Abstract
Changes in the expression of the mismatch repair (MMR) genes hMSH2, hMLH1, hMSH6 and hPMS2 reflect dysfunction of the DNA repair system that may allow the malignant transformation of tissue cells. The aim of the present study was to address the mRNA expression profiles of the mismatch DNA repair system in cancerous and precancerous urothelium. This is the first study to quantify MMR mRNA expression by applying quantitative real-time PCR (qPCR) and translate the results to mRNA phenotypic profiles (r, reduced; R, regular or elevated) in bladder tumors [24 urothelial cell carcinomas (UCCs) and 1 papillary urothelial neoplasm of low malignant potential (PUNLMP)] paired with their adjacent normal tissues (ANTs). Genetic instability analysis was applied at polymorphic sites distal or close to the hMSH2 and hMLH1 locus. Presenting our data, reduced hMSH2, hMSH6 and hPMS2 mRNA expression profiles were observed in cancerous and precancerous urothelia. Significantly, the ANTs of UCCs revealed the highest percentages of reduced hMSH2 (r2), hMSH6 (r6) and hPMS2 (p2) mRNA phenotypes relative to their tumors (P<0.03). In particular, combined r2r6 (P<0.02) presented a greater difference between ANTs of low-grade UCCs vs. their tumors compared with ANTs of high-grade UCCs (P= 0.000). Reduced hMLH1 (r1) phenotype was not expressed in precancerous or cancerous urothelia. The hMSH6 mRNA was the most changed in UCCs (47.8%), while hMSH2, hMLH1 and hPMS2 showed overexpression (47.8, 35 and 30%, respectively) that was associated with gender and histological tumor grading or staging. Genetic instability was rare in polymorphic regions distal to hMLH1. Our data reveal a previously unrecognized hMSH2 and hMSH6 mRNA combined phenotype (r2r6) correlated with a precancerous urothelium and show that hMLH1 is transcriptionally activated in precancerous or cancerous urothelium. In the present study, it is demonstrated that reduction of hMSH6 mRNA is a frequent event in bladder tumorigenesis and reflects a common mechanism of suppression with hMSH2, while alterations of hMSH2 or hMLH1 mRNA expression in UCCs does not correlate with the allelic imbalance of polymorphic regions harboring the genes.
Keywords: bladder tumors, urothelial cell carcinoma, papillary urothelial neoplasm of low malignant potential, normal urothelium, hMSH2, hMLH1, hPMS2, hMSH6, quantitative real-time PCR
Introduction
The most common histological type of bladder cancer is urothelial cell carcinoma (UCC) or transitional cell carcinoma (TCC). Papillary urothelial neoplasm of low malignant potential (PUNLMP) may also arise from urothelium of bladder (1,2). The urothelium of a patient with a bladder cancer is at risk as the cancer often recurrs in the urinary bladder following treatment (1,3). Numerous genetic and epigenetic factors have been implicated in the carcinogenesis of the urinary bladder that involved in its mutator phenotype (4–7). The DNA repair mechanism is essential to prevent DNA mutations that may be lethal for cells (8). Mismatch repair (MMR) genes encode a number of DNA repair enzymes that cooperate to recognize and repair DNA mismatches (8,9). These enzymes act as complexes. A crucial complex that recognizes base-base mismatches is the MutSα, which consists of MSH2 and MSH6 components. MutLα is another MMR complex, consisting of MLH1 and PMS2 components that cooperate with MutSα and other enzymes to repair the damage (10–14). DNA repair dysfunction may allow the generation of a high-risk urothelium for malignant transformation in the urinary bladder. The dysfunction of MMR genes may present as an absence or reduction of MMR gene expression or microsatellite instability (MSI) phenotype (15–17). The protein expression levels of the hMSH2, hMLH1 and hMSH6 MMR genes have been detected in histopathological material of UCC specimens by immunohistochemistry (IHC) with controversial results (17–24). There is little and insufficient literature concerning the expression of the mRNA of MMR genes in bladder cancer specimens (25,26). In the present study, we evaluated for the first time, by a precise quantitative real-time PCR (qPCR) analysis, the mRNA expression levels of the hMSH2, hMLH1, hMSH6 and hPMS2 MMR genes in surgical samples of bladder tumors paired with their corresponding adjacent normal tissues (ANTs). We also present the MMR phenotypes of reduced or elevated mRNA expression that were correlated with a high risk of malignant transformation of urothelium and/or tumor progression in the urinary bladder.
Materials and methods
Tissue collection and patients
Paired surgical specimens from primary bladder tumors and their ANTs were collected from 25 unselected patients who underwent surgery in the University Hospital of Alexandroupolis, Greece, after obtaining informed consent. The Ethics Committees of the University of Thessaly, Department of Pathology, Medical School of Larissa, Larissa, Greece and the Democritus University of Thrace, Departments of Urology and Pathology and University Hospital of Alexandroupolis, Alexandroupolis, Greece approved this study.
The clinical material was frozen at −80°C and further subdivided for standard histological evaluation, DNA and RNA extraction. Tumor content >80% was recorded in all specimens studied. The histological review according to conventional guidelines (WHO/ISUP classification) revealed 24 UCCs and one PUNLMP in our clinical material. The UCCs further revealed 13 low grade tumors (6 in stage pTa and 7 in pT1) and 11 high grade tumors (1 in stage pTa, 9 in pT1 and 1 in pT2; Table I).
Table I.
Quantitative mRNA expression of hMSH2, hMLH1, hMSH6 and hPMS2 in bladder tumors and their adjacent normal tissue.
| Patient no./age (years)/gender | Tumor type | Tumor grade | pTNM classification |
hMSH2/control mRNAa
|
hMLH1/control mRNAa
|
hMSH6/control mRNAa
|
hPMS2/control mRNAa
|
Tumor/ANT MMR mRNA expressionb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | ANT | Tumor | ANT | Tumor | ANT | Tumor | ANT | hMSH2 | hMLH1 | hMSH6 | hPMS2 | ||||
| 1/70/M | PUNLMP | - | pTa | 0.46 | 2.9 | 0.22 | 0.24 | 0.74 | 1.1 | 1.59 | 3.87 | 0.16 | 0.92 | 0.67 | 0.4 |
| 2/50/M | UCC | LG | pTa | 10.2 | 1.58 | 14.7 | 4.6 | 90 | 16 | 24.4 | 7.8 | 6.5 | 3.2 | 5.6 | 3 |
| 3/59/M | UCC | LG | pTa | 3.2 | 0.73 | 5.6 | 1.3 | 8.64 | 15.4 | 13.25 | 26.52 | 4.4 | 0.56 | 0.5 | 4.3 |
| 4/73/M | UCC | LG | pTa | 2 | 0.82 | 12 | 4.45 | 1.7 | 0.88 | 1.7 | 1.67 | 2.4 | 1.9 | 0.8 | 2.7 |
