Abstract
The AgrA transcription factor regulates the quorum-sensing response in Staphylococcus aureus, controlling the production of hemolysins and other virulence factors. AgrA binds to DNA via its C-terminal LytTR domain, a domain not found in humans but common in many pathogenic bacteria, making it a potential target for antimicrobial development. We have determined the crystal structure of the apo AgrA LytTR domain and screened a library of 500 fragment compounds to find inhibitors of AgrA DNA-binding activity. Using NMR, the binding site for five compounds has been mapped to a common locus at the C-terminal end of the LytTR domain, a site known to be important for DNA-binding activity. Three of these compounds inhibit AgrA DNA binding. These results provide the first evidence that LytTR domains can be targeted by small organic compounds.
S. aureus typically causes skin or soft tissue infections at a localized lesion.1 Patients are at risk of more serious life threatening diseases such as pneumonia, osteomyelitis, bacteremia, endocarditis and toxic shock syndrome2–5 if S. aureus is able to cross into the bloodstream. The treatment of S. aureus infections has become increasingly problematic due to the emergence of antibiotic resistant strains. Methicillin resistant S. aureus (MRSA) is now the most common antibiotic resistant pathogen identified in developed world hospitals6 and poses a significant threat to human health.
The pathogenicity of S. aureus requires the coordinated expression of a large number of virulence factors. The agr quorum-sensing system, which allows the pathogen to sense both the density of the local S. aureus population and its degree of confinement, plays a central role in the regulation of the S. aureus virulon. Activation of the agr system represses the production of cell surface adhesins and promotes the secretion of extracellular toxins.
The agr operon consists of four genes, designated agrBDCA. The propeptide, AgrD, is processed and exported by AgrB. The secreted cyclic peptide is the autoinducing peptide (AIP) that functions as the quorum-sensing signal. When the extracellular AIP concentration is high, the histidine kinase, AgrC, is activated, resulting in increased phosphorylation of the cytoplasmic response regulator protein, AgrA. Phosphorylated AgrA activates transcription of the agrBDCA operon and genes encoding phenol soluble modulins7 and the effector RNA molecule, RNAIII.8 In addition to its function as the messenger RNA for hemolysin δ,9 RNAIII uses antisense RNA mechanisms to down-regulate adhesins and activate the transcription of genes encoding hemolysins, Panton-Valentine leukocidin (PVL) and enterotoxins.10–13
Deletion of the agr operon attenuates the S. aureus infection in mouse and rabbit animal models of infection demonstrating the importance of agr quorum sensing for pathogenesis.14–19 Indeed, AIP analogues that inhibit the AgrC histidine kinase are effective in reducing the severity of S. aureus infection.17,19 However, attempts to identify inhibitors of response regulator AgrA have not been reported, despite the attractiveness of AgrA as a target because of the absence of LytTR DNA-binding domain proteins in mammalian proteomes.20
We have determined the high-resolution crystal structure of the AgrA C-terminal LytTR domain (AgrAC) and used a fragment screening approach to search for small molecule binding sites on the DNA-binding surface of this domain. Fragment-screening approaches have been widely used to efficiently screen a broad area of potential chemical space, using a relatively small library of compounds. Although the affinity of small fragment compounds for a target protein is expected to be relatively low, due to the low molecular weight of the compounds used (<300 g·mol−1), fragment screening can identify energetic focal points on a protein surface that can be targeted by small molecule compounds.21
EXPERIMENTAL PROCEDURES
Protein expression and purification
Unlabeled S. aureus AgrAC protein (AgrA residues Asp137 to Ile238) samples were produced in E. coli grown in terrific broth (TB) media as previously described.22 For NMR studies, 15N and 15N/13C isotopically-enriched protein samples of AgrAC were prepared by expressing the AgrAC protein in E. coli BL21 (DE3) pLysS grown in M9 minimal media at 18 °C using 2.5 g L−1 (15NH4)2SO4 and 2 g L−1 [13C]-glucose (Cambridge Isotope Laboratories) as appropriate. The M9 media was supplemented with 50 μg mL−1 kanamycin, 50 μM FeCl2, 2 μM CuCl2, 2 μM Na2MoO4, 2 μM NiCl2, 2 μM CoCl2, 2 μM H3BO3, 10 μM MnCl2, 10 μM ZnSO4, 20 μM CaCl2, and 1 μM of each of the following micronutrients: nicotininc acid, pyridoxine, thiamine, biotin, riboflavin, folic acid, D-pantothenic acid and myo-inositol. Recombinant protein expression was induced using 0.3 mM isopropyl-β-D-thiogalactopyranoside.
All AgrAC protein samples were purified from E. coli lysates following the previously published procedure22 using HiTrap SP HP cation exchange, HiLoad Phenyl Sepharose HP hydrophobic interaction and HiLoad Superdex 75 gel filtration chromatograpy (GE Healthcare). Purified protein was transferred into appropriate buffers by dialysis.
Crystallization of AgrAC
Initial AgrAC crystals were prepared at 4 °C by hanging drop vapor diffusion by mixing 1 μL of 1 mM AgrAC (dissolved in 20 mM Bis Tris, 100 mM NaCl and 10 mM DTT at pH 6.0) with 1 μL of reservoir solution (100 mM Tris, 150 mM LiSO4 and 11% (w/v) PEG 4000 at pH 8.0). The hanging drop was suspended above a 1-mL reservoir. The crystals were improved by streak seeding after 24 h incubation at the same condition except the PEG 4000 concentration was lowered to 8% (w/v).
