Abstract
Background and Aims
Despite differences in morphology, the genera representing ‘true citrus fruit trees’ are sexually compatible, and their phylogenetic relationships remain unclear. Most of the important commercial ‘species’ of Citrus are believed to be of interspecific origin. By studying polymorphisms of 27 nuclear genes, the average molecular differentiation between species was estimated and some phylogenetic relationships between ‘true citrus fruit trees’ were clarified.
Methods
Sanger sequencing of PCR-amplified fragments from 18 genes involved in metabolite biosynthesis pathways and nine putative genes for salt tolerance was performed for 45 genotypes of Citrus and relatives of Citrus to mine single nucleotide polymorphisms (SNPs) and indel polymorphisms. Fifty nuclear simple sequence repeats (SSRs) were also analysed.
Key Results
A total of 16 238 kb of DNA was sequenced for each genotype, and 1097 single nucleotide polymorphisms (SNPs) and 50 indels were identified. These polymorphisms were more valuable than SSRs for inter-taxon differentiation. Nuclear phylogenetic analysis revealed that Citrus reticulata and Fortunella form a cluster that is differentiated from the clade that includes three other basic taxa of cultivated citrus (C. maxima, C. medica and C. micrantha). These results confirm the taxonomic subdivision between the subgenera Metacitrus and Archicitrus. A few genes displayed positive selection patterns within or between species, but most of them displayed neutral patterns. The phylogenetic inheritance patterns of the analysed genes were inferred for commercial Citrus spp.
Conclusions
Numerous molecular polymorphisms (SNPs and indels), which are potentially useful for the analysis of interspecific genetic structures, have been identified. The nuclear phylogenetic network for Citrus and its sexually compatible relatives was consistent with the geographical origins of these genera. The positive selection observed for a few genes will help further works to analyse the molecular basis of the variability of the associated traits. This study presents new insights into the origin of C. sinensis.
Keywords: Phylogeny, evolution, SNP, indel, SSR, Rutaceae, Citrus, Fortunella, Microcitrus, Eremocitrus, Poncirus
INTRODUCTION
Aurantioideae (Rutaceae) are considered to be a monophyletic group (Scott et al., 2000; Groppo et al., 2008; Morton, 2009) and Ruta appears to be sister to Aurantioideae (Scott et al., 2000; Bayer et al., 2009). Furthermore, Groppo et al. (2008) suggest that Aurantioideae should be recognized as a tribe and be included in a subfamily together with Rutoideae, Toddalioideae and Flindersioideae. Although new insights into the circumscription of the tribes of Aurantioideae have been recently released (Bayer et al., 2009; Morton, 2009), it remains unresolved. In the classification of Swingle and Reece (1967), which remains the most used by citrus researchers, Aurantioidae is divided into two tribes, Clauseneae and Citreae. Citreae includes, among others, subtribe Citrinae, which is in turn divided into six genera (Fortunella, Eremocitrus, Poncirus, Clymenia, Microcitrus and Citrus) that comprise the important ‘true citrus fruit trees’ group (Swingle and Reece, 1967; Krueger and Navarro, 2007).
Among these genera, Citrus is by far the most economically important. It is believed to have originated in south-eastern Asia, in an area that includes China, India and the Indochinese peninsula and nearby archipelagos (Krueger and Navarro, 2007). Citrus taxonomy is still controversial due to the large degree of morphological diversity found in the group, sexual compatibility between the species and apomixis in many genotypes. Two major classification systems based on morphological and phenotypic data are currently used, i.e. those of Swingle and Reece (1967) and Tanaka (1977), who recognized 16 and 162 species, respectively. Here we adopt the classification system of Swingle and Reece (1967), which is more in line with the main clustering system derived from the molecular analysis described in this report. More recently, Mabberley (1997) proposed a new classification system for edible citrus that recognizes three species and four hybrid groups. In agreement with a pioneering numerical taxonomic study (Barrett and Rhodes, 1976), the classification system of Mabberley confirms that three main taxa [C. medica L. (citrons), C. maxima (Burm.) Osbeck (pummelos) and C. reticulata Blanco (mandarins)] were the ancestors of cultivated Citrus. However, the subdivision into four hybrid groups remains questionable, and relatively few authors have adopted the classification system of Mabberley. More recent studies involving the diversity of morphological characteristics (Ollitrault et al., 2003) and the analysis of primary metabolites (Luro et al., 2011) and secondary metabolites (Fanciullino et al., 2006) have indicated that the phenotypic diversity of edible citrus species primarily resulted from the initial differentiation between these three basic taxa. Molecular marker studies using restriction fragment length polymorphism (RFLP; Federici et al., 1998), random amplified polymorphic DNA (RAPD) and sequence characterized amplified regions (SCARs; Nicolosi et al., 2000), simple sequence repeats (SSRs; Luro et al., 2001; Barkley et al., 2006), SSR and insertion-deletion (indels; Garcia-Lor et al., 2012a) and single nucleotide polymorphisms (SNPs; Ollitrault et al., 2012a) have confirmed the central role played by these three taxa, but also pointed out that C. micrantha, a member of subgenus Papeda, is a potential parent of some limes (C. aurantifolia Christm.). Swingle and Reece (1967) differentiated between subgenera Papeda and Citrus. The genome of most of the important commercial Citrus spp. (secondary species) can be considered to be a mosaic of large DNA fragments of the ancestral species that resulted from a few interspecific recombination events (Garcia-Lor et al., 2012a).
Fortunella is generally considered to be a separate genus (Swingle and Reece, 1967), but it closely resembles Citrus. According to Swingle (1943), this genus includes four species (F. margarita, F. japonica, F. hindsii and F. polyandra) and Fantz (1988) included two hybrid taxa in Fortunella (F. obovata, F. crassifolia). Its origin is northern China, and it is well adapted to cold areas due to its propensity for prolonged winter dormancy and late flowering. Its fruits, commonly called kumquats, are edible, and Fortunella trees are appreciated for their ornamental qualities. Poncirus is another genus that originated in northern China, and for a long time it was considered to be monotypic (P. trifoliata). However, a new species belonging to this genus, P. polyandra, was found in Yunnan (China) in the 1980s (Ding et al., 1984). Poncirus trifoliata is the only species of ‘true citrus fruit trees’ with deciduous leaves. It is highly tolerant to cold and resistant to several citrus pathogens. It is therefore an important source of germplasm for citrus rootstock breeding.
According to Krueger and Navarro (2007), Microcitrus includes five species that originated in Australia (M. australis, M. australasica, M. inodora, M. garrowayii and M. maindeniana) and two from Papua New Guinea (M. papuana and M. warbugiana). Microcitrus australasica, finger lime, is cultivated on a small scale for its fruit with aromatic, spherical juice vesicles. Eremocitrus is a monospecific genus (E. glauca) that is native to the Australian desert. This genus is cold-tolerant and xerophytic. Eremocitrus and Microcitrus are closely related, morphologically and molecularly (Swingle and Reece, 1967; Bayer et al., 2009). They are graft-compatible with Citrus and other related genera.
Despite considerable morphological differentiation, Citrus, Fortunella, Poncirus, Microcitrus and Eremocitrus are sexually compatible genera (Krueger and Navarro, 2007). Studies based on plastid sequences (Abkenar et al., 2004; Morton, 2009; Bayer et al., 2009) concur that the six genera of tribe Citrinae (Fortunella, Eremocitrus, Poncirus, Clymenia, Microcitrus and Citrus) form a clade. However, these authors did not fully agree on the organization within this clade. Clymenia polyandra appeared in the same subclade as Citrus, Fortunella and Poncirus in the analyses of Abkenar et al. (2004), but Morton (2009) and Bayer et al. (2009) found it in the Eremocitrus and Microcitrus subclade. Moreover, Bayer et al. (2009) included Oxanthera and Feroniella in the clade of the true citrus fruit trees. To analyse the gene pool of true citrus fruit trees, with potential gene flow between sexually compatible taxa, some of which share the same diversification area, phylogenetic analysis based on nuclear sequences should be more informative than the analysis of maternally inherited plastid sequences (Ramadugu et al., 2011). However, the phylogenetic relationships between true citrus fruit trees based on the analysis of nuclear genomes have not been clearly elucidated.
In genetic studies of Citrus, SSR analysis (Gulsen and Roose, 2001; Luro et al., 2001, 2008; Barkley et al., 2006; Ollitrault et al., 2010) is seen as a powerful tool because SSRs are co-dominant, randomly dispersed throughout the plant genome, generally highly polymorphic and locus-specific. However, Barkley et al. (2009) showed that homoplasy might limit the usefulness of SSRs as tags to elucidate the phylogenetic origin of specific DNA fragments in citrus. Moreover, the high mutation rate of SSRs can often lead to an underestimation of subpopulation divergence (Coates et al., 2009). In recent studies, Garcia-Lor et al. (2012a) and Ollitrault et al. (2012b) analysed the value of nuclear indels as genetic markers in Citrus. These studies showed that indels are more suitable than SSRs for differentiating between the three basic taxa of cultivated Citrus. However, the relatively low frequency of indels limits their utility.
SNPs are the most abundant type of DNA sequence polymorphism (Brookes, 1999). Due to the high frequency of occurrence of SNPs and their relatively dense and uniform distribution in genomes, SNPs are an important source of variability and are therefore useful for many applications, including the development of saturated genetic maps, cultivar identification, detection of genotype/phenotype associations and marker-assisted breeding (Botstein and Risch, 2003; Morales et al., 2004; Xing et al., 2005; Lijavetzky et al., 2007). The frequency of occurrence of SNPs in the genomes of eukaryotes depends on the domestication and breeding history, mating system and frequency of mutation, recombination and other features (Buckler and Thornsberry, 2002; Rafalski and Morgante, 2004). Although individual SNPs are less informative than other marker types for population genetic studies because of their biallelic nature, they have several advantages over other marker types due to the high frequency of SNP occurrence, the easy automation of SNP genotyping, the low-scoring error rates and the high levels of reproducibility of SNP analysis results between laboratories (Morales et al., 2004; Helyar et al., 2011).
Many efforts have been made to detect SNPs in plants. SNPs have been used to perform comparative diversity analysis and genotyping, to reveal genetic structures and to assess molecular evolutionary patterns in many plant species including Norway spruce (Heuertz et al., 2006), sunflower (Kolkman et al., 2007), grapevine (Lijavetzky et al., 2007), European aspen (Ingvarsson, 2005) and eucalyptus (Külheim et al., 2009). Some studies have been performed in Citrus, but these studies were generally limited due to narrow genetic basis of the discovery panel. Novelli et al. (2004) searched for SNPs among several sweet orange lines. Terol et al. (2008) identified 6617 putative SNPs from Nules clementine BAC end sequences, from which 622 were successfully transferred to the entire genus using GoldenGate array technology (Ollitrault et al., 2012a). Dong et al. (2010) mined SNPs from sweet orange and satsuma mandarin expressed sequence tag (EST) databases.
The ascertainment bias associated with a low genetic basis of the discovery panel has been widely discussed for humans and animals (Clark et al., 2005; Rosenblum and Novembre, 2007; Albrechtsen et al., 2010), and it was observed for Citrus at the genus level when diversity studies with SNP markers mined in clementine were performed (Ollitrault et al., 2012a). It is therefore important to develop a good sampling strategy for SNP discovery (Garvin et al., 2010; Helyar et al., 2011) that would help to elucidate the true differentiation level between basic taxa and related genera at the nuclear level.
In the present study, we searched for SNP and indel polymorphisms in 45 accessions of Citrus, Poncirus, Fortunella, Microcitrus and Eremocitrus, with Severinia buxifolia employed as outgroup, using Sanger sequencing of amplified DNA fragments from 18 genes involved in primary and secondary metabolite biosynthetic pathways that determine citrus fruit quality (sugars, acids, flavonoids and carotenoids) and nine putative salt tolerance genes. In addition to the identification of useful intra- and interspecific SNP and indel markers, this study addresses the following questions: (1) What are the phylogenetic relationships at the nuclear level between different Citrus spp. and between genera? (2) What is the level of intra- and interspecific diversity between the Citrus taxa at the origin of the cultivated forms? (3) Did the evolution of genes involved in different metabolic pathways and some putative stress adaptation genes follow a similar neutral pattern regarding the history and reproductive biology of Citrus, or did some genes experience selective evolution? (4) What is the phylogenetic inheritance pattern of the analysed genes in secondary Citrus spp.?
MATERIALS AND METHODS
Plant material
Leaf material from 44 true citrus accessions and one relative (Severinia buxifolia) used as the outgroup (Supplementary Data Table S1) was collected, and DNA was extracted using the DNeasy plant mini kit (Qiagen S.A., Madrid, Spain). The samples represented all major Citrus species (seven C. reticulata, five C. maxima, five C. medica and four representatives of subgenus Papeda) and five Fortunella spp., two Microcitrus spp., one Eremocitrus sample and three Poncirus trifoliata, all of which are sexually compatible with Citrus. These eight groups are considered to be ancestral populations. Some representatives of secondary species were added (two diploid clementines and one haploid clementine, two C. sinensis, two C. aurantium, one C. paradisi, one C. limon and one C. aurantifolia) and two hybrids, including one tangor (C. reticulata × C. sinensis) and one tangelo (C. paradisi × C. reticulata). These 12 genotypes are known to be hybrids derived from the ancestral populations and are economically important cultivars. Haploid clementine (Aleza et al., 2009) is currently being used by the International Citrus Genome Consortium to establish the whole genome reference sequence of citrus. It was used in the present study to test whether some genes were duplicated. Forty-two accessions were obtained from the IVIA Citrus Germplasm Bank of pathogen-free plants (Navarro et al., 2002), and three were obtained from the INRA/CIRAD collection. All accessions were used for Sanger sequencing of gene fragments and indel and SSR genotyping.
