Abstract
Recent evidence has shown that transcription is permissible through the purportedly repressive centromere domain, and that this transcriptional activity is of functional consequence. The best-studied example is transcription of the pericentric DNA repeats in the generation of siRNAs required for pericentric heterochromatin assembly in yeast. However, non-siRNA transcripts emanating from both pericentric and centromere core domains have also been detected in a cell cycle and cellular differentiation-dependent manner. Elevated levels of centromeric transcripts have also been detected in some cancers; however, it is still unclear how high levels of centromere transcripts may contribute towards disease progression. More recent studies have demonstrated that careful regulation of the histone modifications and transcription level at the centromere is vital for the recruitment of key centromere proteins and assembly of CENP-A domain. Here, we compare the transcriptional dynamics and function of various transcripts derived from pericentromeric and centromere core regions. We also propose a model in which the chromatin remodelling activity of transcription, and the resultant transcripts, contribute synergistically to perpetuate centromere chromatin identity.
INTRODUCTION
The centromere is a region of specialized chromatin found on all eukaryotic chromosomes, which functions to ensure faithful inheritance of the genome during cell division. This stably transmitted locus directs the assembly of the kinetochore, to mediate spindle capture during mitosis.
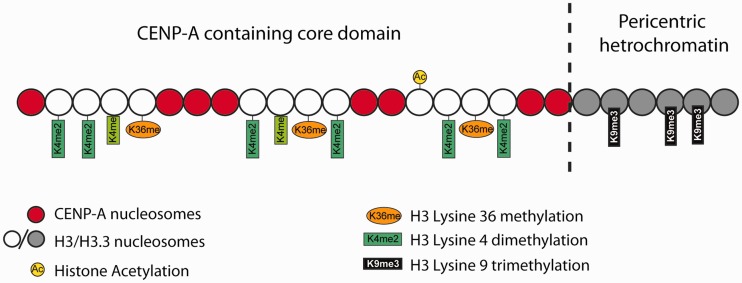
A strict centromere-defining consensus genetic sequence has never been found. Instead, most eukaryotic centromeres (with some exceptions, such as budding yeast Saccharomyces cerevisiae (S. cerevisiae) and Caenorhabditis elegans (C. elegans)) consist of megabases of species-specific repeat DNA, which defy precise sequence definition due to their repetitive nature. Two distinct domains are vital to centromere function, the centromere core domain and its flanking pericentric heterochromatin, which are epigenetically defined by different sets of proteins that dictate their structure and function (Figure 1). The centromere core domain, which specifies kinetochore formation, contains centromere-specific proteins, which form the constitutive centromere-associated network (CCAN) complex (1). The pericentric regions contain typical heterochromatin markers including Heterochromatin Protein 1 (HP1), H3K9 methylation (H3K9me) and DNA methylation.
Figure 1.
Pericentric heterochromatin consists of H3K9 trimethylation, which is vital for HP1 localization to the pericentric domains. The centromere core domain consists of clusters of CENP-A and H3 nucleosomes. In S-phase, both the canonical replication-dependent H3.1 and the replication-independent H3.3 are loaded onto centromere chromatin. The H3.1/H3.3 nucleosomes are enriched for H3K4 dimethylation and H3K36 methylation. No H3K4 trimethylation or H3K9 trimethylation could be detected at the centromere core domain. Although stretched chromatin fibre experiments indicate that histone acetylation is absent at the centromere core domain, but chromatin immunoprecipitation studies have detected a low level of H3 acetylation at the centromere core. However this histone acetylation may be tightly regulated by cell-cycle dynamics. It is currently unknown whether the histone modifications detected so far are carried by H3.1 or H.3 nucleosomes, or if the histone modifications show preferential enrichment at either H3.1 or H3.3.
Due to their inherently heterochromatic nature, it was assumed that centromeres were transcriptionally inert. However, a landmark study showed that small-interfering RNAs (siRNAs) derived from the pericentric domains of the fission yeast, Schizosaccharomyces pombe (S. pombe) centromere, are necessary to propagate and perpetuate their heterochromatic identity (2). This indicated that pericentric heterochromatin transcription was permissible and of functional significance. This discovery prompted a re-evaluation of the traditional definition of heterochromatin, and sparked an interest in non-coding RNA (ncRNA) dynamics in the regulation of hitherto ‘silenced’ chromatin regions, including that of the centromere core domain. To date, there is evidence to suggest that the entire centromeric region is transcriptionally competent. In this article, we will review the different classes of centromere ncRNA arising from transcription of both centromere core and pericentric domains of different model systems. We also discuss evidence that suggest centromere transcription is vital in the maintenance of centromere chromatin identity.
