Summary
Type III secretion systems are used by many Gram-negative pathogens to directly deliver effector proteins into the cytoplasm of host cells. To accomplish this, bacteria secrete translocator proteins that form a pore in the host-cell membrane through which the effector proteins are then introduced into the host cell. Evidence from multiple systems indicates that the pore-forming translocator proteins are exported before effectors, but how this secretion hierarchy is established is unclear. Here we used the P. aeruginosa translocator protein PopD as a model to identify its export signals. The amino-terminal secretion signal and chaperone, PcrH, are required for export under all conditions. Two novel signals in PopD, one proximal to the chaperone-binding site and one at the very C-terminus of the protein, are required for export of PopD before effector proteins. These novel export signals establish the translocator-effector secretion hierarchy, which in turn, is critical for the delivery of effectors into host cells.
Keywords: T3SS, secretion signal, PopB
Introduction
Type III secretion systems are used by many Gram-negative pathogens to promote virulence by delivering effector proteins into targeted host cells (Troisfontaines & Cornelis, 2005, Cornelis, 2006, Galan & Wolf-Watz, 2006). The type III secretion system (T3SS) is a molecular syringe that spans the bacterial envelope. While the apparatus is pre-assembled, effector secretion is triggered in a cell contact-dependent manner (Rosqvist et al., 1994, Vallis et al., 1999, Nordfelth & Wolf-Watz, 2001). Delivery of effector proteins into the host cell requires a specialized set of proteins called translocators. In animal pathogens, two translocator proteins form a pore in the host cell plasma membrane through which the effectors are delivered into the host cell cytoplasm (Sory & Cornelis, 1994, Parsot et al., 1995, Lee et al., 1998, Nordfelth & Wolf-Watz, 2001, Schoehn et al., 2003). A third, hydrophilic translocator protein is situated at the tip of the secretion needle and, beyond its function in delivery of effectors into the host cell, is usually also involved in controlling effector secretion (Menard et al., 1993, Picking et al., 2005, Espina et al., 2006, Nilles et al., 1997, Pettersson et al., 1999, Goure et al., 2004, Lee et al., 2010). The translocon pore in the host cell membrane is believed to consist of 6–8 pore-forming translocator proteins of uncertain stoichiometry (Ide et al., 2001, Schoehn et al., 2003). Several lines of evidence suggest that the translocon pore is docked directly to the needle-tip, forming a continuous conduit from the bacteria cytoplasm to the host cell cytoplasm (Marenne et al., 2003, Ryndak et al., 2005, Lara-Tejero & Galan, 2009). Recent evidence suggests that the translocon may actively contribute to the translocation of effectors into the host cell cytoplasm, rather than simply acting as a pore allowing passive diffusion of the secreted effectors (Akopyan et al., 2011).
The translocator proteins are secreted via the T3SS, but their export is not cell-contact dependent. In P. aeruginosa the translocator proteins that form the pore in the host cell are called PopB and PopD (Frithz-Lindsten et al., 1998). PopB and PopD are required for triggering of effector secretion indicating that they must be exported from the bacterial cell before effector proteins (Cisz et al., 2008). Indeed, export of PopB and PopD under conditions where effector export is prevented can also be demonstrated in vitro (Cisz et al., 2008). Differential export of translocators and effectors has also been observed in Y. enterocolitica. Here, translocator protein secretion could be detected in the presence of serum albumin in the medium (Lee et al., 2001). Similarly, it has been noted that translocator proteins of Shigella are recruited to (or perhaps simply retained at) the needle tip upon addition of bile salts to the growth medium (Olive et al., 2007, Stensrud et al., 2008).
Evidence from several classes of mutations also highlight the differences in export class between translocator and effector proteins. Effector export before triggering of effector secretion is prevented by a family of regulatory proteins including PopN (P. aeruginosa), YopN (Yersinia sp.), MxiC (Shigella sp.), InvE (Salmonella sp.) and SepL (enterohaemorrhagic and eneteropathogenic E. coli)(Pallen et al., 2005). In the case of S. Typhimurium and pathogenic E. coli, deletion of the corresponding gene, invE or sepL, results not only in up-regulation of effector secretion, but also down-regulation of translocator export (Kubori & Galan, 2002, O’Connell et al., 2004). A similar distinction between translocator and effector proteins was discovered when analyzing the integral membrane T3SS component YscU in Y. enterocolitica. YscU belongs to a family of proteins, conserved even in the flagellar T3SS (FlhB), which undergo autocatalytic cleavage to mature (Ferris et al., 2005, Zarivach et al., 2008). Preventing autocleavage of the integral membrane protein YscU in Y. eneterocloitica prevents export of translocator proteins while still allowing effector export (Sorg et al., 2007, Riordan & Schneewind, 2008). Preventing autocatalytic cleavage of SpaS, the YscU-homolog of S. Typhimurium, similarly abrogates translocator export but not effector secretion (Zarivach et al., 2008). In S. Typhimurium, staged access of translocators and effectors to the apparatus was recently linked to three highly conserved components of the T3SS: OrgA, OrgB and SpaO. These three proteins were proposed to form a dynamic sorting platform that controls access of secretion substrates to the T3SS (Lara-Tejero et al., 2011). While translocators SipB/SipC and their cognate secretion chaperone could be pulled down in conjunction with these proteins, effectors were only detected if the translocator genes were deleted.
Several models have been put forward to explain the differential secretion of effectors and translocators. Export of translocators, like all type III secretion substrates, relies on an amino-terminal secretion signal (Lloyd et al., 2001a, Lloyd et al., 2001b, Lloyd et al., 2002, Ramamurthi & Schneewind, 2002, Russmann et al., 2002, Harrington et al., 2003, Sorg et al., 2006, Broms et al., 2007, Arnold et al., 2009, Samudrala et al., 2009). Replacing the amino-terminal export signal of the Y. enterocolitica needle-tip protein LcrV with that of the effector YopE promoted export of the hybrid protein in a yscU autocleavage mutant strain, which interferes with translocator export, suggesting that the identity of translocators and effectors is tied to the amino-terminal secretion signal (Sorg et al., 2007). However, replacing the amino-terminus of the P. aeruginosa translocator proteins PcrV with that of the effector ExoS did not alter its secretion profile, demonstrating that other signals have to be involved in targeting translocators for export before effector secretion has been triggered (Lee et al., 2010). Another hypothesis revolves around the structure of the cognate secretion chaperones of translocators and effectors, which fall into two structural categories (Parsot et al., 2003). Class I chaperones, which promote the export of effectors, contain a mix of alpha-helices and beta-sheets. The chaperone binding domain of the effector wraps around the chaperone (Stebbins & Galan, 2001, Birtalan et al., 2002). In the case of the effector YopE, a patch of amino acids adjacent to the chaperone binding domain is important for export of this protein (Rodgers et al., 2010). Class II chaperones, on the other hand, which promote the export of the pore-forming translocators, are formed form a series of alpha-helical tetratricopeptide repeats that form a palm-shaped structure that cradles the chaperone binding domain of their cognate cargo (Lunelli et al., 2009, Job et al., 2010). The chaperone contributes to secretion efficiency and specificity (Boyd et al., 2000, Birtalan et al., 2002, Letzelter et al., 2006, Lee & Galan, 2004). Analysis of the S. Typhimurium sorting platform highlighted the importance of the chaperone for recruiting substrates, since the effector SptP, lacking its chaperone binding domain, could not associate with this protein complex, even in the absence of the translocator proteins (Lara-Tejero et al., 2011). It has been proposed that the order of export is driven by the affinity of the secretion substrates for the apparatus (Boyd et al., 2000, Birtalan et al., 2002, Lara-Tejero et al., 2011). However, the molecular features driving the export hierarchy are as yet unclear (Stamm & Goldberg, 2011).
Translocator proteins are recruited for export by the T3SS in a manner distinct from effector proteins. Here we use the translocator protein PopD as a model system to characterize the secretion signals governing the export of this translocator protein. As with effector proteins, export of PopD relies on an amino-terminal secretion signal, as well as its cognate chaperone protein, PcrH. In addition, PopD has two translocator-specific export signals that promote export of PopD before effectors, thereby establishing the translocator-effector secretion hierarchy. Disrupting this secretion hierarchy significantly impairs the ability of P. aeruginosa to intoxicate host cells.
Results
The amino-terminal secretion signal is required for secretion of the translocator protein PopD but does not specify export class
Differential secretion of effector and translocator proteins can be assayed quite easily in vitro in P. aeruginosa by virtue of the fact that effector protein secretion is triggered by removing calcium from the medium. Translocator secretion, on the other hand, is detectable in the presence or absence of calcium (Cisz et al., 2008). Export of PopD in the presence of calcium is not due to aberrant export via the flagellar T3SS, akin to the aberrant secretion of SptP via the flagellum in the absence of its chaperone binding domain (Lee & Galan, 2004). In order to rule out this possibility, particularly when analyzing mutants affecting the export signals of PopD, we performed all experiments in a ΔfleQ mutant strain of P. aeruginosa, which lacks the master regulator of flagellar biosynthesis (Arora et al., 1997). Since export of all type III secreted proteins studied to date relies on an amino-terminal secretion signal and evidence from Y. enterocolitica suggested that this signal may help determine the export class of the secretion substrate, we examined the role of the amino-terminal secretion signal in export of the translocator protein PopD.
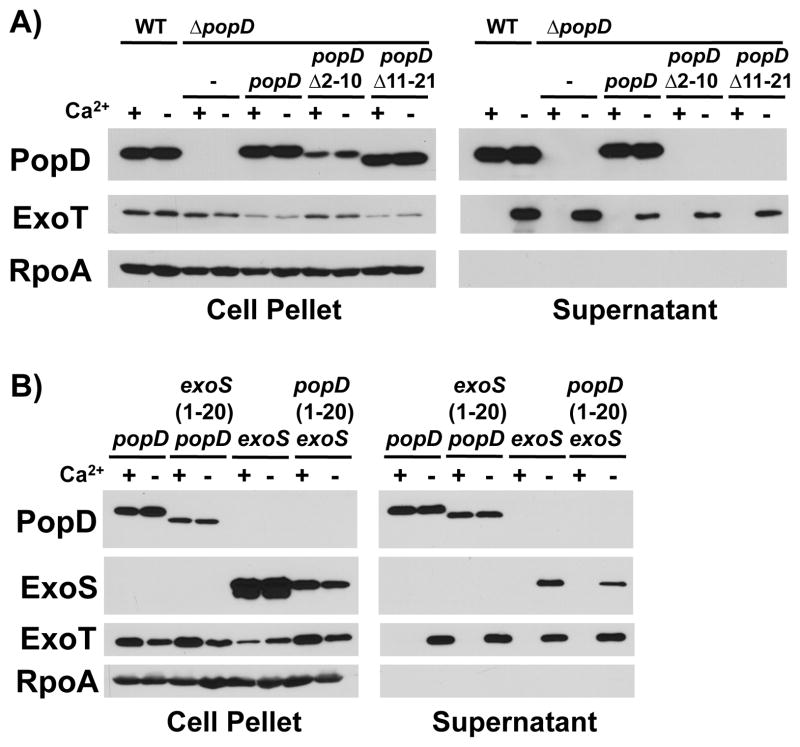
Removing amino acids 2–10 or 11–21 of PopD resulted in a protein that was stably expressed, but failed to be secreted from P. aeruginosa, demonstrating the importance of the amino-terminal secretion signal for PopD export (Fig. 1A). To test the role of the amino-terminal secretion signal in determining the specificity of export, we exchanged the amino-terminal secretion signal of PopD with that of the effector ExoS. Of the three effectors produced by our strain of P. aeruginosa, ExoS and the closely related ExoT are the most highly expressed (unpublished observation). Notably, exchanging the secretion signal of PopD with that of the effector protein ExoS did not abrogate the ability of PopD to be exported in the presence of calcium, the condition where effector secretion is prevented. Similarly, fusing the amino-terminal secretion signal of PopD to the effector ExoS, did not confer the ability to be exported in the presence of calcium to the effector ExoS (Fig. 1B). We therefore conclude that the amino-terminal secretion signal, while critical for secretion of the translocator PopD, does not specify its ability to be exported in the presence of calcium, when effector secretion is prevented.