| 5/75/M | UCC | LG | pTa | 4.12 | 2.9 | 8.29 | 19.25 | 59 | 47 | 8.3 | 16.82 | 1.97 | 0.43 | 1.24 | 0.4 |
| 6/75/M | UCC | LG | pTa | 1.26 | 6.22 | 8 | 7.7 | 21.9 | 64 | 24.3 | 15.8 | 0.2 | 1.04 | 0.34 | 1.5 |
| 7/79/F | UCC | LG | pTa | 1.56 | 1.14 | 2.58 | 2.26 | 2.27 | 1.9 | 4.7 | 1.5 | 1.36 | 1.19 | 3 | 1.13 |
| 8/56/M | UCC | LG | pT1 | 0.4 | 0.54 | 1.89 | 2.4 | 0.44 | 0.55 | 0.86 | 0.83 | 0.73 | 0.8 | 1 | 0.79 |
| 9/69/M | UCC | LG | pT1 | 1.66 | 0.6 | 2.65 | 1.96 | 0.9 | 0.6 | 1.1 | 0.69 | 2.8 | 1.5 | 1.6 | 1.4 |
| 10/69/M | UCC | LG | pT1 | 1.22 | 0.88 | 8.76 | 4.72 | 2.5 | 0.8 | 2.5 | 1.8 | 1.39 | 3.13 | 0.13 | 1.86 |
| 11/73/F | UCC | LG | pT1 | 9.4 | 23.5 | 38 | 38 | 3.2 | 3.4 | 5.9 | 5.9 | 0.4 | 1 | 1 | 1 |
| 12/76/M | UCC | LG | pT1 | 3.97 | 1.43 | 5.4 | 2.49 | 1.73 | 0.76 | 7.29 | 136.27 | 2.8 | 2.28 | 0.5 | 2.2 |
| 13/82/M | UCC | LG | pT1 | 7.5 | 13.8 | 28 | 25 | 1.8 | 1.68 | 17.8 | 7.14 | 0.5 | 1 | 0.2 | 1.12 |
| 14/82/M | UCC | LG | pT1 | 1.8 | 1.45 | 1.87 | 2.36 | 2.2 | 2.4 | 2.29 | 2.78 | 1.3 | 0.9 | 0.8 | 0.79 |
| 15/72/F | UCC | HG | pTa | 0.74 | 0.5 | 2.38 | 4.25 | 0.62 | 0.47 | 0.7 | 0.57 | 1.48 | 1.3 | 1.2 | 0.56 |
| 16/54/M | UCC | HG | pT1 | 3.1 | 1 | 5.58 | 2.26 | 5.8 | 0.79 | 37.8 | 20.39 | 2.6 | 6.8 | 1.85 | 2.5 |
| 17/62/M | UCC | HG | pT1 | 3.78 | 4.29 | 25.73 | 15.78 | 39.7 | 63.4 | 27.4 | 21.4 | 0.87 | 1.6 | 0.6 | 1.6 |
| 18/75/M | UCC | HG | pT1 | 8 | 0.55 | 2.83 | 2.31 | 5.4 | 1 | 47.6 | 16 | 14.55 | 5.4 | 2.9 | 1.22 |
| 19/76/M | UCC | HG | pT1 | 0.49 | 0.7 | 1.65 | 1.8 | 0.73 | 2.2 | 2.3 | 9.8 | 0.57 | 0.33 | 5 | 0.9 |
| 20/83/M | UCC | HG | pT1 | 1.4 | 0.4 | 3 | 5.7 | 2.5 | 2.2 | 3.8 | 5.6 | 3.5 | 1.14 | 0.7 | 0.53 |
| 21/84/M | UCC | HG | pT1 | 0.75 | 0.6 | 1.42 | 1.67 | 0.87 | 0.67 | 1.12 | 6.5 | 1.2 | 1.3 | 0.17 | 0.85 |
| 22/90/F | UCC | HG | pT1 | 0.16 | 1.03 | 1.08 | 2.6 | 0.12 | 0.65 | 0.8 | 104 | 0.15 | 0.19 | 0.001 | 0.4 |
| 23/72/M | UCC | HG | pT1 | 21.66 | 5 | 20 | 12.3 | 130 | 59 | 37 | 24.7 | 4.3 | 14 | 2.2 | 1.5 |
| 24/75/M | UCC | HG | pT2 | 1.63 | 0.7 | 7 | 2.1 | 2.19 | 0.74 | 35.7 | 1.8 | 2.3 | 2.9 | 19.9 | 3.3 |
| 25/68/M | UCC | HG | pT1 | NA | NA | NA | NA | 0.37 | NA | 11.5 | NA | - | - | - | - |
ANT, adjacent normal tissue; MMR, mismatch repair; M, male; F, female; PUNLMP, papillary urothelial neoplasm low malignant potential; UCC, urothelial cell carcinoma; LG, low grade; HG, high grade; pTa, limited to mucosa; pT1, lamina propria invasion; pT2, invasion of the muscularis; NA, non-amplified sample.
Ratios of mRNA expression;
ratios of tumor to ANT mRNA expression.
The cohort of patients included 20 males and 4 females with UCC and 1 male with a PUNLMP with an age range of 50–90 years (median, 74; Table I).
Quantitative analysis of mRNA expression
We used Purescript® RNA isolation and SuperScript First Stand Synthesis System (Invitrogen®, Life Technologies, Paisley, UK) for cDNA synthesis, by reverse transcription (RT), as described previously (27). qPCR analysis of hMSH2 and hMLH1 mRNA was performed as previously described (16). qPCR analysis of hMSH6 and hPMS2 was performed using specific primers: hMSH6 sense, 5′-AACAAGGGGCTGGGTTAG-3′; hMSH6 antisense, 5′-CGTTGCATTGCTCTCAGTATTTC-3′; hPMS2 sense, 5′-GAGTCAAGCAGATGTTTGCCTC-3′; hPMS2 anti-sense, 5′-TGTGTCTCATGGTTGGCCTT-3′; and fluorescent hybridization probes hMSH6 - F L, 5′-TATACA GGTTCAAAATCAAAGGAAGCCC-FL; hMSH6-LC, 5′-LC640-GAAGGGAGGTCATTTTTACAGTGCAAG-PH; hPMS2-FL, 5′-GGGTGATCAGTTTCTTCATCTCGCTTGT-FL; hPMS2-LC, 5′-LC640-TTAAGAGCAGTCCCA ATCATCACCGACTT-PH designed for Light Cycler instrument 3.5 (TIB® Molbiol, Berlin, Germany). All reactions included Porphobilionogen deaminase (hPBGD) housekeeping gene primers as internal controls (Roche Diagnostics, Mannheim, Germany). Following an initial denaturation step at 95°C for 10 min, hMSH6-PCR assays underwent 45 cycles of denaturation at 95°C for 10 sec, annealing at 54.2°C for 15 sec and extension at 72°C for 6 sec and hPMS2-PCR assays underwent 45 cycles of denaturation at 95°C for 10 sec, annealing at 57°C for 10 sec and extension at 72°C for 6 sec. The mRNA expression of each MMR gene was expressed as a ratio of MMR gene mRNA to control hPBGD mRNAs (MMR/control mRNAs) and defined two major phenotypic groups, the reduced (r) for mRNA ratios <1 and the normal or elevated (R) for ratios ≥1, as previously described (16). Additionally, the MMR gene expression of tumor samples was compared with that of the corresponding ANT samples. This value is indicated as relative mRNA expression of MMR genes between tumor and ANT (tumor/ANT) of each patient (Table I).
Genomic instability analysis
Genomic instability analysis was performed for the following polymorphic regions of the hMSH2 and hMLH1 loci: D3S1234 (3p14), D3S1612 (3p21.3–22) and D3S1768 (3p21.3–22) distal to the hMLH1 locus on chromo-some 3p and D2S1788 (2p22.3) proximal to the hMSH2 locus on chromosome 2p, to compare the possible loss of mRNA expression with allelic imbalance of the chromosomal regions that contain the genes (28). The primer sequences for each microsatellite copy were obtained from the National Center for Biotechnology Information database. Nucleotide repeat markers, stretches within non-coding repeats such as BAT26 in intron 5 of hMSH2 and BAT25 in intron 16 of c-kit were used as established mononucleotide markers for determining MSI status (29,30).
MSI analysis was performed as previously described (31). Briefly, following DNA extraction from bladder tissue specimens (Puregene® Cell and Tissue extraction kit; Gentra), genomic DNA samples were stored at −20°C until use. PCR analysis included amplification of the β-globin gene in order to qualify and normalize the amount of DNA in each sample. The primers used for the amplification of the β-globin gene, PCR, qPCR and melting curve analysis conditions were as previously described (31). All samples were run in duplicate and two non-template-controls (NTCs) were included in the reactions. qPCR amplifications and melting curve analyses were repeated twice. The conditions of reactions were 95°C for 15 min, 36 cycles of 95°C for 15 sec, annealing temperature for each set of primers (55°C for D3S1768, D2S1788, BAT25; 56°C for D3S1612; 56.5°C for BAT26; and 58.5°C for D3S1234) for 30 sec and 72°C for 30 sec (acquiring for SYBR). Continuously Melting Curve analysis performed ramping 65–95°C (raising by 0.2°C each step) and finishing at 72°C for 5–10 min. Following completion of the amplification melting curve, analysis was performed as previously described (31).