Gene fragment sequencing
Eighteen genes involved in primary and secondary metabolite biosynthesis pathways that determine citrus fruit quality (sugars, acids, flavonoids and carotenoids) and nine putative salt tolerance genes were selected. Selection of the 27 gene fragments was based on the quality of sequencing chromatograms for all genotypes. Primers were designed using Primer3 (Rozen and Skaletsky, 2000) to amplify fragments ranging from 190 to 941 bp, according to the ESTs available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/; Table 1). PCR amplifications were performed using a Mastercycler Ep Gradient S thermocycler (Eppendorf) in a final volume of 25 µL containing 0·027 U μL−1 Taq DNA polymerase (Fermentas), 1 ng μL−1 of genomic DNA, 10× PCR buffer (Fermentas), 0·2 mmol of each dNTP, 1·5 mm MgSO4 and 0·2 µmol of each primer. The following PCR programme was applied: denaturation at 94 °C for 5 min and 40 cycles of 30 s at 94 °C, 1 min at 55 or 60 °C (according to the melting temperature of the primers), 2 min at 72 °C, and a final elongation step of 4 min at 72 °C. PCR product purification was done using a QIAquick PCR Purification Kit (Qiagen S.A.). Amplicons of the 45 genotypes were sequenced using the Sanger method from the 5′ end using fluorescently labelled dideoxynucleotides (Big Dye Terminator Cycle Sequencing Kit v3·1).
Table 1.
Primer sequences for the studied genes
| Biosyntetic pathway | Gene | Primers | Annealing temperature (°C) | GenBank accession no. |
|---|---|---|---|---|
| Flavonoids | Chalcone isomerase (CHI) | F: TTGTTCTGATGGCCTAATGG | 58 | DY263683 |
| R: AAAGGCTGTCACCGATGAAT | 58 | |||
| Chalcone synthase (CHS) | F: GATGTTGGCCGAGTAATGCT | 60 | CV885475 | |
| R: ATGCCAGGTCCAAAAGCTAA | 59 | |||
| Flavonol synthase (FLS) | F: GGAGGTGGAGAGGGTCCAAG | 59 | AB011796 | |
| R: GGGCCACCACTCCAAGAGC | 61 | |||
| Flavonoid 3'-hydroxylase (F3'H) | F: CTCGAGCCTTCCTCAAAACC | 60 | HQ634392 | |
| R: AACAAGCACAATCCCCATTC | 57 | |||
| Dihydroflavonol 4-reductase (DFR) | F: CTGGGTTTATCGGTTCATGG | 60 | DQ084722 | |
| R: TCCACAGCACCTGTGAACAT | 60 | |||
| Acids | Malic enzyme (EMA) | F: ACATGACGACATGCTTCTGG | 58 | CB417399 |
| R: CGTAGCCACGCCTAGTTCAT | 60 | |||
| Malate dehydrogenase (MDH) | F: ATGGCCGCTACATCAGCTAC | 60 | DQ901430 | |
| R: TGCAACCCCCTTTTCAATAC | 59 | |||
| Aconitase (ACO) | F: AAGCCATGGGTCAAAACAAG | 59 | AF073507 | |
| R: GATTTCCCAGTGTCGGTTGT | 59 | |||
| Vacuolar citrate/H+ symporter (TRPA) | F: GGCGCCACTCCTACCTTCCC | 62 | EF028327 | |
| R: CGGTCATTGAAGAGTGCTCCCC | 60 | |||
| Sugars | Acid invertase (INVA) | F: ATTGCGGATGTGAAGAAAGG | 56 | AB074885 |
| R: TTTGCCATGCTTTGAGTGAG | 56 | |||
| Phosphoenolpyruvate carboxylase (PEPC) | F: AGCCAATGGGATTTCTGACA | 60 | EF058158 | |
| R: GCCAAGCCACACAGGTAAAT | 60 | |||
| Phosphofructokinase (PKF) | F: CGCCGACCTCAGTCCCGTC | 63 | AF095520 | |
| R: GCTGCACGCCCCATAAGCCG | 64 | |||
| Carotenes | 1-Deoxyxylulose 5-phosphate synthase (DXS) | F: GGCGAGGAAGCGACGAAGATGG | 62 | DN959423 |
| R: GGATCAGAACTGGCCCTGGCG | 62 | |||
| Phytoene synthase (PSY) | F: GCTCGTTGATGGGCCTAATGC | 59 | AB037975 | |
| R: CGGGCGTAAGAGGGATTTTGC | 59 | |||
| β-Carotene hydroxylase (HYB) | F: AGCCCTTCTGTCTCCTCACA | 59 | AF315289 | |
| R: CCGTGGAATTTATCCGAGTG | 59 | |||
| Lycopene β-cyclase 2 (LCY2) | F: GCATGGCAACTCTTCTTAGCCCG | 60 | FJ516403 | |
| R: AGCTCGCAAGTAAGGCCATTCC | 61 | |||
| Lycopene β-cyclase (LCYB) | F: GAATTCTTGCCCCAAGTTCA | 60 | AY16696 | |
| R: TATGGGCCACAAATCTTTCC | 59 | |||
| 9-cis-Epoxy hydroxy carotenoid dyoxygenase 3 (NCED3) | F: GCAGTCAAATTCAACAAAGG | 55 | DQ309332 | |
| R: AATCCCAAATCTTGACACCT | 55 | |||
| Aldarate and ascorbate metabolism | Ascorbate oxydase (AOC) | F: TCAGTGAGAACCCTAAAGC | 58 | DY293375 |
| R: CAGTACAACCCCAGTAAGC | 60 | |||
| Ascorbate peroxidase (LAPX) | F: CAGCGGGGACTTATGACG | 58 | EU719653 | |
| R: GCCCTCCGGTAACTTCAAC | 59 | |||
| Cellular Detoxification | MRP-like ABC transporter (MRP4) | F: AGAAGCAGCATGGAAGATGG | 60 | CD574223 |
| R: CCGATCGGTTGGCATACTC | 62 | |||
| Cation chloride cotransporter (CCC1) | F: GCAGCTTGCTACCTACATTGAC | 63 | FN662480 | |
| R: ACTGAACTCCACATCCCAAAAG | 61 | |||
| High-affinity K+ transporter 1 (HKT1) | F: GTCCATGGAGAAAAAGAACC | 58 | DY297409 | |
| R: TGCTAGTGTCCGTGAAGAAG | 60 | |||
| NADH kinase (NADK2) | F: TGCAGAGACAAGATATTCCC | 58 | DN619491 | |
| R: ATGTGAGGTGAGAAATCCC | 58 | |||
| Salt tolerance | Aquaporin PIP1A (PIP1) | F: GACACTCGGCCTGTTCTTG | 62 | CK938271 |
| R: TCCGGTAATTGGGATGGTAG | 60 | |||
| Salt overly sensitive 1 (SOS1) | F: ACCAGTCAGACAACCATTTG | 55 | DN959478 | |
| R: CCAATTAGCACCTCATAGAGAC | 58 | |||
| Sucrose and starch metabolism | Trehalose-6-phosphate synthase (TSC) | F: TGCAGAACCTGTAATGAAGC | 58 | FC875388 |
| R:CTGGTAGGATGCCGACTTAG | 62 |
* AT, annealing temperature (°C).
† GBA, GenBank accession number.
Sequence polymorphism analysis
Sequences were aligned using BioEdit (Hall, 1999), SeqMan version 7·0 (http://www.dnastar.com) and SATé-II (Liu et al., 2012). The homogeneity of the alignment obtained with the three software programs was checked and heterozygosity or homozygosity of all genotypes was verified visually in the chromatogram for all SNP positions. Estimates of nucleotide polymorphisms (segregating sites, S, nucleotide diversity, π) and between-species divergences were obtained using DnaSP v. 5·10·01 (http://www.ub.es/dnasp). The genomic DNA sequences were subjected to blast analysis using the protein databases (blastx) of NCBI to identify the coding and non-coding regions. DnaSP was also used to calculate the statistical test of neutrality, Tajima's D (Tajima, 1989a, b).
For each target gene fragment, the haplotype number and the haplotype diversity were estimated with DnaSP software using coalescent process simulations. Unbiased expected heterozygosity, observed heterozygosity, fixation index value (Fw; Wright, 1978) and the Fstat parameter (Fst) were calculated using genetix v. 4·03 (Belkhir et al., 2002).
Indel marker development
Primer pairs for 12 indel markers are already available for the true citrus fruit trees group (Garcia-Lor et al., 2012a). New primer pairs for genes with indel polymorphisms were designed with Primer3 in conserved regions flanking the indel polymorphism (http://biotools.umassmed.edu/bioapps/primer3_www.cgi; Supplementary Data Table S2) to amplify fragments smaller than 400 bp that were subsequently subjected to fragment size polymorphism analysis in a capillary fragment analyser as described by Garcia-Lor et al. (2012a).
SSR markers
The 50 SSRs markers used for the diversity analysis in Citrus by Garcia-Lor et al. (2012a) were used to complete the genotyping for the accessions of the other genera. The list of primers that were used, the PCR conditions that were employed and the method used for capillary electrophoresis can be found in Garcia-Lor et al. (2012a).
Phylogenetic analysis
Phylogenetic analysis was performed by joining the 27 sequences together for each genotype (eight taxa), creating a sequence with a total length of 16 238 bp. Indels were excluded from the analysis. Several analyses were performed to determine which model best matched our data using the Phylemon 2·0 website (http://phylemon.bioinfo.cipf.es; Sánchez et al., 2011), which integrates different tools for molecular evolution, phylogenetics, phylogenomics and hypothesis testing. PhyML Best AIC Tree (v. 1·02b) software, which uses a model test program (Posada and Crandall, 1998) that performs hierarchical likelihood ratio tests (LRTs) in an ordered way using Akaike's information criteria (AIC), was used to select the model that most closely fitted the data (lowest AIC value), taking into account the nucleotide substitution model, the proportion of invariable sites (I), the nucleotide frequency (F) and the gamma distribution (G).
Construction of the maximum-likelihood (ML) tree was performed using 1000 bootstraps to assess the branch support using the SH-like approximate likelihood ratio test (ranges from 0 to 1), assuming uniform rates among sites and deleting gaps and missing sites. Trees obtained in Phylemon (newick format) were drawn using the TreeDyn 198·3 tool found at www.phylogeny.fr (Dereeper et al., 2008).
Neighbour-joining (NJ) analysis
Population diversity organization based on the SNP data was analysed with DARwin software (Perrier and Jacquemoud-Collet, 2006) as explained by Garcia-Lor et al. (2012a).
Principal coordinate analysis (PCoA)
PCoA was performed using genealex6 software (Peakall and Smouse, 2006). The genomic sequence data were used to obtain a pairwise genetic distance matrix, which was standardized and used for PCoA.
RESULTS
SNP and indel discovery and analysis of polymorphic loci diversity
SNPs
SNPs were not encountered in any of the 27 genetic sequences of haploid clementine. This confirms that there were no duplicated genes in our sample of 27 genes. A total of 16 238 bp were sequenced for each of the accessions analysed, from which 10 427 bp were coding regions and 5811 bp were non-coding sequences (Table 2). A total of 1097 SNPs were found in the true citrus fruit trees samples. Another 262 SNPs were found in the outgroup, Severinia buxifolia. True citrus fruit trees had an average of 52·89 SNPs kb−1 for coding regions and 98·39 SNPs kb−1 for non-coding regions. Considering only Citrus, 28·96 SNPs kb−1 were found in coding regions and 51·45 SNPs kb−1 were found in non-coding regions. In the true citrus fruit trees, most of the SNP loci were biallelic, but 21 (1·86 %) revealed three alleles. Among the polymorphisms described, 59·18 % were transitions (A/G ≈ C/T) and 40·82 % were transversions (A/C ≈ A/T > G/T > C/G). For the true citrus fruit trees, but excluding secondary Citrus spp., the average polymorphism rate was 51·76 SNPs kb−1 for coding regions and 95·43 SNPs kb−1 for non-coding regions, with a total of 1066 SNP loci. Among the basic Citrus taxa, Papeda had 252 polymorphic loci (12·18 SNPs kb−1 in coding and 21·51 SNPs kb−1 in non-coding regions), followed by C. reticulata (236 loci, 15·15 SNPs kb−1 in coding and 13·94 SNPs kb−1 in non-coding regions), C. maxima (107, 4·70 SNPs kb−1 in coding and 9·98 SNPs kb−1 in non-coding regions) and C. medica (70, 2·21 SNPs kb−1 in coding and 8·09 SNPs kb−1 in non-coding regions). Large differences in the number of polymorphic loci were observed among close relatives, including Fortunella (227), Microcitrus (171), Eremocitrus (93) and Poncirus (53). Among the secondary species and hybrids, C. aurantium had 211 polymorphic sites, C. limon had 173, C. sinensis had 162, C. aurantifolia had 158, C. paradisi had 115 and clementine had 119. Among the 31 alleles found exclusively in the secondary species (not present in any other true Citrus spp.), 15 were heterozygous in C. aurantium. Four of these alleles (found in the genes INVA, LCY2, DXS and AOC) were shared with C. limon.
Table 2.
