Abstract
Perifosine belongs to the class of alkylphospholipid analogues, which act primarily at the cell membrane, thereby targeting signal transduction pathways. In phase I/II clinical trials, perifosine has induced tumour regression and caused disease stabilisation in a variety of tumour types. The genetic determinants responsible for its cytotoxicity have not been comprehensively studied, however. We performed a genome-wide analysis to identify genes whose expression levels or genotypic variation were correlated with the cytotoxicity of perifosine, using public databases on the US National Cancer Institute (NCI)-60 human cancer cell lines. For demonstrating drug specificity, the NCI Standard Agent Database (including 171 drugs acting through a variety of mechanisms) was used as a control. We identified agents with similar cytotoxicity profiles to that of perifosine in compounds used in the NCI drug screen. Furthermore, Gene Ontology and pathway analyses were carried out on genes more likely to be perifosine specific. The results suggested that genes correlated with perifosine cytotoxicity are connected by certain known pathways that lead to the mitogen-activated protein kinase signalling pathway and apoptosis. Biological processes such as 'response to stress', 'inflammatory response' and 'ubiquitin cycle' were enriched among these genes. Three single nucleotide polymorphisms (SNPs) located in CACNA2DI and EXOC4 were found to be correlated with perifosine cytotoxicity. Our results provided a manageable list of genes whose expression levels or genotypic variation were strongly correlated with the cytotoxcity of perifosine. These genes could be targets for further studies using candidate-gene approaches. The results also provided insights into the pharmacodynamics of perifosine.
Keywords: perifosine, cytotoxicity, NCI-60, gene expression, genotype
Introduction
Perifosine (NSC639966; Figure 1) belongs to the class of phospholipid analogues or alkylphospholipids, which have anticancer activity in both in vitro (cell culture studies) and in vivo (animal model-based studies) model systems [1,2]. Functionally, perifosine resembles natural phospholipids and acts primarily at the cell membrane, thereby targeting signal transduction pathways. Perifosine has been shown to inhibit, or otherwise modify, signal trans-duction through a number of different pathways, including mitogen-activated protein kinase (MAPK) and Akt [2-5]. Preclinical studies suggest that perifosine inhibits protein kinase B/Akt phosphorylation and induces in vitro and in vivo cytotoxicity in cancer cell lines such as multiple myeloma cells [4], HeLa cells [3] and prostate carcinoma cells [2]. Clinical studies have focused on daily oral dosing (after a loading dose), with two partial responses noted in soft tissue sarcoma (STS) patients, including one patient each with chondrosarcoma and leiomyosarcoma [6,7], as well as patients with renal cell carcinomas [7]. Furthermore, the phase II studies in STS patients were not designed to look for disease stabilisation, a potentially important endpoint for drugs targeting signal transduction pathways [8].
Figure 1.

The chemical structure of perifosine (NSC639966). Molecular formula: C25H52NO4P; Chemical name: piperidinium, 4-[[hydroxy(octadecyloxy)phosphinyl]oxy]-1, 1-dimethyl-, inner salt.
The genetic determinants that are responsible for perifosine's activity have not been comprehensively studied, however. Traditional candidate-gene approaches require a priori knowledge and the selection of a small number of candidate genes for hypothesis testing, while an in silico genome-wide approach could be used to identify any associated genes as potential candidates in an unsupervised way. The US National Cancer Institute (NCI)-60 resources have allowed genome-wide studies using a panel of 60 human cancer cell lines [9]. In addition to the genetic determinants, the NCI-60 resources also provide tools such as COMPARE [10] to identify compounds that show correlated cytotoxic patterns with a particular agent. These compounds, for example, could be potential agents for enhancing the response to a candidate drug or as a substitute for that drug.
The NCI-60 human cancer cell lines have been used in anti-cancer drug screens conducted by the NCI since the late 1980s [9]. The cell lines represent nine distinct tumour types: leukaemia, colon, lung, brain, renal, melanoma, ovarian, breast and prostate. The Developmental Therapeutics Program (DTP) at NCI [11] has maintained a database for the cytotoxicity data, as represented by the GI50 (the concentration required to inhibit cell growth by 50 per cent) on > 40,000 cytotoxic agents, including perifosine [12]. A handful of gene expression datasets using high-throughput platforms such as the Affymetrix oligonucleotide microarrays and cDNA arrays of the untreated NCI-60 cell lines are now publicly available at the DTP/NCI website (Table 1). Recently, the NCI-60 cell lines were genotyped for ~120,000 single nucleotide polymorphism (SNP) markers using the Affymetrix Human 125K Mapping Array manufacturers details [13]. By associating gene expression or SNP genotypes in untreated NCI-60 cell lines, investigators have been able to predict the chemosensitivity of various cytotoxic compounds [14-16]. Here, we report a list of candidate genes whose expression levels or genotypic variation were found to be strongly correlated with the cytotoxicity of perifosine using these publicly available NCI-60 databases. The genes identified could be studied further using a candidate-gene approach. They also could provide new insights into the pharmacodynamics of perifosine.
Table 1.
NCI-60 microarray expression datasets
| Dataset | Institution | Platform | No. of genes on chip | No. of genes analyseda |
|---|---|---|---|---|
| MP-6800 | Millenium Pharmaceuticals, Inc. | Affymetrix Human 6800 | 7,451 | 2,955 |
| GL-U95 | Gene Logic, Inc. | Affymetrix U95 | 63,175 | 23,223 |
| NP-U95 | Novartis Pharmaceuticals, Inc. | Affymetrix U95A | 12,626 | 10,063 |
| NS-cDNA [41,42] | NCI and Stanford University | cDNA array | 9,703 | 5,291 |
aGenes or probe sets that had missing data in more than six cell lines (10 per cent) were not included in the analysis datasets.
NCI; National Cancer Institute.
Materials and methods
Cytotoxicity data
The 60 NCI-60 human cancer cell lines were originally exposed to > 40,000 compounds at NCI/ NIH and outside laboratories. The growth inhibitory effects of each compound were measured for each cell line and reported as the GI50 (for details, see the DTP/NCI website [17]) and maintained in the DTP/NCI online databases. Cytotoxicity data on perifosine (NSC639966) and other agents were obtained as the normalised -log10[GI50] values (released in September 2005). The NSC numbers and common names for the standard agents were retrieved from the DTP/NCI website.