Data collection and structure refinement
AgrAC crystals were soaked for 30 s in 50 mM Tris, 75 mM LiSO4, 8% PEG 4000 and 20% glycerol at pH 8 before flash freezing in liquid nitrogen. A native data set was collected at 100 K using a Rigaku MicroMax-007 HF generator equipped with RAXIS-IV++ detector. Data were processed and scaled with DENZO and SCALEPACK.23 The structure of apo AgrAC was solved by molecular replacement using Phaser 2.024 with the DNA-bound state of AgrAC as the search model (PDB 3BS1). The R factor after rigid body refinement of the best molecular replacement solution was 0.440. From the initial molecular replacement solution, the structure was rebuilt from scratch using RESOLVE25 before iterative refinement using COOT 6.0226 and Phenix.27 The refined model contains two molecules of AgrAC within the asymmetric unit; chains A and B contain residues Glu141 to Ile238 and Ser139 to Ile238, respectively. The model was refined to 1.52 Å with R factor and Rfree values of 0.180 and 0.209 respectively. All residues lie within the allowed regions of the Ramachandran plot and exhibit favorable bond angles and bond lengths. An overview of the data collection and refinement statistics is provided in Table 1.
Table 1.
X-ray data collection and refinement statistics (molecular replacement)
| AgrACa | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 41.4, 45.9, 112.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 25.00-1.52 (1.55-1.52)b |
| Rsymc | 0.045 (0.563) |
| I/σ(I) | 32.9 (2.0) |
| Completeness (%) | 97.2 (89.8) |
| Redundancy | 3.3 (2.7) |
| Refinement | |
| Resolution (Å) | 20.72-1.52 |
| No. reflections | 31964 |
| Data cutoff | σ(F) > 0 |
| Rworkd/Rfreee | 0.180/0.209 |
| No. atoms per asymmetric unit | |
| Protein | 1702 |
| Glycerol | 18 |
| Water | 254 |
| Average B-factors (A2) | |
| Protein | 20.5 |
| Glycerol | 32.3 |
| Water | 32.6 |
| Rms deviations from ideality | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.048 |
A single crystal was used for the data collection.
Values in parentheses are for the highest-resolution shell.
Rsym = Σh|Ih – <I>|/ΣhIh, where Ih and <I> represent the diffraction intensity values of individual measurements and the corresponding mean values, respectively.
Rwork = (Σh||Fo| – |Fc||)/Σh|Fo|, where Fo and Fc are observed and calculated structure factor amplitudes, respectively.
Rfree was calculated for 5% of the randomly selected reflections of the data set that were not used in the refinement.
NMR spectroscopy
All NMR data were acquired on Varian INOVA 600 MHz or Varian 500 MHz spectrometers equipped with triple resonance probes. The NMR spectra were referenced to an internal 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) standard as described previously.28 All spectra were processed using NMRpipe29 and analyzed using CCPNMR analysis 2.1.30
For the backbone NMR assignments, 1H-15N HSQC31, HNCACB, HN(CO)CACB, HNCA, HN(CO)CA and HNCO32 spectra were acquired at 298 K from a sample containing 1 mM AgrAC in 20 mM sodium phosphate, 100 mM NaCl and 5% D2O at pH 5.8. The backbone resonance assignments for AgrAC were established for all residues between Asn138 and Ile238 except for Tyr229.
15N heteronuclear NOE33 spectra were recorded with 3.0 s 1H saturation in the latter part of a 3.5-s preparation period delay, which was also used without radio frequency pulses for the reference 2D spectrum without NOE.
A library of 500 fragment compounds (Maybridge Ro3 500 fragment library) was screened for binding to AgrAC using a WATERGATE W5 LOGSY NMR experiment.34 Each sample contained a mixture of 10 compounds (400 μM of each compound) and 15 μM AgrAC, dissolved in 20 mM sodium phosphate, 100 mM NaCl, 10 mM DTT and 5% D2O at pH 7.0. As a negative control, spectra were acquired for a second sample without AgrAC. Hit compounds were identified by the inversion of the compound spectra when AgrAC was present and validated using single compound repeats of the WATERGATE W5 LOGSY experiment. The WATERGATE W5 LOGSY spectra were acquired at 20 °C using a mixing time of 1.2 s, 8192 complex data points, spectral width of 10000 Hz and number of scans set to 128. The acquisition time was 0.819 s with a relaxation delay of 3.0 s. The total experiment time was 11 min.
To identify the compound-binding sites, 1H- 15N HSQC spectra were acquired for samples containing 300 μM AgrAC dissolved in 20 mM sodium phosphate, 100 mM NaCl, 10 mM DTT and 5% D2O at pH 5.8 after each addition of compound to the protein sample. 200 mM stock solutions of each compound, dissolved in DMSO-d6 were used for the titrations. A control experiment where 3% DMSO-d6 was added to the AgrAC NMR sample in the absence of compound was used to establish which chemical shift perturbations were compound dependent.
The combined chemical shift was calculated for each amide resonance after the addition of compound, using the established method,35 and mapped onto the apo AgrAC crystal structure using Pymol v1.2r2. For each compound the highest spectra obtained with the highest concentration of compound tested in the NMR titrations was used to determine the combined chemical shift perturbation.