TRANSCRIPTION AT THE PERICENTRIC REGION
RNAi-mediated formation of pericentric heterochromatin in S. pombe
RNA interference (RNAi) is a post-transcriptional gene silencing mechanism mediated by siRNA and microRNAs, which regulate gene expression via the inhibition of mRNA translation, or direct degradation of target transcripts (for review, see (3)). However, as shown by Volpe et al., siRNAs can also modify chromatin at specific sites to lead to transcriptional silencing (2).
In S. pombe, the pericentric heterochromatin comprises outermost (otr) repeats called dg and dh. Sequencing of ribonuclease Dicer (dcr1)-generated small RNAs showed sequences that mapped to the pericentric repeats (4). Simultaneously, another study showed that deletion of S. pombe RNAi pathway genes—dcr1, the RNA-binding protein Argonaute (ago1) or the RNA dependent RNA polymerase (RdP1) were susceptible to chromosome missegregation due to defective pericentric heterochromatin formation. These mutants showed loss of repressive chromatin marks H3K9me and swi6 (S. pombe HP1 homologue), and aberrant accumulation of pericentric dg and dh transcripts. These studies indicated that the pericentric heterochromatin is transcribed and the transcripts generated are processed by RNAi machinery to direct heterochromatin assembly.
At the pericentric heterochromatin, the dg/dh transcripts are generated from the conserved cryptic promoters within the otr domain (5,6). The double-stranded RNA (dsRNA) transcripts are cleaved and processed by Dicer into siRNAs, which are then incorporated into the RNA-induced transcriptional silencing complex (RITS complex; consisting of Ago1, Tas3 and Chp1). Through an interaction with Ago1, the siRNAs guide the loading of the RITS complex to the cognate chromatin. Also, the RNA slicing activity of Ago1 helps to recruit the RNA-directed RNA polymerase Complex (RDRC; which consists of the polyA polymerase Cid12 and the helicase Hrr1) to the nascent transcript (siRNA precursor) for further production of dsRNAs (7–10). In addition, the direct association of the RDRC complex with Dicer promotes the production of more siRNAs to further amplify the RNAi signal. Importantly, the RITS complex also recruits Rik1—as part of the CLRC complex (along with Clr4, cullin protein Cul4, WD-40 protein Raf1, Dos1 and Dos2). Clr4 is the methyltransferase crucial for the H3K9me (11) that recruits heterochromatic proteins Swi6 and Chp1 (12). Clr4 is also able to bind existing H3K9me to facilitate the spreading of heterochromatin (13).
RNAPII transcription and RNAi-mediated pericentric heterochromatin assembly in S. pombe
In S. pombe, the pericentric DNA contains TATA-like and putative RNAPII promoter sequences (14), and analysis by 3′-RACE mapping confirmed that the pre-siRNA transcripts were polyadenylated (6)—indicating that RNA Polymerase II (RNAPII) was directly responsible for the generation of the pre-siRNA transcripts, and that RNAi machinery recruitment is coupled to RNAPII transcription. This is further supported by evidence of RNAPII subunits in promoting centromere transcription and RNAi-dependent heterochromatin assembly (6,14). Similar to Dicer, Clr4 and RdRP mutants, the RNAPII subunit Rpb2 mutants showed loss of transcriptional silencing, decreased association of H3K9me and Swi6, at the pericentric dg/dh regions (14). Interestingly, the Rpb2 mutants also showed loss of pericentric-derived siRNAs but elevated levels of corresponding pre-siRNA transcripts. This suggested that the silencing defect did not lie with the transcription of pre-siRNA transcripts, but possibly, with the inefficient downstream processing events that generate the siRNAs. This is in contrast with the mutation of other RNAPII subunit, Rpb7 that also showed loss of transcriptional silencing, accompanied by increased chromosome missegregation (6). In the Rpb7 mutants, there was a decrease in the level of pre-siRNA transcripts although RNAPII recruitment at the pericentric region was not impaired. It was proposed that the loss of transcriptional silencing was caused by a defect in RNAPII transcription initiation (6). Together, these studies suggest that it is likely that siRNA processing is coupled to RNAPII-dependent pericentric transcription, to direct vital chromatin modifying events for heterochromatin maintenance.