COMPARE analysis
The COMPARE software [10,18,19] maintained at the DTP/NCI, was used to screen >40,000 synthetic or natural compounds for agents that showed correlated cell growth (GI50) patterns with that of perifosine. COMPARE generates rank-ordered lists of compounds based on the similarities of cytotoxicity patterns. Every compound from one of several specially prepared databases is ranked for similarity of its in vitro cell growth pattern to the in vitro cell growth pattern of a selected seed or probe compound (ie perifosine). Top-ranking agents based on Pearson correlation coefficient r, whose GI50 patterns correlated with that of perifosine, were reported by the software. To control false correlations due to small sample size, the minimum number of cell lines in common for two compounds to be included in the calculation was 50. We further set the cut-off for COMPARE analysis at |r| = 0.6 (equivalent to nominal p < 0.000001, assuming 40,000 compounds and n = 60, Bonferroni corrected p < 0.05).
NCI-60 microarray expression datasets
The NCI-60 microarray expression datasets (released in August 2005) were downloaded from the DTP/ NCI Molecular Target Databases [20]. These datasets comprise gene expression data on untreated NCI-60 cell lines using different microarray platforms (Table 1). Genes or probe sets that had missing data in more than six cell lines (10 per cent) were not included in the final analysis dataset.
NCI-60 SNP genotyes
The genotype calls for 125,937 SNPs in 58 NCI-60 cell lines were to be downloaded from the DTP/NCI website using the Affymetrix Human 125K Mapping array [13]. We removed uninformative SNPs, such as those with identical genotypes across all cell lines or those with missing data in more than six cell lines (10 per cent). Only SNP markers with at least two data points per genotype were included in the association studies. This left 34,040 highly informative SNPs in the final analysis dataset. Three exploratory genetic models (additive, dominant, recessive) were used to evaluate the association between genotype and cytotoxicity. Given the genotypes of a SNP marker (AA, AB, BB), the genotypes were encoded as (AA = 0, AB = 1, BB = 2) in the additive model, (AA/AB = 1, BB = 0) in the dominant model and (AA/AB = 0, BB = 1) in the recessive model.
Identifying associated copy number alterations
Data on copy number alterations in the NCI-60 cell lines as reported by Garraway and colleagues [13] were downloaded from the DTP/NCI website.
Linear regression model
We performed genome-wide associations between the gene expression (or genotype) and cytotoxicity data. Pearson correlation coefficients and the associated p-values were computed using a linear regression model, which was implemented as the lm function in the R Statistical Package [21]. Specifically, the cytotoxicity, as represented by -log10[GI50], was modelled as dependent on either gene expression or genotype. To adjust for multiple tests, the false discovery rate (FDR) was controlled using the Benjamini and Hochberg step-up FDR procedure [22] (FDRBH). An FDR cut-off of 10 per cent was used to identify candidates for further analyses.
Associations with standard agents
Associations between the identified genes and the cytotoxicity data on the 171 anti-cancer agents in the NCI Standard Agent Database [23] were performed to evaluate perifosine specificity for our gene list. The standard agents cover a variety of mechanisms, besides being phospholipid analogues, and were originally determined by Boyd [24]. The same cut-off (FDRBH < 0.10) was used to determine if an identified gene was associated significantly with any standard agents. The genes that showed no significant associations with any of the 171 standard agents using any dataset were denoted 'perifosine specific'. Genes that showed associations with any of the 171 standard agents using any dataset were denoted 'non-specific'.
Gene ontology and pathway analyses
We used Onto-Express and Pathway-Express [25-27] to search enriched biological processes and known physiological pathways among the perifosine-specific genes from the Gene Ontology (GO)[28] and Kyoto Encyclopaedia of Genes and Genomes (KEGG) databases [29,30]. GO terms or KEGG pathways that were over-represented relative to the corresponding analysis sets (two hits or more, binomial test at FDRBH < 0.05) were called 'enriched' in our gene list.
STS expression database
The identified perifosine-specific genes were queried against a STS expression database, which characterised eight gastrointestinal stromal tumours, eight monophasic synovial sarcomas, four liposarcomas, one myxoid, 11 leiomyosarcomas, eight malignant fibrous histiocytomas and two benign peripheral nerve sheath tumours (Schwannoma) [31]. Genes differentially expressed among different sarcomas were provided by the database using significance analysis of microarrays (SAM) [32].
Results
COMPARE analysis
At p < 0.05 after Bonferroni correction, the COMPARE software [10,18] identified 24 agents with positive correlation with the cytotoxicity pattern of perifosine. By contrast, no agents with significant negative correlation were identified. Table 2 shows some top-ranking agents (r > 0.6) with well-characterised chemical names. Among them, some clearly belong to the same drug class as perifosine: miltefosine (NSC605583, r = 0.81) and edelfosine (NSC324368, r = 0.68). Edelfosine was further used as a representative of phospholipid analogues to verify the associations detected from perifosine (Supplementary Table 1 (Table 5)).
Table 2.
Agents correlated with the GI50 values of perifosine, as reported by COMPARE (r > 0.6)
| NSC# | r | Chemical name |
|---|---|---|
| 605583 | 0.81 | Miltefosin C; choline, hexadecyl hydrogen phosphate, inner salt |
| 643826 | 0.75 | Choline, hydroxide, 3-methoxy-2-[methyl(octadecyl)amino] propyl hydrogen phosphate, inner salt |
| 643828 | 0.68 | Choline, hydroxide, 2-methoxy-3-[methyl(octadecyl)amino] propyl hydrogen phosphate, inner salt |
| 324368 | 0.68 | Edelfosine; 1-octadecyl-2-methylphosphorylcholine |
| 643827 | 0.68 | Choline, hydroxide, 3-methoxy-1-[methyl(octadecyl)amino]-2-propyl hydrogen phosphate, inner salt |
| 18268 | 0.65 | Actinomycin D |
| 678144 | 0.62 | 4H-l,3,6,2-dioxazaphosphocinium, 4-hexadecyltetrahydro- 2,6,6-trimethyl-, bromide, 2-oxide |
| 337591 | 0.62 | ES 12H; choline, hydroxide, 3-(dodecyloxy)propyl hydrogen phosphate, inner salt |
| 87222 | 0.62 | Actinomycin C3 |
| 266763 | 0.61 | 2-Propenamide, N-[2-(decylsulfinyl)-1-(hydroxymethyl)ethyl]-3-(1,2,3,4-tetrahydro-6-methyl-2,4-dioxo-5-pyrimidinyl)- |
| 207895 | 0.60 | Benzofurazan, 4-(4-methyl-l-piperazinyl)-7-nitro-, 3-oxide |
Supplementary Table 1.