Fig. 1. The N-terminal secretion signal does not direct export of translocators before effectors.
A) Export of PopD lacking amino acid residues 2–10 or 11–21 was assayed in the presence and absence of calcium by complementing a popD deletion mutant (ΔpopD: PAO1F ΔfleQ ΔpopD) with plasmids expressing either wild-type PopD, or the corresponding mutant protein and compared to export of PopD expressed from its native chromosomal location (WT: PAO1F ΔfleQ). Cell pellet and supernatant fractions were probed with antibodies directed against PopD, the effector ExoT, as well as the RNA polymerase alpha subunit (RpoA, fractionation control).
B) PopD, ExoS, as well as proteins in which the secretion signals of the two proteins had been exchanged [ExoS(1-20)-PopD(21-295) and PopD(1-20)-ExoS(21-453)] were expressed in a strain lacking the chromosomal copies of exoS, popB and popD (PAO1F ΔfleQ ΔexoS ΔpopBD). Export was assayed in the presence and absence of calcium. Cell pellet and supernatant fractions were probed with antibodies directed against PopD, RpoA, as well as the effectors ExoS and ExoT.
The C-terminus of PopD and PopB is required for translocator export in the presence of calcium
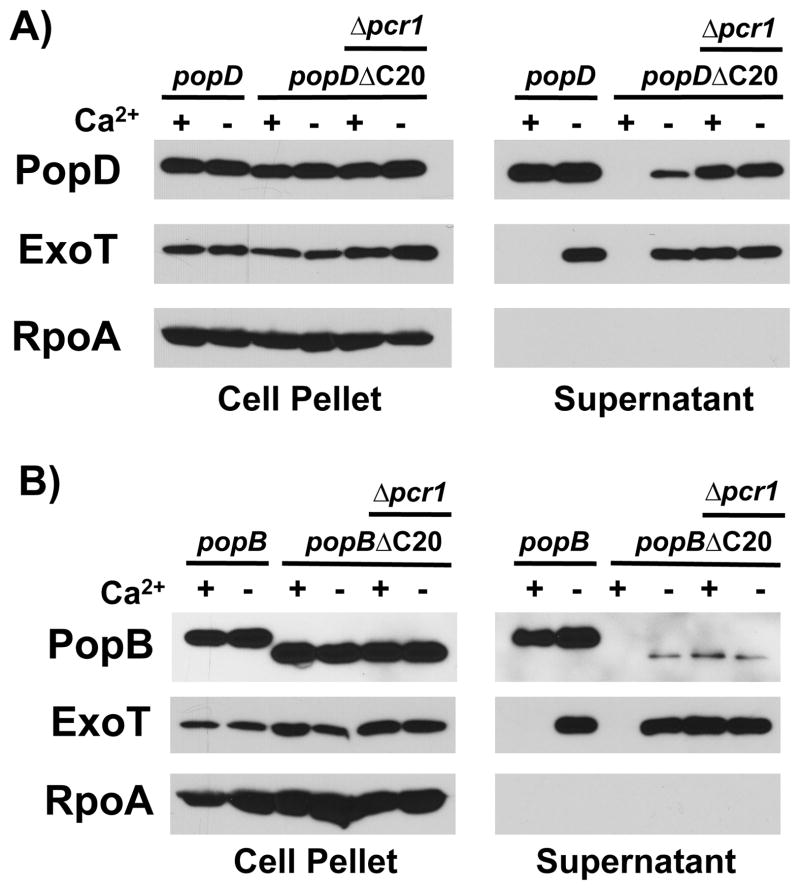
In order to characterize the determinants of PopD that facilitate its export before effector proteins, we generated a series of C-terminally truncated proteins fused to beta-lactamase, a common reporter of protein export via the T3SS (Charpentier & Oswald, 2004). Interestingly, even the full-length fusion demonstrated a severe secretion defect in the presence of calcium, which was not alleviated by exchanging the bulky beta-lactamase tag with the small VSV-G protein epitope (data not shown). This observation suggested that the C-terminus of PopD is important for export of the translocator in the presence of calcium. To test this hypothesis directly, we expressed PopD lacking the C-terminal 20 amino acids, PopDΔC20, and compared its export to wild-type PopD (Fig. 2A). The protein was stably expressed in P. aeruginosa, however, export of PopDΔC20 in the presence of calcium was completely abrogated, and export after effector secretion had been triggered, in the absence of calcium, was severely reduced (23% of total PopD secreted, as opposed to 56% in the case of wild type PopD). We observed a similar secretion defect when we assayed the export the second pore-forming translocator, PopB, using a similarly, C-terminally truncated protein, PopBΔC20 (Fig. 2B). Here 9% of total PopBΔC20 was exported, compared to 52% in the case of wild-type PopB.
Fig. 2. PopD and PopB harbor a C-terminal export signal that is required for export before effectors and efficient secretion overall.
A) PopD or PopDΔC20 (PopDΔ276-295) were expressed in trans from a plasmid in PAO1F ΔfleQ ΔpopD or PAO1F ΔfleQ ΔpopD Δpcr1 (indicated as Δpcr1) and export of these proteins was assayed in the presence or absence of calcium. Cell pellet and supernatant fractions were probed with antibodies directed against PopD, the effector ExoT, as well as RpoA.
B) PopB or PopBΔC20 (PopBΔ371-390) were expressed in trans from a plasmid in PAO1F ΔfleQ ΔpopB or ΔfleQ ΔpopB Δpcr1 (Δpcr1) and export of these proteins was assayed in the presence or absence of calcium. Cell pellet and supernatant fractions were probed with antibodies directed against PopB, the effector ExoT, as well as RpoA.
We next determined if the absence of PopDΔC20 and PopBΔC20 in the supernatants of bacteria grown in the presence of calcium was due to an inability of the protein to be routed for export in the presence of calcium or an unrelated effect, such as reduced stability of the secreted protein in the presence of calcium. To this end, we expressed either protein in a strain lacking a negative regulator of effector secretion, Pcr1 (Fig 2A, B). Effector secretion is constitutive in a pcr1 mutant and no longer controlled by the presence of calcium in the medium as demonstrated by the export of the effector ExoT in the presence of calcium (Fig. 2A, B)(Yang et al., 2007). Both PopDΔC20 and PopBΔC20 were exported in the presence of calcium in the Δpcr1 mutant, demonstrating that the these truncated proteins have effectively changed secretion class and have lost the ability to be exported in the presence of calcium in wild type bacteria. While expression of PopDΔC20 in the Δpcr1 mutant strain somewhat restored the overall export of PopDΔC20 (moving from 23% of total protein in a pcr1+ strain to 43% in the Δpcr1 background), export of PopBΔC20 remained low (8% of total protein). This difference between PopB and PopD could be due to a difference in the degree to which either translocator relies on this C-terminal export signal for overall secretion efficiency. The difference could also indicate that removing the last 20 amino acids of PopB impairs the C-terminal export signal to a greater degree in this protein, than it does in the case of PopD.
The C-terminal translocator export signal is required for the delivery of effector proteins on cell-contact
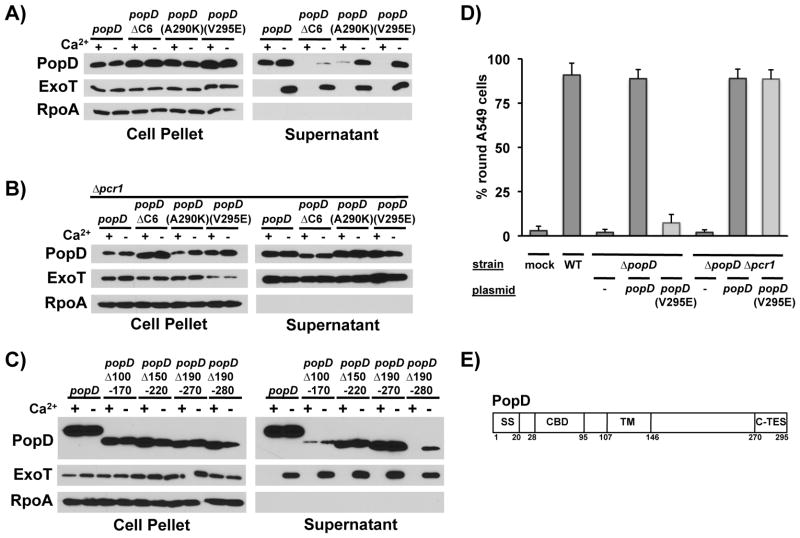
To better define the extent of the C-terminal translocator export signal (C-TES), we generated a popD mutant in which a smaller portion of the open reading frame was truncated, popDΔC6, removing the C-terminal six codons of popD. PopDΔC6 was only exported in the absence of calcium, indicating that even removing six amino acids from the C-terminus of PopD interfered with export in the presence of calcium (Fig. 3A). As with PopDΔC20, removal of the last 6 amino acids of PopD also resulted in a general secretion defect (Fig. 3A), which was detectable even in a strain in which effector secretion had been deregulated (Δpcr1, Fig. 3B). Since the C-terminus of PopD is hydrophobic in nature, we probed the functional significance of the six residues deleted in PopDΔC6 by substituting them individually with charged residues. Of the six mutants assayed, point mutations at codon 290(A290K) and 295(V295E), the last codon of popD, similarly prevented export of PopD in the presence of calcium (Fig. 3A). Export of the mutant protein in the presence of calcium could be restored by deregulating effector secretion (Fig. 3B), indicating that these mutations specifically abrogated export of PopD in the presence of calcium. Taken together, these data demonstrate that the C-TES extends to the very C-terminus of PopD. Interestingly, unlike PopDΔC6, however, neither of the point mutant proteins was defective in its export in the absence of calcium. This finding suggests that determining the secretion hierarchy and promoting efficient export are separable TES functions.
Fig. 3. The C-terminal translocator export signal (C-TES) of PopD spans residues 270-295 and is required for delivery of effectors into host cells.
A) PopD, PopDΔC6 (PopDΔ290-295) as well as point mutants PopD(A290K) and PopD(V295E) were expressed from a plasmid in PAO1F ΔfleQ ΔpopD and secretion was monitored in the presence and absence of calcium. PopD, ExoT and RpoA were detected in cell pellet and supernatant fractions as indicated.