Compound docking
In-silico docking of the compounds binding to AgrAC was performed using Autodock Vina.36
Inhibition Assays
The activities of five compounds as inhibitors of AgrAC DNA binding were tested using an electrophoretic mobility shift assay performed in the presence of increasing amounts of each compound (from 0 mM to 5 mM) to test whether the presence of compound inhibits the DNA-binding activity of AgrAC. A 19-bp DNA duplex was prepared by annealing 5′-ATTTAACAGTTAAGTATTT-3′ with its complementary oligonucleotide in 21 mM sodium phosphate, 105 mM NaCl, 10.5 mM DTT at pH 5.8 and purified by size exclusion chromatography using a Superdex 75 HR column. Solutions were prepared containing 2 μM DNA duplex, 20 μM AgrAC and various compound concentrations in 20 mM sodium phosphate, 100 mM NaCl, 10 mM DTT and 2.5% DMSO-d6 at pH 5.8. After incubation at room temperature for 30 min, 12% (w/v) Ficoll 400 was added to each sample to give a 2% (w/v) final concentration before running the samples on an 8% acrylamide/bis-acrylamide 40 mM Tris·HCl, 20 mM acetic acid, 1 mM EDTA at pH 8.0 (TAE) gel at 70 V and 4 °C. The gels were stained in TAE supplemented with 0.1 mg mL−1 ethidium bromide for 10 min prior to destaining in TAE for 10 min. DNA was visualized with UV light and images were captured using a CCD camera imager (Alpha Innotech).
RESULTS
Backbone Resonance Assignments of AgrAC
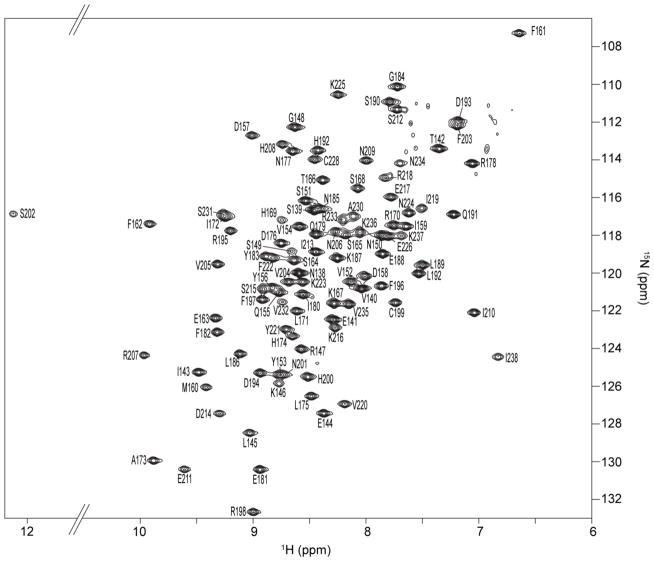
Backbone resonance assignments were determined for AgrAC using a 15N,13C-labeled sample of AgrA LytTR domain (Asp137 to Ile238). The protein sample gave excellent quality NMR spectra with well-dispersed chemical shifts (Fig. 1.) allowing for assignment of the backbone resonances. Assignments could be made for all of the residues between Asn138 and Ile238 except for Tyr229 whose resonances could not be confidently assigned, probably due to overlapping crosspeaks with another amide crosspeak. The unusual downfield shift of the serine 202 backbone amide proton resonance is caused by the hydrogen bonding of this proton with the imidazole ring of His200, as observed in the crystal structure of AgrAC. A similar downfield shift has been described for the hydrogen bonding of Arg7 backbone amide to His106 side chain in B. subtilis chorismate mutase.37
Figure 1.
2D 1H-15N HSQC spectrum of AgrAC. Residue specific cross-peak assignments for the backbone amides are indicated.
AgrAC in the Absence of DNA
The structure of AgrAC bound to its target DNA duplex has been reported previously,22 but any structural changes that occur within this domain upon DNA binding were unknown. To establish if there are structural differences between the apo and DNA-bound states of AgrAC both NMR and X-ray crystallographic analyses were used to investigate the structure of AgrAC protein in the absence of DNA.
AgrAC was crystallized in its apo state, forming P212121 space group crystals that diffracted to 1.5-Å resolution. The structure of apo AgrAC was solved by molecular replacement. To remove model bias from the molecular replacement solution, the AgrAC protein was rebuilt from scratch using RESOLVE and then iteratively refined. The final structure contains two molecules of AgrAC in the asymmetric unit of the crystals: chain A contains residues Glu141 to Ile238 and chain B contains Ser139 to Ile238).