Supplementary Table 1a. A majority of perifosine-specific gene expression levels are associated with the cytotoxicity of edelfosine
| Symbol | Perifosine (p-value) | Edelfosine (p-value) | Perifosine (r-value) | Edelfosine (r-value) | Notes |
|---|---|---|---|---|---|
| NS-cDNA | |||||
| ATF2 | 0.0000 | 0.0075 | -0.560 | -0.348 | Significant |
| TRA2A | 0.0008 | 0.0005 | -0.438 | -0.456 | Significant |
| ETS2 | 0.0005 | 0.0004 | -0.438 | -0.449 | Significant |
| UBE2D3 | 0.0009 | 0.0026 | -0.419 | -0.382 | Significant |
| VEGFB | 0.0006 | 0.0001 | 0.443 | 0.502 | Significant |
| ANP32A | 0.0002 | 0.0003 | 0.492 | 0.469 | Significant |
| GL-U95 | |||||
| REG4 | 0.0000 | 0.0004 | -0.579 | -0.443 | Significant |
| SLCO4A1 | 0.0000 | 0.0060 | -0.518 | -0.351 | Significant |
| RPL18A | 0.0000 | 0.0093 | -0.500 | -0.333 | Significant |
| OAZ2 | 0.0000 | 0.0018 | 0.527 | 0.394 | Significant |
| DZIP3 | 0.0000 | 0.0001 | 0.580 | 0.478 | Significant |
| NP-U95 | |||||
| STK39 | 0.0001 | 0.0004 | -0.479 | -0.449 | Significant |
| FAM32A | 0.0002 | 0.0529 | -0.460 | -0.253 | Marginal |
| MAPKAPK3 | 0.0003 | 0.0920 | -0.454 | -0.221 | Marginal |
| RAB8A | 0.0005 | 0.0018 | -0.441 | -0.399 | Significant |
| STK17B | 0.0006 | 0.0243 | -0.435 | -0.293 | Significant |
| TCF3 | 0.0006 | 0.0054 | -0.435 | -0.358 | Significant |
| PARP4 | 0.0006 | 0.0000 | -0.433 | -0.519 | Significant |
| PSMA2 | 0.0006 | 0.0060 | -0.432 | -0.354 | Significant |
| DGKE | 0.0007 | 0.0019 | -0.429 | -0.396 | Significant |
| PVT1 | 0.0010 | 0.0018 | 0.418 | 0.398 | Significant |
| ELOVL2 | 0.0009 | 0.0005 | 0.419 | 0.438 | Significant |
| SMARCA3 | 0.0009 | 0.0038 | 0.420 | 0.371 | Significant |
| USP6 | 0.0007 | 0.0094 | 0.428 | 0.335 | Significant |
| NFATC4 | 0.0005 | 0.0305 | 0.437 | 0.282 | Significant |
| IGF1R | 0.0005 | 0.0570 | 0.437 | 0.249 | Marginal |
| POU4F1 | 0.0005 | 0.0023 | 0.439 | 0.390 | Significant |
| PDLIM3 | 0.0005 | 0.0058 | 0.440 | 0.355 | Significant |
| CBS | 0.0004 | 0.0094 | 0.443 | 0.336 | Significant |
| ARMCX2 | 0.0004 | 0.0008 | 0.447 | 0.425 | Significant |
| OPHN1 | 0.0003 | 0.0194 | 0.459 | 0.304 | Significant |
| ZNF609 | 0.0002 | 0.0091 | 0.462 | 0.337 | Significant |
| ATN1 | 0.0001 | 0.0754 | 0.474 | 0.233 | Marginal |
| DZIP3 | 0.0001 | 0.0093 | 0.479 | 0.336 | Significant |
| PPBPL2 | 0.0001 | 0.0081 | 0.487 | 0.342 | Significant |
| MPDZ | 0.0000 | 0.0001 | 0.534 | 0.498 | Significant |
| SKIV2L | 0.0000 | 0.0007 | 0.557 | 0.427 | Significant |
| GABRG3 | 0.0000 | 0.0000 | 0.601 | 0.533 | Significant |
| Supplementary Table 1b. The perifosine-specific SNPs are associated with the cytotoxicity of edelfosine | |||||
| dbSNP | Perifosine (p-value) | Perifosine (r-value) | Edelfosine (p-value) | Edelfosine (r-value) | Notes |
| rs4236669 | 2.80E-07 | 0.64 | I.75E-03 | 0.42 | Significant |
| rsI468400 | 8.80E-07 | 0.62 | 6.27E-04 | 0.46 | Significant |
| rsI345938 | 2.60E-06 | 0.58 | 3.66E-05 | 0.52 | Significant |
Genes with expression associated with perifosine cytotoxicity and GO and pathway analyses
Table 3a lists the perifosine-specific genes identified from the microarray expression datasets. The non-specific genes are listed in Supplementary Table 2a (Table 6). The GO and pathway analyses were then carried out to find any enriched biological processes and known KEGG pathways among the perifosine-specific genes (Table 4).
Supplementary Table 2.