B) PopD, PopDΔC6 (PopDΔ290-295) as well as point mutants PopD(A290K) and PopD(V295E) were expressed from a plasmid in PAO1F ΔfleQ ΔpopD Δpcr1 (Δpcr1) and secretion was monitored in the presence and absence of calcium. PopD, ExoT and RpoA were detected in cell pellet and supernatant fractions as indicated.
C) PopD, as well as internal, in-frame deletion mutants of PopD lacking amino acids 100-170 (Δ100-170), 150-220 (Δ150-220), 190-270(Δ190-270) and 190-280(Δ190-280) were expressed in PAO1F ΔfleQ ΔpopD and their export was assayed in the presence and absence of calcium. PopD, ExoT and RpoA were detected in cell pellet and supernatant fractions as indicated.
D) A549 lung epithelial cells were infected at an MOI of 50, incubated for 3.5 hours at 37°C while expression of popD was induced in trans. Delivery of effectors was assayed by evaluating ExoS and ExoT-mediated depolymerization of actin, which results in cell-rounding. The relevant genotype of the infecting strain is indicated in the top row (mock = no bacteria added). The open reading frame encoded in the complementing plasmid is indicated below (- = vector control).
E) Schematic representation of PopD indicating the amino-terminal secretion signal (SS), chaperone binding domain (CBD), transmembrane domain (TM) and C-terminal translocator export signal (C-TES). The amino acids indicating the beginning and end of each domain are indicated below.
We next determined the amino-terminal boundary of the C-TES by introducing internal, in-frame deletions into the popD open reading frame and assaying their effect on export of the truncated protein. While deletions covering codons 100–170, 150–220 and 190–270 did not interfere with the export of PopD in the presence of calcium, deleting codons 190–280 did, suggesting that the N-terminal boundary of the C-TES lies between residues 270 and 280 (Fig. 3C).
Mutation of the C-TES abolishes the translocator-effector secretion hierarchy, thereby allowing us to test if this hierarchy is important for T3SS function. We assayed delivery of effectors into host cells by analyzing effector-dependent depolymerization of the host-cell actin cytoskeleton, which results in cell-rounding. The TES-mutant popD(V295E) was unable to complement a popD null mutant strain of P. aeruginosa (Fig. 3D). This defect was not due to a defect in translocon function, since restoring calcium-independent export of PopD(V295E) through deletion of pcr1 also restored delivery of effectors. Taken together, these data indicate that the C-TES spans residues 270–295 of PopD (Fig. 3E). It is required for establishing the translocator-effector secretion hierarchy, which, in turn, is critical for the efficient intoxication of host cells.
The C-terminus of PopD interacts with the chaperone PcrH
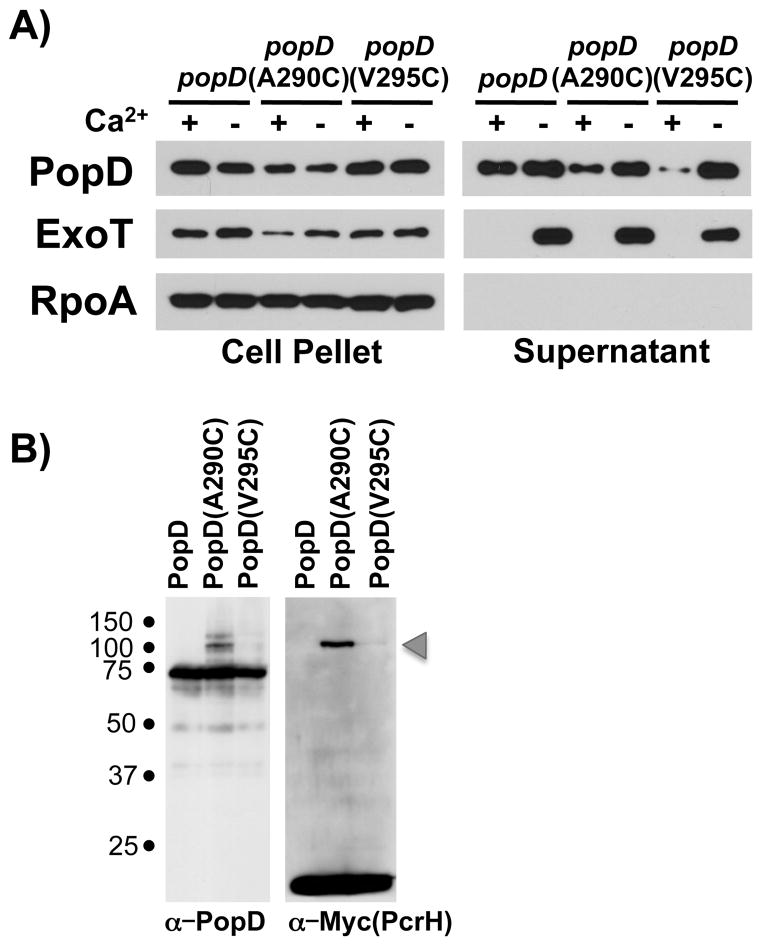
We used a crosslinking approach to identify the interaction partner of the PopD C-TES. PopD does not contain any cysteine residues, which allowed us to easily introduce unique cysteine substitutions at positions 290 and 295 of the C-TES. While these cysteine substitutions reduced the export of the corresponding protein in the presence of calcium, neither cysteine substitution prevented export of PopD in the presence of calcium, indicating that the C-TES still retained its ability to direct export of PopD (Fig. 4A). We next fused these PopD cysteine mutant proteins to maltose binding protein, thereby preventing their export from the cell and providing a tag for purification of cross-linked protein complexes. Interacting proteins were cross-linked to the MBP-PopD fusion proteins using the membrane-permeable, hetero-bifunctional crosslinkers succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC) or LC-SMCC. SMCC and LC-SMCC are sulfhydryl to amine crosslinker that harbors maleimide- and NHS-ester functional groups separated by a 8.3 angstrom (SMCC) or 16.2 angstrom (LC-SMCC) spacer. The fusion proteins were expressed in P. aeruginosa lacking popD and interacting proteins were purified following crosslinking. While we detected little crosslinking in the case of the MBP-PopD(V295C) mutant, crosslinking of MBP-PopD(A290C) resulted in multiple crosslinked complexes. Mass-spectrometric analysis determined that the only type III secretion-related protein thus crosslinked to the MBP-PopD(A290C) fusion protein was the chaperone PcrH (data not shown). We confirmed this cross-linking result by expressing the fusion proteins in a strain in which the chromosomal copy of pcrH had been modified to express PcrH tagged with two copies of the Myc epitope tag at its C-terminus. Crosslinking of MBP-PopD(A290C) in this strain background resulted in a higher molecular weight complex which reacted both with the anti-PopD and anti-Myc antibody. The formation of this complex depended on the presence of the A290C mutation (Fig. 4B). The C-terminus of PopD therefore interacts with PcrH. The fact that PcrH could be cross-linked to PcrH via a cysteine substituted at residue 290 and not 295, suggests that this interaction is specific and not simply a function of the normal PopD-PcrH binding interaction via the more amino-terminal chaperone-binding domain of PopD. The secretion defect of the PopD(A290K) mutant furthermore suggests that this interaction is important for C-TES function. This result was somewhat surprising, given the fact that prior analysis of the PopD-PcrH interaction by pull-downs and limited proteolysis had been unable to detect an interaction between the C-terminus of PopD and PcrH (Faudry et al., 2007). However, we were similarly unable to detect an interaction between the C-terminal half of PopD alone and PcrH by two-hybrid analysis (Fig. S1A), and none of our TES mutants interfered with the PcrH-PopD interaction when assayed in our two-hybrid system (Fig. S1B), indicating that the interaction is weak.
Fig. 4. PcrH interacts with the C-TES of PopD.
A) PopD, PopD(A290C) and PopD(V295C) were expressed in trans in PAO1F ΔfleQ ΔpopD and secretion was monitored in the presence and absence of calcium. PopD, ExoT and RpoA were detected in cell-pellet and supernatant fractions as indicated.
B) PopD and PopD cysteine mutants A290C and V295C were expressed as His tagged MBP fusions in PAO1F ΔfleQ ΔexsE ΔpopD pcrH-Myc and crosslinked in vivo using the site specific heterobifunctional cysteine crosslinker SMCC. Total cell lysates were probed with antibodies directed against PopD and Myc (PcrH). The MBP-PopD-PcrH complex is indicated by an arrowhead.
The chaperone PcrH is required for export of PopD before effector secretion is triggered
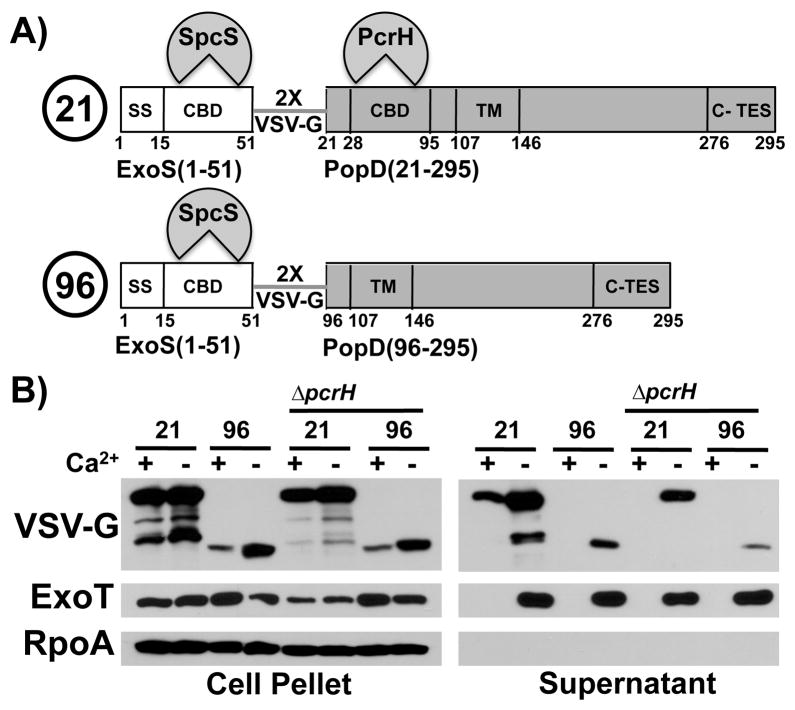
Translocator and effector chaperones belong to different structural classes. The chaperone is therefore a reasonable candidate for helping to establish the translocator-effector secretion hierarchy. PcrH, however, is required for stabilizing PopB and PopD prior to export (Broms et al., 2006), this is also the case in our strain of P. aeruginosa (Fig. S2). We hypothesized that we could stabilize PopD in the absence of PcrH by fusing PopD to a secretion competent protein, such as an effector, which is stabilized by an unrelated chaperone. Such a fusion protein should then allow us to examine the export properties of PopD in the absence of PcrH. We therefore generated hybrid proteins in which the first 51 amino acids of ExoS, comprising its secretion signal and chaperone binding site (Epaulard et al., 2006), are fused to either aminoacids 21–295 of PopD (still able to bind to the chaperone PcrH) or amino acids 96–295 (lacking the PcrH binding site) (Fig. 5A). The export chaperone of ExoS is SpcS (Frithz-Lindsten et al., 1997, Shen et al., 2008). The fusion partners in these hybrid proteins are linked by two copies of the VSV-G epitope tag, allowing detection of all fusions using an anti-VSV-G antibody.