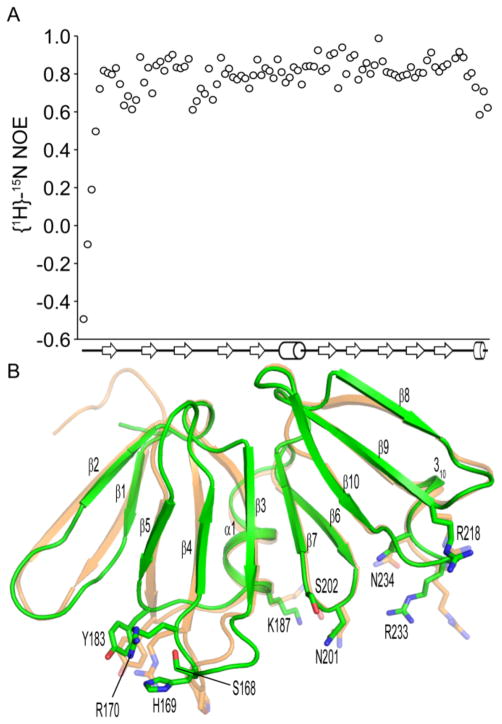
For both AgrAC protomers, the N-terminal residues are not observed in the electron density map, suggesting that this region is disordered, consistent with the negative {1H}15N heteronuclear NOE values observed for these amino acids (Fig. 2A) under solution conditions. The apo AgrAC protein adopts a β-β-β-sandwich fold as observed previously for the DNA-bound state. The fold is stabilized by a set of conserved salt bridges: Glu141-Arg195, Asp157-His208, Asp193-Arg195, Asp157-Arg195 and His174-Glu226. The domain is very stable with heteronuclear NOE values for the backbone amide bond vectors between 0.7 and 1.0 for the majority of the protein. However, the β1-β2 loop, the β3-β4 loop and the C terminus have 1H15N-heteronuclear NOE values between 0.6 and 0.7, suggesting greater conformational flexibility for these regions relative to the rest of the LytTR domain (Fig. 2A). In addition, alternate rotamer conformations are observed in the electron density map for several amino acid sidechains within the AgrAC crystal structure (Ser164, Ser215 and Glu217 from protomer chain A and Ile143, Ser149 and Asp176 from chain B), indicative of localized conformational flexibility within the domain. When the two protomer chains in the AgrAC crystal are superposed, the only differences in the protein backbone are observed for the N and C termini as well as the β1-β2, and β3-β4 loops, in excellent agreement with the heteronuclear NOE data. The root mean square (RMS) deviation for the two protomer chains in the apo AgrAC crystal is 0.19 Å for all backbone atoms, but a much greater difference is seen when the DNA-bound state is compared with the apo AgrAC protein chains A or B (RMS deviations of 0.41 or 0.46 respectively), suggesting that a conformational change takes place upon DNA binding.
Figure 2.
Conformational differences between apo and DNA-bound AgrAC. A, Steady-state heteronuclear {1H}15N-NOE values for each residue of AgrAC plotted versus the secondary structure topology of the protein. β strands and α helices are depicted as arrows and cylinders respectively. B, Superposition of the apo AgrAC crystal structure (green) with AgrAC in its DNA-bound state (orange).22 The sidechains of amino acid residues within the DNA-binding surface that adopt different conformations between the apo and DNA-bound states are shown in stick format.
When the apo and DNA-bound structures of AgrAC are compared, changes in the conformation of the DNA-binding surface are observed for residues that are known to interact with the DNA backbone (Ser 168, Arg170, Tyr183, Lys187, Ser202, Arg218, Asn234) as well as for three residues that make direct, base-specific contacts (His169, Asn201 and Arg233)22 (Fig. 2B). The largest change between the DNA-bound and apo states of the AgrAC protein is observed for the β3-β4 loop (Ser165 to Arg170). There are no crystal contacts for this region for the chain B protomer in the apo protein structure, so crystal lattice packing does not explain the different configuration of the β3-β4 loop relative to that in the DNA-bound state of AgrAC. The conformational differences between the apo and DNA-bound structures suggest that DNA binding traps the protein into a conformation that represents only one conformation from the population of states adopted by the apo AgrAC protein. The conformational freedom of the DNA-binding surface would allow some flexibility for AgrA binding to its DNA target sequences, albeit with an entropic penalty for binding.
Fragment Screening for Ligands of AgrAC
LytTR domains are considered attractive drug targets for the development of new antibiotics because these domains are not present in humans and are often found to activate virulence pathways in bacterial pathogens.20 NMR fragment screening methods were chosen because they allow the entire molecular surface of the target protein to be probed by a fragment compound library, in contrast to X-ray diffraction screening where some surfaces are occluded by lattice contacts. Fragment screening by NMR has proven to be a powerful tool for identifying regions of a protein that can be targeted by low complexity, small molecule ligands (fragment compounds), which subsequently can be used as starting points for drug development.38,39 A library of 500 compounds was screened for binding to AgrAC using a WATERGATE W5 LOGSY NMR experiment. Five compounds, 4-phenoxyphenol, 9H-xanthene-9-carboxylic acid, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, [5-(2-thienyl)-3-isoxazolyl]methanol and 4-hydroxy-2,6-dimethylbenzonitrile, were identified as binding to AgrAC as indicated by inversion of the compound spectrum when AgrAC was present in the solution.