Statistical analysis
We used the paired Student’s t-test to compare ratios of hMSH2, hMLH1, hMSH6 and hPMS2 alterations between tumor and matched ANT for different patient characteristics, including age, gender and clinicohistopathological parameters such as tumor type, grade and stage. The correlation between the mRNA expression ratios of hMSH2, hMLH1, hMSH6 and hPMS2 in bladder tumors and their ANTs for different patient and tumor characteristics was examined by Pearson test. The χ2 test was also used to examine the distribution of MMR mRNA phenotypes (r, R, rr, rR, Rr and RR) in tumor and ANT specimens at different tumor histopathological grades or stages. Statistical significance was considered for values of p<0.05.
Results
hMSH2, hMLH1, hMSH6 and hPMS2 mRNA quantification in bladder tumors
We evaluated hMSH2, hMLH1, hMSH6 and hPMS2 mRNA levels in primary UCCs and their corresponding ANTs relative to the reference hPBGD control gene by qPCR (Fig. 1). These data are summarized in Table I with patient age, gender, tumor type, stage and grade.
Figure 1.
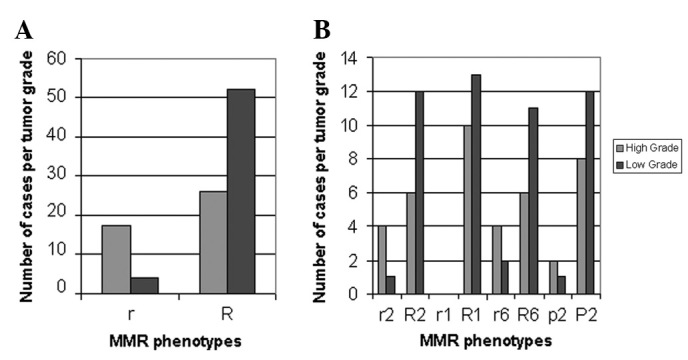
(A) Relative distribution of reduced (r/p) to normal or elevated (R/P) MMR phenotypes to histological tumor grades in UCCs. (B) Relative distribution of each hMSH2 (r2/R2), hMLH1 (R1), hMSH6 (r6/R6) and hPMS2 (p2/P2) mRNA phenotypes to histological tumor grades in UCCs. MMR, mismatch repair; UCC, urothelial cell carcinoma.
The urothelium adjacent to UCCs revealed reduced mRNA ratios (<1) of the hMSH2, hMSH6 and hPMS2 genes in 47.8 (11/23), 43.5 (10/23) and 13% (3/23) of samples, respectively, compared with 21.7 (5/23), 29.2 (7/24) and 12.5 (3/24) of UCC tumors (P= 0.027284). We observed a statistically significant difference between the proportions of reduced mRNA ratios of hMSH2/control and hMSH6/control genes observed in ANTs of low grade UCCs relative to their tumors (P=0.025347) that was more pronounced than those observed between ANTs and high grade UCCs (P= 0.000; Table I). Moreover, high grade UCCs exhibited higher proportions of reduced mRNA ratios of hMSH2/control, hMSH6/control and hPMS2/control (40, 40 and 20%, respectively) compared with low grade tumors (7.6, 15.3 and 7.6%, respectively; P=0.000).
The hMLH1/control mRNA ratios were ≥1 in UCCs and their ANTs, contrary to PUNLMP and its corresponding ANT that exhibited reduced mRNA ratios (<1). PUNLMP also showed reduced (<1) hMSH2/control and hMSH6/control mRNA ratios, while its corresponding ANT exhibited elevated or normal (≥1) ratios.
hMSH2, hMLH1, hMSH6 and hPMS2 mRNA relative expression
Calculation of tumor/ANT MMR mRNA ratios from quantification data (Table I) revealed different proportions of UCC ratios ≤0.8 with 26.1% (6/23) for hMSH2, 21.7% (5/23) for hMLH1, 47.8% (11/23) for hMSH6 and 26% (6/23) for hPMS2, respectively. Calculation of PUNLMP/ANT MMR mRNA revealed ratios ≤0.8 of hMSH2, hMSH6 and hPMS2 (Table I). The reduction in hMSH2, hMLH1, hMSH6 and hPMS2 mRNA expression between primary bladder tumors and their matched ANTs, male and female, pTa stage and pT1, high and low grade UCC was not statistically significant by paired Student’s t-test analysis (Table II).
Table II.
Alterations of hMSH2, hMLH1, hMSH6 and hPMS2 mRNA levels between paired bladder tumor and adjacent normal tissues relative to their clinicopathological parameters.
| Relative copies of hMSH2 mRNA
|
Relative copies of hMLH1 mRNA
|
Relative copies of hMSH6 mRNA
|
Relative copies of hPMS2 mRNA
|
Tumor/ANT MMR mRNA gene expression
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | n | Tumor | ANT | Tumor | ANT | Tumor | ANT | Tumor | ANT | hMSH2 | hMLH1 | hMSH6 | hPMS2 |
| All patients | 24 | 3.77a,b | 3.06 | 8.69e | 7 | 16 | 12 | 12.9j | 18.34 | 2.4n | 2.4 | 2.19 | 1.52 |
| Gender | |||||||||||||
| Male | 20 | 3.93c | 2.37 | 8.23f | 6 | 19 | 14 | 14.9k | 16.41 | 2.75o | 2.79 | 2.43 | 1.68 |
| Female | 4 | 2.96 | 6.54 | 11 | 12 | 1.6 | 1.6 | 3.03 | 28 | 0.85p,v | 0.9w,y | 1.3z | 0.77 |
| Tumor type | |||||||||||||
| PUNLMP | 1 | 0.46 | 2.9 | 0.22 | 0.2 | 0.74 | 1.1 | 1.59 | 3.87 | 0.16 | 0.67 | 0.4 | 0.9 |
| UCC | 23 | 3.9 | 3.07 | 9.06 | 7.3 | 16.7 | 12.5 | 13.42 | 18.97 | 2.5q | 2.66 | 2.33 | 1.55 |
| Tumor stage | |||||||||||||
| Ta | 7 (6LG+1HG) | 3.3d | 1.98 | 7.65 | 6.26 | 26.3 | 20.8 | 11.05 | 10.1 | 2.62r | 1.73x | 1.5 | 2.8 |
| T1 | 15 (7LG+8HG) | 4.35 | 3.73 | 9.86g | 8.09 | 13.13 | 9.34 | 13.04l | 24.25 | 2.51s | 1.89 | 1.24 | 2.07 |
| T2 | 1 (HG) | 1.63 | 0.7 | 7 | 2.1 | 2.19 | 0.74 | 35.7 | 1.8 | 2.3 | 2.9 | 19.9 | 3.3 |
| Tumor grade | |||||||||||||
| LG | 13 (6Ta+7T1) | 3.71 | 4.27 | 10.6h | 8.96 | 15.1 | 12 | 8.8 | 17.35 | 2.06t | 1.64 | 1.11 | 2.17 |
| HG | 10 (1Ta+9T1) | 4.17 | 1.5 | 7.07i | 5.08 | 18.79 | 13.1 | 19.42m | 21.08 | 3.15u | 2.21 | 3.45 | 2.59 |
Mean age of the patients was 72 years. ANT, adjacent normal tissue; MMR, mismatch repair; PUNLMP, papillary urothelial neoplasm of low malignant potential; UCC, urothelial cell carcinoma; LG, low grade; HG, high grade.
r=0.795, by Pearson test; correlation between hMSH2/hPBGD and hMSH6/hPBGD mRNA ratios.
P=0.019165,
P=0.020265,
P=0.048544,
P=0.000,
P=0.000366,
P=0.012455,
P=0.012,
P=0.032,
P=0.005896,
P=0.005159,
P=0.038387,
P=0.028778; by Student’s t-test.
r=0.415684,
r=0.37,
r=0.873155,
r=0.44,
r=0.778713,
r=0.613262,
r=0.700724,
r=0.598678; by Pearson test, correlation between hMSH2 and hMLH1 mRNA ratios of tumor/ANT.
r=0.735679 by Pearson test, correlation between hMSH2 and hMSH6 mRNA ratios of tumor/ANT.
r=0.728012,
r=0.567703 by Pearson test, correlation between hMLH1 and hMSH6 mRNA ratios of tumor/ANT.
r=0.560746 Pearson test, correlation between hMLH1 and hPMS2 mRNA ratios of tumor/ANT.
r=0.801165 Pearson test, correlation between hMSH6 and hPMS2 mRNA ratios of tumor/ANT.