Polymorphisms of nucleotide sequences of genes for all samples analysed
| Gene | CS | TS | GS | SC | SNC | SNPc | Freq. | SNPnc | Freq. | πnonsyn/πsyn | indelc | Freq. | indelnc | Freq. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHI | 652 | 721 | 721 | 206 | 446 | 11 | 53·40 | 68 | 152·47 | 1·38 | 0 | 0 | 8 | 17·94 |
| CHS | 565 | 659 | 659 | 574 | 0 | 20 | 35·40 | – | – | 0·06 | 0 | 0 | – | – |
| FLS | 473 | 763 | 763 | 419 | 54 | 41 | 97·85 | 6 | 111·11 | 0·12 | 0 | 0 | 3 | 55·56 |
| F3'H | 783 | 1000 | 1400 | 569 | 214 | 40 | 70·30 | 20 | 93·46 | 0·55 | 0 | 0 | 3 | 14·02 |
| DFR | 421 | 1017 | 1650 | 171 | 250 | 7 | 40·94 | 26 | 104·00 | 0·25 | 0 | 0 | 3 | 12·00 |
| EMA | 428 | 166 | 450 | 131 | 297 | 7 | 53·44 | 27 | 90·91 | 2·27 | 1 | 7·63 | 4 | 13·47 |
| MDH | 712 | 1209 | 1250 | 712 | 0 | 28 | 39·33 | – | – | 1·06 | 0 | 0 | – | – |
| ACO | 695 | 1196 | 2000 | 250 | 445 | 5 | 20·00 | 39 | 87·64 | 0·02 | 0 | 0 | 2 | 4·49 |
| TRPA | 795 | 987 | 1300 | 657 | 138 | 40 | 60·88 | 15 | 108·70 | 0·43 | 0 | 0 | 1 | 7·25 |
| INVA | 908 | 679 | 1100 | 515 | 393 | 36 | 69·90 | 38 | 96·69 | 0·23 | 0 | 0 | 1 | 2·54 |
| PEPC | 694 | 1201 | 2000 | 61 | 633 | 2 | 32·79 | 51 | 80·57 | 0·00 | 0 | 0 | 4 | 6·32 |
| PKF | 775 | 807 | 1650 | 406 | 369 | 16 | 39·41 | 31 | 84·01 | 0·88 | 0 | 0 | 3 | 8·13 |
| DXS | 722 | 935 | 1500 | 327 | 395 | 13 | 39·76 | 37 | 93·67 | 0·29 | 0 | 0 | 3 | 7·59 |
| PSY | 606 | 727 | 2100 | 97 | 509 | 5 | 51·55 | 40 | 78·59 | 0·39 | 0 | 0 | 2 | 3·93 |
| HYB | 680 | 787 | 1600 | 379 | 301 | 19 | 50·13 | 27 | 89·70 | 0·91 | 1 | 2·638 | 2 | 6·64 |
| LCY2 | 738 | 850 | 850 | 738 | 0 | 65 | 88·08 | – | – | 0·27 | 5 | 6·77 | – | – |
| LCYB | 941 | 1206 | 1500 | 941 | 0 | 37 | 39·32 | – | – | 0·13 | 0 | 0 | – | – |
| NCED3 | 560 | 650 | 650 | 560 | 0 | 22 | 39·29 | – | – | 0·39 | 0 | 0 | – | – |
| AOC | 675 | 801 | 800 | 675 | 0 | 37 | 54·81 | – | – | 0·12 | 0 | 0 | – | – |
| MRP4 | 774 | 782 | 900 | 363 | 411 | 14 | 38·57 | 24 | 58·39 | 0·29 | 0 | 0 | 1 | 2·43 |
| CCC1 | 762 | 805 | 850 | 762 | 0 | 33 | 43·31 | – | – | 0·06 | 0 | 0 | – | – |
| HKT1 | 238 | 1003 | 1200 | 116 | 122 | 10 | 86·21 | 9 | 73·77 | 0·17 | 0 | 0 | 1 | 8·20 |
| LAPX | 282 | 321 | 400 | 145 | 137 | 11 | 75·86 | 8 | 58·39 | 0·19 | 0 | 0 | – | – |
| NADK2 | 339 | 787 | 1200 | 65 | 274 | 3 | 46·15 | 25 | 91·24 | 2·12 | 0 | 0 | 1 | 3·65 |
| PIP1 | 190 | 346 | 500 | 103 | 87 | 5 | 48·54 | 21 | 241·38 | 0·01 | 0 | 0 | 0 | 0·00 |
| SOS1 | 495 | 579 | 1000 | 358 | 137 | 22 | 61·45 | 12 | 87·59 | 0·18 | 0 | 0 | 1 | 7·30 |
| TSC | 335 | 505 | 800 | 136 | 199 | 7 | 51·47 | 17 | 85·43 | 0·58 | 0 | 0 | 0 | 0·00 |
| Total | 16238 | 10427 | 5811 | 556 | 52·89 | 541 | 98·39 | 7 | 0·66 | 43 | 7·58 |
CS, cleaned sequence (bp); TS, theoretical size of EST (bp); GS, genomic size (bp); SC, sequence coding region (bp); SNC, sequence non-coding region (bp); SNPc, SNPs in the coding region; Freq, SNP frequency per kb; SNPnc, SNPs in the non-coding region; πnonsyn/πsyn, average non-synonymous/synonymous substitution rate; indelc, indels in coding region; indelnc, indels in non-coding region. See Table 1 for gene abbreviations.
The average rate of heterozygosity observed in the eight ancestral taxa was low (Ho = 0·051), and 27·79 % of the SNPs detected were homozygous in all individuals (Ho = 0). The most heterozygous site was at locus F3′H (SNP51), with an Ho = 0·39.
We estimated the average rates of inter-accession polymorphism (SNPs kb−1) within and between the ancestral taxa (Table 3). Considering only Citrus spp., the average rates of intra- and inter-taxon polymorphisms were 1·76 and 11·31 SNPs kb−1, respectively. Intra-taxon SNP rates varied from 0·65 for C. maxima to 3·37 for Papeda (C. hystrix, C. inchangensis, C. micrantha). Interspecific rates in Citrus varied from 8·56 between C. reticulata and Papeda to 14·43 between C. medica and Papeda. The SNP rate between C. reticulata and C. maxima, the two species believed to have given rise to C. sinensis, C. aurantium, C. paradisi and clementine, was 10·16 SNPs kb−1. Comparing genera, the lowest density of SNPs was found in Poncirus trifoliata (0·55 SNPs kb−1), but the highest level of inter-species differentiation was found between the latter and C. medica (18·18 SNPs kb−1).
Table 3.
Inter-accession polymorphism levels within and between taxa, and frequency of SNPs found in only a single taxon
| SNP per kb | C. reticulata | C. maxima | C. medica | Papeda | Fortunella | Microcitrus | Eremocitrus | Poncirus |
|---|---|---|---|---|---|---|---|---|
| C. reticulata | 1·54 | |||||||
| C. maxima | 10·16 | 0·65 | ||||||
| C. medica | 13·92 | 11·13 | 1·50 | |||||
| Papeda | 8·56 | 9·66 | 14·43 | 3·37 | ||||
| Fortunella | 8·70 | 7·95 | 12·27 | 5·71 | 6·04 | |||
| Microcitrus | 9·99 | 10·09 | 13·77 | 9·74 | 8·74 | 2·41 | ||
| Eremocitrus | 9·62 | 9·96 | 13·17 | 10·24 | 8·82 | 2·85 | – | |
| Poncirus | 13·37 | 13·17 | 18·18 | 13·85 | 13·00 | 14·90 | 14·98 | 0·55 |
| Specific SNPs | 6·77 | 4·25 | 6·28 | 6·47 | 6·65 | 4·93 | 3·33 | 6·84 |
Diagonal: average dissimilarities between two accessions within taxa (SNP per kb). Intersection: average dissimilarities between two accessions between taxa (SNP per kb). Last lane: frequency of SNPs found only in one taxon (SNP per kb).
The average number of SNPs per kb that were specific to one taxon (observed at least in one genotype of the considered taxon but not in other taxa) was similar for C. reticulata, C. medica, Papeda, Fortunella and Poncirus, with an average of 6·6, but lower rates were observed for Microcitrus (4·93), C. maxima (4·25) and Eremocitrus (3·3). No polymorphisms were observed between accessions of the same secondary species when two cultivars per species were studied (clementine, C. sinensis, C. aurantium).
Indels
Fifty indel polymorphisms were found. The average indel frequency in coding regions was 0·66 per kb, and the non-coding regions contained an average of 7·58 per kb. The most frequent indel was a mononucleotide (20 out of 50), but di-, tri-, tetra- and hexa-nucleotides were also abundant (20 out of 50 in total). Larger indels were less common. The largest indel, 56 bp long, was found in the PKF gene.
Comparison of diversity revealed at the intra- and inter-taxa level by SNPs, indels and SSRs
We compared the diversity structures revealed by the identification of SNPs, indel markers defined from mined indel polymorphisms and 50 SSR markers [previously used by Garcia-Lor et al. (2012b) to describe genetic structure in Citrus]. Among the 50 indel sites identified, 25 were selected to develop indel markers. Twelve indel markers were published by Garcia-Lor et al. (2012b), and the primers for the 13 remaining markers can be found in Supplementary Data Table S2.
Averaged data for all of the SNP, indel and SSR loci analysed in this study are presented in Table 4. The lowest average number of alleles (n) and the observed (Ho) and expected heterozygosity (He) in the combined eight taxa were revealed in the SNP markers (n = 2·008, Ho = 0·045, He = 0·173). SSR markers had the highest values (n = 11·080, Ho = 0·486, He = 0·822) and indel markers displayed intermediate values (n = 3·308, Ho = 0·125, He = 0·317). At the interspecific level in Citrus, an increasing order of He values was observed for C. medica, C. maxima and C. reticulata in all marker types (SNP, indel, SSR). However, the relative values were variable. For example, the ratios between C. maxima and C. reticulata were 0·54 or 0·92 for SNPs and SSRs, respectively.
Table 4.
Statistical summary of the diversity of SNP, indel and SSR markers
| SNP |
Indel |
SSR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| He | Ho | Fw | N | He | Ho | Fw | N | He | Ho | Fw | N | |
| C. reticulata | 0·067 | 0·061 | 0·091 | 1·212 | 0·225 | 0·245 | –0·093 | 1·615 | 0·586 | 0·569 | 0·029 | 3·680 |
| C. maxima | 0·036 | 0·034 | 0·050 | 1·097 | 0·083 | 0·096 | –0·155 | 1·231 | 0·540 | 0·549 | –0·016 | 2·900 |
| C. medica | 0·022 | 0·006 | 0·737 | 1·059 | 0·027 | 0·031 | –0·124 | 1·077 | 0·268 | 0·179 | 0·331 | 1·860 |
| Papeda | 0·088 | 0·048 | 0·450 | 1·223 | 0·113 | 0·051 | 0·545 | 1·308 | 0·775 | 0·480 | 0·380 | 3·520 |
| Fortunella | 0·075 | 0·065 | 0·140 | 1·207 | 0·260 | 0·231 | 0·112 | 1·923 | 0·616 | 0·575 | 0·067 | 3·674 |
| Microcitrus | 0·082 | 0·069 | 0·163 | 1·150 | 0·077 | 0·077 | 0·000 | 1·077 | 0·713 | 0·610 | 0·145 | 2·700 |
| Eremocitrus | 0·085 | 0·085 | 0·000 | 1·085 | 0·000 | 0·000 | 0·000 | 1·000 | 0·563 | 0·563 | 0·000 | 1·563 |
| Poncirus | 0·024 | 0·034 | –0·416 | 1·049 | 0·046 | 0·077 | –0·665 | 1·077 | 0·309 | 0·440 | –0·423 | 1·660 |
| Total AT | 0·173 | 0·045 | 0·741 | 2·008 | 0·317 | 0·125 | 0·605 | 3·308 | 0·822 | 0·486 | 0·409 | 11·080 |
| Whole dataset | 0·166 | 0·072 | 0·568 | 2·036 | 0·317 | 0·172 | 0·457 | 4·154 | 0·814 | 0·554 | 0·320 | 11·560 |
Mean values are represented in the table. He, unbiased expected heterozygosity; Ho, observed heterozygosity; Fw, Wright fixation index; N, allele number; AT, ancestral taxa.
Average Fw values (excluding secondary species) for the three types of markers showed that there was a large deficit of heterozygous individuals observed in the population (Fw,SNP = 0·741, Fw,indel = 0·605, Fw,SSR = 0·409), suggesting a high level of differentiation between the taxa. Fst values of the differentiation between taxa (excluding secondary species) (Fst,SNP = 0·644; Fst,indel = 0·596; Fst,SSR = 0·392) were similar to Fw values, indicating that the taxon subdivision represents most of the genetic stratification. SNPs and indels revealed a higher inter-taxon structure than SSRs. At the intraspecific level, the only taxon that showed a consistently higher level of heterozygosity than was expected for all three marker types was Poncirus trifoliata.
