the manufacturer’s protocol, to allow the amine groups on the proteins to react with the NHS esters. Finally, the slide was passivated with bovine serum albumin (BSA) for 1 hr, rinsed with PBS buffer, and dried.
Figure 1.
a) Schematic illustration of the PPL patterning process used for making multiplexed protein arrays. Fluorescent images of: b) a Si mould inked with 3 proteins by inkjet printing; c) a polymer pen array dipped into the Si mould in b); d) multiplexed proteins arrays made by PPL with the polymer pen array in c). Yellow: TRITC conjugated anti-mouse IgG; Green: Alexa Fluor 488 conjugated anti-prostate specific antigen (anti-PSA); Red: Alexa Fluor 647 conjugated anti-cholera toxin beta (anti-CTβ).
The wells in the mould are inverted-pyramids with an average depth of 86 μm, edge length of 120 μm, and centre-to-centre distance of 240 μm. As a proof-of-concept, we loaded 1600 inkwells with three different dye-labelled proteins, and by fluorescence microscopy one can see that they have been properly addressed with the inkjet printer (Figure 1b). By making the well surface hydrophobic with 1H,1H,2H,2H-perfluorodecyltrichlorosilane, the ink is driven by gravity into the wells. A PPL array was then levelled, aligned, and brought into contact with the ink-filled mould by the optical microscope of the NSCRIPTOR. Importantly, because the polymer pen array is transparent, one can readily confirm inking optically (Movie 1, Supporting Information). The PPL array was allowed to absorb ink for 10 min at 90% relative humidity, imaged by fluorescence microscopy (Figure 1c), and then used for patterning experiments. As a proof-of-concept, each pen in an array was used to make a 5 × 5 protein dot array with 5 μm spacing between the dots (Figure 1d). As shown in the inset image, the sizes of the protein features from left column to right column are 0.63 ± 0.06, 1.94 ± 0.02, 3.09 ± 0.1, 3.94 ± 0.09, 4.83 ± 0.08 μm, respectively. There is no apparent cross-contamination, a consequence of the one-step, top-down writing attribute of PPL. Finally, the inkwells can be used repeatedly to ink more than five pen arrays with very similar results and less than 10% variation in feature size across the studied length scale.
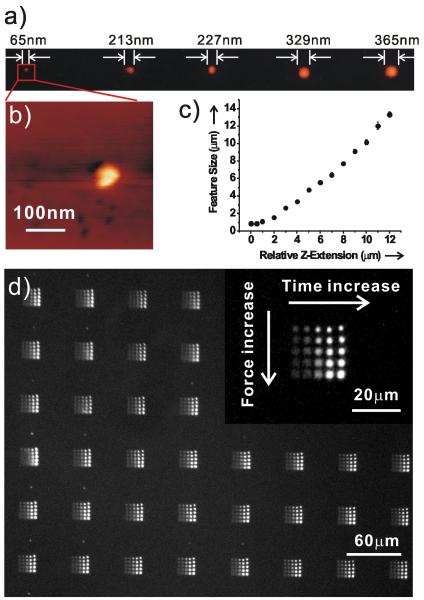
Importantly, one can control feature size over the sub-100 nm to many μm length scale by varying both the tip-substrate contact time and contact force (Figure 2). When the tip makes initial contact with the substrate, 65 nm features are made at 0.01 s contact time (Figure 2a and b). One sees the feature area dependence upon the tip-substrate contact time typical of DPN and PPL (Figure S1). Because feature size in a PPL experiment is also dependent upon contact force, one can rapidly access larger feature sizes by controlling Z-piezo extension (Figure 2c). For example, with a 500 nm extension (relative to initial contact) and a fixed contact time of 10 s, the resulting protein feature size is 860 ± 40 nm. Further extending the Z-piezo results in a quasi-linear increase in feature size. For example, 13.32 ± 0.32 μm dots were generated with a 12 μm Z-piezo extension in the same pen array configuration. This feature of PPL allows one to not only multiplex, but also span the sub-100 nm to many μm length scale in a single patterning experiment.
Figure 2.
a) Tapping mode atomic force microscopy (AFM, topographic mode) of CTβ/glycerol patterned on a Codelink slide by PPL. b) A zoom in AFM topography of a). c) Feature size of patterned protein arrays as a function of tip-substrate contact force. d) Fluorescent image of PSA arrays labelled with Alexa Fluor 488 conjugated anti-PSA at different tip-substrate contact time and contact force. The inset is a magnified fluorescence image.