Heterochromatin is normally inaccessible to transcriptional machinery; thus, it seems paradoxical that the pericentric heterochromatin is transcribed to recruit the heterochromatin apparatus. The levels of siRNAs peak in S-phase, coincident with the localization of CLRC and RITS complex subunits at the pericentric repeats (15,16). RNAPII is also preferentially recruited to transcribe both forward and reverse strands of the pericentric repeats during S-phase—presumably when silencing is briefly alleviated as heterochromatin markers are distributed onto newly replicated strands (15,16). However, to date, it remains unknown how RNAPII is recruited to the repressive pericentric region. It has been proposed that Epe1, a JmjC domain protein is recruited by Swi6, possibly to facilitate RNAPII transcription of heterochromatic repeats (17). However, other work has suggested that Epe1 regulate heterochromatin assembly independently of the RNAi pathway and transcription (18). Thus, the factors that recruit RNAPII to heterochromatic regions are still unclear.
Dynamics of pericentric heterochromatin transcription in vertebrate systems
Although it is clear that RNAi pathway is important to pericentric heterochromatin assembly in S. pombe, the role of pericentric transcription and RNAi-mediated heterochromatin assembly in vertebrates is less straightforward. There have been conflicting reports in terms of the size of the transcripts (Table 1), cell-cycle expression pattern and their role in heterochromatin assembly.
Table 1.
Listing of transcripts detected emanating from pericentric heterochromatin and centromere core domain in different model systems
| Organism | Derived from | RNA characteristics | References |
|---|---|---|---|
| Schizosaccharomyces pombe | Pericentric heterochromatin | ||
| • Dg and dh sequences from the outermost repeat domains | RNAPII generated | 14 | |
| Double stranded, 21–25 nt siRNAs | 2 | ||
| Centromere core | |||
| • Cnt and imr repeats | ∼0.5 kb transcripts | 45 | |
| • tRNA genes found within the imrI region | RNAPIII transcribed to generate functional tRNAs | 49, 50 | |
| Saccharomyces cerevisiae | Centromere core | RNAPII generated | 41 |
| Zea mays (Maize) | Centromere core | Single stranded, 40–900 bp | 67 |
| • Centromeric retroelements (CRs) | |||
| • Satellite repeat CentC | |||
| Human | Endogenous centromeres | ||
| Centromere core | RNAPII generated | ||
| • α-satellite | Single stranded | 66, 71 | |
| Pericentric heterochromatin | |||
| • α-Satellite | |||
| • Satellite II | 2–5 kb | ||
| • Satellite III | 33–38 | ||
| Neocentromeres | |||
| Centromere core | |||
| • Mardel(10): L1 retroelement | Single stranded | 72 | |
| • Genes found within the 10q25 neocentromere | 64 | ||
| Rice | CRR retrotrasposon | dsRNA, 23–24 nt | 62, 63 |
| Active genes | |||
| Tammar Wallaby | Centromeric | ||
| • sat23 | dsRNA, 34–42 nt, 60 nt and 100 nt | 68 | |
| • KERV-1 | |||
| Mouse | Centromere core | ||
| • Minor satellite | 120 nt, 2 kb, 4 kb transcripts | 69, 70 | |
| Pericentric heterochromatin | 22, 23, 25–30 | ||
| • Major satellite | 25–30 nt, 150 nt, >1 kb + transcripts |
Dicer is an essential gene in vertebrates—Dicer knockout mice and zebrafish are embryonic lethal (19,20). Thus, the study of RNAi pathway in heterochromatin assembly in vertebrate cells has been limited to cell-culture-based systems. In human–chicken hybrid cells (chicken DT40 cells carrying a human chromosome), ablation of Dicer led to mitotic defects as a result of premature sister chromatid separation. This was attributed to the loss of HP1 at the pericentric regions, misregulation of cohesin and the mitotic checkpoint protein BubR1. Notably, as in S. pombe, depletion of Dicer caused an accumulation of pericentric transcripts, as evidenced by the detection of long α-satellite and satellite III transcripts (more discussion on Satellite III transcription below). This study was the first to demonstrate the importance of Dicer/RNAi machinery for heterochromatin assembly in vertebrate cells (21).