Bladder tumors revealed mRNA overexpression ratios (≥1.8) of hMSH2, hMLH1, hMSH6 and/or hPMS2 compared with their corresponding ANTs in 47.8 (11/23), 35 (8/23), 30 (7/23) and 30% (7/23) of specimens, respectively (Table I).
The statistically significant difference of hMSH2 transcriptional levels between UCC tumors and their matched ANTs (P=0.019165; Student’s t-test) was more pronounced in males compared with females (males, P=0.020265; females, P= 0.169501; Student’s t-test) and was independent of stage (pTa, P=0.127745; pT1, P=0.089642; Student’s t-test; Table II). Low grade UCCs exhibited a statistically significant elevation of hMSH2 mRNA expression relative to their matched ANTs (low grade, P=0.048544; high grade, P=0.130441; Student’s t-test; Table II).
We observed significant difference in hMLH1 transcriptional activation between bladder tumors and their matched ANTs (P= 0.000; Student’s t-test; Table II). Notably, males showed a higher hMLH1 transcriptional activation in tumors compared with females (males, P=0.000366; females, P=0.5; Student’s t-test; Table II). We also observed statistically significantly higher levels of hMLH1 mRNA expression between UCC tumors and their matched ANTs of pT1 stages than pTa (pT1, P= 0.012455; pTa, P=0.081273; Student’s t-test; Table II). Low grade UCCs compared with their matched ANTs exhibited relatively higher mRNA expression levels of hMLH1 than high grade UCCs (low grade, P=0.012; high grade, P=0.032; Student’s t-test; Table II).
The statistically significantly elevated mRNA expression of hPMS2 observed between UCC tumors and their matched ANTs (P= 0.005896; Student’s t-test) was also identified in males compared with females (males, P=0.005159; females, P=0.474152; Student’s t-test), pT1 UCCs (pT1, P= 0.038387; pTa, P=0.143226; Student’s t-test) and was pronounced in high grade compared with low grade tumors (high grade, P=0.028778; low grade, P=0.05819; Student’s t-test; Table II).
Correlation between mRNA expression of the hMSH2, hMLH1, hMSH6 and hPMS2 MMR genes in bladder tumors
We observed a statistically significant correlation between hMSH2/hPBGD and hMSH6/hPBGD mRNA ratios in bladder tumors (r=0.795; Pearson test; Table II).
We also observed a statistically significant association between the relative mRNA expression of hMSH2 and hMLH1 (tumor/ANT) expression (r=0.415684; Pearson test) that was more pronounced in females compared with males (females, r= 0.873155; males, r= 0.37; Pearson test), slightly more intense in pTa than pT1 stages (pTa, r= 0.778713; pT1, r=0.61326; Pearson test) and in low grade than high grade UCCs (low grade, r=0.700724; high grade, r=0.598678; Pearson test; Table II). Only females exhibited a statistically significant association of relative mRNA expression between hMSH2 and hMSH6 (r=0.735679, Pearson test), hMLH1 and hPMS2 (r= 0.560746; Pearson test) and hMSH6 and hPMS2 (r= 0.801165; Pearson test; Table II). In addition, females and pTa UCCs exhibited a significant association between hMLH1 and hMSH6 relative (tumor/ANT) levels of mRNA expression (female, r=0.728012; pTa, r=0.567703; Pearson test; Table II).
Phenotyping MMR sorting
We used the ratio of MMR mRNA expression relative to reference mRNA control to adopt a functional unified assessment for our findings, as previously described (16). We classified our specimens into two major phenotypic groups, one with reduced (r) and the other with regular or enhanced (R) ratios of expression (Materials and methods) and subdivided our study group into eight phenotypes, r2 and R2 for hMSH2, r1 and R1 for hMLH1, r6 and R6 for hMSH6 and finally p2 and P2 for hPMS2 DNA repair system components (Table III) or their combined phenotypes R2R1, R2R6, R2P2, R1R6, R1P2, R6P2 and R2r1, R2r6, R2p2, R1r6, R1p2, R6p2 and r2R1, r2R6, r2P2, r1R6, r1P2, r6P2 and r2r1, r2r6, r2p2, r1r6, r1p2, r6p2 by descending MMR system activity.
Table III.
Distribution of individual hMSH2, hMLH1, hMSH6 and hPMS2 mRNA phenotypes in UCCs and their ANTs.
| MMR mRNA phenotype | UCC (n) | UCC observed phenotypic frequency | Grade (n)
|
Stage (n)
|
ANT (n) | ANT observed phenotypic frequency | Samples (n)
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| LG | HG | pTa | pT1–2 | PUNLMP | ANT | |||||
| hMSH2 | ||||||||||
| r2 | 5 | 0.2174 | 1 | 4 | 1 (HG) | 4 (1LG+3HG) | 11 | 0.4783 | 1 | |
| R2 | 18 | 0.7826 | 12 | 6 | 6 (LG) | 12 (6LG+6HG) | 12 | 0.5217 | 1 | |
| hMLH1 | ||||||||||
| r1 | 0 | 0.000 | 0 | 0 | 0 | 0 | 0 | 0.000 | 1 | 1 |
| R1 | 23 | 1.000 | 13 | 10 | 7 (1LG+6HG) | 16 (7LG+9HG) | 23 | 1.000 | ||
| hMSH6 | ||||||||||
| r6 | 6 | 0.2609 | 2 | 4 | 1 (HG) | 5 (2LG+3HG) | 10 | 0.4348 | 1 | |
| R6 | 17 | 0.7391 | 11 | 6 | 6 (LG) | 11 (5LG+6HG) | 13 | 0.5652 | 1 | |
| hPMS2 | ||||||||||
| p2 | 3 | 0.1304 | 1 | 2 | 1 (HG) | 2 (1LG+1HG) | 3 | 0.1304 | ||
| P2 | 20 | 0.8696 | 12 | 8 | 6 (LG) | 14 (6LG+8HG) | 20 | 0.8696 | 1 | 1 |
ANT, adjacent normal tissue; MMR, mismatch repair; UCC, urothelial cell carcinoma; LG, low grade; HG, high grade; PUNLMP, papillary urothelial neoplasm of low malignant potential; pTa, limited to mucosa; pT1, lamina propria invasion; pT2, invasion of the muscularis; r/p, reduced, mRNA ratio <1; R/P, normal/elevated, mRNA ratio ≥1; r/R2, hMSH2; r/R1, hMLH1; r/R6, hMSH6; p/P2, hPMS2.
Clinical and biological evaluation of single and combined MMR phenotypic distributions
We tested the ability of our phenotypes to distinguish our study group into distinct bladder tumors and ANTs subtypes. We thus examined the significance of the differences between the subgroups created according to our phenotypic criteria (Table III, Fig. 1).
The distribution of individual r and R MMR mRNA phenotypes was significantly different between ANTs and UCCs (P=0.021751; χ2 test), particularly r2 vs. R2 (P= 0.012261; χ2 test). Additionally, a marked difference of r and R phenotypic distribution was observed between high and low grade UCCs (P= 0.00013; χ2 test; Fig. 1A), particularly r2 vs. R2 (P=0.00053, χ2 test) and r6 vs. R6 (P<0.04, χ2 test; Fig. 1B). The frequencies of independent r2, r1, r6, p2, R2, R1, R6 and P2 phenotypes in UCCs and their corresponding ANTs are shown in Table III. The reduced r1 phenotype was not identified in any UCC or corresponding ANTs, in contrast to the PUNLMP and its ANT (Table III).