Genes whose expression levels were associated with the cytotoxicity of perifosine (FDRBH < 0.10) but were not perifosine specific
| Symbol | Gene title | r | p | Control totala |
|---|---|---|---|---|
| GL-U95 | ||||
| FNBP3 | Formin-binding protein 3 | -0.53 | 3.5E-05 | 75 |
| MOBKL2A | MOBI, Mps One Binder kinase activator-like 2A (yeast) | -0.50 | 5.5E-05 | 43 |
| TP53INP2 | Tumour protein p53 inducible nuclear protein 2 | 0.51 | 3.9E-05 | 24 |
| FBXO44 | F-box protein 44 | 0.52 | 2.4E-05 | 53 |
| TMF1 | TATA element modulatory factor 1 | 0.53 | 1.7E-05 | 57 |
| NP-U95 | ||||
| HNRPDL | Heterogeneous nuclear ribonucleoprotein D-like | -0.49 | 6.9E-05 | 3 |
| DDX39 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 | -0.49 | 7.3E-05 | 22 |
| MRPL23 | Mitochondrial ribosomal protein L23 | -0.49 | 8.7E-05 | 2 |
| RPS24 | Ribosomal protein S24 | -0.47 | 1.9E-04 | 21 |
| LBR | Lamin B receptor | -0.47 | 2.0E-04 | 73 |
| ERCC5 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 | -0.47 | 2.0E-04 | 11 |
| HDAC1 | Histone deacetylase 1 | -0.46 | 2.2E-04 | 2 |
| GTF3A | General transcription factor IIIA | -0.45 | 3.IE-04 | 11 |
| EEF1B2 | Eukaryotic translation elongation factor 1 beta 2 | -0.44 | 4.3E-04 | 68 |
| ICAM3 | Intercellular adhesion molecule 3 | -0.44 | 5.IE-04 | 43 |
| SNRPF | Small nuclear ribonucleoprotein polypeptide F | -0.44 | 5.2E-04 | 83 |
| SH2D1A | SH2 domain protein IA, Duncan's disease (lymphoproliferative syndrome) | -0.44 | 5.8E-04 | 61 |
| KIR3DL1 | Killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 | -0.43 | 7.3E-04 | 76 |
| RPL35 | Ribosomal protein L35 | -0.42 | 8.IE-04 | 47 |
| NUPL2 | Nucleoporin-like 2 | -0.42 | 8.8E-04 | 1 |
| PTMA | Prothymosin, alpha (gene sequence 28) | -0.42 | 9.0E-04 | 33 |
| CORO1A | Coronin, actin-binding protein, IA | -0.42 | 9.3E-04 | 104 |
| LCNI | Lipocalin 1 (tear prealbumin) | -0.42 | 9.3E-04 | 21 |
| POLE3 | Polymerase (DNA directed), epsilon 3 (pl7 subunit) | -0.42 | 9.2E-04 | 31 |
| RPS27A | Ribosomal protein S27a | -0.42 | 9.4E-04 | 79 |
| TRIMI4 | Tripartite motif-containing 14 | -0.42 | 9.3E-04 | 60 |
| LHFPL2 | Lipoma HMGIC fusion partner-like 2 | 0.42 | l.0E-03 | 99 |
| DOK5 | Docking protein 5 | 0.42 | 9.5E-04 | l |
| EIF4GI | Eukaryotic translation initiation factor 4 gamma, l | 0.42 | 9.6E-04 | 28 |
| RGSI9 | Regulator of G-protein signalling 19 interacting protein l | 0.42 | 9.2E-04 | 116 |
| COPB2 | Coatomer protein complex, subunit beta 2 (beta prime) | 0.42 | 8.9E-04 | l |
| TLE2 | Transducin-like enhancer of split 2 (E(spl) homologue, Drosophila) | 0.42 | 9.0E-04 | 4 |
| ITGA7 | Integrin, alpha 7 | 0.42 | 8.6E-04 | 13 |
| SEMA3C | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | 0.42 | 8.2E-04 | 22 |
| SI00AI3 | S100 calcium binding protein A13 | 0.43 | 7.8E-04 | 88 |
| FLOTI | Flotillin 1 | 0.43 | 6.5E-04 | 2 |
| MLFI | Myeloid leukemia factor 1 | 0.43 | 6.0E-04 | 11 |
| ARHGEFII | Rho guanine nucleotide exchange factor (GEF) 11 | 0.44 | 5.5E-04 | 9 |
| COLI5AI | Co11agen, type XV, alpha l | 0.44 | 4.9E-04 | l |
| DAGI | Dystroglycan l (dystrophin-associated glycoprotein l) | 0.44 | 4.9E-04 | 118 |
| IFNAI4 | Interferon alpha l4 | 0.44 | 4.8E-04 | 2 |
| PHLDBI | Pleckstrin homology-like domain, family B, member l | 0.44 | 4.5E-04 | 12 |
| PTPRS | Protein tyrosine phosphatase, receptor type S | 0.45 | 4.0E-04 | 2 |
| SASHI | SAM and SH3 domain containing l | 0.45 | 4.lE-04 | 52 |
| ACVRIB | Activin A receptor, type IB | 0.45 | 3.2E-04 | 18 |
| CTNNAI | Catenin (cadherin-associated protein), alpha 1, 102 kDa | 0.46 | 3.0E-04 | 30 |
| IL6ST | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | 0.46 | 2.9E-04 | 68 |
| ATP6VID | ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D | 0.46 | 2.5E-04 | 2 |
| SUOX | Sulphite oxidase | 0.46 | 2.1E-04 | 58 |
| TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | 0.48 | 1.3E-04 | 51 |
| GRINA | Glutamate receptor, ionotropic, N-methyl D-asparate-associated protein 1 (glutamate binding) | 0.49 | 9.3E-05 | 1 |
| ABCB6 | ATP-binding cassette, sub-family B (MDR/ TAP), member 6 | 0.49 | 8.2E-05 | 8 |
| CTSF | Cathepsin F | 0.49 | 7.4E-05 | 9 |
| VEGFB | Vascular endothelial growth factor B | 0.50 | 4.6E-05 | 6 |
| GGCX | Gamma-glutamyl carboxylase | 0.52 | 2.1E-05 | 23 |
| LAPTM4B | Lysosomal-associated protein transmembrane 4 beta | 0.53 | 1.6E-05 | 84 |
| FYN | Sialidase 1 (lysosomal sialidase) | 0.55 | 6.3E-06 | 59 |
| NS-cDNA | ||||
| LBR | Lamin B receptor | -0.47 | 1.3E-04 | 1 |
| TRA2A | Transformer-2 alpha | -0.44 | 8.3E-04 | 13 |
| ETS2 | V-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | -0.44 | 5.3E-04 | 30 |
| UBE2D3 | Ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homologue, yeast) | -0.42 | 8.5E-04 | 4 |
| RRMI | Ribonucleotide reductase M1 polypeptide | -0.41 | 9.8E-04 | 1 |
| ATP6VICI | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C, isoform 1 | 0.42 | 7.7E-04 | 51 |
| VEGFB | Vascular endothelial growth factor B | 0.44 | 6.3E-04 | 57 |
| RDX | Radixin | 0.45 | 4.2E-04 | 29 |
| APOD | Apolipoprotein D | 0.45 | 3.1E-04 | 3 |
| PTMS | Parathymosin | 0.47 | 1.8E-04 | 2 |
aThe total number of associated standard agents (see Methods).
Table 4.