Fig. 5. PcrH and the chaperone binding domain are required for export of PopD before effectors.
A) Schematic representation of the two fusion proteins. ExoS(1-51, white) is fused to PopD (21-295, grey, “21”) or PopD (96-295, grey, “96”). The two fusion partners are joined by a linker consisting of two copies of the VSV-G protein epitope tag. The secretion signal of ExoS (SS) as well as the chaperone binding domains of ExoS and PopD (CBD) are indicated. The chaperone that binds to each of the cognate chaperone binding domains is also indicated (SpcS, the export chaperone of ExoS and ExoT as well as PcrH, the export chaperone of PopB and PopD). The location of the C-TES (TES) is noted as well. Amino acid numbers corresponding do the domain boundaries in the native protein are indicated below.
B) Two fusion proteins, ExoS(1-51)-VSV-G-PopD(21-295) (labeled 21), and ExoS(1-51)-VSV-G-popD(96-295) (labeled 96) were expressed in trans in PAO1F ΔfleQ ΔexoS ΔpopBD and PAO1F ΔfleQ ΔexoS ΔpcrHpopBD(ΔpcrH). Export was assayed in the presence and absence of calcium. Cell pellet and supernatant fractions were probed with antibodies directed against VSV-G(ExoS-PopD hybrid), RpoA, as well as the effector ExoT.
Both hybrid proteins were stable in P. aeruginosa and therefore allowed us to assess the role of the chaperone and chaperone binding site of PopD in routing this translocator for export. The full-length fusion protein, ExoS(1-51)-VSV-G-PopD(21-295), was exported both in the presence and absence of calcium, demonstrating that the translocator export signals in PopD override the export signals inherent in the ExoS(1-51) fragment and SpcS (Fig. 5B). Deletion of pcrH or expression of the shorter, ExoS(1-51)-VSV-G-PopD(96-295), hybrid protein, which lacks the PcrH binding site, prevented export of the hybrid protein in the presence of calcium. PcrH, a signal in PopD in the vicinity of the chaperone binding site, or both, are therefore required for routing PopD for export before effectors.
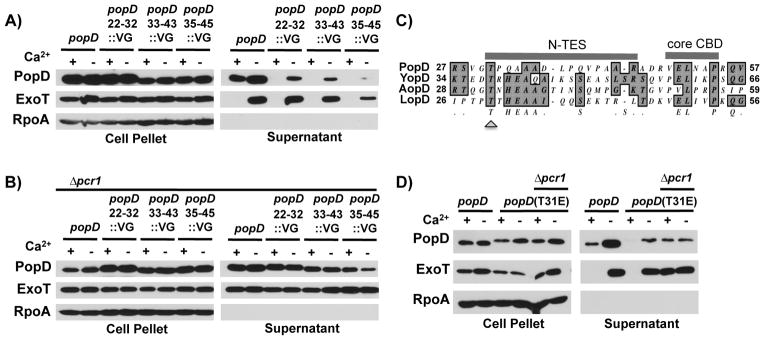
PopD harbors a second translocator export signal adjacent to the chaperone binding site
The preceding result raised the possibility that PopD harbors a second translocator export signal in the vicinity of the chaperone binding domain. Indeed, residues adjacent to the SycE binding site of the Y. pseudotuberculosis effector YopE were recently shown to be important for directing this effector for export via the T3SS (Rodgers et al., 2010). While the chaperone binding domain of PopD was initially determined to lie between residues 28–95 by limited proteolysis (Faudry et al., 2007), subsequent crystallographic analysis of the interaction demonstrated that the core chaperone binding domain of PopD resides at residues 49–54 (Job et al., 2010). To probe whether residues flanking the chaperone binding site of PopD are important for export of the translocator, we assayed the export of a series of amino-terminally truncated PopD proteins fused to ExoS(1-51) akin to the fusion proteins assayed in the preceding section. Fusion of PopD(21-295), PopD(27-295) and PopD(31-295) but not PopD(43-295) resulted in a hybrid protein that was exported both in the presence or absence of calcium (Fig. S3). Replacing residues 22–32, 33–43 and 35–45 of native PopD with the 11 amino-acid VSV-G epitope tag resulted in a TES mutant defect. Export in the presence of calcium was abrogated (Fig. 6A), but could be restored by deleting pcr1, thereby deregulating effector secretion (Fig. 6B). Mutation of the conserved threonine residue at position 31 of PopD (Fig. 6C) resulted in a TES mutant phenotype, abrogating export of PopD in the presence of calcium (Fig. 6D). Export of PopD before effector secretion has been triggered therefore relies on a second, amino-terminal translocator export signal (N-TES) that is adjacent to the chaperone binding domain. Notably, as was the case for C-TES deletions above, mutation of the N-TES resulted in a drop in overall secretion. This was particularly evident for the PopD variant in which residues 35–45 were replaced by the VSV-G epitope tag.
Fig. 6. PopD residues adjacent to the CBD encode a translocator export signal (N-TES).
A) PopD or mutants of PopD in which amino acids 22-32, 33-43 or 35-45 had been replaced by the VSV-G epitope tag were expressed in strain PAO1F ΔfleQ ΔpopD and assayed for export in the presence or absence of calcium. Cell pellet and supernatant fractions were probed for the presence of PopD, ExoT and RpoA.
B) PopD or mutants of PopD in which amino acids 22-32, 33-43 or 35-45 had been replaced by the VSV-G epitope tag were expressed in strain PAO1F ΔfleQ ΔpopD Δpcr1 (Δpcr1) and assayed for export in the presence or absence of calcium. Cell pellet and supernatant fractions were probed for the presence of PopD, ExoT and RpoA.
C) T-COFFEE alignment of residues surrounding the core chaperone binding site in PopD as well as its homologs from Yersinia pseudotuberculosis (YopD), Aeromonas hydrophila (AopD) and Photorhabdus luminescens (LopD). The core chaperone binding domain (core CBD), as well as the N-TES and the conserved threonine residue (T31 in PopD, arrowhead) are indicated.
D) PopD or PopD(T31E) were expressed in trans from a plasmid in PAO1F ΔfleQ ΔpopD or PAO1F ΔfleQ ΔpopD Δpcr1 (Δpcr1) and export of these proteins was assayed in the presence or absence of calcium. Cell pellet and supernatant fractions were probed with antibodies directed against PopD, ExoT or RpoA.
Residue A49 defines a region important for the export function of PcrH
The chaperone PcrH could either contribute directly to the export of PopB and PopD by interacting with an apparatus component and mediating recruitment of the chaperone-translocator complex to the T3SS, or indirectly, by presenting its cargo in a manner that the export signals can be recognized by the T3SS. To distinguish between these possibilities we carried out a genetic screen for mutations in pcrH that prevent export of PopD without preventing binding of PcrH to PopD.
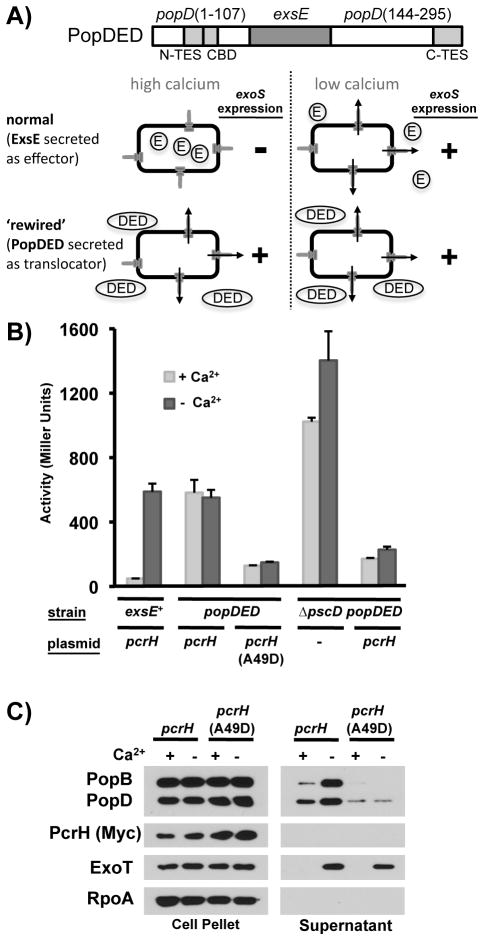
In order to isolate pcrH mutants that meet these criteria we exploited the fact that expression of the type III secretion apparatus and effector genes in P. aeruginosa is linked to effector export (Vallis et al., 1999, Wolfgang et al., 2003, Yahr & Wolfgang, 2006). In wild-type P. aeruginosa, ExsE, a small, secreted protein, serves as a negative regulator of gene expression. When effector secretion is triggered, ExsE is exported via the T3SS from the bacterial cell, thereby precipitating a series of events that lead to the activation of the master regulator of T3SS gene expression, ExsA (Rietsch et al., 2005, Urbanowski et al., 2005)(Fig. 7A). This change in gene expression can be easily assayed using a lacZ reporter inserted in the exoS open reading frame. We adapted this regulatory system to act as a reporter of PopD export by replacing the chromosomal copy of exsE with a sandwich fusion in which the exsE open reading frame replaces codons 108–143 of popD (popDED). Export of the PopDED fusion protein is controlled by the export signals in PopD, resulting in elevated T3SS gene expression both in the presence and absence of calcium (Fig. 7A, B). Expression of the exoS reporter was significantly reduced in the absence of a functional T3SS (ΔpscD), indicating that the elevated level of exoS transcription in the PopDED expressing strain was due to export of PopDED via the T3SS. The basal level of exoS expression in the PopDED expressing pscD null mutant strain was still somewhat elevated compared to the strain expressing wild type ExsE in the presence of calcium, suggesting that the PopDED fusion protein is either somewhat unstable, or slightly defective in its interaction with ExsC. ExsE has to interact with ExsC to repress T3SS gene expression (Dasgupta et al., 2004, Rietsch et al., 2005, Urbanowski et al., 2005). As with wild-type PopD, stability of the PopDED fusion protein relies on its interaction with PcrH, since exoS expression was high in the absence of the pcrH complementation plasmid, even in a strain lacking the T3SS (Fig. 7B).
Fig. 7. Alanine 49 of PcrH defines an epitope on PcrH that is required for the secretion of PopB and PopD.
A) Diagram representing the PopDED sandwich fusion protein in which amino acids 108-143 of PopD are replaced by ExsE (dark grey). Location of the N-TES, C-TES and core chaperone binding domain (CBD) are also indicated (light grey). In wild-type P. aeruginosa, the negative regulator ExsE is only exported via the T3SS when effector secretion is triggered (e.g. in vitro, by removing calcium from the medium). PopDED export, on the other hand, is controlled by the export determinants of PopD, resulting in secretion both in the presence and absence of calcium. Accordingly, exoS expression is elevated even in the presence of calcium in a strain in which exsE has been replaced by popDED.