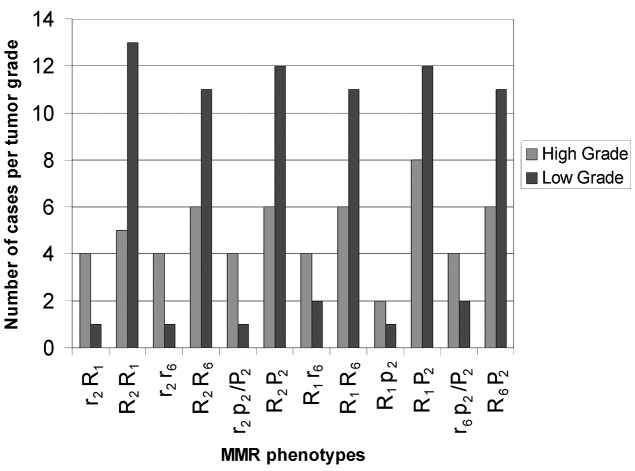
The distribution of combined MMR mRNA phenotypes in UCCs and their matched ANTs are shown in Table IV. We observed two r2R1 and R2R1 combined hMSH2 and hMLH1 mRNA phenotypes, two R1p2 and R1P2 combined hMLH1 and hPMS2 phenotypes and two R1r6 and R1R6 combined hMLH1 and hMSH6 mRNA phenotypes in UCCs and/or their ANTs with same frequencies between observed and calculated frequencies of combined loci. We also observed four R2R6, r2R6, R2r6 and r2r6 combined hMSH2 and hMSH6, three R2P2, r2P2 and r2p2 combined hMSH2 and hPMS2 and three R6P2, r6P2 and r6p2 combined hMSH6 and hPMS2 mRNA phenotypes in UCCs and/or their ANTs with different frequencies between observed and calculated frequencies of combined loci. There was a statistically significant difference between high and low grade tumors in r2R1 vs. R2R1 (P=0.00019; χ2 test), r2r6 vs. R2R6 (P=0.000786; χ2 test), r2p2/P2 vs. R2P2 (P=0.00053, χ2 test) and less marked in R1r6 vs. R1R6 (P<0.04; χ2 test) and in r6p2/P2 vs. R6P2 (P<0.04, χ2 test). The histogram in Fig. 2 shows the association of reduced (homozygous or heterozygous) r2R1 or r2r6 or r2p2/P2 or R1r6 or r6p2/P2 and normal or elevated (homozygous) R2R1 or R2R6 or R2P2 or R1R6 or R6P2 to high and low grades, respectively (Table IV and Fig. 2).
Table IV.
Distribution of combined hMSH2, hMLH1, hMSH6 and hPMS2 mRNA phenotypes in UCCs and their ANTs.
| Tissue | R2R1 | r2R1 | R2r1 | r2r1 | R6P2 | r6P2 | r6p2 | R6p2 | R2R6 | r2R6 | R2r6 | r2r6 |
|
| ||||||||||||
| UCCs | 18 | 5 | 0 | 0 | 17 | 3 | 3 | 0 | 17 | 0 | 1 | 5 |
| Observed frequencies | 0.7826 | 0.2714 | 0.000 | 0.000 | 0.7391 | 0.1304 | 0.1304 | 0.0000 | 0.7391 | 0.000 | 0.0435 | 0.2174 |
| Calculated phenotypic frequencies of combined loci | 0.7826 | 0.2714 | 0.000 | 0.000 | 0.6427 | 0.2269 | 0.0340 | 0.0964 | 0.5784 | 0.2006 | 0.2042 | 0.0708 |
| Grade | ||||||||||||
| LG | 13 | 1 | 0 | 0 | 11 | 1 | 1 | 0 | 11 | 0 | 1 | 1 |
| HG | 5 | 4 | 0 | 0 | 6 | 2 | 2 | 6 | 0 | 0 | 4 | |
| Stage | ||||||||||||
| pTa | 6 | 1 | 0 | 0 | 6 | 0 | 1 | 0 | 6 | 0 | 0 | 1 |
| pT1–2 | 12 | 4 | 0 | 0 | 11 | 3 | 2 | 0 | 11 | 0 | 1 | 4 |
| ANT | 14 | 9 | 0 | 0 | 14 | 6 | 3 | 0 | 10 | 4 | 4 | 5 |
| Observed frequencies | 0.6087 | 0.3913 | 0.000 | 0.000 | 0.6087 | 0.2609 | 0.1304 | 0.000 | 0.4348 | 0.1739 | 0.1739 | 0.2174 |
|
| ||||||||||||
| R2P2 | r2P2 | R2p2 | r2p2 | R1P2 | r1P2 | R1p2 | r1p2 | R1R6 | r1R6 | R1r6 | r1r6 | |
|
| ||||||||||||
| UCCs | 18 | 2 | 0 | 3 | 20 | 0 | 3 | 0 | 17 | 6 | 0 | |
| Observed frequencies | 0.7826 | 0.087 | 0.0000 | 0.1304 | 0.8696 | 0.000 | 0.1304 | 0.000 | 0.7391 | 0.000 | 0.2609 | 0.000 |
| Calculated phenotypic frequencies of combined loci | 0.6806 | 0.236 | 0.1021 | 0.0354 | 0.8696 | 0.000 | 0.1304 | 0.000 | 0.7391 | 0.000 | 0.2609 | 0.000 |
| Grade | ||||||||||||
| LG | 12 | 0 | 0 | 1 | 12 | 0 | 1 | 0 | 11 | 0 | 2 | 0 |
| HG | 6 | 2 | 0 | 2 | 8 | 0 | 2 | 0 | 6 | 0 | 4 | 0 |
| Stage | ||||||||||||
| pTa | 6 | 0 | 0 | 1 | 6 | 0 | 1 | 0 | 6 | 0 | 1 | 0 |
| pT1–2 | 12 | 2 | 0 | 2 | 14 | 0 | 2 | 0 | 11 | 0 | 5 | 0 |
| ANT | 14 | 6 | 0 | 3 | 20 | 0 | 3 | 0 | 14 | 0 | 9 | 0 |
| Observed frequencies | 0.6087 | 0.2609 | 0.000 | 0.1304 | 0.8696 | 0.000 | 0.1304 | 0.000 | 0.6087 | 0.000 | 0.3913 | 0.000 |
UCC, urothelial cell carcinoma; ANT, adjacent normal tissue; LG, low grade; HG, high grade; pTa, limited to mucosa; pT1, lamina propria invasion; pT2, invasion of the muscularis; r/p, reduced, mRNA ratio <1; R/P, normal/elevated, mRNA ratio ≥1; r/R2, hMSH2; r/R1, hMLH1; r/R6, hMSH6; p/P2, hPMS2.
Figure 2.
Relative distribution of combined hMSH2, hMLH1, hMSH6 and hPMS2 mRNA phenotypes to histological tumor grades in UCCs. r/p, reduced, mRNA ratio <1; R/P, normal/elevated, mRNA ratio ≥1; r/R2, hMSH2; r/R1, hMLH1; r/R6, hMSH6; p/P2, hPMS2; MMR, mismatch repair; UCC, urothelial cell carcinoma.
A statistically significant difference was also observed between R2R6 and r2r6 phenotypic distribution in UCCs relative to their ANTs (P<0.02; χ2 test) that was pronounced in low compared with high grade tumors (P=0.0000006; χ2 test; Table IV and Fig. 3).
Figure 3.
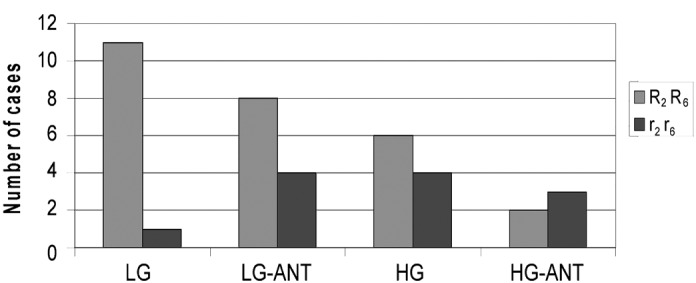
Relative distribution of cases with combined reduced (r2r6) or normal or elevated (R2R6) hMSH2 and hMSH6 mRNA phenotypes to low or high grade UCCs (LG, HG) and their matched ANTs (LG-ANT, HG-ANT). UCC, urothelial cell carcinoma; ANT, adjacent normal tissue.
Genomic instability
We examined 6 genetic markers for genomic instability [MSI and/or loss of heterozygosity (LOH)] in bladder tumors (Table V) distal or close to hMLH1 and hMSH2 to determine the correlation between possible loss of mRNA expression and allelic imbalance of the chromosomal regions that harbor the genes. D2S1788 (2p22.3) and BAT26 are located distal to and in the hMSH2 locus, respectively, while D3S1612 (3p21.3–22), D3S1768 (3p21.3–22) and D3S1234 (3p14) are distal to the hMLH1 locus. Additionally, BAT25 stretches within the c-kit gene and was included as it has been previously correlated with a DNA repair mechanism (29,30).