Enriched Gene Ontology biological processes among the perifosine-specific genes
| GO ID | Process | p | Gene symbol |
|---|---|---|---|
| NP-U95 | |||
| GO:0006950 | Response to stress | 3.8E-04 | STK39 MAPKAPK3 |
| GO:0006511 | Ubiquitin-dependent protein catabolism | 7.9E-04 | PSMA2 USP6 |
| GO:0006954 | Inflammatory response | 4.3E-03 | NFATC4 PARP4 |
| GO:0006512 | Ubiquitin cycle | 6.5E-03 | DZIP3 USP6 |
| GO:0006366 | Transcription from RNA polymerase II promoter | 7.8E-03 | PPBPL2 NFATC4 |
| NS-cDNA | |||
| GO:0006355 | Regulation of transcription, DNA-dependent | 5.6E-03 | EST2 ATF2 |
At FDRBH < 0.10, no genes were associated with perifosine cytotoxicty using the MP-6800 dataset, although at a more lenient cutoff (FDRBH < 0.25), one gene, FABP5 (encoding fatty acid binding protein 5), could be described as being significantly correlated with the sensitivity response to perifosine. The expression of FABP5 was denoted as non-specific, as it was also associated with one standard agent. For the two Affymetrix U95 series of microarray datasets (GL-U95 and NP-U95), one gene, DZIP3 (encoding zinc finger DAZ-interacting protein 3), was correlated with the resistance response to perifosine using both datasets (FDRBH < 0.10). DZIP3 was denoted as perifosine specific, as it showed no associations with any standard agents. In total, ten genes were found to be correlated with perifosine cytotoxicity (FDRBH < 0.10) using the GL-U95 dataset: five each with sensitivity and resistance. Of these, five did not show associations with any standard agents. The GO biological process 'ubiquitin cycle' was enriched among all ten genes (two hits or more, binomial test at FDRBH < 0.05); however, it was not significant among the five perifosine-specific genes. No KEGG pathways were enriched among the identified genes. By contrast, 79 genes were found to be correlated with perifosine cytotoxicity (FDRBH < 0.10) in the NP-U95 dataset. Among them, 30 genes were correlated with sensitivity, while 49 genes were correlated with resistance. Five GO biological processes were enriched among the 27 perifosine-specific genes (two hits or more, binomial test, FDRBH < 0.05). No KEGG pathways were enriched among the identified genes. Using the NS-cDNA dataset, 23 genes were identified, with significant associations with perifosine cytotoxicity (FDRBH < 0.10). Among them, 12 genes were correlated with sensitivity and 11 genes were correlated with resistance. One GO biological process, 'DNA-dependent regulation of transcription', was enriched among the five perifosine-specific genes. No KEGG pathways were enriched among the identified genes.
SNPs associated with perifosine cytotoxicity
Three SNPs under the recessive model were found to be significantly correlated with the resistance response to perifosine (FDRBH < 0.10; Table 3b, Figure 2). These included two SNPs located in the introns of CACNA2D1 (calcium channel, voltage-dependent, alpha 2/delta subunit 1). The third SNP is located in an intron of EXOC4 (exocyst complex component 4). Using both additive and dominant models, these three SNPs did not show significant associations with any standard agents. By contrast, rs1468400 in CACNA2D1 was correlated with one standard agent under the recessive model.
Table 3.
Table 3a. Genes with Gene symbol expression levels specifically associated with the cytotoxicity of perifosine (FDRBH < 0.10)
| Gene symbol | Gene title | ra | p | Response | |
|---|---|---|---|---|---|
| GL-U95 | |||||
| REG4 | Regenerating islet-derived family, member 4 | -0.58 | I.3E-06 | Sensitivity | |
| SLCO4A1 | Solute carrier organic anion transporter family, member 4A1 | -0.52 | 2.3E-05 | Sensitivity | |
| RPL18A | Ribosomal protein L18a | -0.50 | 4.7E-05 | Sensitivity | |
| OAZ2 | Ornithine decarboxylase antizyme 2 | 0.53 | I.6E-05 | Resistance | |
| DZIP3 | Zinc finger DAZ-interacting protein 3 | 0.58 | I.5E-06 | Resistance | |
| NP-U95 | |||||
| STK39 | Serine threonine kinase 39 (STE20/ SPSI homologue, yeast) | - 0.48 | I.2E-04 | Sensitivity | |
| FAM32A | Family with sequence similarity 32, member A | - 0.46 | 2.5E-04 | Sensitivity | |
| MAPKAPK3 | Mitogen-activated protein kinase-activated protein kinase 3 | - 0.45 | 3.0E-04 | Sensitivity | |
| RAB8A | RAB8A, member Ras oncogene family | - 0.44 | 4.7E-04 | Sensitivity | |
| STK17B | Serine/threonine kinase I7b (apoptosis-inducing) | - 0.44 | 5.8E-04 | Sensitivity | |
| TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors EI2/E47) | - 0.44 | 5.8E-04 | Sensitivity | |
| PARP4 | Poly (ADP-ribose) polymerase family, member 4 | - 0.43 | 6.IE-04 | Sensitivity | |
| PSMA2 | Proteasome (prosome, macropain) subunit, alpha type, 2 | - 0.43 | 6.3E-04 | Sensitivity | |
| DGKE | Diacylglycerol kinase, epsilon 64 kDa | - 0.43 | 6.9E-04 | Sensitivity | |
| PVT1 | Pvtl oncogene homologue, MYC activator (mouse) | 0.42 | I.0E-03 | Resistance | |
| ELOVL2 | Elongation of very long chain fatty acids (FENI/Elo2, SUR4/Elo3, yeast)-like 2 | 0.42 | 9.5E-04 | Resistance | |
| SMARCA3 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 3 | 0.42 | 9.3E-04 | Resistance | |
| USP6 | TLI32 protein | 0.43 | 7.IE-04 | Resistance | |
| IGF1R | Insulin-like growth factor I receptor | 0.44 | 5.3E-04 | Resistance | |
| NFATC4 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | 0.44 | 5.4E-04 | Resistance | |
| POU4F1 | POU domain, class 4, transcription factor I | 0.44 | 5.IE-04 | Resistance | |
| PDLIM3 | PDZ and LIM domain 3 | 0.44 | 5.0E-04 | Resistance | |
| CBS | Cystathionine beta-synthase | 0.44 | 4.4E-04 | Resistance | |
| ARMCX2 | Armadillo repeat containing, X-linked 2 | 0.45 | 3.9E-04 | Resistance | |
| OPHN1 | Oligophrenin I | 0.46 | 2.5E-04 | Resistance | |
| ZNF609 | Zinc finger protein 609 | 0.46 | 2.3E-04 | Resistance | |
| ATN1 | Atrophin I | 0.47 | I.5E-04 | Resistance | |
| DZIP3 | Zinc finger DAZ-interacting protein 3 | 0.48 | I.3E-04 | Resistance | |
| PPBPL2 | Pro-platelet basic protein-like 2 | 0.49 | 9.3E-05 | Resistance | |
| MPDZ | Multiple PDZ domain protein | 0.53 | I.3E-05 | Resistance | |
| SKIV2L | Superkiller viralicidic activity 2-like (Saccharomyces cerevisiae) | 0.56 | 4.6E-06 | Resistance | |
| GABRG3 | Gamma-aminobutyric acid (GABA) A receptor, gamma 3 | 0.60 | 4.9E-07 | Resistance | |
| NS-cDNA | |||||
| ATF2 | Activating transcription factor 2 | - 0.56 | 4.8E-06 | Sensitivity | |
| TRA2A | Transformer-2 alpha | - 0.44 | 8.3E-04 | Sensitivity | |
| ETS2 | V-ets erythroblastosis virus E26 oncogene homologue 2 (avian) | - 0.44 | 5.3E-04 | Sensitivity | |
| UBE2D3 | Ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homologue, yeast) | - 0.42 | 8.5E-04 | Sensitivity | |
| ANP32A | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | 0.49 | I.6E-04 | Resistance | |
| Table 3b. SNPs associated with the cytotoxicity of perifosine (FDRBH < 0.10) | |||||
| dbSNPa | Gene locus | Location | r | p | Model |
| rs4236669 | CACNA2DI | Intron | 0.64 | 2.8E-07 | Recessive |
| rsl468400 | CACNA2DI | Intron | 0.62 | 8.8E-07 | Recessive |
| rs1345938 | EXOC4 | Intron | 0.58 | 2.6E-06 | Recessive |
Table 3a. aPearson correlation coefficients were calculated by linear regression in which cytotoxicity (-log10[GI50]) was dependent on gene expression. A positive r-value indicates that a gene is correlated with resistance, while a negative r-value indicates that a gene is correlated with sensitivity.