B) Expression of an exoS-lacZ reporter was linked to translocator export by replacing the exsE open reading frame with a hybrid gene encoding popD in which codons 108-143 have been replaced with the exsE open reading frame (popD(1-107)-exsE(2-81)-popD(144-295), popDED). Expression of the exoS-lacZ reporter was assayed either in the presence or absence of calcium. The relevant genotype of the strain is noted in the first line [WT: PAO1F ΔpcrHpopBD ΔexoS::GL3(GFP-lacZ), popDED: PAO1F ΔpcrHpopBD ΔexoS::GL3 (GFP-lacZ) pcrG(Δ30-40Δ60-70) popDED(ΔexsE::popD(1-107)-exsE(2-81)-popD(144-295)) or popDED ΔpscD: PAO1F ΔpcrHpopBD ΔexoS::GL3 (GFP-lacZ) pcrG(Δ30-40Δ60-70) popDED(ΔexsE::popD(1-107)-exsE(2-81)-popD(144-295)) ΔpscD]. The pcrH deletion was complemented with vector alone (-) or a plasmid encoding pcrH or pcrH(A49D), indicated in the second line below the graph.
C) PcrH-Myc or PcrH(A49D)-Myc were expressed in strain PAO1F ΔfleQ ΔpcrH(Δ46-71), expressing the wild type translocator proteins (and wild type ExsE). Export of PopB and PopD was monitored in the presence or absence of calcium. Cell pellet and supernatant fractions were probed for the presence of PopB, PopD, PcrH(Myc), ExoT and RpoA.
We next mutagenized pcrH by error-prone PCR and screened the resultant library for mutants that stabilize PopDED but prevent its export (pale blue on LB X-Gal plates). The screen identified one mutant, pcrH(G50D) which resulted in a weak PopD export defect (data not shown). In order to probe this region of PcrH more carefully, we used mutagenic primers to specifically randomize codons 45, 46, 49, 50, 51, 53 and 54 of pcrH and assayed the ability of the resultant PcrH mutants to promote export of the PopDED fusion protein. This screen identified the pcrH(A49D) allele of pcrH. PcrH(A49D) strongly inhibits PopDED export while still able to stabilize the fusion protein inside the cell (Fig. 7B). To assess the ability of PcrH(A49D) to promote export of native PopB and PopD, we complemented a pcrH null mutant with a Myc-tagged version of pcrH(A49D). Expression of PcrH(A49D) severely reduces the export of PopB and PopD both in the presence and absence of calcium, while still stabilizing both proteins in the cytoplasm of P. aeruginosa (Fig. 7C). The corresponding Myc-tagged, wild-type, PcrH was able to promote export of PopB and PopD in the presence and absence of calcium. The A49 residue therefore identifies a region of PcrH that is specifically required for the export of PopB and PopD both before and after triggering of effector secretion has occurred.
Discussion
Evidence from several bacteria suggests that translocators are exported before effector proteins. Work from our own lab demonstrated that the pore-forming translocators PopB and PopD are needed to trigger effector secretion on cell-contact, suggesting that the implied secretion hierarchy, translocators preceding effectors, is important for bringing the T3SS to bear. Here we used the translocator PopD as a model protein to define the signals governing its export. We present evidence that export is controlled by three signals in PopD: the secretion signal and two translocator export signals (N- and C-TES). Moreover, the translocator chaperone PcrH actively contributes to secretion as well. Two of these signals, the amino-terminal secretion signal, as well as a region defined by residue A49 of PcrH are needed for export of PopD under all conditions. The N- and C-terminal TES, on the other hand, are required for routing PopD for export before effector proteins. Disrupting the export hierarchy by mutating the C-TES of PopD results in a defect in contact-dependent delivery of effectors into epithelial cells, highlighting the importance of the translocator-effector secretion sequence for T3SS function.
The two classes of export mutants suggest that targeting the PopD/PcrH complex for export requires at least two steps. As with all type III secreted proteins, the amino-terminus of PopD harbors a secretion signal that is required for export (Lloyd et al., 2001b, Lloyd et al., 2002). Similarly, the secretion chaperone is important for promoting the export of type III secretion substrates (Boyd et al., 2000, Birtalan et al., 2002, Letzelter et al., 2006, Lee & Galan, 2004). This is also the case for PopD. Here we used a genetic screen to identify a chaperone residue, A49, that defines a region on PcrH that is required for promoting the export of both PopB and PopD. This region is specifically required for the export function of the chaperone, since it does not abrogate the ability of PcrH to bind to and stabilize PopB and PopD in the cytoplasm of P. aeruginosa. The mutation also demonstrates that the chaperone plays an active role in promoting the export of type III secretion substrates, rather than only promoting export indirectly by presenting the secretion signals inherent in its substrate to the T3SS. Having said that, PcrH also plays a role in specifically promoting the export of PopD before effector secretion has been triggered. In a pcrH null mutant background, the ExoS(1-51)-PopD(21-295) fusion protein was only exported after effector secretion had been triggered. PcrH therefore either directly interacts with the apparatus to deliver translocators to the T3SS before effector secretion has been triggered, or does so indirectly, by presenting the TES in PopD. In this context, it is interesting that mutation of residue A290 of PopD, which, based on our crosslinking experiments is in close proximity to PcrH, results in a TES-mutant phenotype. One possible interpretation of these data is that the two TES domains act in concert and that PcrH serves as a scaffold to bring the two export signals together.
We identified two translocator specific export signals that are required for secretion of PopD before triggering of effector secretion has occurred. One signal is adjacent to the chaperone binding site. Several substitutions in which residues 22–45 were replaced with VSV-G epitope tags, as well as a point mutation, in which we mutated a conserved threonine residue at position 31 of PopD, prevented export of PopD before effector secretion had been triggered. Interestingly, a similar chaperone-proximal export signal was recently discovered in the effector YopE (Rodgers et al., 2010). While not a secretion hierarchy determinant, this export signal suggests that chaperone-binding domain proximal residues are generally important for the export of both effectors and translocators.
Deletions and point mutations at the C-terminus of PopD also prevented export of PopD before triggering of effector secretion. Depending on the mutation, some of the TES mutants (e.g. replacing codons 35–45 with the VSV-G epitope tag, or removing the C-terminal 6 amino acids of PopD) also resulted in an overall secretion defect, demonstrating that the TES, while responsible for targeting PopD for export before effectors also helps improve the overall efficiency with which translocators are exported. Importantly, an intact TES is required for efficient delivery of effectors into host cells, demonstrating that export of translocators before effectors is an important feature of type III secretion. This observation is consistent with our previous finding that the pore forming translocators are required for triggering of effector secretion on cell contact. Notably, the export hierarchy is not needed to allow the assembly of a functional translocon, since a pcr1 mutant can intoxicate cells despite secreting effectors and translocators simultaneously. The primary function of the export hierarchy therefore appears to be to allow the translocon to participate in sensing host-cell contact and triggering of effector secretion.
Interactions of the C-terminus of PopD homologs with their cognate export chaperone have been documented in two related secretion systems, those of Y. pseudotuberculosis and A. hydrophila (Francis et al., 2000, Tan et al., 2009). In the case of Y. pseudotuberculosis, a small deletion near the C-terminus of the protein also interfered with export of YopD, akin to the general secretion defect we have observed with certain TES mutants (Francis et al., 2000). At the time, these deletions were not, however, linked to establishing the translocator-effector secretion hierarchy. Previous attempts to demonstrate an interaction of the C-terminus of PopD with PcrH by pull-down were unsuccessful (Faudry et al., 2007). As noted above, we were similarly not able to detect an interaction of the C-terminus of PopD with PcrH by two-hybrid analysis and none of the TES mutants diminished the PopD-PcrH interaction in our two-hybrid system. The interaction of the PopD C-terminus with PcrH is therefore likely weak and transient in nature.
PopB export appears to be governed by similar export signals since truncation of the C-terminal 20 amino acids of PopB results in a similar TES-mutant phenotype. Indeed, the fact that the translocon operons of Y. pseudotuberculosis and Aeromonas hydrophila can be used to complement a P. aeruginosa pcrGVHpopBD null mutant (A. Rietsch, unpublished observation) demonstrates that the signals governing the translocator-effector secretion hierarchy are likely a conserved feature of this class of proteins.
In the flagellar system, which relies on a type III secretion apparatus for export of flagellar subunits, late substrates (e.g. flagellin and the flagellar cap protein, FliD) bind to their respective export chaperones via their C-termini (Auvray et al., 2001, Bennett et al., 2001, Ozin et al., 2003, Muskotal et al., 2006). An argument could be made that these late substrates of the flagellar T3SS are analogous to translocator proteins, since both are exported in a manner dependent on a conserved specificity switch, either upon completion of the hook basal body (flagellum) or the needle structure (virulence associated T3SS) (Zarivach et al., 2008, Riordan & Schneewind, 2008, Edqvist et al., 2003, Lavander et al., 2002, Ferris et al., 2005, Williams et al., 1996, Hirano et al., 1994). However, many differences exist, including the fact that the primary site of contact between translocator proteins and their cognate chaperone resides close to the amino-terminus of the translocator protein (Job et al., 2010, Faudry et al., 2007, Lunelli et al., 2009), as opposed to the C-terminus of flagellin and FliD in the flagellar system. Moreover, the flagellar chaperones fall into a different structural class (Imada et al., 2010) and, from a functional standpoint, seem to be more closely related to the chaperones of type III secretion needle-proteins, which prevent premature polymerization of the needle (Quinaud et al., 2005), akin to the manner in which FliS prevents polymerization of flagellin (Auvray et al., 2001), and also interact with their cognate secretion substrate via the C-terminus (Quinaud et al., 2005, Sun et al., 2008).
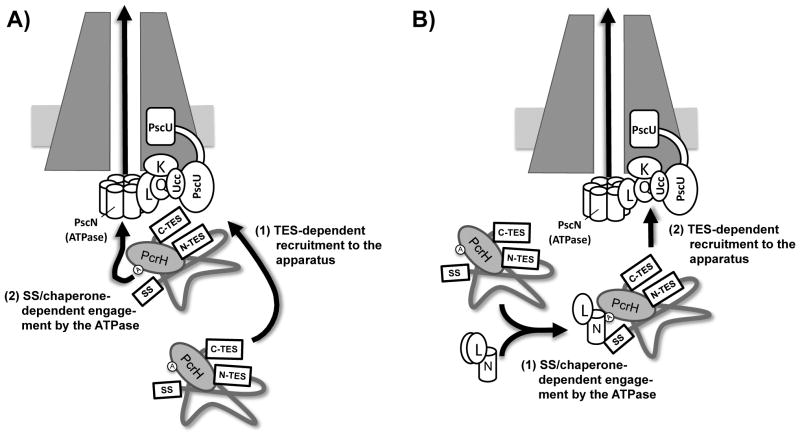
It is as yet unclear how the export signals we have identified are parsed by the T3SS. The simplest interpretation of our data is that export of PopD involves two steps. One step involves the TESs and is required for export before effector secretion has been triggered. The second step involves the N-terminal secretion signal and chaperone PcrH and is required regardless of whether or not effector secretion has been triggered. Since secretion substrates have been demonstrated to associate with the T3SS ATPase in a secretion signal-dependent manner, it would seem that the N-terminal secretion signal, and likely the chaperone interaction, are required for interfacing with the ATPase (Sorg et al., 2006, Akeda & Galan, 2004, Akeda & Galan, 2005). What components of the T3SS the TES interacts with is less clear. Data from several systems suggests that export of translocators depends on auto-cleavage of homologs of P. aeruginosa PscU (Sorg et al., 2007, Riordan & Schneewind, 2008, Zarivach et al., 2008). This is also the case in P. aeruginosa (P.C. Lee and A. Tomalka, unpublished observation). One candidate protein therefore is PscU. Recent evidence from S. Typhimurium, suggests that the effector and translocator proteins are routed for export via a cytoplasmic sorting platform comprised of OrgA, OrgB and SpaO (P. aeruginosa PscK, PscL and PscQ)(Lara-Tejero et al., 2011). Interestingly, YscK, YscL and YscQ of Y. enterocolitica bind to the C-terminal cytoplasmic domain of YscU (YscUc). Co-purification of these three components is either diminished or abolished if autoproteolytic cleavage of YscU is prevented by mutation (Riordan & Schneewind, 2008). Cleavage of PscU could therefore present a surface to which the appropriate sorting platform component is recruited once it has bound to the PcrH-PopD complex. The order in which these two steps occur is unclear. On the one hand, the PopD-PcrH complex could be recruited to the T3SS via the TESs and subsequently routed to the ATPase for export (Fig. 8A). On the other hand, in the flagellar system, it has been proposed that the chaperone-substrate complex associates with ATPase monomers in the cytoplasm and the resulting complex is then recruited to the flagellar T3SS export apparatus (Minamino & Namba, 2008). In this scenario, the signal-sequence/chaperone-dependent interaction with the ATPase occurs first and subsequent recruitment to the T3SS is mediated by the TES (Fig. 8B). More work is needed to distinguish between these possibilities.