Mapping the Compound-Binding Site
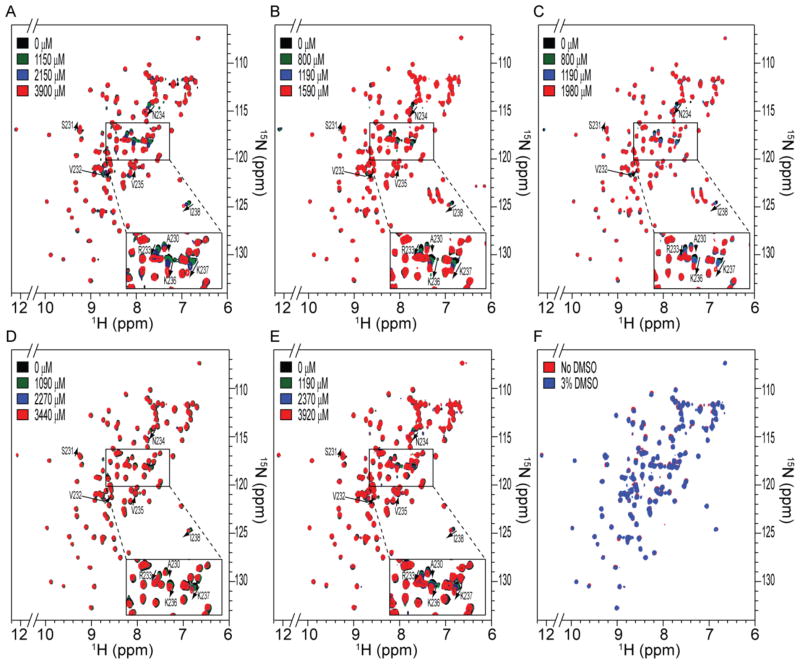
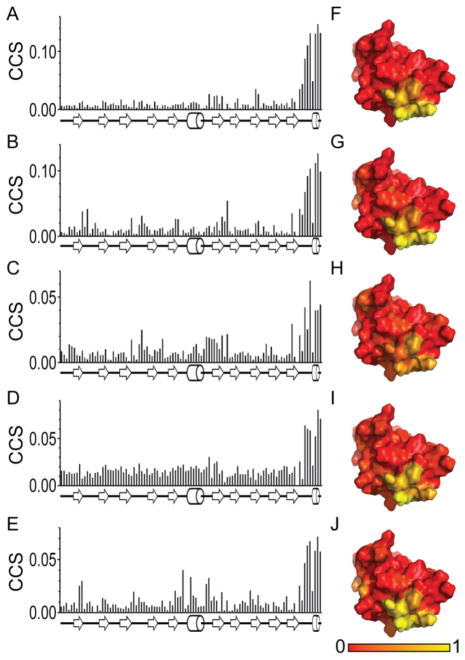
Having identified five compounds that interacted with AgrAC in the initial screen, increasing amounts of the each compound were titrated into a 15N-labeled sample of AgrAC and chemical shift perturbations in the 15N-1H-HSQC spectrum of the protein were monitored (Fig. 3.). Addition of each of the five compounds resulted in the perturbation of the same chemical shifts corresponding to the backbone amides for residues Ser231, Val232, Arg233, Asn234, Lys236, Lys237 and Ile238, at the C-terminal end of the protein (Fig. 4). A control experiment confirmed that the chemical shift perturbations were dependent on the presence of the compounds and not caused by the addition of DMSO-d6 to the NMR sample (Fig. 4F). Mapping the chemical shift perturbations onto the surface of AgrAC revealed a surface corresponding to a shallow groove formed by the β10-α2 loop and the α2 helix. Interestingly, the β10-α2 loop is known to be important for DNA binding by AgrA22 so ligands that occlude this site would be expected to reduce the DNA-binding activity of AgrA.
Figure 3.
Chemical shift perturbations observed upon binding of fragment compounds to AgrAC. 1H-15N HSQC spectra of AgrAC recorded in the presence of the indicated concentrations of A, 4-phenoxyphenol, B, 9H-xanthene-9-carboxylic acid, C, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, D, [5-(2-thienyl)-3-isoxazolyl]methanol and E, 4-hydroxy-2,6-dimethylbenzonitrile. The residue specific assignments for backbone amide cross-peaks that shift in a compound concentration dependent manner are highlighted. F, 1H-15N HSQC spectra of 15N-labeled AgrAC before and after the addition of 3% DMSO-d6.
Figure 4.
Mapping the compound dependent chemical shift perturbations. Combined chemical shift perturbations of backbone amide resonances plotted versus the amino acid residue number for the addition of 4-phenoxyphenol A, 9H-xanthene-9-carboxylic acid, B, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, C, [5-(2-thienyl)-3-isoxazolyl]methanol, D, and E, 4-hydroxy-2,6-dimethylbenzonitrile. The normalized combined chemical shift perturbations for F, 4-phenoxyphenol, G, 9H-xanthene-9-carboxylic acid, H, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, I, [5-(2-thienyl)-3-isoxazolyl]methanol and J, 4-hydroxy-2,6-dimethylbenzonitrile are mapped onto the surface of the crystal structure of apo AgrAC with the region showing the greatest combined chemical shift perturbation colored in yellow.
Compound Binding Predictions
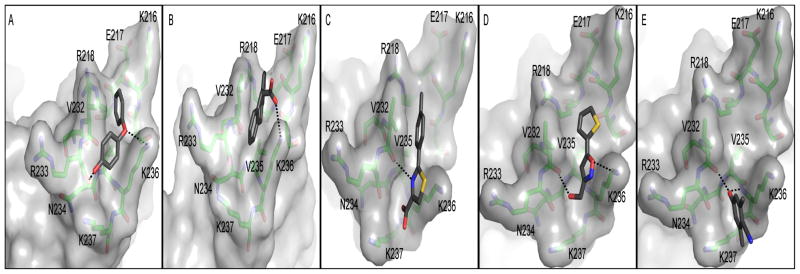
In silico docking was used to predict the binding mode of the compounds using AutoDock Vina.36 The search box was restricted to the region at the C-terminal end of AgrAC, identified by NMR as the binding site for the compounds (Fig. 5). All five compounds docked into a shallow groove located between Val232 and Lys236. 4-Phenoxyphenol is predicted to form hydrogen bond interactions with Lys236 and the backbone carbonyl of Arg233 and bends around Val232 to make tight Van der Waals interactions with this amino acid sidechain. 9H-xanthene-9-carboxylic acid binds with the planar xanthene ring structure between Val232 and Lys236. The predicted interaction is further stabilized by a salt bridge between the carboxylic acid group of the compound and the sidechain amine of Lys236. 2-(4-Methylphenyl)-1,3-thiazole-4-carboxylic acid binding is predicted to be stabilized by a hydrogen bond between the thiazole nitrogen and the carbonyl group of Val232 and a salt bridge between Lys237 and the carboxylic acid group in the compound. [5-(2-thienyl)-3-isoxazolyl]-methanol is predicted to bind with the isoxazolyl ring oxygen forming a hydrogen bond with Lys236 amine. The putative compound binding mode is further stabilized by a hydrogen bond between Val232 backbone carbonyl group and the hydroxyl group of the compound. The 4-hydroxy-2,6-dimethylbenzonitrile compound is predicted to bind in the shallow groove formed between Val232, Arg233, Lys236 and Lys237, with the hydroxyl group of the compound making hydrogen bond interactions with the backbone of Val232 and Lys236. The calculated binding energies for the best docking poses for 4-phenoxyphenol, 9H-xanthene-9-carboxylic acid, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, [5-(2-thienyl)-3-isoxazolyl]methanol and 4-hydroxy-2,6-dimethylbenzonitrile were −4.1 kcal mol−1, −4.1 kcal mol−1, −4.0 kcal mol−1, −3.5 kcal mol−1 and −3.6 kcal mol−1, respectively, in agreement with the millimolar affinity observed by NMR.