Table 3b. adbSNP Build 126 (May, 2006).
Figure 2.

SNPs specifically associated with the cytotoxicity of perifosine in the recessive model. AA/AB = 0; BB = 1. (A) Genotypes of rs4236669 in CACNA2DI were associated with the cytotoxicity of perifosine. (B) Genotypes of rs1345938 in EXOC4 were associated with the cytotoxicity of perifosine.
Copy number alterations and perifosine cytotoxicity
At FDRBH < 0.10, no copy number alterations or gene amplifications were found to be correlated with perifosine cytotoxicity.
Querying gene expression patterns in STS cells
Perifosine-specific genes in Table 3a were queried against the STS expression database [31]. Genes that are either up- or downregulated in each type of tumour are listed in Supplementary Table 3 (Table 7). Six genes (STK17B, IGF1R, POU4F1, CBS, MPDZ, EST2) were included in the database. With the exception of EST2, the other five genes were found to be up-or downregulated in certain STS cells.
Supplementary Table 3.
Perifosine-specific genes whose expression levels are up- or downregulated in STS
| Gene | ra | Calponin-positive leiomyosarcoma1b | Calponin-negative leiomyosarcoma1b | GIST1b | Synovial sarcoma1b | Liposarcomab | MFHb | Schwannoma1[3] |
|---|---|---|---|---|---|---|---|---|
| STKI7B | -0.44 | Down regulated | Upregulated | |||||
| IGFIR | 0.44 | Up regulated | ||||||
| P0U4FI | 0.44 | Down regulated | Down regulated | Upregulated | Down regulated | |||
| CBS | 0.44 | Up regulated | Down regulated | |||||
| MPDZ | 0.53 | Up regulated | Down regulated | |||||
| EST2 | -0.44 | |||||||
aCorrelation coefficients (see Table 3 in the text).
bSTS type (see Nielsen et al. 2002).31
Discussion
We performed a genome-wide analysis to identify genes whose expression levels were significantly associated with perifosine's activity, as represented by its cytotoxicity (GI50). Four independent gene expression datasets of untreated NCI-60 cancer cell lines (Table 1), using different microarray platforms, were used to evaluate the association between cytotoxcity and gene expression. We further focused on the identified genes that are more likely to be perifosine specific (Table 3). Previous studies, using traditional candidate-gene approaches, have suggested that perifosine inhibits, or otherwise modifies, signal transduction through a number of different pathways, including MAPK and Akt [2-4]. An in silico genome-wide scan without a priori knowledge in this work provided more candidate genes in an unsupervised way.
The use of COMPARE [10,18] allowed us to identify compounds that have similar cell growth patterns with perifosine (Table 2). To limit the effects due to factors such as small sample size and multiple comparisons, we took measures to control potential false positives. Compounds including those belonging to the same drug class as perifosine (such as miltefosine and edelfosine) were among the top-ranking agents with strong positive correlation coefficients (r > 0.6, p < 0.05 after Bonferroni correction). Not surprisingly, a majority of the perifosine-specific genes were also significantly associated (nominal p < 0.05) with edelfosine, which was used to represent phospholipid analogues (Supplementary Table 1 (Table 5). The remaining few genes showed at least marginal associations (nominal p < 0.10) with edelfosine. This suggests that our list of perifosine-specific genes also contains a set of common genes that determines the pharmacodynamics of this drug class. To our knowledge, this is the most comprehensive list of associated genes for phospholipid analogues. The COMPARE program also retrieved drugs acting through different mechanisms (Table 2). The shared cytotoxicity profiles could be explained by the common pathways between these drugs and perifosine. For example, the correlation with actinomycin, which inhibits transcription by binding DNA at the transcription initiation complex and preventing elongation by RNA polymerase [33], could be explained via general transcriptional modulation (Table 4).