Table V.
Genetic alterations in UCCs using melting curve analysis.
| Case no.,T/N | Polymorphic markers at the 3p loci (distal to hMLH1)
|
Polymorphic markers at the 2p loci (within or distal to hMSH2)
|
Polymorphic marker related to MSI BAT25
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
D3S1234
|
D3S1768
|
D3S1612
|
BAT26
|
D2S1788
|
||||||
| aPeak | Peak 1 | Peak 2 | Peak 1 | Peak 2 | Peak 1 | Peak 1 | Peak 2 | Peak 3 | Peak 1 | |
| 1 | ||||||||||
| T | 77.7 | 73.5 | 75.6 | 76.2 | 73.8 | 73.5 | 72.5 | |||
| N | 77.8 | 73.7 | 75.6 | 76.2 | 73.8 | 73.5 | 72.5 | |||
| 2 | ||||||||||
| T | NA | NA | NA | NA | NA | NA | ||||
| N | NA | NA | NA | NA | NA | NA | ||||
| 3 | ||||||||||
| T | 77 | 73.3 | 76.4 | 73.7 | NA | 72.6 | ||||
| N | 77 | 73.3 | NA | NA | NA | 72.5 | ||||
| 4 | ||||||||||
| T | 76.8 | 73 | 76.4 | 73.7 | 72.3 | 73.5 | NA | |||
| N | 76.7 | 73.3 | 75.5 | 76.2 | 73.9 | NA | NA | NA | ||
| 5 | ||||||||||
| T | NA | NA | NA | NA | NA | NA | ||||
| N | NA | NA | NA | NA | NA | NA | ||||
| 6 | ||||||||||
| T | NA | NA | NA | NA | NA | NA | ||||
| N | NA | NA | NA | NA | NA | NA | ||||
| 7 | ||||||||||
| T | 77.5 | 73.5 | 75.7 | 76.3 | 73.7 | 73.5 | 72.75 | |||
| N | 77.5 | 73.5 | 75.7 | 76.3 | 73.7 | 73.5 | 72.75 | |||
| 8 | ||||||||||
| T | 77.4 | 73.3 | 75.6 | 73.7 | 73.4 | 72.8 | ||||
| N | 77.5 | 73.5 | 75.6 | 73.7 | 73.4 | 72.8 | ||||
| 9 | ||||||||||
| T | 77.6 | 73.2 | 76.4 | 73.8 | 73.5 | 74.5 | 72.7 | |||
| N | 77.5 | 73.5 | 76.4 | 73.8 | 73.5 | 74.5 | 72.6 | |||
| 10 | ||||||||||
| T | 76.8 | 73.66 | NA | NA | 73 | 74.2 | 73 | |||
| N | 76.8 | 73.70 | NA | NA | 73 | 74.2 | 73.2 | |||
| 11 | ||||||||||
| T | 77.7 | 73.4 | 77.1 | 73.3 | 73.4 | 76.8 | NA | |||
| N | 77.7 | NA | 77.2 | NA | NA | NA | NA | |||
| 12 | ||||||||||
| T | 76.7 | NA | NA | 73.7 | NA | NA | ||||
| N | 76.5 | NA | NA | 73.7 | NA | NA | ||||
| 13 | ||||||||||
| T | 77.5 | 73.5 | 76.4 | 73.8 | 73.4 | 76.8 | NA | |||
| N | 77.5 | NA | 76.4 | NA | NA | NA | NA | |||
| 14 | ||||||||||
| T | 77.4 | 73.2 | 76.9 | 73.7 | 73.4 | 76.6 | 72.55 | |||
| N | 77.4 | 73.2 | 77 | 73.7 | 73.4 | 76.6 | 72.65 | |||
| 15 | ||||||||||
| T | 78.1 | NA | 75.7 | 76.8 | 73.7 | 73.5 | 76.6 | 72.8 | ||
| N | 78.1 | 73.5 | 75.7 | 76.9 | 73.7 | 73.4 | 76 | 72.9 | ||
| 16 | ||||||||||
| T | 77.4 | 73.5 | 75.5 | 76.7 | 73.9 | NA | 72.4 | |||
| N | 77.3 | 73.64 | 75.5 | 76.7 | 73.9 | NA | 72.8 | |||
| 17 | ||||||||||
| T | NA | NA | NA | NA | NA | NA | ||||
| N | NA | NA | NA | NA | NA | NA | ||||
| 18 | ||||||||||
| T | 77.8 | 73.6 | 76.5 | 73.7 | NA | 72.3 | ||||
| N | 77.7 | NA | 76.5 | 73.7 | NA | 72.6 | ||||
| 19 | ||||||||||
| T | 77.5 | 73.4 | 75.6 | 76.9 | 73.7 | 73.4 | 75.7 | 72.7 | ||
| N | 77.5 | 73.7 | 75.6 | 76.9 | 73.7 | NA | NA | 73.2 | ||
| 20 | ||||||||||
| T | 77.3 | 73.5 | 76.5 | 73.7 | 73.5 | 72.5 | ||||
| N | 77.2 | NA | 75.6 | 76.5 | 73.7 | 73.5 | 72.5 | |||
| 21 | ||||||||||
| T | 78.2 | 73.5 | 76.6 | NA | NA | NA | ||||
| N | NA | NA | 76.5 | NA | NA | NA | ||||
| 22 | ||||||||||
| T | 77 | 73.76 | NA | NA | NA | NA | ||||
| N | 77 | 73.70 | NA | NA | NA | 73.2 | ||||
| 23 | ||||||||||
| T | NA | NA | NA | NA | NA | NA | ||||
| N | NA | NA | NA | NA | NA | NA | ||||
| 24 | ||||||||||
| T | 77.2 | NA | 76.6 | 73.7 | 73.3 | 76 | 72.5 | |||
| N | 77 | NA | 76.4 | 73.8 | 73.5 | 76 | 72.6 | |||
| 25 | ||||||||||
| T | 77.5 | 72.7 | 73.7 | 77.2 | 73.8 | 73.4 | 76.6 | 72.9 | ||
| N | 77.5 | 73.7 | 77.1 | 73.8 | NA | NA | 72.9 | |||
Melting temperature peaks of polymorphic markers. Genotyping, heterozygous samples (two peaks), homozygous (one peak). NA, non-amplified sample. Genetic alterations: LOH, loss of heterozygosity, shown as loss of a melting peak temperature in tumor tissue sample (cases 4 and 20, loss of peak 1/D3S1612 locus); MSI, microsatellite instability, shown as creation of a new melting peak in tumor tissue sample (case 25, new peak 1/D3S1768 locus). UCC, urothelial cell carcinoma; T, tumor; N, normal.
We observed genetic instability (MSI and/or LOH) in 3 (15%) of the 20 analyzed bladder tumors vs. their ANTs. Two polymorphic regions at the 3p loci had been affected (distal to hMLH1). D3S1768 locus exhibited MSI in one of the 12 (8.3%) analyzed cases, which was characterized as MSI-L (one unstable marker), while the remaining cases were not informative for LOH (Table V). D3S1612 locus was affected by LOH in two out of 7 (28.57%) informative cases (Table V). None of the MSI-analyzed cases were informative for LOH at the D3S1234, D3S1788, BAT25 or BAT26 polymorphic regions (Table V).
The MSI-L bladder tumor was a high grade UCC with stage pT1, while LOH was noted in a low grade pTa stage and a high grade pT1 stage UCC. The 2 UCCs which showed LOH at the D3S1612 (3p21.3–22) locus exhibited normal or elevated MMR phenotypes but reduced (≤0.8) hMSH6 mRNA tumor/ANT ratios (Tables I and V).
Discussion
To date, a series of studies have attempted to determine the expression of MMR proteins, mainly MSH2 and MLH1, in bladder cancer, the majority using IHC methods (17–24). Only two previous studies have determined MMR mRNA levels in bladder cancer by qPCR analysis and even in a few series of clinical specimens with different percentages from IHC analysis (25,26). The current study presents for the first time a quantification analysis of MMR mRNA transcripts in paired bladder tumors and their ANTs.