Statistical test of neutrality and haplotype structure in the true citrus fruit trees excluding secondary cultivated Citrus spp. and hybrid cultivars
The nucleotide variation observed for the gene sequences analysed is summarized for each taxon in Table 5, and the data presented for each gene are provided in Supplementary Data Table S3. Average total nucleotide diversity (πT) was 0·012 for the entire sample set, ranging from 0·003 for citron to 0·009 for the Papeda group. Nucleotide diversity in silent and synonymous substitution sites was similar between the taxa and for the entire population, but non-synonymous nucleotide diversity was 3·52 times lower than the synonymous one (average πnonsyn = 0·006). The non-synonymous substitution rate varied from 0·000 (PEPC, ACO and PIP1) to 0·010 (CHI, PSY, NADK), and the ratio of non-synonymous to synonymous diversity ranged from 0·000 at PEPC (high conservative selection) to 2·273 at the EMA locus, which suggests that selective constraints and/or the history of adaptive evolution vary between genes. The average non-synonymous/silent substitution rate was 0·345 for all of the genes and the entire population, indicating purifying selection. Within taxa, only the C. reticulata group at the HYB locus (πnonsyn/πsyn = 1·421) and the F3′H locus (πnonsyn/πsyn = 1·767) displayed higher non-synonymous than synonymous diversity. There were some groups with null synonymous mutations in the exons, so the πnonsyn/πsyn ratio was not possible to calculate. In the entire sample set, several loci displayed a non-synonymous/synonymous ratio >1, including CHI (πnonsyn/πsyn = 1·377), EMA (πnonsyn/πsyn = 2·273) and NADK2 (πnonsyn/πsyn = 2·117). Taking into account only the basic taxa (excluding secondary species and recent hybrids), four loci showed values >1, including CHI (πnonsyn/πsyn = 1·381), EMA (πnonsyn/πsyn = 1·511), PSY (πnonsyn/πsyn = 3·533) and NADK2 (πnonsyn/πsyn = 2·043). The PKF locus had a πnonsyn/πsyn value of 0·883 for the entire population and 1·072 for the ancestral taxa group. For the entire population MDH and HYB loci had a πnonsyn/πsyn value of 1·065 and 0·914, respectively.
Table 5.
Summary of nucleotide diversity and divergence within and between species.
| Taxa | S | πT | πsil | πsyn | πnonsyn | πnonsyn/πsyn | Nh | Hd | Hd (s.d.) |
|---|---|---|---|---|---|---|---|---|---|
| C. reticulata | 8·926 | 0·005 | 0·008 | 0·010 | 0·003 | 0·411 | 4·407 | 0·593 | 0·096 |
| C. maxima | 3·926 | 0·004 | 0·005 | 0·004 | 0·001 | 0·191 | 3·222 | 0·521 | 0·116 |
| C. medica | 2·815 | 0·003 | 0·004 | 0·004 | 0·001 | 0·256 | 2·037 | 0·296 | 0·068 |
| Fortunella | 8·481 | 0·006 | 0·009 | 0·008 | 0·003 | 0·285 | 5·185 | 0·683 | 0·097 |
| Papeda | 9·630 | 0·009 | 0·015 | 0·014 | 0·003 | 0·292 | 4·519 | 0·871 | 0·126 |
| Microcitrus | 5·889 | 0·006 | 0·009 | 0·011 | 0·003 | 0·184 | 2·926 | 0·760 | 0·198 |
| Eremocitrus | 3·407 | 0·006 | 0·009 | 0·013 | 0·004 | 0·154 | 1·778 | 0·772 | 0·380 |
| Poncirus | 2·407 | 0·003 | 0·005 | 0·003 | 0·000 | 0·088 | 2·148 | 0·469 | 0·099 |
| Main taxa | 39·667 | 0·013 | 0·021 | 0·020 | 0·006 | 0·555 | 23·074 | 0·926 | 0·016 |
| Whole population | 40·926 | 0·012 | 0·021 | 0·020 | 0·005 | 0·495 | 28·333 | 0·901 | 0·015 |
| Max | 9·630 | 0·009 | 0·015 | 0·014 | 0·004 | 0·411 | 5·185 | 0·871 | 0·380 |
| Min | 2·407 | 0·003 | 0·004 | 0·003 | 0·000 | 0·088 | 1·778 | 0·296 | 0·068 |
S, segregating sites; πT, nucleotide diversity total; πsil, nucleotide diversity silent sites; πsyn, nucleotide diversity synonymous sites; πnonsyn/syn, ratio nucleotide diversity non-synonymous/synonymous sites; Nh, number of haplotypes; Hd, haplotype diversity. Max and min: maximum and minimum values within the basic taxa.
The level of differentiation between the taxa (evaluated by Fst; Supplementary Data Table S3) was relatively homogeneous among the genes. Highest and lowest values were found for SOS1 (Fst = 0·814) and PIP1 (Fst = 0·438), respectively, with an average of 0·644 ± 0·036.
No significant Tajima's D value was found in any of the genes in the entire population (Supplementary Data Table S3).
The average number of haplotypes per locus in the entire population was 28·33, with a maximum value of 5·185 haplotypes in Fortunella and a minimum value of 1·778 in Eremocitrus. Regarding the four main ancestors in Citrus, Papeda had the highest number of haplotypes (4·519), followed by C. reticulata(4·407), C. maxima (3·222) and C. medica (2·037). At the intra-taxon level, haplotype diversity ranged from 0·871 for Papeda to 0·296 for C. medica (Table 5).
Phylogenetic analysis
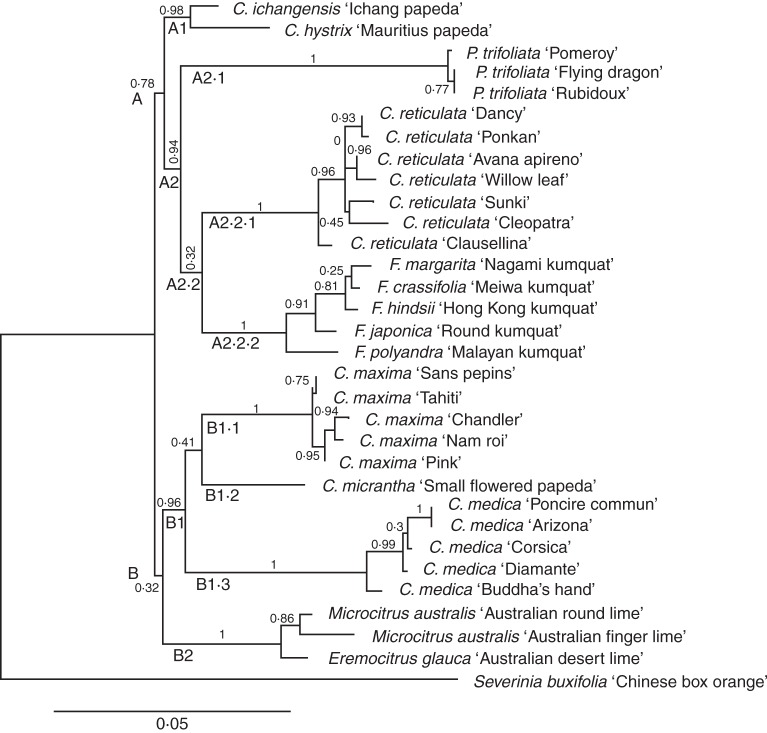
Among all of the models tested via the Phylemon website, the model with the best fit was TVM + I + G + F (with SH-like branch supports alone). This model takes into account the nucleotide substitution model TVM ‘transitional model’ (five substitution classes: AC, AT, CG, GT, AG = CT), the proportion of invariable sites (I), the nucleotide frequency (F) and the gamma distribution (G). The phylogenetic relationships between Citrus species and their relatives inferred from the ML method using this model are shown in Fig. 1. Branch support (BS) is given for all branches. The different true citrus fruit tree genotypes were rooted using Severinia buxifolia as outgroup. The first two clades (A and B) are each divided in two subclades. Clade A has a medium BS (0·78), joining a subclade A1 (BS = 0·98) of two Papeda species (C. hystrix and C. ichangensis) and a strong subclade A2 (BS = 0·94) including all Poncirus trifoliata (monospecific subclade A2·1, BS = 1), all C. reticulata accessions (monospecific subclade A2·2·1, BS = 1) and all Fortunella accessions (monogeneric subclade A2·2·2, BS = 1). Fortunella and C. reticulata are joined in a subclade A2·2 with a low BS (0·32). In the other part of the tree, clade B (low BS = 0·32) includes two groups. The first group, B1 (BS = 0·96), is divided into three highly supported specific subclades, C. maxima accessions (B1·1; BS = 1), C. micrantha (B1·2; only one accession) and C. medica (B1·3; BS = 1) accessions. The second subclade, B2 (BS = 1), includes Microcitrus and Eremocitrus, two strongly associated genera of Australian origin. Papeda is the only group that does not display a monophyletic structure; the accessions of each of the other groups (Poncirus, C. reticulata, Fortunella, C. maxima and C. medica, Microcitrus and Eremocitrus) are all joined in specific clades clearly differentiated from the other taxa.
Fig. 1.
Phylogenetic relationship between Citrus ancestral taxa (C. reticulata, C. maxima, C. medica, Papeda) and relatives (Fortunella, Microcitrus, Eremocitrus, Poncirus trifoliata). Phyml Best AIC Tree (v. 1·02b), model TVM + I + G + F (with SH-like branch supports alone).
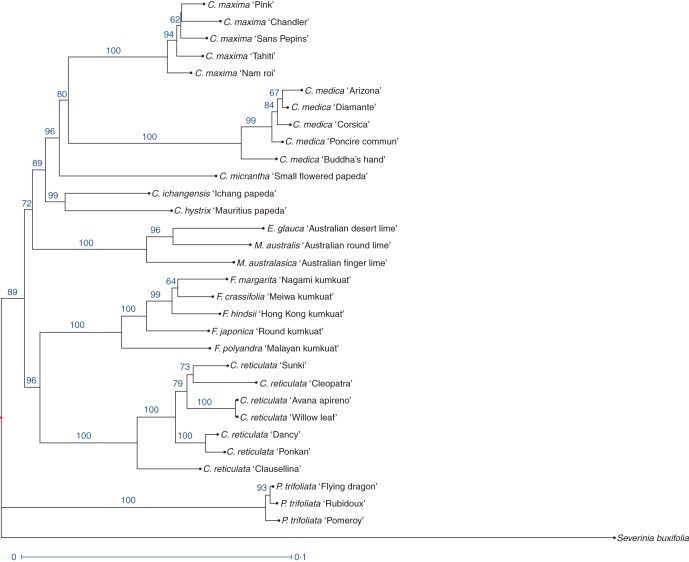
This phylogenetic structure is similar, for several strong groupings, to the structure observed using NJ analysis based on SNP data (Fig. 2). In the NJ tree, the association between C. reticulata and Fortunella (BS = 0·96) is maintained, as are the C. maxima/C. medica (BS = 0·8) and Microcitrus/Eremocitrus (BS = 1) associations. The Papeda group is shifted from one group to the other. Poncirus trifoliata appears as the most distant species, i.e. it is the first one that separates from the others. This in agreement with the high level of differentiation of Poncirus with all other taxa (Table 3).
Fig. 2.
NJ tree with 1097 SNP markers in the ancestral Citrus species and relatives (1000 bootstraps performed). Branch support values of over 50 % are shown.
When the secondary species and interspecific hybrids were added to the analysis (Supplementary Data Fig. S1), the NJ representation was modified and the relationships described before are not maintained. Citrus reticulata appears to be more closely related to C. maxima than to Fortunella, and C. medica is not as closely related to C. maxima, as was suggested by the Phylemon and Darwin analysis that excluded the hybrid genotypes.
Genome structure of citrus secondary species and hybrids
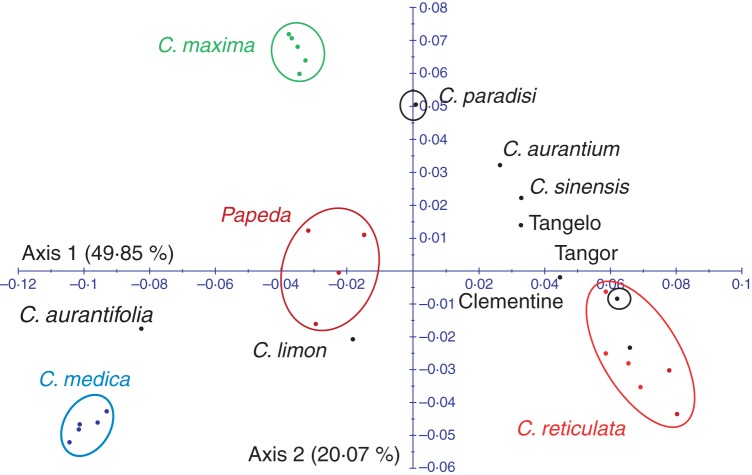
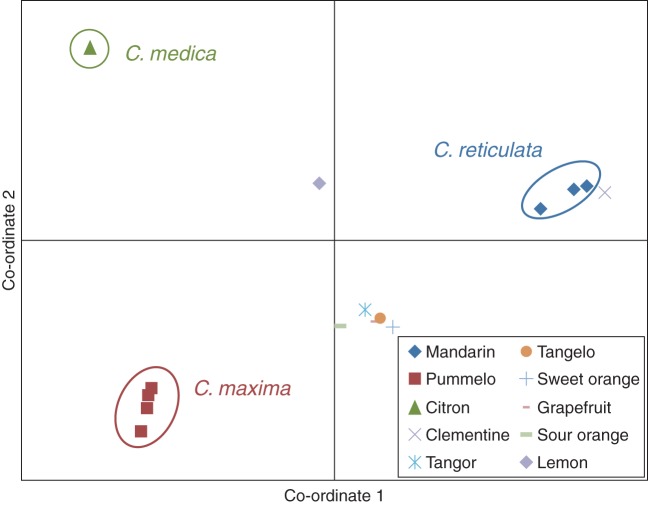
We used factorial analysis to examine the potential contribution of the ancestral species to the inheritance of 27 genes in secondary cultivated species (Table 6). For the SNPs of these 27 genes, almost 70 % of the diversity in Citrus spp. is explained by the first two axes (Fig. 3). The basic Citrus taxa are clearly distinguished. Secondary species are positioned between their putative parental gene pools: C. sinensis between C. maxima and C. reticulata, C. paradisi between C. sinensis and C. maxima, C. limon between C. aurantium and C. medica, and C. aurantifolia between C. medica and C. micrantha (Fig. 3). With the goal of performing a gene-by-gene analysis of the phylogenetic inheritance in the secondary species, we performed a PCoA for each gene using the basic taxa of cultivated citrus as active individuals, and we projected the secondary species genotypes onto the defined axes. The phylogenetic inheritance was inferred from the position of the secondary species in the PCoA relative to the ancestral species and the analysis of SNP allelic locus configurations. The genetic structure of the FLS locus (Fig. 4) is presented as an example of phylogenetic assignation. Grapefruit, sweet orange, sour orange, tangor ‘King’ and tangelo ‘Orlando’ are in an intermediate position between the C. reticulata (mandarin; M) and C. maxima (pummelo; P) groups. It was therefore assumed that these species should have inherited one allele of this gene from each of these ancestral groups (interspecific heterozygosity MP). This was confirmed by examining the allelic configuration at each SNP locus. Using the same approach, lemon appears to be heterozygous (MC) for the C. reticulata and C. medica (citron; C) alleles, whereas clementine appears to have inherited two C. reticulata alleles (MM).