We wanted to know the interactions among the perifosine-specific genes with known biological processes or pathways. Searches against the GO and KEGG databases identified six biological processes that were enriched among the perifosine-specific genes (Table 4). Among them, the biological process of the ubiquitin cycle was identified with DZIP3 and USP6. Notably, DZIP3 was significantly associated with resistance to perifosine, using two of the Affymetrix U95 series of arrays (Table 3a). The function of DZIP3, a ubiquitin ligase [34], in the pharmacodynamics of perifosine has not been investigated, although, given the potential of ubiquitin ligases as anti-cancer targets [35,36], DZIP3 and the role of ubiquitin-dependent protein degradation could be an interesting candidate for further studies. The perifosine-specific genes also over-represented such biological processes as 'response to stress' and 'inflammatory response', which are more evidently related to drug response. Although no particular known KEGG pathways were found to be enriched among the perifosine-specific genes, many of these genes could be connected by a network of known physiological pathways (Figure 3) which have interactions with perifosine through known mechanisms that lead to the MAPK signalling pathway and apoptosis. For example, perifosine can affect the phosphatidylinositol signalling pathway, Akt signalling pathway and MAPK signalling pathway [37,38]. Some of our identified perifosine-specific genes are known to be involved in these pathways; for example, DGKE (the phosphatidylinositol signalling pathway) and MAPKAPK3 (the MAPK signalling pathway). Furthermore, the gene product of DGKE is involved in the phosphatidylinositol signalling system pathway and interacts with the phosphatidylinositol 3-kinase/phosphatase and tensin homologue deleted on chromosome 10 (PTEN)/ Akt pathways [30], suggesting its potential role in the perifosine response. The connected pathways can be divided into three categories:[29] cell communication (tight junction, adherens junction and focal adhesion); immune systems (T/B cell receptor signalling pathways); and signal transduction (MAPK, Wnt, vascular endothelial growth factor and phosphatidylinositol signalling pathways). Given the fact that perifosine, as well as edelfosine, significantly affects the pathway of extrinsic apoptosis [38-40], our findings showed that while perifosine was involved in such pathways as the MAPK and phosphatidylinositol signalling pathways that can lead to apoptosis [2-4], it could also influence other interconnected pathways, such as those in cell communication.
Figure 3.
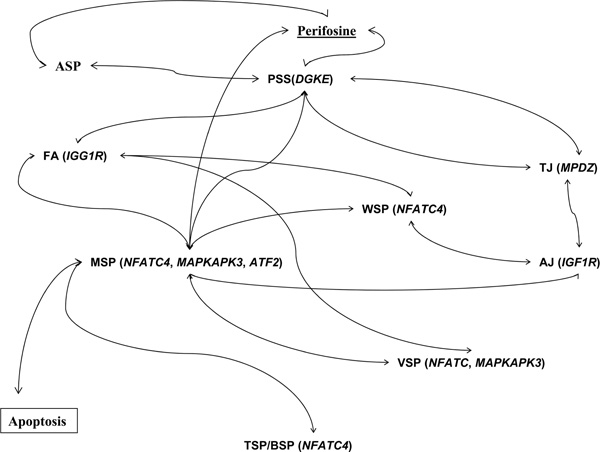
Some perifosine-specific genes are connected by common pathways leading to MAPK signalling pathway and apoptosis. Signal transduction pathways: ASP Akt signalling pathway; PSS, phosphatidylinositol signalling system; WSP: Wnt signalling pathway; VSP vascular endothelial growth factor signalling pathway; MSP, MAPK signalling pathway. Cell communication pathways: TJ, tight junction; AJ, adherens junction; FA, focal adhesion. Immune systems: TSP, T-cell receptor signalling pathway; BSP, B-cell receptor signalling pathway. The relationships among pathways were retrieved from the KEGG database (Release 45.0, January 1, 2008).
Variation in DNA sequence is partially responsible for gene expression; [41,42] therefore, we performed an association test between SNP [13] genotypes and the cytotoxicity of perifosine. Different models (additive, dominant and recessive) were used to explore the genetic relationships between genotypes and cytotoxicity. Two SNPs (rs4236669 in CACNA2D1 and rs1345938 in EXOC4) showed strong perifosine-specific associations under the recessive model (Figure 2). Since the expression of CACNA2D1 was not found to be significantly correlated with perifosine cytotoxicity, the relationship between gene expression and its genotypes is not straightforward. Given that CACNA2D1 is involved in the MAPK signalling pathway [29], however, these SNPs could be interesting candidates for further studies.
Studies have shown that alkylphospholipids are a class of anti-cancer agents that perturb signal transduction pathways through inhibition of MAPK [2-4] and Akt phosphorylation. These drugs have shown consistent clinical anti-cancer activity, but their systemic application has been limited by toxicity. Therefore, one impact of our list of genes could be to help to identify better targeted cancer types for perifosine. One potential candidate, for example, could be multiple myeloma, given the fact that the PSMA2 gene (associated with the sensitivity response to perifosine; Table 3a) was found to be highly upregulated in multiple myeloma cells [43]. In fact, perifosine activity has been reported in myeloma preclinically [4,39]. A recent multicentre phase II study of perifosine alone and in combination with dexamethasone for patients with relapsed or relapsed/refractory multiple myeloma suggested promising activity (eg stabilisation of disease) as combination therapy, with manageable toxicity [44]. Our results thus warrant further clinical trials for this tumour type. There is some evidence of perifosine having activity in STS, with responses reported in chondrosarcoma and leiomyosarcoma [6,7]. Based on these studies, continued assessment of perifosine in STS also appears to be warranted. Given the heterogeneity of STS, it is a plausible hypothesis that there is an identifiable subset of tumours that will respond to this agent [45]. A search against a STS expression database [31] further indicated that a type of leiomyosarcomas that does not express calponin showed the best correlated pattern of gene expression with our perifosine-specific genes (Supplementary Table 4). For example, STK17B (associated with the sensitivity response to perifosine; Table 3a) is significantly upregulated in this tumour type, while POU4F1 and MPDZ (associated with the resistance response to perifosine; Table 3a) are significantly downregulated in this tumour type, suggesting that this type of leiomyo-sarcoma could be a better target for perifosine. As the available STS expression dataset contains only ~5,000 genes [31], a more comprehensive dataset could provide more insights.
In summary, we used the public NCI-60 resources to identify a list of genes potentially relevant to the cytotoxicity of perifosine. Although there were some limitations; such as the gene coverage of the current microarray platforms, relatively small sample size of 60 cell lines and severity of multiple comparisons, our results not only confirmed that perifosine is involved in some known pathways (eg MAPK signalling) that can lead to apoptosis, but also suggested that it could influence some new candidate genes and pathways. Our unsupervised in silico analyses, therefore, could provide targeted candidates that are globally associated with the perifosine response for further studies.
Acknowledgements
This research was supported by a grant from Keryx Biopharmaceuticals, Inc., New York. The authors declare competing financial interests. E.P is an employee of Keryx Biopharmaceuticals, Inc., New York. His employment with Keryx could be construed as a conflict of interest because he may indirectly benefit from sales of their products.