Fig. 8. Model for targeting PopD for export.
Two possible models for PopD export are depicted [K, PscK; L, PscL; Q, PscQ; N, PscN; Ucc, C-terminal autoproteolytic fragment of PscU; SS, amino-terminal secretion signal; C- and N-TES, translocator export signals).
A) The PcrH-PopD complex is targeted to the apparatus via the concerted action of the two TES domains (C-TES and N-TES) that are held in proximity to one another by virtue of the interaction of the C-terminus of PopD with PcrH. Subsequent to this targeting step, PopD is delivered to the T3SS ATPase (PscN) through interactions with the secretion signal and the domain defined by A49 of PcrH(circled A).
B) The PcrH-PopD complex binds to an ATPase component in the cytoplasm (likely bound to PscL) in an interaction that relies on the amino-terminal secretion signal and the PcrH domain around alanine 49 (circled A). The PcrH-PopD-ATPase complex is then targeted to the T3SS in a TES-dependent manner.
Our work has identified two sets of signals that govern export of PopD. Export proceeds via a two-step process and establishes the translocator-effector secretion hierarchy that is crucial for T3SS function.
Experimental procedures
Media and culture conditions
E. coli strains were routinely grown at 37°C in LB medium containing 10g/l NaCl. P. aeruginosa was grown at 37°C in a modified LB medium (LB-MC) containing 200mM NaCl, 0.5mM CaCl2 and 10mM MgCl2. Strains and plasmids used in this study are listed in Table 1. A549 cells were obtained from the American Type Culture Collection (lung epithelial cell line, ATCC CCL-185) and propagated in RPMI1640 medium with 10% fetal bovine serum.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| KDZif1ΔZ | E. coli two-hybrid analysis strain lacking rpoZ (encoding ω) and harboring a test promoter-lacZ fusion to detect Zif-dependent two- hybrid interactions | (Vallet-Gely et al., 2005) |
|
| ||
| BN469 | E. coli two-hybrid analysis strain harboring a test promoter-lacZ fusion to detect lambda cI-dependent two-hybrid interactions | (Nickels, 2009) |
| RP1831 | PAO1F, wild type P. aeruginosa PAO1 | (Bleves et al., 2005) |
| RP1865 | PAO1F ΔfleQ | This study |
| RP2937 | PAO1F Δfle Q ΔpopD | This study |
| RP3013 | PAO1F Δfle Q Δpop B Δpop D | This study |
| RP3674 | PAO1F Δfle Q Δpop B | This study |
| RP4043 | PAO1F Δfle Q Δpop D Δpcr1 | This study |
| RP4042 | PAO1F Δfle Q Δpop B Δpcr1 | This study |
| RP3711 | PAO1F Δfle Q ΔexoS Δpop B Δpop D | This study |
| RP4039 | PAO1F Δfle Q ΔexoS ΔpcrH Δpop B Δpop D | This study |
| RP5985 | PAO1F Δfle Q ΔexsE Δpop D pcrH-Myc | This study |
| RP6800 | PAO1F ΔpcrH Δpop B Δpop D ΔexoS::GL3 | This study |
| RP7655 | PAO1F ΔpcrH Δpop B Δpop D ΔexoS::GL3 ΔexsE::popD(1-107)-exsE(2- 81)-popD(144-295) pcrGΔ30-40Δ60-70 | This study |
| RP7656 | PAO1F ΔpcrH Δpop B Δpop D ΔexoS::GL3 ΔpscD ΔexsE::popD(1-107)- exsE(2-81)-popD(144-295) pcrGΔ30-40Δ60-70 | This study |
| RP5795 | PAO1F Δfle Q pcrHΔ46-71 | This study |
| Plasmid | Relevant Features | Reference |
|
| ||
| pPSV37 | colE1 oriR, gentR, PA origin, oriT, lacUV5 promoter, lacIq, stops in every reading frame preceding the MCS and T7 terminator following the MCS relative to the lacUV5 promoter | (Lee et al., 2010) |
| pHMAL | pPSV37 encoding an (8X) His tag fused to a signal-sequenceless malE gene (codons 27–396) lacking a stop codon followed by a polylinker to create N-terminal His-MBP fusions, gentR | This study |
| pBRω-GP | two-hybrid plasmid to create fusions to the C-terminus of the ω subunit of E. coli RNAP, carb R, ColE1 oriR, lacUV5 promoter | (Castang & Dove, 2010) |
| pACtr-VSVG-ZifAP | two-hybrid plasmid to create fusions to the C-terminus of Zif (zinc-finger DNA-binding domain of murine Zif268 protein), VSV-G epitope tag fused to the N-terminus of Zif, tetR, p15A oriR, lacUV5 promoter | (Castang & Dove, 2010) |
| pEXG2 | allelic exchange vector, colE1 origin, oriT, gentR, sacB | (Rietsch et al., 2005) |
| pP37-popD | popD under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ2-10 | popD lacking codons 2-10 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ11-21 | popD lacking codons 11-21 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-20)-popD(21-295) | codons 1–20 of exoS fused to codons 21–295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(1-20)-exoS(21-453) | codons 1–20 of popD fused to codons 21–453 of exoS, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔC20 | popD lacking codons 276-295 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popB | popB under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popBΔC20 | popB lacking codons 371-390 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔC6 | popD lacking codons 290-295 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(A290K) | popD(A290K) under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(V295E) | popD(V295E) under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(A290C) | popD(A290C) under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(V295C) | popD(V295C) under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popD(T31E) | popD(T31E) under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ100-170 | popD lacking codons 100-170 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ150-220 | popD lacking codons 150-220 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ190-270 | popD lacking codons 190-270 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ190-280 | popD lacking codons 190-280 under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ22- 32::VSVG | popD codons 22-32 are replaced with a (1X) VSV-G tag under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ33- 43::VSVG | popD codons 33-43 are replaced with a (1X) VSV-G tag under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-popDΔ35- 45::VSVG | popD codons 35-45 are replaced with a (1X) VSV-G tag under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-51)- VSVG-popD(21-295) | codons 1–51 of exoS fused to a (2X) VSV-G tag, fused to codons 21 – 295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-51)- VSVG-popD(27-295) | codons 1–51 of exoS fused to a (2X) VSV-G tag, fused to codons 27 – 295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-51)- VSVG-popD(31-295) | codons 1–51 of exoS fused to a (2X) VSV-G tag, fused to codons 31 – 295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-51)- VSVG-popD(43-295) | codons 1–51 of exoS fused to a (2X) VSV-G tag, fused to codons 43 – 295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-exoS(1-51)- VSVG-popD(96-295) | codons 1–51 of exoS fused to a (2X) VSV-G tag, fused to codons 96 – 295 of popD, expressed under the control of the lacUV5 promoter in pPSV37 | This study |
| pP37-pcrH-Myc | pcrH fused to a (2X) Myc tag at the C-terminus under control of the lacUV5 promoter in pPSV37 | This study |
| pP37-pcrH(A49D)- Myc | pcrH(A49D) fused to a (2X) Myc tag at the C-terminus under control of the lacUV5 promoter in pPSV37 | This study |
| pHMAL-popD | plasmid encoding His-MBP-PopD fusion protein under control of a lacUV5 promoter | This study |
| pHMAL-popD(A290C) | plasmid encoding His-MBP-PopD(A290C) fusion protein under control of a lacUV5 promoter | This study |
| pHMAL-popD(V295C) | plasmid encoding His-MBP-PopD(V295C) fusion protein under control of a lacUV5 promoter | This study |
| pBRω-GP-pcrH | two-hybrid plasmid encoding ω-PcrH fusion protein | This study |
| pACtr-VSVG-ZifAP-popD | two-hybrid plasmid encoding a VSV-G-Zif-PopD fusion protein | This study |
| pACtr-VSVG-ZifAP-popDΔ22-32::VSVG | two-hybrid plasmid encoding a VSV-G-Zif-PopD fusion protein, codons 22-32 of popD are replaced with a (1X) VSV-G tag | This study |
| pACtr-VSVG-ZifAP-popDΔ33-43::VSVG | two-hybrid plasmid encoding a VSV-G-Zif-PopD fusion protein, codons 33-43 of popD are replaced with a (1X) VSV-G tag | This study |
| pACtr-VSVG-ZifAP-popDΔ35-45::VSVG | two-hybrid plasmid encoding a VSV-G-Zif-PopD fusion protein, codons 35-45 of popD are replaced with a (1X) VSV-G tag | This study |
| pACtr-VSVG-ZifAP-popDΔ58-68::VSVG | two-hybrid plasmid encoding a VSV-G-Zif-PopD fusion protein, codons 58-68 of popD are replaced with a (1X) VSV-G tag | This study |
| pACtr-VSVG-ZifAP-popD(T31E) | two-hybrid plasmid encoding a VSV-G-Zif-PopD(T31E) fusion protein | This study |
| pACtr-VSVG-ZifAP-popDΔC6 | two-hybrid plasmid encoding a VSV-G-Zif-PopDΔC6 fusion protein | This study |
| pACtr-VSVG-ZifAP-popD(V295E) | two-hybrid plasmid encoding a VSV-G-Zif-PopD(V295E) fusion protein | This study |
| pACtr-VSVG-ZifAP-popD(A290K) | two-hybrid plasmid encoding a VSV-G-Zif-PopD(A290K) fusion protein | This study |
| pACλcI35 | two-hybrid plasmid to create fusions to the C-terminus of lambda cI | (Dove & Hochschild, 2004) |
| pBRα35 | two-hybrid plasmid to create fusions to the C-terminus of RNAP alpha | (Dove & Hochschild, 2004) |
| pACλCI35-popD | two-hybrid plasmid encoding a cI-PopD fusion protein | This study |
| pACλCI35-popD(1- 146) | two-hybrid plasmid encoding a cI-PopD(1-146) fusion protein | This study |
| pACλCI35-popD(58- 295) | two-hybrid plasmid encoding a cI-PopD(58-295) fusion protein | This study |
| pBRα35-pcrH | two-hybrid plasmid encoding an alpha-PcrH fusion protein | This study |
| pEXG2-ΔexoS::GFP- lacZ | allelic exchange vector which deletes exoS and inserts translationally coupled versions of GFP and lacZ in its place | (Rietsch et al., 2004) |
| pEXG2-Δfle Q | allelic exchange vector designed to delete codons 4-487 of fleQ | This study |
| pEXG2-Δpop D | allelic exchange vector designed to delete codons 3-268 of popD | (Cisz et al., 2008) |
| pEX-Δpop B | pEXGW plasmid with popB deletion allele (deletes codons 22-364 of popB) | (Sundin et al., 2002) |