It is known that MSH2/MSH6 proteins form heterodimers that act as a complex (MutSα). This complex function is to detect single base-base mismatches and insertion-deletion loops and bind to the side of the DNA error (11,12,14). Our data showed that unaffected urothelia adjacent to UCC tumors (mainly adjacent to low grade UCC tumors) express low ratios of hMSH2 and hMSH6 mRNA levels (r2r6 phenotype), implying a low activity of DNA damage recognition of single mismatches and insertion-deletion loops errors. The ANTs of high grade UCCs also exhibited a reduced r2r6 phenotype, leaving the urothelium at high risk of cancer. Moreover, urothelia adjacent to high grade UCCs showed statistically higher percentages of the reduced r2r6 phenotype, approaching the levels of high grade tumors, in contrast to ANT of low grade UCCs, which showed a significant difference between the corresponding tumors. However, the hMLH1 gene was found to have elevated mRNA ratios (R1 phenotype) both in UCCs and their ANTs, indicating either high requirements for DNA repair of the progressively increasing errors in cancerous or precancerous urothelium or the involvement of hMLH1 in another tumorigenesis pathway (33). The counterpart of hMLH1, hPMS2, was also overexpressed in a percentage of pT1–2 and high grade UCCs, to cooperate with MLH1 as complex (MutLα) due to the demanding repair or another function (10,14,34). Nevertheless, a percentage of UCCs presented reduced levels of hMLH1 and hPMS2 mRNA expression relative to their ANTs which indicates low DNA repair activity in a large proportion of UCCs and therefore accumulation of replication errors in the abnormal proliferating malignant cells.
The unbalanced mRNA levels of MMR genes, including overexpression of hMSH2, hMLH1 and hPMS2 and reduction of mRNA levels of hMSH6, in the urothelium of UCC, mainly in males, was correlated with tumor progression. A recent study implicates MutLα as a general stimulating factor for miRNA biogenesis, giving the complex an additional function in tumorigenesis (34). In our cohort of specimens we observed that a proportion of tumors exhibited mRNA overexpression of hMSH2, hMLH1 and hPMS2. For hMSH2 this was more frequent in low grade pTa tumors; for hMLH1 in low grade pT1 tumors; and for hPMS2 for high grade pT1–2 tumors relative to the ANTs that may indicate the tumor progression. An explanation may be that from low to high grade tumors or from pTa to pT3 histological stages additional DNA errors take place, e.g., small and larger insertion-deletion loops (12,13). The need for recognition of these errors by other MMR complexes and enzymes, such as MutSβ (MSH2-MSH3), is indicated by the significant reduction of the hMSH6 counterpart of hMSH2(25).
We analyzed a case of PUNLMP and its ANT for MMR mRNA expression. The normal urothelium adjacent to PUNLMP revealed regular or elevated (≥1) mRNA levels of MutSα complex which detects single base-base mismatches and insertion-deletion loops while the mRNA levels of hMLH1, a crucial component of MutLα that is responsible for repairing the DNA errors (10,14), were <1. This is in agreement with the results of a previous study which showed that MLH1 is expressed at a lower level than MSH2 and MSH6 in human cells (35), suggesting a regular proliferation of urothelium cells and a limited DNA repair requirement. hMSH2, hMSH6 and hPMS2 mRNAs were reduced in PUNLMP compared with its ANT, probably due to a low rate of apoptosis (36).
The correlation of our results with clinical data revealed the statistically significant association of hMSH2 and hMLH1, hMSH2 and hMSH6, hMSH6 and hPMS2, hMLH1 and hMSH6 tumor/ANT mRNA expression ratios in females. We derive the conclusion that the urothelium of females has a better balance in the expression DNA MMR genes compared with males, who exhibited imbalance. Most likely, the MMR mechanisms are biologically differently regulated in the two genders. Additionally, a significant association was also found between the changes in hMSH2 and hMLH1 mRNA expression levels in UCCs compared with their ANTs, indicating that hMSH2 and hMLH1 cooperation in DNA repair (10,11,14) requires an associated mechanism for regulating hMSH2 and hMLH1 gene expression.
The biological significance of these findings is indicated by the association between hMSH2, hMLH1, hMSH6 and hPMS2 mRNA expression in our tissue cohort. We identified a significant association between reduced mRNA expression levels of hMSH2/control and hMSH6/control, indicating a common mechanism of hMSH2 and hMSH6 suppression of transcriptional activation that is in accordance with their biological function, as components of the MutSα complex act cooperatively (11–13). The interdependence of the four genetic loci was shown by the observed and calculated frequencies of their combined phenotypes (Table IV) (31). hMSH2 and hMSH6 revealed different frequencies and were considered as depended loci, as were hMSH2 and hPMS2 or hMSH6 and hPMS2(37). Besides, hMSH2 and hMLH1, hMHL1 and hMSH6 or hMHL1 and hPMS2 exhibited identical observed and calculated frequencies in UCCs and/or their ANTs and were considered as independent loci (16).
The identification of MSI in bladder tumors vs. their ANTs and correlation with MMR mRNA expression or MMR phenotypes showed that MSI-H was absent, MSI-L was rare in our study group and LOH was found in a small proportion of informative UCC samples. This result is in agreement with those of previous studies which reported the absence or low frequencies of MSI in bladder cancer (32,38). LOH and MSI-L were observed in a region distal to the hMLH1 locus. The two UCCs affected by LOH at 3p loci exhibited regular or elevated MMR phenotypes. Therefore, allelic imbalance at these chromosomal regions which harbor hMLH1 was not correlated with loss of hMLH1 mRNA expression; this is in agreement with a previous study of non-small cell lung tumors (28). However, the UCCs affected by LOH showed reduced hMSH6 mRNA tumor/ANT ratios, which may mean that genetic instability in the bladder, distal to the hMLH1 locus, is correlated with a reduced expression of hMSH6.
In conclusion, this is the first study to quantify MMR mRNA expression in bladder tumors and adjacent normal urothelium. Reduced (r) mRNA phenotypes of hMSH2, hMSH6 and hPMS2 were found to be correlated with precancerous or cancerous urothelium and a previously unrecognized reduced r2r6 (hMSH2 and hMSH6) phenotype with a precancerous urothelium. Additionally, we did not identify a reduced r1 phenotype of hMLH1, a crucial component of MutLα complex, in UCCs or their ANTs and hMLH1 was overexpressed in a significant proportion of UCCs. Therefore, the hMLH1-elevated (R1) mRNA phenotype and mRNA overexpression was correlated with urothelium with malignant potential. The correlation of our results with clinical data revealed that in males the MMR mechanism appears to be unbalanced relative to females and gradually elevated mRNAs expression levels of hMSH2, hMLH1 and hPMS2 in males show a progression from low to high grade and from pTa to pT1–2 tumors. Biologically, we demonstrated that hMSH2, hMSH6 and hPMS2 are interdependent loci; particularly, hMSH2 and hMSH6 were indicated to have a common mechanism of suppressing transcriptional activation. hMLH1 was independent, but requires an association with a hMSH2 mechanism, frequently in low grade tumors, for regulation of mRNA expression. Finally, reduction of hMSH2 and hMLH1 mRNA expression in UCCs is unlikely to be correlated with allelic imbalance at polymorphic regions which harbor the genes; however, LOH distal to hMLH1 may be correlated with hMSH6 reduction.