Figure 5.
In silico compound docking predictions. Autodock Vina predicted binding modes are shown for A, 4-phenoxyphenol, B, 9H-xanthene-9-carboxylic acid, C, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, D, [5-(2-thienyl)-3-isoxazolyl]methanol and E, 4-hydroxy-2,6-dimethylbenzonitrile. Orientations of the protein are slightly different in each panel to optimize viewing of the predicted hydrogen bonds between the compound and protein, indicated by dashed lines.
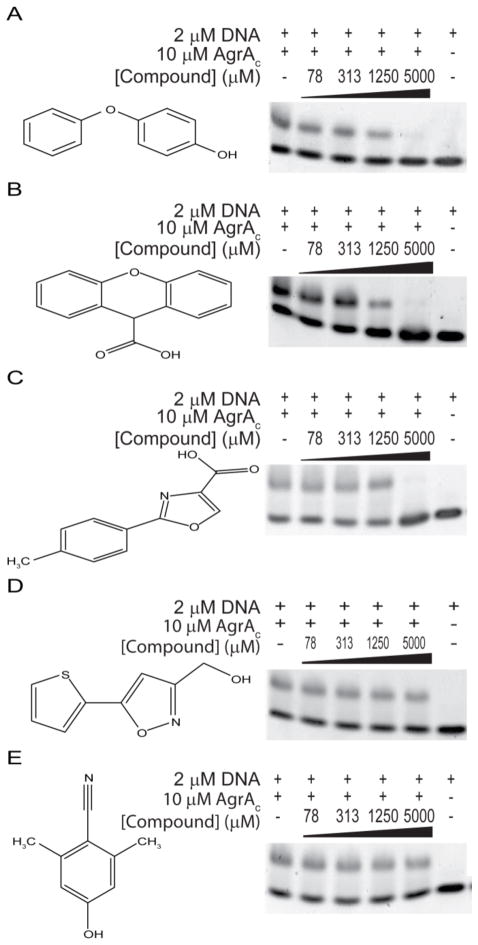
Three Fragment Compounds Inhibit the DNA-binding Activity of AgrAC
The five compounds identified by fragment screening all target the C-terminal end of AgrAC, a region known to be involved in DNA binding.22 It was therefore important to establish whether the compounds alter the DNA-binding activity of AgrA. The DNA-binding activity of AgrAC was tested using electrophoretic mobility shift assays in the presence of various compound concentrations from 75 μM to 5000 μM (Fig. 6). For three compounds (4-phenoxyphenol, 9H-xanthene-9-carboxylic acid and 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid), the intensity of the band corresponding to the AgrAC:DNA complex decreases as the compound concentration is increased; no AgrAC:DNA complex is observed when 5 mM compound is present. Two other compounds ([5-(2-thienyl)-3-isoxazolyl]methanol and 4-hydroxy-2,6-dimethylbenzonitrile) had no apparent effect on the DNA-binding activity of AgrAC at concentrations up to 5 mM. The DMSO concentration in all binding reactions was constant so the disruption to the DNA-binding activity of AgrAC is dependent only on the presence of the compound. In the control experiments where no AgrAC was present, there is no effect of the compounds on the migration of DNA through the gel. Some of the compounds that were identified to bind at the C terminus of AgrAC can disrupt AgrA DNA-binding activity.
Figure 6.
Effects of compounds on AgrA DNA-binding activity. Electrophoretic mobility shift assays were performed with AgrAC, its target DNA duplex and the indicated concentrations of A, 4-phenoxyphenol, B, 9H-xanthene-9-carboxylic acid, C, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, D, [5-(2-thienyl)-3-isoxazolyl]methanol and E, 4-hydroxy-2,6-dimethylbenzonitrile. DNA incubated with or without AgrAC in the absence of compound are shown as positive and negative controls, respectively.