| pEXG2-Δpop BΔpop D | allelic exchange vector designed to delete from popB codon 3 to popD codon 268 | (Sun et al., 2010) |
| pEXG2-Δpcr1 | allelic exchange vector designed to delete codons 10-81 of pcr1 | This study |
| pEX-ΔexoS | pEXGW plasmid with exoS deletion allele | (Vance et al., 2005) |
| pEXG2-ΔpcrHΔpop BΔpop D | allelic exchange vector designed to delete from pcrH codon 5 to popD codon 268 | This study |
| pEXG2-ΔexsE | allelic exchange vector designed to delete codons 3-80 of exsE | (Rietsch et al., 2005) |
| pEXG2-pcrH-Myc | allelic exchange vector designed to introduce a (2X) Myc tag at the C- terminus of pcrH | This study |
| pEXG2-pcrGΔ30- 40Δ60-70 | allelic exchange vector designed to delete codons 30-40 and 60-70 of pcrG | This study |
| pEXG2-ΔexsE::popD(1-107)- exsE(2-81)- popD(144-295) | allelic exchange vector designed to make a functional sandwich fusion of popD (codons 1-107, 144-295) and exsE (2-81) | This study |
| pEXG2-ΔpscD | allelic exchange vector designed to delete codons 39-410 of pscD | (Sun et al., 2010) |
| pEXG2-pcrHΔ46-71 | allelic exchange vector designed to delete codons 46-71 of pcrH | This study |
Plasmid construction
Escherichia coli strain DH5α was used for cloning, and E. coli strain Sm10λpir was used to mate plasmids into P. aeruginosa (both E. coli strains were derived from laboratory stocks). pHMAL was constructed by amplifying malE from pMAL (Lee et al 2010) with a primer encoding an (8X) His tag (HisMBP-5R) and malE-3K and cloned into pMAL to replace malE with his-malE using EcoRI and KpnI. Primers used in this study are listed in Table S1. Constructs designed to introduce mutations on the chromosome (cloned into the allelic exchange vector pEXG2) were created using splicing by overlap extension (SOE) PCR (Warrens, 1997). Internal primers (defining the mutation) and external primers (defining the ends of the two flanking regions amplified, as well as the restriction sites used to cloned the spliced cross-over PCR product) are listed in Table S1. PCR products were cloned into pPSV37 and pEXG2 with EcoRI and HindIII, with the exception of popDΔ22-32::VSVG which was cloned into existing pP37-popD with PvuI and HindIII. PCR products were cloned into pHMAL, pBRω-GP and pACtr-VSVG-ZifAP as NotI and HindIII fragments.
Constructs to fuse popD, codons 21–295, to the C-terminus of exsE on the chromosome were constructed by amplifying and fusing three PCR products. The 5′ flank specifying exsE was amplified using primers 1711-5-1 and ED21exsE-5-2. The central portion specifying popD was amplified using primers ED21popD-5-1, paired with EDpopD-3-2. The 3′ flank was amplified using primers 1711-3-2 paired with EDexsE-3-1. All three products were combined by splicing by overlap extension (SOE) PCR and cloned as an EcoRI/HindIII fragment into plasmid pEXG2. These constructs were used to replace the wild-type exsE gene with the exsE-popD(21-295) fusion. To create the popDED allele in which the exsE open reading frame replaces codons 108-143 of popD, four PCR products were fused. Primers 1711-5-1 and EinsDflank-5-2 were used to amplify the 5′ flanking region. Primers EinsD-5-1 and EinsD-D5-2 were used to amplify popD codons 1-107. Primers EinsD-E5 and EinsD-E3 were used to amplify the exsE open reading frame and primers EinsDflank-3-1 and EinsDflank-3-2 were used to amplify the 3′ end of popD (codons 144-289). The four PCR products were fused by two rounds of SOE PCR and cloned as an EcoRI/HindIII fragment into plasmid pEXG2. The resultant plasmid, pEXG2-popDED, was used to convert the chromosomal exsE-popD(21-295) fusion alleles into the corresponding popDED alleles. Two internal deletions (Δ30-40 and Δ60-70) were introduced into the pcrG open reading frame to increase export of the PopDED fusion protein without altering low-calcium dependent triggering of effector secretion (P.C. Lee, unpublished data).
To generate pEXG2-pcrH-CMyc2, flanks were amplified using primers pcrH5R and pcrH2Myc-5-2 (5′ flank) as well as pcrH2Myc-3-1 and pcrH3-2 (3′ flank). The two flanks were combined by SOE PCR and cloned as an EcoRI/HindIII fragment into plasmid pEXG2.
T-COFFEE (Notredame et al., 2000) alignment of PopD homologs was performed in MacVector (Rastogi, 2000).
RECC Assay
The assay to Re-Establish Calcium Control, a modified secretion assay, was performed as previously described (Cisz et al 2008, Lee et al 2010). Bacteria were grown in LB-MC with the addition of EGTA (5mM final concentration) to induce effector secretion and up-regulate expression of the T3SS and its secretion substrates. Cultures were removed at an OD600 of 0.4–0.6, bacteria were pelleted and either resuspended in pre-warmed LB-MC (+ calcium condition) or LB-MC with 5mM EGTA (- calcium condition). The cultures were then incubated for an additional 25 minutes and chilled on ice for 5 minutes before pelleting the bacteria from 1 ml of culture and removing 0.5 ml of the supernatant into a separate tube. The remaining supernatant was discarded and the bacteria pellet was resuspended in 1x SDS sample buffer, normalized to an OD600 of 10. The supernatant proteins were precipitated with trichloroacetic acid (10% final concentration), pelleted by centrifugation, washed once with acetone, dried and resuspended in 1x SDS sample buffer to match the concentration of the cognate cell-pellet sample. Complementation plasmids were induced to WT levels with IPTG.
Immunoblotting
Protein was run on a 10% SDS-PAGE, transferred to PVDF membrane, blocked with 5% milk and probed with specific antisera. Primary antibodies were detected using HRP conjugated secondary antibodies and a chemiluminescent detection reagent (SuperSignal West Pico (Pierce)). Exposure times for cell and supernatant fractions vary between experiments and antibodies, but corresponding pellet and supernatants probed with the same antibody are of the same exposure time. Antibodies to PopD, PopB and ExoT were generated in rabbits using a commercial service (Covance). The antisera were further purified using an affinity purification protocol. Antibodies directed against RpoA (Neoclone), VSV-G tag (Sigma) and Myc tag (Novus Biologicals) were obtained commercially. Quantitation of the western blot in Fig. 2 was performed using ImageJ software (NIH).
Cytotoxicity assay
A549 cells were infected in duplicate with the indicated P. aeruginosa strains at an MOI of 50 in RPMI with 40μM IPTG and incubated at 37°C with 5% CO2 for 3.5 hours. The media was then removed, cells were fixed with 3.7% formaldehyde in PBS for 20 minutes, washed with PBS and visualized with a Nikon Eclipse TE200 inverted microscope. Delivery of effectors was detected by visualizing ExoS/T-mediated cell rounding and expressed as the percent of round vs. flat cells in a field of view. The results from four biological replicates, 3 fields of view each, were averaged.
Cysteine crosslinking
P. aeruginosa strains indicated were grown in 3 mL high salt LB with 40 uM IPTG until OD600 = 0.3–0.4. Cells were spun down by centrifugation, resuspended in PBS with 0.5 mM CaCl2, 5 mM MgCl2 and 500 μM SMCC crosslinker (Succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate, Thermo Scientific). Proteins were crosslinked at room temperature, in the dark, for 30 minutes. Cells were pelleted, resuspended in 1x SDS sample buffer and boiled for 10 minutes to create cell lysates.
pcrH mutant library screen
A silent MfeI site at codons 38–39 of pcrH was introduced by amplifying a 5′ flank with PcrH5R-opt and H-Mfe1-5-2 and a 3′ flank with CMyc2-3H and H-Mfe1-3-1 using pP37-pcrH-Myc as template. The two flanks were combined by SOE PCR and cloned into pPSV37 as an EcoRI/HindIII fragment to create pP37-pcrH-Myc-MfeI. Seven individual pcrH-Myc mutant libraries were constructed in which the following codons were randomized by PCR: 45, 46, 49, 50, 51, 53, 54. Fragments were amplified with Hmut45, Hmut46, Hmut49, Hmut50, Hmut51, Hmut53 and Hmut54 paired with CMyc2-3H and cloned into pP37-pcrH-Myc-MfeI with MfeI and HindIII. Libraries were the following sizes: pP37-pcrH(45*)-Myc-MfeI 710 clones with 0% religation, pP37-pcrH(46*)-Myc-MfeI 690 clones with 0% religation, pP37-pcrH(49*)-Myc-MfeI 720 clones with 0% religation, pP37-pcrH(50*)-Myc-MfeI 1180 clones with 0% religation, pP37-pcrH(51*)-Myc-MfeI 1160 clones with 0% religation, pP37-pcrH(53*)-Myc-MfeI 1970 clones with 0% religation, pP37-pcrH(54*)-Myc-MfeI 2460 clones with 0% religation. Each library was transformed individually into PAO1F ΔpcrH ΔpopBD ΔexoS::GL3 popDED pcrGΔ30-40Δ60-70 and plated on LB with 200 mM NaCl, 10 mM MgCl2, 0.5 mM CaCl2, 30 μg/mL gentamycin, 100 μM IPTG and 75 μg/mL X-gal. Mutations in pcrH that failed to secrete popDED were identified as pale blue colonies. These plasmids were miniprepped, re-transformed to assure that the mutant phenotype is linked to the plasmid and sequenced.
β-galactosidase assay
P. aeruginosa strains were grown overnight in LB-MC with 15μg/ml gentamicin. The overnight cultures were diluted 1:300 into fresh LB-MC with or without the addition of 5mM EGTA (- calcium) and grown to mid-logarithmic phase. Cells were permeabilized with chloroform and SDS, β-galactosidase activity was assayed as previously described (Miller, 1992). For two-hybrid analysis, E. coli KDZif1ΔZ was co-transformed with plasmids containing Zif and ω fusions. Overnight cultures were diluted 1:300 in LB containing 0 or 25 μM IPTG and grown to mid-log phase and processed as noted for P. aeruginosa above. All assays were performed in triplicate with separate overnight cultures inoculated from separate colonies.