References
- 1.Lee TK, Chaux A, Karram S, Miyamoto H, Miller JS, Fajardo DA, Epstein JI, Netto GJ. Papillary urothelial neoplasm of low malignant potential of the urinary bladder: clinicopathologic and outcome analysis from a single academic center. Hum Pathol. 2011;42:1799–1803. doi: 10.1016/j.humpath.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 2.MacLennan GT, Kirkali Z, Cheng L. Histologic grading of noninvasive papillary urothelial neoplasms. Eur Urol. 2007;51:889–898. doi: 10.1016/j.eururo.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 3.Chaux A, Karram S, Miller JS, Fajardo DA, Lee TK, Miyamoto H, Netto GJ. High-grade papillary urothelial carcinoma of the urinary tract: a clinicopathologic analysis of a post-World Health Organization/International Society of Urological Pathology classification cohort from a single academic center. Hum Pathol. 2012;43:115–120. doi: 10.1016/j.humpath.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Cheng L, Davidson DD, Maclennan GT, Williamson SR, Zhang S, Koch MO, Montironi R, Lopez-Beltran A. The origins of urothelial carcinoma. Expert Rev Anticancer Ther. 2010;10:865–880. doi: 10.1586/era.10.73. [DOI] [PubMed] [Google Scholar]
- 5.Cordon-Cardo C, Cote RJ, Sauter G. Genetic and molecular markers of urothelial premalignancy and malignancy. Scand J Urol Nephrol. 2000;(Suppl 205):82–93. doi: 10.1080/003655900750169338. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SM. Urinary bladder carcinogenesis. Toxicol Pathol. 1998;26:121–127. doi: 10.1177/019262339802600114. [DOI] [PubMed] [Google Scholar]
- 7.Vageli D, Kiaris H, Delakas D, Anezinis P, Cranidis A, Spandidos DA. Transcriptional activation of H-ras, K-ras and N-ras proto-oncogenes in human bladder tumors. Cancer Lett. 1996;107:241–247. doi: 10.1016/0304-3835(96)04372-8. [DOI] [PubMed] [Google Scholar]
- 8.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 10.Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 11.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 13.Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa T, Datta A, Kolodner RD. Multiple functions of MutS- and MutL-related heterocomplexes. Proc Natl Acad Sci USA. 1999;96:14186–14188. doi: 10.1073/pnas.96.25.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruszkiewicz A, Bennett G, Moore J, Manavis J, Rudzki B, Shen L, Suthers G. Correlation of mismatch repair genes immunohistochemistry and microsatellite instability status in HNPCC-associated tumours. Pathology. 2002;34:541–547. doi: 10.1080/0031302021000035965-2. [DOI] [PubMed] [Google Scholar]
- 16.Vageli D, Daniil Z, Dahabreh J, Karagianni E, Vamvakopoulou DN, Ioannou MG, Scarpinato K, Vamvakopoulos NC, Gourgoulianis KI, Koukoulis GK. Phenotypic mismatch repair hMSH2 and hMLH1 gene expression profiles in primary non-small cell lung carcinomas. Lung Cancer. 2009;64:282–288. doi: 10.1016/j.lungcan.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 18.Mylona E, Zarogiannos A, Nomikos A, Giannopoulou I, Nikolaou I, Zervas A, Nakopoulou L. Prognostic value of microsatellite instability determined by immunohistochemical staining of hMSH2 and hMSH6 in urothelial carcinoma of the bladder. APMIS. 2008;116:59–65. doi: 10.1111/j.1600-0463.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto Y, Matsuyama H, Kawauchi S, Furuya T, Liu XP, Ikemoto K, Oga A, Naito K, Sasaki K. Biological characteristics in bladder cancer depend on the type of genetic instability. Clin Cancer Res. 2006;12:2752–2758. doi: 10.1158/1078-0432.CCR-05-0805. [DOI] [PubMed] [Google Scholar]
- 20.Catto JW, Xinarianos G, Burton JL, Meuth M, Hamdy FC. Differential expression of hMLH1 and hMSH2 is related to bladder cancer grade, stage and prognosis but not microsatellite instability. Int J Cancer. 2003;105:484–490. doi: 10.1002/ijc.11109. [DOI] [PubMed] [Google Scholar]
- 21.Rubio J, Blanes A, Sanchez-Carrillo JJ, Diaz-Cano SJ. Microsatellite abnormalities and somatic down-regulation of mismatch repair characterize nodular-trabecular muscle-invasive urothelial carcinoma of the bladder. Histopathology. 2007;51:458–467. doi: 10.1111/j.1365-2559.2007.02795.x. [DOI] [PubMed] [Google Scholar]
- 22.Ericson KM, Isinger AP, Isfoss BL, Nilbert MC. Low frequency of defective mismatch repair in a population-based series of upper urothelial carcinoma. BMC Cancer. 2005;5:23. doi: 10.1186/1471-2407-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saetta AA, Goudopoulou A, Korkolopoulou P, Voutsinas G, Thomas-Tsagli E, Michalopoulos NV, Patsouris E. Mononucleotide markers of microsatellite instability in carcinomas of the urinary bladder. Eur J Surg Oncol. 2004;30:796–803. doi: 10.1016/j.ejso.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Kassem HS, Varley JM, Hamam SM, Margison GP. Immunohistochemical analysis of expression and allelotype of mismatch repair genes (hMLH1 and hMSH2) in bladder cancer. Br J Cancer. 2001;84:321–328. doi: 10.1054/bjoc.2000.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thykjaer T, Christensen M, Clark AB, Hansen LR, Kunkel TA, Ørntoft TF. Functional analysis of the mismatch repair system in bladder cancer. Br J Cancer. 2001;85:568–575. doi: 10.1054/bjoc.2001.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leach FS, Hsieh JT, Molberg K, Saboorian MH, McConnell JD, Sagalowsky AI. Expression of the human mismatch repair gene hMSH2: a potential marker for urothelial malignancy. Cancer. 2000;88:2333–2341. [PubMed] [Google Scholar]
- 27.Vageli D, Ioannou MG, Koukoulis GK. Transcriptional activation of hTERT in breast carcinomas by the Her2-ER81-related pathway. Oncol Res. 2009;17:413–423. doi: 10.3727/096504009788912507. [DOI] [PubMed] [Google Scholar]
- 28.Wang YC, Lu YP, Tseng RC, Lin RK, Chang JW, Chen JT, Shih CM, Chen CY. Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. J Clin Invest. 2003;111:887–895. doi: 10.1172/JCI15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou XP, Hoang JM, Cottu P, Thomas G, Hamelin R. Allelic profiles of mononucleotide repeat microsatellites in control individuals and in colorectal tumors with and without replication errors. Oncogene. 1997;15:1713–1718. doi: 10.1038/sj.onc.1201337. [DOI] [PubMed] [Google Scholar]
- 30.Hoang JM, Cottu PH, Thuille B, Salmon RJ, Thomas G, Hamelin R. BAT-26, an indicator of the replication error phenotype in colorectal cancers and cell lines. Cancer Res. 1997;57:300–303. [PubMed] [Google Scholar]
- 31.Vageli D, Daniil Z, Dahabreh J, Karagianni E, Liloglou T, Koukoulis G, Gourgoulianis K. Microsatellite instability and loss of heterozygosity at the MEN1 locus in lung carcinoid tumors: a novel approach using real-time PCR with melting curve analysis in histopathologic material. Oncol Rep. 2006;15:557–564. [PubMed] [Google Scholar]
- 32.Ericson KM, Isinger AP, Isfoss BL, Nilbert MC. Low frequency of defective mismatch repair in a population-based series of upper urothelial carcinoma. BMC Cancer. 2005;5:23. doi: 10.1186/1471-2407-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shcherbakova PV, Hall MC, Lewis MS, Bennett SE, Martin KJ, Bushel PR, Afshari CA, Kunkel TA. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao G, Lee S, Ortega J, Gu L, Li GM. Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 2012;22:973–985. doi: 10.1038/cr.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem. 2000;275:18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- 36.Köster F, Schröer A, Fischer D, Greweldinger T, Diedrich K, Friedrich M. Correlation of DNA mismatch repair protein hMSH2 immunohistochemistry with p53 and apoptosis in cervical carcinoma. Anticancer Res. 2007;27:63–68. [PubMed] [Google Scholar]
- 37.Hayes AP, Sevi LA, Feldt MC, Rose MD, Gammie AE. Reciprocal regulation of nuclear import of the yeast MutSalpha DNA mismatch repair proteins Msh2 and Msh6. DNA Repair (Amst) 2009;8:739–751. doi: 10.1016/j.dnarep.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnal C, Ravery V, Toublanc M, Bertrand G, Boccon-Gibod L, Henin D, et al. Absence of microsatellite instability in transitional cell carcinoma of the bladder. Urology. 2000;55:287–291. doi: 10.1016/s0090-4295(99)00399-4. [DOI] [PubMed] [Google Scholar]