Table 6.
Phylogenetic origins of genes of secondary species and hybrids
| Gene | Clementine |
C. sinensis | C. paradisi | C. aurantium | C. aurantifolia | C. limon | Tangor ‘King’ | Tangelo ‘Orlando’ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHI | M | P | M | P | P | P | M | P | C | C | C | M/P? | M | M | M | P |
| CHS | M | M | M | M | M | P | M | P | C | PAP | C | M | M | M | M | M |
| FLS | M | M | M | P | M | P | M | P | C | PAP | C | M | M | P | M | P |
| F3'H | M | M | M | M | P | P | M | P | C | PAP | C | M | M | M | M | M |
| DFR | M | P | M | P | P | P | M | P | C | PAP | C | M | M | M | M | P |
| EMA | M | M | M | M | M | P | M | P | C | PAP | C | M | M | M | M | M |
| MDH | M | M | M | P | P | P | M | P | C | PAP | C | P | M | P | M | P |
| ACO | M | M | M | M | M | P | M | P | ? | ? | C | M | M | M | M | M |
| TRPA | M | P | P | P | P | P | M | P | C | PAP | C | M | M | P | M | P |
| INVA | M | M | M | P | P | P | M | P | C | PAP | C | P | M | P | M | P |
| PEPC | M | M | M | P | P | P | M | P | C | PAP | C | P | M | M | M | P |
| PKF | M | P | M | P | P | P | ? | P | C | PAP | C | M | M | P | M | P |
| DXS | M | M | M | P | M | P | M | P | C | PAP | C | M | M | P | M | P |
| PSY | M | M | M | P | M | P | M | P | C | PAP | C | M | M | M | M | M |
| HYB | M | M | M | M | M | P | M | P | ? | ? | C | M | M | M | M | M |
| LCY2 | M | P | M | P | M | P | M | P | C | PAP | C | M | M | P | M | M |
| LCYB | M | P | M | P | M | P | M | P | C | PAP | C | M | M | M | M | M |
| NCED3 | M | P | P | P | P | P | M | P | C | PAP | C | P | M | P | M | P |
| AOC | M | M | M | M | M | P | M | P | C | PAP | C | M | M | M | M | M |
| MRP4 | M | M | M | M | M | P | M | P | C | PAP | C | M | M | M | M | M |
| CCC1 | M | P | P | P | M | P | M | P | C | PAP | C | M | M | P | P | P |
| HKT1 | M | P | M | P | M | P | M | P | C | PAP | C | M | M | P | M | M |
| LAPX | M | M | M | P | M | P | M | P | C | PAP | C | M | ? | P | M | P |
| NADK2 | M | M | M | P | P | P | M | P | C | PAP | C | M | M | M | P | P |
| PIP1 | M | M | M | M | M | P | ? | P | ? | ? | C | P | M | M | M | M |
| SOS1 | M | M | M | P | P | P | M | P | C | PAP | C | P | M | P | M | P |
| TSC | M | M | M | P | M | P | M | P | C | PAP | C | M | M | M | M | P |
M, mandarin; P, pummelo; C, citron; PAP, Papeda; ?, origin not known. See Table 1 for gene abbreviations.
Fig. 3.
Genetic relationship between secondary Citrus species and basic taxa (factorial analysis; axes 1/2).
Fig. 4.
Genetic organizational analysis (principal co-ordinates) of secondary species and recent hybrids (flavonoid synthase gene).
For most genes (18/27) clementines appear to have inherited C. reticulata alleles in phylogenetic homozygosity. However, nine genes appear to be heterozygous between C. reticulata and C. maxima. For all the genes analysed, the estimated contribution of C. reticulata was 83·3 %, and the estimated contribution of C. maxima was 16·7 %.
Citrus sinensis appears to contain more alleles from C. reticulata (59·3 %) than from C. maxima (40·7 %). It inherited two alleles from C. maxima (PP) for three genes and two alleles from C. reticulata (MM) for eight genes. The remainder of the genes are phylogenetically heterozygous with alleles from both gene pools (MP).
Citrus paradisi has 11 genes solely inherited from C. maxima, whereas the remaining genes were heterozygously inherited from C. maxima and C. reticulata. The contributions from the parental lines were therefore 70·4 % for C. maxima and 29·6 % for C. reticulata.
Citrus aurantium contains two loci with parental origins that were not possible to define due to the presence of specific alleles at the SNP loci. The other loci were heterozygous for C. maxima/C. reticulata alleles (MP). Therefore, for the loci with complete phylogenetic assignation, the contributions of C. maxima and C. reticulata were each 50 %.
Citrus aurantifolia contains three genes with phylogenetic origins that were not possible to infer. Most of the other genes showed interspecific heterozygosity between C. medica and Papeda. However, CHI appeared to be homozygous for C. medica alleles (CC). Therefore, for the 24 genes that could be analysed, the contributions of C. medica and Papeda were 53 and 47 %, respectively.
Citrus limon showed the most diverse combination of parental contribution patterns. Twenty genes resembled a combination of C. medica and C. reticulata genes, six genes resembled a combination of C. medica and C. maxima genes, and one locus could not clearly be identified. For the genes that could be identified, C. medica contributed 50 %, C. reticulata contributed 38·5 % and C. maxima contributed 11·5 % to the C. limon genome.
‘King’, which is assumed to be a tangor (C. reticulata × C. sinensis), and tangelo ‘Orlando’ (C. paradisi × C. reticulata) contained some genes that exhibited interspecific heterozygosity (C. reticulata and C. maxima; MP) and some that displayed monospecific inheritance (MM or PP). The relative contributions of the C. reticulata and C. maxima gene pools were, respectively, 75·93 and 24·07 % for ‘King’ and 66·67 and 33·33 % for ‘Orlando’.
DISCUSSION
SNP and indel discovery and analysis of the relative utility of these markers compared with SSRs for use in diversity and phylogenetic studies
In true citrus fruit trees, the average number of SNPs per kb in non-coding regions is almost two times higher than in coding regions. This value is high compared with the value obtained for Eucalyptus spp. (1·5 times higher; Külheim et al., 2009). The mean frequency of SNPs kb−1 found in exons was 28·96 for Citrus, which is higher than in other species such as Populus tremula, with 16·7 SNPs kb−1 (Ingvarsson, 2005), and in maize, with 23·25 SNPs kb−1 (Yamasaki et al., 2005). Regarding the SNP frequency in Citrus spp. the values were lower [C. reticulata (15·15 SNPs kb−1), C. maxima (4·70 SNPs kb−1), C. medica (2·21 SNPs kb−1)]. Moreover, the value is lower than that found in Quercus crispula, with 40 SNPs kb−1 (Quang et al., 2008) and Eucalyptus camaldulensis, with 47·62 SNPs kb−1 (Külheim et al., 2009). The percentages of transition and transversion events are similar to those found in other species, such as oil palm (0·58 and 0·42, respectively; Riju et al., 2007). In Citrus, these results are in agreement with results reported by Dong et al. (2010), Terol et al. (2008) and Novelli et al. (2004). In contrast, the transition fraction was found to be substantially higher in poplar (70 %; Tuskan et al., 2006).
The nucleotide diversity value observed in the true citrus fruit trees and in C. reticulata (π = 0·005) was similar to the values observed in grapevine (π = 0·005; Lijavetzky et al., 2007), maize (π = 0·006; Ching et al., 2002) and rye (π = 0·006; Li et al., 2011), but the value was approximately five times higher than those observed in soybean (π = 0·00097; Zhu et al., 2003) and human (π = 0·001; Sachidanandam et al., 2001). Compared with the diversity data in Citrus obtained with SNPs mined in clementine (Ollitrault et al., 2012a), it appears that the relative diversity levels of the three basic taxa were quite different. Indeed, the Nei diversity values (He) of C. maxima and C. medica over C. reticulata were 0·23 (0·063/0·279) and 0·20 (0·057/0·279), respectively, whereas the values obtained in the present study were 0·53 (0·036/0·067) and 0·33 (0·022/0·067), respectively, confirming the conclusion of Ollitrault et al. (2012a) that the ascertainment bias due to the scarcity and specificity of the discovery panel of the SNPs mined in clementine resulted in an over-estimation of the relative diversity within C. reticulata. Analysis of the average inter-accession polymorphism within and between species reveals that for the three basic taxa of cultivated Citrus (C. reticulata, C. maxima, C. medica), the ratios between and within species were high. For example, within C. reticulata and between C. reticulata and C. maxima, the ratio was close to 6·6 (10·16/1·54). Therefore, the analysis of SNP density along the genome should help differentiate between genomic regions with interspecific heterozygosity (MP for example) and those that result from intraspecific inheritance (MM or PP, for example) in the genomes of secondary species.
The information obtained by studying the allelic diversity of the analysed genes will allow us to optimize molecular tools for both genomic and transcriptomic studies. The identification of conserved areas can be used to develop primers or hybridization sequences to limit sources of bias such as null alleles or differential allelic PCR competition or hybridization. Identification of the different alleles of these genes also opens the way for allele-specific expression studies.
The frequencies of indels per kb in the true citrus fruit trees species were 0·66 and 7·58 in exon and intron sequences, respectively. These frequencies are comparable to values reported for other species such as maize (18 genes studied, 6935 bp), where 0·43 and 11·76 indels kb−1 were found in coding and non-coding regions, respectively (Ching et al., 2002), and Brassica (557 clone sequences, 1396 498 bp), with 0·45 and 7·42 indels kb−1 in coding and non-coding regions, respectively (Park et al., 2010). In melon (34 ESTs sequenced, ±15 000 bp), indels occurred less frequently in introns (approx. 0·60 kb−1), and no indels were found inside coding regions (Morales et al., 2004). In grapevine (230 gene fragments sequenced, >1 Mb), low levels of indel polymorphism were found, with 0·11 and 2·25 indel kb−1 in coding and non-coding regions, respectively (Lijavetzky et al., 2007).
Considering the eight basic taxa together, the fixation index (Fw) values and the differentiation index values (Fst) between taxa obtained using three types of markers (SSRs, SNPs, indels) confirmed the high degree of stratification in differentiated taxa with limited gene flows. However, the levels of diversity revealed by the three types of markers were quite different. The indel markers developed in this study confirmed that indels are efficient tools for interspecific differentiation, as demonstrated by Garcia-Lor et al. (2012a) and Ollitrault et al. (2012b). The indel markers developed in this study had an average Fst value of 0·596, similar to that obtained using SNP markers (Fst = 0·644), whereas with 50 SSR markers analysed for the same accessions, the Fst value was only 0·392. In contrast, the SNP loci and indels mined from our much diversified interspecific panel appeared, on average, to be less polymorphic to describe intraspecific polymorphism. However, in our study, which includes several genotypes for each species, we also identified numerous SNP loci that revealed intraspecific diversity that should be useful for germplasm characterization and management. Unlike SSRs and indel sequences, SNPs can be employed in high-throughput screening and in relatively low-cost genotyping methods. Their utility is limited, however, due to the fact that they are usually present only as diallelic polymorphisms.
Evolution of citrus genes
In true citrus fruit trees, the average ratio of non-synonymous to silent SNP rates per site (πnonsyn/πsil) was 0·345. In Citrus spp. similar values were found in C. reticulata (0·385) and C. medica (0·339), but a higher value in C. maxima (0·577). This is higher than the 0·17 and 0·21 ratios observed in white spruce (Pavy et al., 2006) and in Arabidopsis thaliana (in a study of 242 genes; Zhang et al., 2002), respectively. These relatively low values indicate that, on average, white spruce open reading frames and nuclear genes in A. thaliana are probably under higher purifying selection pressure than the genes of true citrus fruit trees. This can probably be attributed to the wide diversity encompassed by true citrus fruit trees and the high genetic and phenotypic differentiation between the different taxa that have experienced allopatric evolution (even if they are still sexually compatible). The minimum value of πnonsyn/πsil in our entire data set was 0 at the PEPC locus, and the maximum value was 1·09 at the NADK2 locus. The non-synonymous substitution rate varied from 0·000 in PEPC to 0·010 in CHI, which suggests that selective constraints vary between loci (Fu et al., 2010).