Supplementary Material
Acknowledgments
We would like to thank Dr. Simon Dove for the components of the Zif-omega and alpha-cI two hybrid systems, Dr. Susann Brady-Kalnay for the use of her inverted microscope, as well as Dr. Cathleen Carlin for the use of her tissue-culture microscope. This work was funded by the Pulmonary Host Defense training grant from the National Institutes of Health (T32 HL083823)(AGT) as well as an American Cancer Society Research Scholar Grant (RSG-09-198-01-MPC) and Pilot and Feasibility grant from Cystic Fibrosis Foundation program project grant (R447-CR07) to AR.
Footnotes
The authors have no conflicts of interest to declare with regard to the published work.
References
- Akeda Y, Galan JE. Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J Bacteriol. 2004;186:2402–2412. doi: 10.1128/JB.186.8.2402-2412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeda Y, Galan JE. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- Akopyan K, Edgren T, Wang-Edgren H, Rosqvist R, Fahlgren A, Wolf-Watz H, Fallman M. Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci U S A. 2011;108:1639–1644. doi: 10.1073/pnas.1013888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes HW, Horn M, Rattei T. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5:e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. Journal of molecular biology. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JC, Thomas J, Fraser GM, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Molecular microbiology. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–980. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187:3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd AP, Lambermont I, Cornelis GR. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J Bacteriol. 2000;182:4811–4821. doi: 10.1128/jb.182.17.4811-4821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Edqvist PJ, Forsberg A, Francis MS. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol Lett. 2006;256:57–66. doi: 10.1111/j.1574-6968.2005.00099.x. [DOI] [PubMed] [Google Scholar]
- Broms JE, Francis MS, Forsberg A. Diminished LcrV secretion attenuates Yersinia pseudotuberculosis virulence. J Bacteriol. 2007;189:8417–8429. doi: 10.1128/JB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang S, Dove SL. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Molecular microbiology. 2010;78:916–931. doi: 10.1111/j.1365-2958.2010.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X, Oswald E. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisz M, Lee PC, Rietsch A. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J Bacteriol. 2008;190:2726–2738. doi: 10.1128/JB.01553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2004;53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- Dove SL, Hochschild A. A bacterial two-hybrid system based on transcription activation. Methods Mol Biol. 2004;261:231–246. doi: 10.1385/1-59259-762-9:231. [DOI] [PubMed] [Google Scholar]
- Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, Wolf-Watz H, Lloyd SA. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epaulard O, Toussaint B, Quenee L, Derouazi M, Bosco N, Villiers C, Le Berre R, Guery B, Filopon D, Crombez L, Marche PN, Polack B. Anti-tumor immunotherapy via antigen delivery from a live attenuated genetically engineered Pseudomonas aeruginosa type III secretion system-based vector. Mol Ther. 2006;14:656–661. doi: 10.1016/j.ymthe.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faudry E, Job V, Dessen A, Attree I, Forge V. Type III secretion system translocator has a molten globule conformation both in its free and chaperone-bound forms. Febs J. 2007;274:3601–3610. doi: 10.1111/j.1742-4658.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- Francis MS, Aili M, Wiklund ML, Wolf-Watz H. A study of the YopD-lcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol Microbiol. 2000;38:85–102. doi: 10.1046/j.1365-2958.2000.02112.x. [DOI] [PubMed] [Google Scholar]
- Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- Frithz-Lindsten E, Holmstrom A, Jacobsson L, Soltani M, Olsson J, Rosqvist R, Forsberg A. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol Microbiol. 1998;29:1155–1165. doi: 10.1046/j.1365-2958.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect Immun. 2004;72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington AT, Hearn PD, Picking WL, Barker JR, Wessel A, Picking WD. Structural characterization of the N terminus of IpaC from Shigella flexneri. Infect Immun. 2003;71:1255–1264. doi: 10.1128/IAI.71.3.1255-1264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Laarmann S, Greune L, Schillers H, Oberleithner H, Schmidt MA. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-5822.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- Imada K, Minamino T, Kinoshita M, Furukawa Y, Namba K. Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8812–8817. doi: 10.1073/pnas.1001866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job V, Mattei PJ, Lemaire D, Attree I, Dessen A. Structural basis of chaperone recognition of type III secretion system minor translocator proteins. J Biol Chem. 2010;285:23224–23232. doi: 10.1074/jbc.M110.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galan JE. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol. 2002;184:4699–4708. doi: 10.1128/JB.184.17.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Stopford CM, Svenson AG, Rietsch A. Control of effector export by the P. aeruginosa type III secretion proteins PcrG and PcrV. Mol Microbiol. 2010;75:924–941. doi: 10.1111/j.1365-2958.2009.07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- Lee VT, Anderson DM, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- Lee VT, Mazmanian SK, Schneewind O. A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J Bacteriol. 2001;183:4970–4978. doi: 10.1128/JB.183.17.4970-4978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, Kuhn M, Callebaut I, Cornelis GR. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SA, Forsberg A, Wolf-Watz H, Francis MS. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 2001a;9:367–371. doi: 10.1016/s0966-842x(01)02100-x. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Norman M, Rosqvist R, Wolf-Watz H. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol Microbiol. 2001b;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Sjostrom M, Andersson S, Wolf-Watz H. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol Microbiol. 2002;43:51–59. doi: 10.1046/j.1365-2958.2002.02738.x. [DOI] [PubMed] [Google Scholar]
- Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci U S A. 2009;106:9661–9666. doi: 10.1073/pnas.0812900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenne MN, Journet L, Mota LJ, Cornelis GR. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog. 2003;35:243–258. doi: 10.1016/s0882-4010(03)00154-2. [DOI] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics : a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press; Plainview, NY: 1992. p. 2. [Google Scholar]
- Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- Muskotal A, Kiraly R, Sebestyen A, Gugolya Z, Vegh BM, Vonderviszt F. Interaction of FliS flagellar chaperone with flagellin. FEBS letters. 2006;580:3916–3920. doi: 10.1016/j.febslet.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Nickels BE. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods. 2009;47:53–62. doi: 10.1016/j.ymeth.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nilles ML, Williams AW, Skrzypek E, Straley SC. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordfelth R, Wolf-Watz H. YopB of Yersinia enterocolitica is essential for YopE translocation. Infect Immun. 2001;69:3516–3518. doi: 10.1128/IAI.69.5.3516-3518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, Albert MJ, Kaper JB, Frankel G, Donnenberg MS. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol. 2004;52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozin AJ, Claret L, Auvray F, Hughes C. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS microbiology letters. 2003;219:219–224. doi: 10.1016/S0378-1097(02)01208-9. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Beatson SA, Bailey CM. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 2005;5:9. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C, Hamiaux C, Page AL. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol. 2003;6:7–14. doi: 10.1016/s1369-5274(02)00002-4. [DOI] [PubMed] [Google Scholar]
- Parsot C, Menard R, Gounon P, Sansonetti PJ. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- Pettersson J, Holmstrom A, Hill J, Leary S, Frithz-Lindsten E, von Euler-Matell A, Carlsson E, Titball R, Forsberg A, Wolf-Watz H. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol Microbiol. 1999;32:961–976. doi: 10.1046/j.1365-2958.1999.01408.x. [DOI] [PubMed] [Google Scholar]
- Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, Pastor A, Elsen S, Dessen A, Attree I. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Schneewind O. Yersinia enterocolitica type III secretion: mutational analysis of the yopQ secretion signal. J Bacteriol. 2002;184:3321–3328. doi: 10.1128/JB.184.12.3321-3328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi PA. MacVector. Integrated sequence analysis for the Macintosh. Methods in molecular biology. 2000;132:47–69. doi: 10.1385/1-59259-192-2:47. [DOI] [PubMed] [Google Scholar]
- Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A, Wolfgang MC, Mekalanos JJ. Effect of Metabolic Imbalance on Expression of Type III Secretion Genes in Pseudomonas aeruginosa. Infect Immun. 2004;72:1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008;68:1485–1501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers L, Mukerjea R, Friedberg D, Birtalan S, Ghosh P. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. J Bacteriol. 2010;192:3114–3122. doi: 10.1128/JB.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R, Magnusson KE, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russmann H, Kubori T, Sauer J, Galan JE. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol Microbiol. 2002;46:769–779. doi: 10.1046/j.1365-2958.2002.03196.x. [DOI] [PubMed] [Google Scholar]
- Ryndak MB, Chung H, London E, Bliska JB. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect Immun. 2005;73:2433–2443. doi: 10.1128/IAI.73.4.2433-2443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudrala R, Heffron F, McDermott JE. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 2009;5:e1000375. doi: 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. Embo J. 2003;22:4957–4967. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DK, Quenee L, Bonnet M, Kuhn L, Derouazi M, Lamotte D, Toussaint B, Polack B. Orf1/SpcS chaperones ExoS for type three secretion by Pseudomonas aeruginosa. Biomed Environ Sci. 2008;21:103–109. doi: 10.1016/S0895-3988(08)60014-8. [DOI] [PubMed] [Google Scholar]
- Sorg I, Wagner S, Amstutz M, Muller SA, Broz P, Lussi Y, Engel A, Cornelis GR. YscU recognizes translocators as export substrates of the Yersinia injectisome. Embo J. 2007;26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA, Blaylock B, Schneewind O. Secretion signal recognition by YscN, the Yersinia type III secretion ATPase. Proc Natl Acad Sci U S A. 2006;103:16490–16495. doi: 10.1073/pnas.0605974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Goldberg MB. Microbiology. Establishing the secretion hierarchy. Science. 2011;331:1147–1148. doi: 10.1126/science.1203195. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 2008;283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. Journal of molecular biology. 2008;377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Karmakar M, Roy S, Ramadan RT, Williams SR, Howell S, Shive CL, Han Y, Stopford CM, Rietsch A, Pearlman E. TLR4 and TLR5 on Corneal Macrophages Regulate Pseudomonas aeruginosa Keratitis by Signaling through MyD88-Dependent and -Independent Pathways. J Immunol. 2010;185:4272–4283. doi: 10.4049/jimmunol.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin C, Wolfgang MC, Lory S, Forsberg A, Frithz-Lindsten E. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb Pathog. 2002;33:265–277. doi: 10.1006/mpat.2002.0534. [DOI] [PubMed] [Google Scholar]
- Tan YW, Yu HB, Sivaraman J, Leung KY, Mok YK. Mapping of the chaperone AcrH binding regions of translocators AopB and AopD and characterization of oligomeric and metastable AcrH-AopB-AopD complexes in the type III secretion system of Aeromonas hydrophila. Protein Sci. 2009;18:1724–1734. doi: 10.1002/pro.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–339. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- Urbanowski ML, Lykken GL, Yahr TL. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci U S A. 2005;102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:11082–11087. doi: 10.1073/pnas.0502663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis AJ, Yahr TL, Barbieri JT, Frank DW. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67:914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Rietsch A, Mekalanos JJ. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens AN, Jones MD, Lechler RI. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997;186:29–35. doi: 10.1016/s0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, V, Lee T, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Shan Z, Kim J, Wu W, Lian W, Zeng L, Xing L, Jin S. Regulatory role of PopN and its interacting partners in type III secretion of Pseudomonas aeruginosa. J Bacteriol. 2007;189:2599–2609. doi: 10.1128/JB.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.