We patterned 5 × 5 PSA dot arrays by PPL onto a Codelink slide with increasing tip-substrate contact times and contact forces. The distances between neighbouring dots (in one array) and neighbouring arrays were 5 μm and 60 μm, respectively. This protein chip was labelled by its corresponding antibody by immersion in a PBS (pH 7.4) solution containing 100 nM Alexa Fluor 488 conjugated anti-PSA for 1 hr, followed by rinsing, drying and imaging with fluorescence microscopy. As shown in Figure 2d, anti-PSA binds selectively onto the PSA regions with undetectable background. The feature size increases from 1.1 to 3.2 μm with increasing contact force. Interestingly, the fluorescence intensity increases with increasing tip-substrate contact time, most likely because of higher PSA densities at longer contact times. In addition, we demonstrated that PPL-patterned proteins retain their structural integrity by patterning the PSA antibody onto Codelink slides and incubating an Alexa Fluor 488 labelled PSA with the surface-bound antibodies. Fluorescence microscopy results demonstrate that the PPL-patterned antibodies bound their antigens (Figure S2, Supporting Information).
In conclusion, we have demonstrated a novel way of using a PPL array mould to localize different inks on the pens of a PPL array. This new strategy for localizing the respective inks on the nanoscale tips of a two-dimensional PPL array allows for the multiplexed patterning of protein nano and micro arrays in a high throughput and low-cost manner. The resulting protein features retain their structure and can be prepared with no evidence of cross-contamination over very large areas. This novel method is a general approach, which in principle can be applied to large scale, multiplexed nano- and micropatterning of many biomolecules and other libraries of small molecules, catalysts, and essentially any set of structures which can be transported by PPL.
Experimental Section
Materials
Si wafers <100> with 500 nm of thermally deposited SiO2 were purchased from Silicon Quest International. Codelink slides were purchased from SurModics. Shipley1805 photoresist and MF319 developing solution were purchased from MicroChem. 1H,1H,2H,2H-perfluorodecyltrichlorosilane was purchased from Gelest. TRITC conjugated anti-mouse IgG, bovine serum albumin (BSA), and prostate specific antigen (PSA) proteins were purchased from Sigma-Aldrich. Anti-PSA was purchased from R and D Systems. Alexa Fluor 488 and 647 monoclonal antibody labelling kits and anti-cholera toxin beta (anti-CTβ) antibodies were purchased from Invitrogen. The antibodies were labelled with the Alexa Fluor dyes following the manufacturer’s instructions. Buffered HF etching solution was purchased from Transene Company. Isopropanol and acetone were purchased from Fisher.
Antibody labelling
After the antigens were bound to the slides, they were rinsed with 0.15 M PBS supplemented with 0.1% Tween 20. Then, the labelled antibodies were each diluted to a final concentration of 100 nM in 0.15 M PBS with 0.025% Tween 20 and 0.1% BSA and incubated with the surface bound antigens for 1 hr. The slide was then rinsed with the 0.15 M PBS and Tween 20 solution, briefly rinsed with water, and spun dry.