In the carotenoid biosynthetic pathway, different key steps have been found to be associated with differentiation between cultivated Citrus spp. (Kato et al., 2004; Fanciullino et al., 2006, 2007). Several studies have tried to clarify the regulation of carotenoid biosynthesis (Rodrigo et al., 2004; Kato et al., 2004; Kim et al., 2001), but this regulation has not yet been fully elucidated.
PSY drives the formation of phytoene, the first product in the carotenoid biosynthetic pathway and a major step in the differentiation between cultivated basic taxa (Fanciullino et al., 2006, 2007). Considering the eight taxa studied, it appears that PSY is under positive selection (πnonsyn/πsyn = 3·533) and is associated with a high level of allelic differentiation between the taxa (Fst = 0·750), which is higher than the average. There were nine sites with SNP polymorphisms between C. reticulata and the other taxa that produced changes in the amino acid composition that may be responsible for their differentiation. In contrast, in C. reticulata, no changes were found (excepted for one heterozygous change in the cultivar ‘Ponkan’). Further functional analysis of the different alleles of this gene should provide insights into the molecular basis of phenotypic differentiation.
LCYB is a key enzyme required for the conversion of lycopene into β-carotenoids (Fanciullino et al., 2006; Alquézar et al., 2009). Fanciullino et al. (2007) proposed that allelic variation at this locus should strongly limit this biosynthetic step in C. maxima. The numerous amino acid changes observed in C. maxima compared with C. reticulata might be associated with this limitation due to changes in the functionality of the pummelo allele.
HYB also plays a major role in the carotenoid biosynthetic pathway (Fanciullino et al., 2006) by catalysing the transformation of β-carotene into β-cryptoxanthin and zeaxanthin. Citrus reticulata produces these compounds, whereas C. maxima does not convert β-carotene into β-cryptoxanthin and zeaxanthin and C. medica only convert β-carotene into β-cryptoxanthin. In C. reticulata, the ratio between non-synonymous/synonymous substitutions was higher than one (positive selection) at the HYB locus, which might be related to the significant variation in β-cryptoxanthin levels found among C. reticulata cultivars (Fanciullino et al., 2006). The β-cryptoxanthin content greatly enhances fruit colour and has probably been under human-induced selection during domestication.
In the flavonoid pathway, positive selection was found to occur in C. reticulata at the F3′H locus, which belongs to the cytochrome P450 family and catalyses the hydroxylation of flavonoids at the 3′ position of the B-ring, leading to the production of hydroxylated flavonols, proanthocyanidins (condensed tannins) and anthocyanins (Winkel-Shirley, 2001). This gene plays an important role in flavonoid biosynthesis in Arabidopsis (Schoenbohm et al., 2000) and grapevine (Bogs et al., 2006) and was previously isolated in clementine by Garcia-Lor et al. (2012b). Schoenbohm et al. (2000) demonstrated that, in yeast, this enzyme could convert naringenin or dihydrokaempferol into eriodictyol or dihydroquercetin, respectively. Therefore, the changes in non-synonymous amino acid composition in the mandarin group (C. reticulata) may be associated with the different flavonol compositions found in some studies (Gattuso et al., 2007). At the CHI locus, a greater number of non-synonymous vs. synonymous substitutions were not found to have occurred in the eight subpopulations studied, but at the interspecific level, the ratio was higher than 1, meaning that the gene was probably subject to positive selection during the interspecific differentiation process. This gene controls the second step of the flavonoid biosynthetic pathway (Winkel-Shirley, 2001), and it was shown that it can alter flavonoid levels in citrus leaves (Koca et al., 2009). Understanding F3′H and CHI regulation and allelic functionality could be important for the analysis of molecular determinants of flavonoid composition in citrus fruits.
In the biosynthesis of acidic compounds, EMA displayed non-synonymous/synonymous ratios greater than 1 (πnonsyn/πsyn = 2·273) and evidenced positive selection at the interspecific level. EMA is involved in the last steps of the citric acid cycle, catalysing the transformation of malate into pyruvate, the precursor of citrate formation (Kay and Weitzman, 1987). Malic enzyme is activated by the accumulation of citric acid cycle intermediates, allowing excess intermediates to leave the cycle and re-enter as acetyl groups, producing more citric acid. Citric acid content is strongly differentiated between Citrus taxa and ranges from 0·005 mol L−1 for oranges and grapefruits to 0·30 mol L−1 for lemons and limes (Penniston et al., 2008).
None of the sugar biosynthesis genes exhibited positive selection. It is well known that the total concentration of sugars increases throughout maturation in all Citrus spp. (Albertini et al., 2006). The null level of non-synonymous divergence at PEPC is consistent with strong selection for conserved amino acid sequences in this gene, which plays a crucial role in such important processes as C4 and crassulacean acid metabolism (CAM) photosynthesis.
In the entire sample set, taking into account only the eight ancestral taxa (excluding secondary species and recent hybrids), NADK2 displayed a non-synonymous/synonymous ratio greater than 1 (πnonsyn/πsyn = 2·117 and 2·043, respectively). NADK (NAD kinase) catalyses the ATP-dependent phosphorylation of NAD(H) (Berrin et al., 2005). In A. thaliana, there are three isoforms of NADK. Two isoforms, NADK1 and NAD(H)K3, are cytosolic and one, NADK2, is found in the plastid (Turner et al., 2004, 2005; Chai et al., 2005, 2006). These isoforms play an essential role in the phosphorylation of NAD(H) and have been linked to plant stress response. Chai et al. (2005) showed that manipulation of AtNADK2 levels affected plastid NADPH levels, and null mutants were stunted, with a pale yellow colour, and were hypersensitive to abiotic stress.
Differences found in the coding regions of NADK2, and thus variations in amino acid sequences between the taxa, might affect the responses of these genotypes to abiotic stresses. Full sequencing of this gene and functional analysis of the different alleles could greatly increase our understanding of the role that this gene plays in increasing stress tolerance in Citrus and its relatives.
For all of the genes discussed here, the sequence data highlight amino acid variability of corresponding proteins that were probably subject to selection. Therefore, these genes are good candidates for further complete sequencing studies (including promoter sequencing) and allelic functional studies to decipher the molecular basis of the phenotypic variability in the species examined.
Despite the previous discussion concerning the possible selective pressure exerted on some of the genes studied, the genetic organization of Citrus obtained from the SNP data (Fig. 1) is similar to the genetic organization elucidated in previous SSR studies (Ollitrault et al., 2010; Garcia-Lor et al., 2012a). This suggests that the same basic type of evolutionary components led to the diversity structures of both types of markers. Therefore, a predominantly neutral selection pattern can be assumed for most of the current SNP markers. The minimum Fst value was 0·438 at the PIP1 locus and the maximum value was 0·814 at the SOS1 locus for the differentiation of the eight taxa analysed in this work, i.e. C. reticulata, C. maxima, C. medica, Papeda, Fortunella, Microcitrus, Eremocitrus and Poncirus trifoliata. This study sheds light on the important differentiation between the taxa and demonstrates that SNP markers are efficient tools for phylogenetic studies and inheritance analysis of secondary species.
Phylogenetic relationships
For a biologically complex crop such as citrus, information obtained from nuclear gene sequences is more useful than the information from maternally inherited plastid sequences (Ramadugu et al., 2011; Puritz et al., 2012) due to the possibility of gene flow between sexually compatible species and the fact that the species belong to the same area of diversification. Previous phylogenetic molecular analyses using plastid markers showed that all true citrus fruit tree species constitute a clade that is differentiated from other genera (de Araújo et al., 2003; Bayer et al., 2009).
In our study, all accessions of the same species form a clade with mainly high branch support values. Two species in the Papeda group, C. hystrix and C. ichangensis, are closely related. The other species of subgenus Papeda, C. micrantha, is separated from the two previous ones, possibly due to its geographical origin and distribution. The origin of C. micrantha is believed to be in the Philippines, whereas C. hystrix and C. ichangensis are of continental origin, in Burma, Thailand and Indo-China (Tanaka, 1954). Therefore, Swingle and Reece's (1967) subdivision of the genus into subgenera Papeda and Citrus seems to be inadequate.
An important observation maintained through the ML phylogenetic trees and the NJ cluster analysis is that C. reticulata and Fortunella form a cluster clearly differentiated from another cluster including C. maxima, C. medica and C. micrantha. The close relationship between C. reticulata and Fortunella matches the results obtained by Penjor et al. (2010) based on the analysis of rbcL plastid gene sequences, but it differs from the results obtained from the analysis of amplified fragment length polymorphism (AFLP) molecular markers (Pang et al., 2007) and SSR markers (Barkley et al., 2006) and Swingle and Reece's (1967) treatment of Fortunella. In the ML phylogenetic analysis, Poncirus trifoliata was found to belong to the same clade as C. reticulata and Fortunella with strong branch support (0·94). However, in the NJ analysis, P. trifoliata appears as the most distant to all the true citrus fruit tree taxa analysed, in agreement with our estimation of the inter-taxon differentiations. The strongly supported clade (B1; BS = 0·96) including C. medica, C. maxima and C. micrantha of subgenus Papeda is also observed in the NJ analysis. However, our results are in contrast to information derived from other studies, including the analysis of nine plastid markers by Bayer et al. (2009), the analysis of SSR, sequence-related amplified polymorphism (SRAP) and cleaved amplified polymorphic sequences (CAPS)-SNP markers (Amar et al., 2011), SSRs (Barkley et al., 2006), and RAPD, SCAR and plastid DNA markers (Nicolosi et al., 2000). All of these studies suggested that C. maxima and C. reticulata share a clade and are separated from C. medica. The inconsistency with previous nuclear studies may be due to the inclusion of secondary species of interspecific origin in these previous studies, which might have led to the artefactual clustering of the C. maxima and C. reticulata gene pools due to the numerous accessions resulting from hybridization between these gene pools. Our phylogenetic ML analysis (Fig. 1) and the NJ analysis done with the SNPs in the absence of secondary species (Fig. 2) are consistent, whereas the NJ tree that includes the secondary species (Supplementary Data Fig. S1) shows clustering of C. maxima and C. reticulata with low branch support. This illustrates the bias associated with the inclusion of genotypes of inter-taxon origin in NJ cluster analyses. Another source of bias in molecular studies might be the choice of molecular marker type and the genotype panel used for its development. In our study, using Sanger sequencing, all SNPs from all accessions are revealed, so there was no bias towards any of the ancestral species.
The consistent clades observed in the ML phylogenetic study are in agreement with the geographical distribution of species divided by the ‘Tanaka line’ (Tanaka, 1954). Fortunella, Poncirus and C. reticulata (clade A2) share the same area of diversification, where subgenus Metacitrus predominates (East Asiatic floral zone) (Tanaka, 1954), whereas the C. medica and C. maxima clade (B1) is in agreement with the area of distribution where the subgenus Archicitrus, described by Tanaka (1954), predominates (Indo-Malayan floral zone). Some phenotypic traits differentiate these two clades. For example, Fortunella, Poncirus and C. reticulata are facultatively apomictic with high carotenoid contents, whereas C. maxima and C. medica are monoembryonic non-apomictic species, which have strong limitations in the carotenoid pathway. The speciation between Fortunella, Poncirus and C. reticulata might be explained by their different flowering periods (precocious in Poncirus and late in Fortunella). However, gene flow probably occurred by accidental, out-of-time flowering. Despite sharing the Indo-Malayan floral zone (Tanaka, 1954), C. maxima and C. medica are geographically separated, with a more intertropical specialization for C. maxima.
Eremocitrus and Microcitrus were found to be associated in all our analyses. This result is consistent with the conclusions of Barrett and Rhodes (1976), based on morphological traits, and also with previous molecular phylogenetic analyses (e.g. Bayer et al., 2009). The phylogenetic placement of these Australian genera among the true citrus fruit trees remains unclear, due to the lack of branch support for the deeper branches in the phylogenetic trees.
Secondary species structure
The origin of secondary species and many recent hybrids formed by interspecific hybridization between the basic Citrus taxa (C. maxima, C. reticulata, C. medica and C. micrantha) has been well documented in several molecular studies (Nicolosi et al., 2000; Barkley et al., 2006; Garcia-Lor et al., 2012a; Ollitrault et al., 2012a), and the relative contribution of the ancestral taxa to their genomes was estimated by Barkley et al. (2006) and Garcia-Lor et al. (2012a). However, these two studies were based on SSRs and these estimations could be biased by the frequent homoplasy observed for these markers (Barkley et al., 2009). The genomes of secondary species can be considered to be mosaics of large DNA fragments of ancestral species that resulted from a few interspecific recombination events (Garcia-Lor et al., 2012a). However, the phylogenetic structures of secondary species in concrete points of the genome remain obscure. For C. sinensis, C. aurantium, C. paradisi and clementine, previous molecular studies (Nicolosi et al., 2000; Barkley et al., 2006; Garcia-Lor et al., 2012a; Ollitrault et al., 2012a) also showed that intra-taxon diversity resulted only from mutation and/or epigenetic variation without further sexual recombination events. Therefore, these species generally present low or null molecular intercultivar diversity for genetic markers such as SSRs or SNPs. Such low molecular diversity was confirmed in this work for secondary taxa for which two cultivars were sequenced (C. sinensis, C. aurantium and clementine). Due to this intra-secondary taxon diversification history, most of the conclusions about the mosaic structure inferred from one or two genotypes should be extended to other cultivars of the same secondary species.
Clementine is believed to have resulted from a cross between mandarin ‘Willow Leaf’ and sweet orange (Nicolosi et al., 2000; Ollitrault et al., 2012a), which means that there were contributions from both the C. reticulata and the C. maxima gene pools (Garcia-Lor et al., 2012a). From the analysis of 27 genes, the observation that there was a majority of mandarin/mandarin phylogenetic homozygosity and very little mandarin/pummelo heterozygosity is in agreement with this hypothesis. The proportion of the pummelo genome estimated from these 27 sequences (16·7 %) is higher than that estimated from SSR markers (7 %) by Garcia-Lor et al. (2012a).