DISCUSSION
The agr quorum sensing system has been suggested as a possible drug target because deletion of the agr system attenuates S. aureus virulence in animal models of infection.14–16,18 Furthermore, a functional agr system has been found to be important for bacterial survival in the early stages of abscess formation in animal models.19 On the other hand, isolation of agr-deficient mutants from patients with chronic S. aureus infections40 suggests that agr is not important for maintaining the infection once it has been established. Thus substantial controversy surrounds the validity of the agr system as a target for antimicrobial drug development. agr inhibitors would be especially useful tools in addressing the role of the agr system during the course of infection in animal models of infection. Furthermore, agr inhibitors might be effective for prophylactic treatment. Indeed, co-inoculation of S. aureus with an agr inhibitor was sufficient to reduce virulence in a mouse skin infection model.19
To date, research on inhibitors of S. aureus agr quorum sensing has focused on analogues of the autoinducing peptide as inhibitors of AgrC activation.17,19,41–43 Targeting agr quorum sensing using AIP analogues is complicated by the existence of several Staphylococcal AgrC types that are inhibited or activated by different AIP molecules.17,44,45
The results presented here represent the first attempt to inhibit the agr quorum sensing pathway by targeting the DNA-binding surface of the AgrA LytTR domain. Fragment screening can be used to identify an energetic focal point on the surface of a protein that is favorable to compound binding.21 While small fragments are efficient in covering large areas of chemical space and have advantages as starting points for drug development, their binding affinities are typically very low. Indeed, only three of the five compounds that were identified to bind to AgrAC by NMR analyses disrupted DNA binding in electrophoretic mobility shift assays. For these compounds, inhibition was detected only at millimolar concentrations. The low affinity of ligand binding provides a likely explanation for the lack of inhibition observed with the other two compounds.
The five compounds identified by fragment screening as AgrAC inhibitors all bind to a highly conserved region that forms a short helix at the C terminus of the AgrA protein. To date, there are 211 AgrA protein sequences across all Staphylococcal strains in the Uniprot knowledgebase database. Conservation of 100% sequence identity is observed for Ser231, Val232, Arg233, Asn234, Lys236 and Lys237 that are identified as residues in the compound-binding site. Val235 is conserved in 97% of all Staphylococcal sequences, although isoleucine or leucine amino acids are observed at this position in S. siminae, S. pseudintermedius, S. intermedius and S. carnosus strains. The C-terminal isoleucine residue (Ile238) is conserved in 94% of the AgrA protein sequences but is replaced by a lysine in S. aureus Mu3 and Mu50 strains or by valine in S. lugdunensis or S. saprophyticus strains. Other modifications at the AgrA C terminus give rise to short extensions. However, these extensions result in a reduced or completely deficient agr phenotype.40 Given the high degree of sequence conservation of the compound-binding site, it is reasonable to expect that any compound binding to this surface would be able to inhibit agr quorum sensing across all Staphylococcal strains.
The fragment compounds identified in this study bind with millimolar affinity to the AgrAC protein. The low affinity reflects the small size of the compounds that populate the library used in the screen. Despite the low affinity of initial hits, fragment screening has previously been shown to provide a useful starting point for the development of higher affinity inhibitors of protein:DNA interactions.21,46 It is commonly observed that features of the original low affinity fragment compound, identified by the initial screen, are maintained in the compounds that ultimately enter clinical development47–49. Further research is required to expand the compounds that have been identified as AgrA inhibitors, to improve their affinity and specificity. The apo AgrAC crystal structure and NMR assignments that have been established in this research will provide valuable information to guide future compound development.
Acknowledgments
We thank G. V. T. Swapna for her assistance setting up the NMR experiments and G. T. Montelione for use of the NMR facility within the CABM Structural Bioinformatics Laboratory.
Funding Sources
This work was supported in part by National Institutes of Health grant R37 GM047958 to A.M.S.
ABBREVIATIONS
- AIP
autoinducing peptide
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- NMR
nuclear magnetic resonance
- NOE
Nuclear Overhauser Effect
- PEG
polyethylene glycols
- rms
root mean square
Footnotes
The authors declare no competing financial interest.
Structural Data
The atomic coordinates and structure factors for apo AgrAC have been deposited in the Protein Data Bank. (PDB accession code: 4G4K) and NMR resonance assignments have been deposited into the Biological Magnetic Resonance Data Bank (BMRB accession number 18598).