Fabrication of Si moulds
Shipley1805 photoresist was spun-coated onto Si wafers with a 500 nm thick top layer of SiO2. Square well arrays were fabricated by photolithography using a chrome mask. The photoresist patterns were developed in an MF319 developing solution and then exposed to O2 plasma for 30 s (200 mTorr) to remove the residual organic layer. Subsequently, the substrates were placed in a buffered HF etching solution for 6 min. The photoresist was then removed with acetone to expose the SiO2 pattern. The SiO2 patterned substrate was placed in a KOH etching solution (30% KOH in H2O:isopropanol (4:1 v/v)) at 75 °C for ~2.5 hr with vigorous stirring. The uncovered areas of the Si wafer were etched anisotropically, resulting in the formation of recessed pyramids. The remaining SiO2 layer was removed by exposure to HF etching solution for 1 min. Copious rinsing with MilliQ water was required after each etching step to clean the surface. Finally, the pyramid inkwell/master was modified with 1H,1H,2H,2H-perfluorodecyltrichlorosilane by gas phase silanization.[13, 28]
Supplementary Material
Footnotes
C.A.M. acknowledges the AFOSR, DARPA/SPAWAR, and NSF for support of this research. C.A.M. is also grateful for the NIH Director’s Pioneer Award and a NSSEF Fellowship from the DoD. L.R.G. acknowledges the NSF for a graduate research fellowship.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- [1].Mirkin CA. ACS Nano. 2007;1:79. doi: 10.1021/nn700228m. [DOI] [PubMed] [Google Scholar]
- [2].Balboni I, Chan SM, Kattah M, Tenenbaum JD, Butte AJ, Utz PJ. Annu. Rev. Immuno. 2006;24:391. doi: 10.1146/annurev.immunol.24.021605.090709. [DOI] [PubMed] [Google Scholar]
- [3].Jonkheijm P, Weinrich D, Schröder H, Niemeyer CM, Waldmann H. Angew. Chem. 2008;120:9762. doi: 10.1002/anie.200801711. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:9618. [Google Scholar]
- [4].Christman KL, Enriquez-Rios VD, Maynard HD. Soft Matter. 2006;2:928. doi: 10.1039/b611000b. [DOI] [PubMed] [Google Scholar]
- [5].Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- [6].Yousaf MN, Houseman BT, Mrksich M. Proc. Natl. Acad. Sci. 2001;98:5992. doi: 10.1073/pnas.101112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee K-B, Park S-J, Mirkin CA, Smith JC, Mrksich M. Science. 2002;295:1702. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- [8].Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nat. Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- [9].Walter N, Selhuber C, Kessler H, Spatz JP. Nano Lett. 2006;6:398. doi: 10.1021/nl052168u. [DOI] [PubMed] [Google Scholar]
- [10].Huo F, Zheng Z, Zheng G, Giam LR, Zhang H, Mirkin CA. Science. 2008;321:1658. doi: 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E. Adv. Mater. 2000;12:1067. [Google Scholar]
- [12].Duan X, Sadhu VB, Perl A, Peter M, Reinhoudt DN, Huskens J. Langmuir. 2008;24:3621. doi: 10.1021/la702975q. [DOI] [PubMed] [Google Scholar]
- [13].Zheng Z, Jang J-W, Zheng G, Mirkin CA. Angew. Chem. 2008;120:10099. doi: 10.1002/anie.200803834. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:9951. [Google Scholar]
- [14].Coyer SR, Garcia AJ, Delamarche E. Angew. Chem. 2007;119:6961. doi: 10.1002/anie.200700989. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:6837. [Google Scholar]
- [15].Hoff JD, Cheng LJ, Meyhofer E, Guo LJ, Hunt AJ. Nano Lett. 2004;4:853. [Google Scholar]
- [16].Christman KL, Schopf E, Broyer RM, Li RC, Chen Y, Maynard HD. J. Am. Chem. Soc. 2009;131:521. doi: 10.1021/ja804767j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Loh OY, Ho AM, Rim JE, Kohli P, Patankar NA, Espinosa HD. Proc. Natl. Acad. Sci. 2008;105:16438. doi: 10.1073/pnas.0806651105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pavlovic E, Oscarsson S, Quist AP. Nano Lett. 2003;3:779. [Google Scholar]
- [19].Kenseth JR, Harnisch JA, Jones VW, Porter MD. Langmuir. 2001;17:4105. [Google Scholar]
- [20].Wadu-Mesthrige K, Xu S, Amro NA, Liu G.-y. Langmuir. 1999;15:8580. [Google Scholar]
- [21].Tinazli A, Piehler J, Beuttler M, Guckenberger R, Tampé B. Nat. Nanotech. 2007;2:220. doi: 10.1038/nnano.2007.63. [DOI] [PubMed] [Google Scholar]
- [22].Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. Science. 1999;283:661. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- [23].Ginger DS, Zhang H, Mirkin CA. Angew. Chem. 2004;116:30. doi: 10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2004;43:30. [Google Scholar]
- [24].Salaita K, Wang YH, Mirkin CA. Nat. Nanotech. 2007;2:145. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- [25].Lenhert S, Sun P, Wang Y, Fuchs H, Mirkin CA. Small. 2007;3:71. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- [26].Lee KB, Lim JH, Mirkin CA. J. Am. Chem. Soc. 2003;125:5588. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- [27].Wang YH, Giam LR, Park M, Lenhert S, Fuchs H, Mirkin CA. Small. 2008;4:1666. doi: 10.1002/smll.200800770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pallandre A, Glinel K, Jonas AM, Nysten B. Nano Lett. 2004;4:365. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.