Several hypotheses have been proposed for the origin of C. sinensis. According to Barrett and Rhodes (1976), Torres et al. (1978), Scora (1988), Nicolosi et al. (2000) and Moore (2001), sweet orange should be a direct interspecific hybrid between a pummelo (C. maxima) and a mandarin (C. reticulata), whereas Roose et al. (2009) and Garcia-Lor et al. (2012a) suggested that C. sinensis resulted from a backcross 1 (BC1) [(C. maxima × C. reticulata) × C. reticulata)]. The identification of interspecific phylogenetic heterozygosity MP and phylogenetic homozygosity PP and MM (Table 6) in the C. sinensis genome contradicts these two models. Indeed, the presence of both types of phylogenetic homozygosity (reported for the first time for pummelo homozygosity) implies that both parents of sweet orange were of interspecific origin. The presence of intraspecific heterozygous SNPs for some genes in phylogenetic homozygosity (EMA and HYB; data not shown) also contradicts the hypothesis that C. sinensis resulted from an F2 interspecific hybrid (self-fecundation of an interspecific F1).
Sour orange (C. aurantium) is thought by some authors to be a natural hybrid of a mandarin and a pummelo (Scora, 1975; Barrett and Rhodes, 1976; Nicolosi et al., 2000; Uzun et al., 2009). The interspecific heterozygosity (MP, Table 6) observed for all interpretable loci is in agreement with this hypothesis. However, specific SNP alleles were found in C. aurantium, indicating that the parental pummelo or mandarin was not part of the germplasm analysed and that sweet orange and sour orange were not related as considered by some authors.
Grapefruit (C. paradisi) is thought to have arisen from natural hybridization between C. maxima and C. sinensis in the Caribbean after the discovery of the New World by Columbus (Barrett and Rhodes, 1976; de Moraes et al., 2007; Ollitrault et al., 2012a). The results obtained in this study help to confirm this theory, as many loci were homozygous for the C. maxima genome and other loci showed interspecific heterozygosity (MP, Table 6). Nicolosi et al. (2000) proposed that Mexican lime (C. aurantifolia) is a hybrid between C. medica and C. micrantha. This theory fits with our data for 23 out of 27 genes. For three genes, it was not possible to decipher the mosaic structure and for the gene leading to a CC conclusion it should be supposed that PCR competition resulted in an apparent Papeda null allele (C0). The tri-hybrid origin (C. medica, C. reticulata, C. maxima) accepted for C. limon (Nicolosi et al., 2000; Barkley et al., 2006; Garcia-Lor et al., 2012a) was confirmed by our sequence data for lemon ‘Eureka’, which has contributions from its ancestors (C. medica 50 %, C. reticulata 38·46 % and C. maxima 11·54 %, Table 6) that are similar to those described by Garcia-Lor et al. (2012a). Moreover, the systematic presence of a C. medica allele and the fact that lemon shares heterozygosity with some rare sour orange alleles support the hypothesis proposed by Nicolosi et al. (2000) that lemon resulted from direct hybridization between C. medica and C. aurantium.
Both tangors (C. reticulata × C. sinensis) and tangelos (C. paradisi × C. reticulata) were bred from recombination between the C. reticulata and C. maxima gene pools. The SNP pattern for tangelo ‘Orlando’ originating from a controlled cross between a grapefruit and mandarin ‘Dancy’ (Hodgson, 1967) with both mandarin and pummelo allele inheritance is logical. Our results also confirm that the tangor ‘King’ classified by Tanaka (1977) as C. nobilis is most probably a tangor with at least one mandarin allele for each gene and MP heterozygosity inheritance for some genes.
With the next release of the pseudo-chromosome sequence assembly of the reference haploid clementine genome (Gmitter, 2012), the assignation of the phylogenetic origin of these 27 genes will contribute to deciphering of the interspecific mosaic genome structure of the secondary species. Moreover, this allelic assignation in genotypes of interspecific origin, coupled with further analysis of functionality of the alleles of the different ancestral species, will provide a promising pathway for understanding the molecular basis of phenotypic variability in this highly stratified gene pool in which the organization of phenotypic and molecular diversity is closely linked.
CONCLUSIONS AND PERSPECTIVES
Sanger sequencing of 27 nuclear gene fragments for 45 genotypes resulted in the identification of a large number of molecular polymorphisms (1097 SNPs and 50 indels). For the indels, half of the mined polymorphisms have been used to define new markers. A significant number of the mined SNP loci could be converted into efficient markers to perform high-throughput genotyping studies that will be important for the management of Citrus collections and marker/trait association studies. The nuclear phylogenetic analyses of Citrus and its sexually compatible relatives showed coherence with the geographical distribution and differentiation proposed by Tanaka (1954), with C. reticulata and Fortunella appearing to be closely related. A cluster that joins C. medica, C. maxima and the Papeda species C. micrantha was consistently revealed.
In the near future, by using the entire Citrus genome as a reference and resequencing data from the main secondary species, the resulting estimations of the relative levels of within- and between-taxon differentiation will be useful for deciphering the interspecific mosaic structure of Citrus secondary cultivated species and modern cultivars. The present study has allowed us to assign a phylogenetic inheritance of the genes that were examined for most of the genotypes of interspecific origin under study. One of our major results concerns C. sinensis, which has alleles of three genes that appear to have been inherited solely from the C. maxima gene pool and alleles of eight genes that appear to have been inherited from C. reticulata. This result contradicts the hypothesis that C. sinensis originated directly from F1 or by BC1 hybridization between the C. maxima and C. reticulata gene pools. However, our study confirms previous hypotheses concerning the origins of the other secondary species.
Positive selection was observed for a few genes within or between the species studied, suggesting that these genes may play a key role in phenotypic differentiation. These genes are therefore major candidates for future studies, including complete gene sequencing and functional analysis of different alleles to analyse the molecular basis of the phenotypic variability of corresponding traits.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was jointly financed by the Ministerio de Ciencia e Innovación [AGL2008-00596-MCI] and Generalidad Valenciana [Prometeo 2008/121] projects, by the European Commission, under the CIBEWU project [FP6-2003- INCO-DEV-2–015453] and the French Bureau des Resources Génétiques project [BRG no. 34].
LITERATURE CITED
- Abkenar AA, Isshiki S, Tashiro Y. Phylogenetic relationships in the ‘true citrus fruit trees’ revealed by PCR-RFLP analysis of cpDNA. Scientia Horticulturae. 2004;102:233–242. [Google Scholar]
- Albertini MV, Carcouet E, Pailly O, Gambotti C, Luro F, Berti L. Changes in organic acids and sugars during early stages of development of acidic and acidless citrus fruit. Journal of Agricultural and Food Chemistry. 2006;54:8335–8339. doi: 10.1021/jf061648j. [DOI] [PubMed] [Google Scholar]
- Albrechtsen A, Nielsen FC, Nielsen R. Ascertainment biases in SNP chips affect measures of population divergence. Molecular Biology and Evolution. 2010;27:2534–2547. doi: 10.1093/molbev/msq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquézar B, Zacarias L, Rodrigo MJ. Molecular and functional characterization of a novel chromoplast-specific lycopene beta-cyclase from Citrus and its relation to lycopene accumulation. Journal of Experimental Botany. 2009;60:1783–1797. doi: 10.1093/jxb/erp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleza P, Roldan J, Hernández M, Ollitrault P, Navarro L. Obtention and characterization of Citrus clementina Hort. ex Tan ‘Clemenules’ haploid lines to establish the reference whole Citrus genome sequence. BMC Plant Biology. 2009;9(110) doi: 10.1186/1471-2229-9-110. http:dx.doi.org/10.1186/1471-2229-9-110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar MH, Biswas MK, Zhang Z, Guo W. Exploitation of SSR, SRAP and CAPS-SNP markers for genetic diversity of Citrus germplasm collection. Scientia Horticulturae. 2011;128:220–227. [Google Scholar]
- de Araújo EF, de Queiroz LP, Machado MA. What is Citrus? Taxonomic implications from a study of cp-DNA evolution in the tribe Citreae (Rutaceae subfamily Aurantioideae) Organisms Diversity & Evolution. 2003;3:55–62. [Google Scholar]
- Barkley NA, Roose ML, Krueger RR, Federici CT. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs) Theoretical and Applied Genetics. 2006;112:1519–1531. doi: 10.1007/s00122-006-0255-9. [DOI] [PubMed] [Google Scholar]
- Barkley NA, Krueger RR, Federici CT, Roose ML. What phylogeny and gene genealogy analyses reveal about homoplasy in Citrus microsatellite alleles. Plant Systematics and Evolution. 2009;282:71–86. [Google Scholar]
- Barrett HC, Rhodes AM. A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Systematic Botany. 1976;1:105–136. [Google Scholar]
- Bayer RJ, Mabberley DJ, Morton C, et al. A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. American Journal of Botany. 2009;96:668–685. doi: 10.3732/ajb.0800341. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Goudet J, Chikhi L, Bonhomme F. Laboratoire Génome et Population. GENETIX v. 4·03, Logiciel sous Windows pour la génétique des populations. 2002 Université de Montpellier 2, Montpellier, France. http://www.univ-montp2.fr . [Google Scholar]
- Berrin JG, Pierrugues O, Brutesco C, et al. Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) kinase in Arabidopsis thaliana. Molecular Genetics and Genomics. 2005;273:10–19. doi: 10.1007/s00438-005-1113-1. [DOI] [PubMed] [Google Scholar]
- Bogs J, Ebadi A, McDavid D, Robinson SP. Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiology. 2006;140:279–291. doi: 10.1104/pp.105.073262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nature Genetics. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- Buckler ES, Thornsberry JM. Plant molecular diversity and applications to genomics. Current Opinion in Plant Biololgy. 2002;5:107–111. doi: 10.1016/s1369-5266(02)00238-8. [DOI] [PubMed] [Google Scholar]
- Chai MF, Chen QJ, An R, Chen YM, Chen J, Wang XC. NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Molecular Biology. 2005;59:553–564. doi: 10.1007/s11103-005-6802-y. [DOI] [PubMed] [Google Scholar]
- Chai MF, Wei PC, Chen QJ, et al. NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant Journal. 2006;47:665–674. doi: 10.1111/j.1365-313X.2006.02816.x. [DOI] [PubMed] [Google Scholar]
- Ching A, Caldwell KS, Jung M, et al. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genetics. 2002;3(19) doi: 10.1186/1471-2156-3-19. http://dx.doi.org/10.1186/1471-2156-3-19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Research. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates BS, Sumerford DV, Miller NJ, et al. Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. Journal of Heredity. 2009;100:556–564. doi: 10.1093/jhered/esp028. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. http://dx.doi.org/10.1093/nar/gkn180 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SQ, Zhang XN, Bao ZR, Ling MQ. A new species of Poncirus from China. Acta Botanica Yunnanica. 1984;6:292–293. [Google Scholar]
- Dong J, Qing-liang Y, Fu-sheng W, Li C. The mining of Citrus EST-SNP and its application in cultivar discrimination. Agricultural Sciences in China. 2010;9:179–190. [Google Scholar]
- Fanciullino AL, Dhuique-Mayer C, Luro F, Casanova J, Morillon R, Ollitrault P. Carotenoid diversity in cultivated Citrus is highly influenced by genetic factors. Journal of Agricultural and Food Chemistry. 2006;54:4397–4406. doi: 10.1021/jf0526644. [DOI] [PubMed] [Google Scholar]
- Fanciullino AL, Dhuique-Mayer C, Luro F, Morillon R, Ollitrault P. Carotenoid biosynthetic pathway in the citrus genus: number of copies and phylogenetic diversity of seven genes. Journal of Agricultural and Food Chemistry. 2007;55:7405–7417. doi: 10.1021/jf070711h. [DOI] [PubMed] [Google Scholar]
- Fantz PR. Nomenclature of the Meiwa and Changshou kumquats, intrageneric hybrids of Fortunella. HortScience. 1988;23:249–250. [Google Scholar]
- Federici CT, Fang DQ, Scora RW, Roose ML. Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theoretical and Applied Genetics. 1998;96:812–822. [Google Scholar]
- Fu B, Chen M, Zou M, Long M, He S. The rapid generation of chimerical genes expanding protein diversity in zebrafish. BMC Genomics. 2010;11(657) doi: 10.1186/1471-2164-11-657. http://dx.doi.org/10.1186/1471-2164-11-657 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lor A, Luro F, Navarro L, Ollitrault P. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity; a perspective for genetic association studies. Molecular Genetics and Genomics. 2012;287:77–94. doi: 10.1007/s00438-011-0658-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Lor A, Garcia-Martinez J, Perez-Amador M. Identification of ovule and seed genes from Citrus clementina. Tree Genetics & Genomes. 2012;8:227–235. [Google Scholar]
- Garvin MR, Saitoh K, Gharret AJ. Application of single nucleotide polymorphisms to non-model species: a technical review. Molecular Ecology Resources. 2010;10:915–934. doi: 10.1111/j.1755-0998.2010.02891.x. [DOI] [PubMed] [Google Scholar]