References
- Berdel WE, Bausert WR, Fink U. et al. Anti-tumor action of alkyl-lysophospholipids (Review) Anticancer Res. 1981;1:345–352. [PubMed] [Google Scholar]
- Kondapaka SB, Singh SS, Dasmahapatra GP. et al. Perifosine, a novel alkylphospholipid, inhibits protein kinase B activation. Mol Cancer Ther. 2003;2:1093–1103. [PubMed] [Google Scholar]
- Ruiter GA, Zerp SF, Bartelink H. et al. Anti-cancer alkyl-lysophospholipids inhibit the phosphatidylinositol 3-kinase-Akt/PKB survival pathway. Anticancer Drugs. 2003;14:167–173. doi: 10.1097/00001813-200302000-00011. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Catley L, Yasui H. et al. Perifosine, an oral bio-active novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood. 2006;107:4053–4062. doi: 10.1182/blood-2005-08-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiter GA, Zerp SF, Bartelink H. et al. Alkyl-lysophospholipids activate the SAPK/JNK pathway and enhance radiation-induced apoptosis. Cancer Res. 1999;59:2457–2463. [PubMed] [Google Scholar]
- Bailey HH, Mahoney MR, Ettinger DS. et al. Phase II study of daily oral perifosine in patients with advanced soft tissue sarcoma. Cancer. 2006;107:2462–2467. doi: 10.1002/cncr.22308. [DOI] [PubMed] [Google Scholar]
- Van Ummersen L, Binger K, Volkman J. et al. A phase I trial of perifosine (NSC 639966) on a loading dose/maintenance dose schedule in patients with advanced cancer. Clin Cancer Res. 2004;10:7450–7456. doi: 10.1158/1078-0432.CCR-03-0406. [DOI] [PubMed] [Google Scholar]
- Michaelis LC, Ratain MJ. Measuring response in a post-RECIST world: From black and white to shades of grey. Nat Rev Cancer. 2006;6:409–414. doi: 10.1038/nrc1883. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Zaharevitz DW, Holbeck SL, Bowerman C. et al. COMPARE: A web accessible tool for investigating mechanisms of cell growth inhibition. J Mol Graph Model. 2002;20:297–303. doi: 10.1016/S1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- http://dtp.nci.nih.gov
- Paull KD, Shoemaker RH, Hodes L. et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA. et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Staunton JE, Slonim DK, Coller HA. et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AC, Feulner JA, Reilly A. Evidence for functionally significant polymorphism of human glutamate cysteine ligase catalytic subunit: Association with glutathione levels and drug resistance in the National Cancer Institute tumor cell line panel. Toxicol Sci. 2001;61:218–223. doi: 10.1093/toxsci/61.2.218. [DOI] [PubMed] [Google Scholar]
- Moisan F, Longy M, Robert J. et al. Identification of gene polymorphisms of human DNA topoisomerase I in the National Cancer Institute panel of human tumour cell lines. Br J Cancer. 2006;95:906–913. doi: 10.1038/sj.bjc.6603361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://dtp.nci.nih.gov
- Fang X, Shao L, Zhang H. Web-based tools for mining the NCI databases for anticancer drug discovery. J Chem Inf Comput Sci. 2004;44:249–257. doi: 10.1021/ci034209i. [DOI] [PubMed] [Google Scholar]
- http://dtp.nci.nih.gov/docs/compare/compare.html
- http://dtp.nci.nih.gov/mtargets/download.html
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2006. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc [Ser B] 1995;57:289–300. [Google Scholar]
- http://dtp.nci.nih.gov/docs/cancer/searches/standard_agent.html
- Boyd MR. In: Cancer: Principles and Practice of Oncology. 3. DeVita, VT, Jr, Hellman, S, Rosenberg, SA, editor. Lippincott, Philadelphia, PA; 1989. Status of the NCI preclinical antitumor drug discovery screen; pp. 1–12. [Google Scholar]
- Draghici S, Khatri P, Bhavsar P. et al. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Martins RP. et al. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/S0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Khatri P, Bhavsar P, Bawa G. et al. Onto-Tools: An ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA. et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia ofgenes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S. et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC. et al. Molecular characterisation of soft tissue tumours: A gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci USA. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft SG, Nassal M. hRUL138, a novel human RNA-binding RING-H2 ubiquitin-protein ligase. J Cell Sci. 2003;116:605–616. doi: 10.1242/jcs.00261. [DOI] [PubMed] [Google Scholar]
- Pray TR, Parlati F, Huang J. et al. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5:249–258. doi: 10.1016/S1368-7646(02)00121-8. [DOI] [PubMed] [Google Scholar]
- Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geroldi D, Aldeghi A, Emanuele E. Perifosine may be of therapeutic usefulness in morbid obese females via inhibition of the PI3K/Akt signaling pathway. Med Hypotheses. 2007;68:236. doi: 10.1016/j.mehy.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Elrod HA, Lin YD, Yue P. et al. The alkylphospholipid perifosine induces apoptosis of human lung cancer cells requiring inhibition of Akt and activation of the extrinsic apoptotic pathway. Mol Cancer Ther. 2007;6:2029–2038. doi: 10.1158/1535-7163.MCT-07-0004. [DOI] [PubMed] [Google Scholar]
- Gajate C, Mollinedo F. Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood. 2007;109:711–719. doi: 10.1182/blood-2006-04-016824. [DOI] [PubMed] [Google Scholar]
- Nyakern M, Cappellini A, Mantovani I. et al. Synergistic induction of apoptosis in human leukemia T cells by the Akt inhibitor perifosine and etoposide through activation of intrinsic and Fas-mediated extrinsic cell death pathways. Mol Cancer Ther. 2006;5:1559–1570. doi: 10.1158/1535-7163.MCT-06-0076. [DOI] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO. et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–640. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ratain MJ, Dolan ME. The HapMap Resource is providing new insights into ourselves and its application to pharmacogenomics. Bioinform Biol Insights. 2008;2:15–23. doi: 10.4137/bbi.s455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi NC, Hideshima T, Carrasco D. et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- Richardson P, Lonial S, Jakubowiak A. et al. Multi-center phase II study of perifosine (KRX-0401) alone and in combination with dexamethasone (dex) for patients with relapsed or relapsed/refractory multiple myeloma (MM): Promising activity as combination therapy with manageable toxicity. ASH Annual Meeting Abstracts. 2007;110:1164. [Google Scholar]
- Skubitz KM, Skubitz AP. Characterization of sarcomas by means of gene expression. J Lab Clin Med. 2004;144:78–91. doi: 10.1016/j.lab.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Scherf U, Ross DT, Waltham M. et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB. et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]