References
- 1.Talan DA, Krishnadasan A, Gorwitz RJ, Fosheim GE, Limbago B, Albrecht V, Moran GJ. Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin Infect Dis. 2011;53:144–149. doi: 10.1093/cid/cir308. [DOI] [PubMed] [Google Scholar]
- 2.Dohin B, Gillet Y, Kohler R, Lina G, Vandenesch F, Vanhems P, Floret D, Etienne J. Pediatric bone and joint infections caused by Panton-Valentine leukocidin-positive Staphylococcus aureus. Pediatr Infect Dis J. 2007;26:1042–1048. doi: 10.1097/INF.0b013e318133a85e. [DOI] [PubMed] [Google Scholar]
- 3.Kollef MH, Micek ST. Staphylococcus aureus pneumonia: a “superbug” infection in community and hospital settings. Chest. 2005;128:1093–1097. doi: 10.1378/chest.128.3.1093. [DOI] [PubMed] [Google Scholar]
- 4.Petti CA, Fowler VG., Jr Staphylococcus aureus bacteremia and endocarditis. Infect Dis Clin North Am. 2002;16:413–435. x–xi. doi: 10.1016/s0891-5520(01)00003-4. [DOI] [PubMed] [Google Scholar]
- 5.Todd J, Fishaut M, Kapral F, Welch T. Toxic-shock syndrome associated with phage-group-I Staphylococci. Lancet. 1978;2:1116–1118. doi: 10.1016/s0140-6736(78)92274-2. [DOI] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) Stockholm: ECDC; 2011. Antimicrobial resistance surveillance in Europe 2010. [Google Scholar]
- 7.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol. 2004;186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janzon L, Lofdahl S, Arvidson S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol Gen Genet. 1989;219:480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- 10.Bronner S, Stoessel P, Gravet A, Monteil H, Prevost G. Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl Environ Microbiol. 2000;66:3931–3938. doi: 10.1128/aem.66.9.3931-3938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth MC, Atkuri RV, Nanda SK, Iandolo JJ, Gilmore MS. Accessory gene regulator controls Staphylococcus aureus virulence in endophthalmitis. Invest Ophthalmol Vis Sci. 1995;36:1828–1836. [PubMed] [Google Scholar]
- 16.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright JS, 3rd, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galperin MY. Telling bacteria: do not LytTR. Structure. 2008;16:657–659. doi: 10.1016/j.str.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J Med Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 22.Sidote DJ, Barbieri CM, Wu T, Stock AM. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 31.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 32.Grzesiek S, Dobeli H, Gentz R, Garotta G, Labhardt AM, Bax A. 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry. 1992;31:8180–8190. doi: 10.1021/bi00150a009. [DOI] [PubMed] [Google Scholar]
- 33.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 34.Furihata K, Shimotakahara S, Tashiro M. An efficient use of the WATERGATE W5 sequence for observing a ligand binding with a protein receptor. Magn Reson Chem. 2008;46:799–802. doi: 10.1002/mrc.2264. [DOI] [PubMed] [Google Scholar]
- 35.Mulder FA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 36.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eletsky A, Heinz T, Moreira O, Kienhofer A, Hilvert D, Pervushi K. Direct NMR observation and DFT calculations of a hydrogen bond at the active site of a 44 kDa enzyme. J Biomol NMR. 2002;24:31–39. doi: 10.1023/a:1020697627485. [DOI] [PubMed] [Google Scholar]
- 38.Fejzo J, Lepre CA, Peng JW, Bemis GW, Ajay Murcko MA, Moore JM. The SHAPES strategy: an NMR-based approach for lead generation in drug discovery. Chem Biol. 1999;6:755–769. doi: 10.1016/s1074-5521(00)80022-8. [DOI] [PubMed] [Google Scholar]
- 39.Lepre CA, Peng J, Fejzo J, Abdul-Manan N, Pocas J, Jacobs M, Xie X, Moore JM. Applications of SHAPES screening in drug discovery. Comb Chem High Throughput Screen. 2002;5:583–590. doi: 10.2174/1386207023329950. [DOI] [PubMed] [Google Scholar]
- 40.Traber K, Novick R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ- and α-haemolysins. Mol Microbiol. 2006;59:1519–1530. doi: 10.1111/j.1365-2958.2006.04986.x. [DOI] [PubMed] [Google Scholar]
- 41.Fowler SA, Stacy DM, Blackwell HE. Design and synthesis of macrocyclic peptomers as mimics of a quorum sensing signal from Staphylococcus aureus. Org Lett. 2008;10:2329–2332. doi: 10.1021/ol800908h. [DOI] [PubMed] [Google Scholar]
- 42.Mansson M, Nielsen A, Kjaerulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine photobacterium. Mar Drugs. 2011;9:2537–2552. doi: 10.3390/md9122537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto M, Sussmuth R, Vuong C, Jung G, Gotz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 44.Geisinger E, George EA, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–309. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 46.Hajduk PJ, Dinges J, Miknis GF, Merlock M, Middleton T, Kempf DJ, Egan DA, Walter KA, Robins TS, Shuker SB, Holzman TF, Fesik SW. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J Med Chem. 1997;40:3144–3150. doi: 10.1021/jm9703404. [DOI] [PubMed] [Google Scholar]
- 47.Artis DR, Lin JJ, Zhang C, Wang W, Mehra U, Perreault M, Erbe D, Krupka HI, England BP, Arnold J, Plotnikov AN, Marimuthu A, Nguyen H, Will S, Signaevsky M, Kral J, Cantwell J, Settachatgull C, Yan DS, Fong D, Oh A, Shi S, Womack P, Powell B, Habets G, West BL, Zhang KY, Milburn MV, Vlasuk GP, Hirth KP, Nolop K, Bollag G, Ibrahim PN, Tobin JF. Scaffold-based discovery of indeglitazar, a PPAR pan-active anti-diabetic agent. Proc Natl Acad Sci USA. 2009;106:262–267. doi: 10.1073/pnas.0811325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park CM, Bruncko M, Adickes J, Bauch J, Ding H, Kunzer A, Marsh KC, Nimmer P, Shoemaker AR, Song X, Tahir SK, Tse C, Wang X, Wendt MD, Yang X, Zhang H, Fesik SW, Rosenberg SH, Elmore SW. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51:6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 49.Wyatt PG, Woodhead AJ, Berdini V, Boulstridge JA, Carr MG, Cross DM, Davis DJ, Devine LA, Early TR, Feltell RE, Lewis EJ, McMenamin RL, Navarro EF, O’Brien MA, O’Reilly M, Reule M, Saxty G, Seavers LC, Smith DM, Squires MS, Trewartha G, Walker MT, Woolford AJ. Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a novel cyclin dependent kinase inhibitor using fragment-based X-ray crystallography and structure based drug design. J Med Chem. 2008;51:4986–4999. doi: 10.1021/jm800382h. [DOI] [PubMed] [Google Scholar]