- Gattuso G, Barreca D, Gargiulli C, Leuzzi U, Caristi C. Flavonoid composition of Citrus juices. Molecules. 2007;12:1641–1673. doi: 10.3390/12081641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmitter FG. New Citrus genome sequence resources. Plant and Animal Genome Conference XX. 2012 January 14–18, San Diego, USA. Abstract W160. https://pag.confex.com/pag/xx/webprogram/Paper2097.html . [Google Scholar]
- Groppo M, Pirani JR, Salatino MLF, Blanco SR, Kallunki JA. Phylogeny of Rutaceae based on two noncoding regions from cpDNA. American Journal of Botany. 2008;95:985–1005. doi: 10.3732/ajb.2007313. [DOI] [PubMed] [Google Scholar]
- Gulsen O, Roose ML. Lemons: diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. Journal of the American Society for Horticultural Science. 2001;126:309–317. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Helyar SJ, Hemmer-Hansen J, Bekkevold D, et al. Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Molecular Ecology Resources. 2011;11:123–136. doi: 10.1111/j.1755-0998.2010.02943.x. [DOI] [PubMed] [Google Scholar]
- Heuertz M, De Paoli E, Källman T, et al. Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce [Picea abies (L.) Karst] Genetics. 2006;174:2095–2105. doi: 10.1534/genetics.106.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson RW. Horticultural varieties of Citrus. In: Reuther W, Webber HJ, Batchelor LD, editors. The citrus industry. Berkeley, CA: University of California; 1967. pp. 431–589. [Google Scholar]
- Ingvarsson PK. Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae) Genetics. 2005;169:945–953. doi: 10.1534/genetics.104.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiology. 2004;134:824–837. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J, Weitzman PD. Krebs' citric acid cycle: half a century and still turning. London: Biochemical Society; 1987. [Google Scholar]
- Kim IJ, Ko KC, Kim CS, Chung WI. Isolation and characterization of cDNAs encoding β-carotene hydroxylase in Citrus. Plant Science. 2001;161:1005–1010. [Google Scholar]
- Koca U, Berhow MA, Febres VJ, Champ KI, Carrillo-Mendoza O, Moore GA. Decreasing unpalatable flavonoid components in Citrus: the effect of transformation construct. Physiologia Plantarum. 2009;137:101–114. doi: 10.1111/j.1399-3054.2009.01264.x. [DOI] [PubMed] [Google Scholar]
- Kolkman JM, Berry ST, Leon AJ, et al. Single nucleotide polymorphisms and linkage disequilibrium in sunflower. Genetics. 2007;177:457–468. doi: 10.1534/genetics.107.074054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RR, Navarro L. In: Citrus germplasm resources. Citrus genetics, breeding, and biotechnology. Khan IA, editor. Wallingford, UK: CAB International; 2007. pp. 45–140. [Google Scholar]
- Külheim C, SuatHui Y, Maintz J, Foley WJ, Moran GF. Comparative SNP diversity among four Eucalyptus species for genes from secondary metabolite biosynthetic pathways. BMC Genomics. 2009;10(452) doi: 10.1186/1471-2164-10-452. http://dx.doi.org/10.1186/1471-2164-10-452 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Haseneyer G, Schön C, et al. High levels of nucleotide diversity and fast decline of linkage disequilibrium in rye (Secale cereale L.) genes involved in frost response. BMC Plant Biology. 2011;11(6) doi: 10.1186/1471-2229-11-6. http://dx.doi.org/10.1186/1471-2229-11-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky D, Cabezas J, Ibáñez A, Rodríguez V, Martínez-Zapater J. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics. 2007;8(424) doi: 10.1186/1471-2164-8-424. http://dx.doi.org/10.1186/1471-2164-8-424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Warnow TJ, Holder MT, et al. SATé-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Systematic Biology. 2012;61:90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- Luro F, Rist D, Ollitrault P. Evaluation of genetic relationships in Citrus genus by means of sequence tagged microsatellites. Acta Horticulturae. 2001;546:237–242. [Google Scholar]
- Luro F, Costantino G, Terol J, et al. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics. 2008;9(287) doi: 10.1186/1471-2164-9-287. http://dx.doi.org/10.1186/1471-2164-9-287 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luro F, Gatto J, Costantino G, Pailly O. Analysis of genetic diversity in Citrus. Plant Genetic Resources. 2011;9:218–221. [Google Scholar]
- Morales M, Roig E, Monforte AJ, Arús P, Garcia-Mas J. Single-nucleotide polymorphisms detected in expressed sequence tags of melon (Cucumis melo L.) Genome. 2004;47:352–360. doi: 10.1139/g03-139. [DOI] [PubMed] [Google Scholar]
- Mabberley DJ. A classification for edible Citrus (Rutaceae) Telopea. 1997;7:167–172. [Google Scholar]
- Moore GA. Oranges and lemons: clues to the taxonomy of Citrus from molecular markers. Trends in Genetics. 2001;17:536–540. doi: 10.1016/s0168-9525(01)02442-8. [DOI] [PubMed] [Google Scholar]
- de Moraes A, dos Santos Soares Filho W, Guerra M. Karyotype diversity and the origin of grapefruit. Chromosome Research. 2007;15:115–121. doi: 10.1007/s10577-006-1101-2. [DOI] [PubMed] [Google Scholar]
- Morton CM. Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpB-rbcL spacer, rps16, and trnL-trnF. Organisms, Diversity & Evolution. 2009;9:52–68. [Google Scholar]
- Navarro L, Pina JA, Juárez J, et al. The Citrus Variety Improvement Program in Spain in the period 1975–2001. In: Duran-Vila N, Milne RG, da Graça JV, editors. Proceedings of the 15th Conference of the International Organization for Citrus Virology. Riverside, CA: IOCV; 2002. pp. 306–316. [Google Scholar]
- Nicolosi E, Deng ZN, Gentile A, Malfa S, Continella G, Tribulato E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theoretical and Applied Genetics. 2000;100:1155–1166. [Google Scholar]
- Novelli VM, Takita MA, Machado MA. Identification and analysis of single nucleotide polymorphisms (SNPs) in Citrus. Euphytica. 2004;138:227–237. [Google Scholar]
- Ollitrault P, Jacquemond C, Dubois C, Luro F. Citrus. In: Hamon P, Seguin M, Perrier X, Glaszmann J-C, editors. Genetic diversity of cultivated tropical plants. Montpellier/Enfield, NH: CIRAD/Science Publishers, Inc; 2003. pp. 193–217. [Google Scholar]
- Ollitrault F, Terol J, Pina JA, Navarro L, Talon M, Ollitrault P. Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. American Journal of Botany. 2010;97:e124–9. doi: 10.3732/ajb.1000280. [DOI] [PubMed] [Google Scholar]
- Ollitrault P, Terol J, Garcia-Lor A, et al. SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics. 2012;13(13) doi: 10.1186/1471-2164-13-13. http://dx.doi.org/10.1186/1471-2164-13-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault F, Terol J, Alonso Martin A, et al. Development of InDel markers from Citrus clementina (Rutaceae) BAC-end sequences and interspecific transferability in Citrus. American Journal of Botany. 2012;99:268–273. doi: 10.3732/ajb.1100569. [DOI] [PubMed] [Google Scholar]
- Pang X, Hu C, Deng X. Phylogenetic relationships within Citrus and its related genera as inferred from AFLP markers. Genetic Resources and Crop Evolution. 2007;54:429–436. [Google Scholar]
- Park S, Yu H, Mum J, Lee S. Genome-wide discovery of DNA polymorphism in Brassica rapa. Molecular Genetics and Genomics. 2010;283:135–145. doi: 10.1007/s00438-009-0504-0. [DOI] [PubMed] [Google Scholar]
- Pavy N, Parsons L, Paule C, MacKay J, Bousquet J. Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics. 2006;7(174) doi: 10.1186/1471-2164-7-174. http://dx.doi.org/10.1186/1471-2164-7-174 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penjor T, Anai T, Nagano Y, Matsumoto R, Yamamoto M. Phylogenetic relationships of Citrus and its relatives based on rbcL gene sequences. Tree Genetics and Genomes. 2010;6:931–939. [Google Scholar]
- Penniston KL, Nakada SY, Holmes RP, Assimos DG. Quantitative assessment of citric acid in lemon juice, lime juice, and commercially-available fruit juice products. Journal of Endourology. 2008;3:567–570. doi: 10.1089/end.2007.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. DARwin software. 2006 http://darwin.cirad.fr/darwin . [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Puritz JB, Addison J, Toonen RJ. Next-generation phylogeography: a targeted approach for multilocus sequencing of non-model organisms. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034241. http://dx.doi.org/10.1371/journal.pone.0034241 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang N, Ikeda S, Harada K. Nucleotide variation in Quercus crispula Blume. Heredity. 2008;101:166–174. doi: 10.1038/hdy.2008.42. [DOI] [PubMed] [Google Scholar]
- Rafalski A, Morgante M. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends in Genetics. 2004;20:103–111. doi: 10.1016/j.tig.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Ramadugu C, Keremane LM, Lee RF, Roose M. Single nucleotide polymorphisms in Citrus and members of Aurantioideae. Plant & Animal Genomes XIX Conference. 2011 Absract W147. http://www.intl-pag.org/19/abstracts/W21_PAGXIX_147.html . [Google Scholar]
- Riju A, Chandrasekar A, Arunachalam V. Mining for single nucleotide polymorphisms and insertions/deletions in expressed sequence tag libraries of oil palm. Bioinformation. 2007;2:128–131. doi: 10.6026/97320630002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo M, Marcos J, Zacarías L. Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. Journal of Agricultural and Food Chemistry. 2004;52:6724–6731. doi: 10.1021/jf049607f. [DOI] [PubMed] [Google Scholar]
- Roose ML, Federici CT, Mu L, Kwok K, Vu C. Map-based ancestry of sweet orange and other citrus variety groups. In: Gentile A, Tribulato E, editors. Second International Citrus Biotechnology Symposium. Vol. 28. Tremestieri Etneo, Italy: Emme Erre Grafica; 2009. [Google Scholar]
- Rosenblum EB, Novembre J. Ascertainment bias in spatially structured populations: a case study in the eastern fence lizard. The Journal of Heredity. 2007;98:331–336. doi: 10.1093/jhered/esm031. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1·42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Sánchez R, Serra F, Tárraga J, et al. Phylemon 2·0: a suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Research. 2011;39:1–5. doi: 10.1093/nar/gkr408. http://dx.doi.org/10.1093/nar/gkr408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biological Chemistry. 2000;381:749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- Scora RW. On the history and origin of Citrus. Bulletin of the Torrey Botanical Club. 1975;102:369–375. [Google Scholar]
- Scora RW. Goren R, Mendel K, editors. Biochemistry, taxonomy and evolution of modern cultivated Citrus. Proceedings of the 6th International Citrus Congress. 1988:277–289. Philadelphia/Rehovot: Balaban Publishers; Weikersheim, Germany: Margraf Scientific Books. [Google Scholar]
- Scott KD, McIntyre CL, Playford J. Molecular analyses suggest a need for a significant rearrangment of Rutaceae subfamilies and a minor reassessment of species relationships within Flindersia. Plant Systematics and Evolution. 2000;223:15–27. [Google Scholar]
- Swingle WT. The botany of Citrus and its relatives of the orange sub-family. In: Webber HJ, Batchelor DL, editors. The citrus industry, vol. 1. History, world distribution, botany and varieties. Berkeley, CA: University of California; 1943. pp. 129–474. [Google Scholar]
- Swingle W, Reece P. The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD, editors. The citrus industry, vol. 1. The botany of Citrus and its wild relatives. Berkeley, CA: University of California; 1967. pp. 190–430. [Google Scholar]
- Tajima F. DNA polymorphism in a subdivided population: the expected number of segregating sites in the two-subpopulation model. Genetics. 1989;123:229–240. doi: 10.1093/genetics/123.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. Species problem in Citrus (Revisio Aurantiacearum IX). Tokyo: Japanese Society for Promotion of Science; 1954. [Google Scholar]
- Tanaka T. Fundamental discussion of Citrus classification. Study in Citrologia, Osaka. 1977;14:1–6. [Google Scholar]
- Terol J, Naranjo MA, Ollitrault P, Talon M. Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46,000 BAC end sequences. BMC Genomics. 2008;9(423) doi: 10.1186/1471-2164-9-423. http://dx.doi.org/10.1186/1471-2164-9-423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AM, Soost RK, Diedenhofen U. Leaf isozymes as genetic markers in Citrus. American Journal of Botany. 1978;65:869–881. [Google Scholar]
- Turner WL, Waller JC, Vanderbeld B, Snedden WA. Cloning and characterization of two NAD kinases from Arabidopsis: identification of a calmodulin binding isoform. Plant Physiology. 2004;135:1243–1255. doi: 10.1104/pp.104.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Snedden WA. Identification, molecular cloning and functional characterization of a novel NADK kinase from Arabidopsis thaliana (thale cress) Biochemical Journal. 2005;385:217–223. doi: 10.1042/BJ20040292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Uzun A, Yesiloglu T, Aka-Kacar Y, Tuzcu O, Gulsen O. Genetic diversity and relationships within Citrus and related genera based on sequence related amplified polymorphism markers (SRAPs) Scientia Horticulturae. 2009;121:306–312. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of population, variability within and among natural populations. Chicago: The University of Chicago Press; 1978. [Google Scholar]
- Xing C, Schumacher F, Xing G, Lu Q, Wang T, Elston R. Comparison of microsatellites, single-nucleotide polymorphisms (SNPs) and composite markers derived from SNPs in linkage analysis. BMC Genetics. 2005;6(29) doi: 10.1186/1471-2156-6-S1-S29. http://dx.doi.org/10.1186/1471-2156-6-S1-S29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Bi IV, et al. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Molecular Biology and Evolution. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.