Dicer-deficient mouse embryonic stem (ES) cells, although viable in culture, were defective in cellular differentiation (22,23). Similar to S. pombe and chicken cells, Dicer deficiency in mouse ES cells caused an accumulation of pericentric Major satellite transcripts, ranging from 40 nt to over 200 nt in size (22,23), along with other normally repressed DNA repeats such as L1 and IAP elements (23). This indicated a role for Dicer in the repression of pericentric regions and other normally silenced genetic elements in ES cells. However, no aneuploidy or genomic instability was observed in Dicer-depleted mouse ES cells—although, the cells were significant less proliferative than wildtype ES cells (22,23). In the literature, there is a difference of opinion as to whether the Dicer/RNAi pathway is essential for the regulation of heterochromatin assembly in mouse ES cells. Kanellopoulou et al. reported loss of DNA methylation and H3K9me3 at the pericentric regions of Dicer-deficient cells. Whereas, Murchison et al. concluded that Dicer function was dispensable for the maintenance of pericentric heterochromatin, as no significant loss of DNA methylation or H3K9me3 was detected.
Heterogeneity of pericentric heterochromatin transcripts
It is well documented that an RNA component is necessary for the binding of HP1 at pericentric heterochromatin (24,25), and it was thought that the Dicer-generated siRNAs could be this RNA component. However, other than in chicken cells (21), canonically sized (21–25 nt) siRNAs derived from the pericentric domain have been difficult to detect in vertebrates. In mouse ES cells, a 150 nt centromeric transcript, as well as a 25–30 nt RNA species have been detected (23). Although these were not the canonical sized 21–25 nt siRNAs, the generation of the 25–30 nt transcripts were Dicer-dependent, which led to the suggestion that these RNA molecules maybe analogous to heterochromatic siRNAs found in S. pombe (23).
In mouse cells, non-siRNA-sized pericentric-derived transcripts have also been detected (26,27). A study in NIH 3T3 cells showed two distinct RNA species derived from the pericentric region; a G2/M-specific 150 nt RNA, and a G1-phase 1 Kb+ transcript, which was undetectable by mid-S-phase, coincident with centromere DNA replication. The significance of this cell-cycle-regulated pericentric transcription remains unknown, but the transcriptional activity was independent of Suv39h1/2 H3K9 methyltransferase activity (26). This is in contrast with S. pombe, in which S-phase pericentric transcription is linked to the maintenance of H3K9me and HP1 at the pericentric heterochromatin (15,16). However, a more recent study reported a functional link between ncRNA and heterochromatin assembly in mammalian cells. In mouse cells, large Major satellite transcripts (several repeat lengths in size; not siRNA transcripts) were detected (27) and interestingly, HP1α showed a greater association with the forward strand Major satellite RNA over the reverse transcript. This interaction between the forward RNA strand and HP1α was dependent on SUMO (Small Ubiquitin-like Modifier)-ylation of HP1α at the hinge domain. It was proposed that the tendency of the purine-rich forward strand towards secondary structures could positively regulate the RNA–HP1α interaction and heterochromatin assembly (27). The cell-cycle dynamics of the Major satellite RNA–HP1 association is currently undefined. It is not known if the previously reported G1 expression of Major satellite transcripts (26) is linked to HP1 re-loading during G1. In future work, it would be interesting to investigate the factors regulating pericentric transcription during the cell cycle, and its link with HP1 recruitment in mammalian cells.
Major satellite transcripts have also been implicated in developmental progression in mouse embryos. In the 2-cell embryo, there is an initial peak in the production of forward Major satellite transcripts, followed by a peak in the level of the reverse strand. However, by the 4-cell stage, both strands of Major satellite transcripts were rapidly down-regulated. This burst of transcriptional activity overlapped with the reorganization of pericentric region into heterochromatic chromocentres in the early embryo. Disruption of these transcripts led to developmental arrest at the 2-cell stage and failed chromocentre formation—indicating a role for these transcripts in the de novo heterochromatin formation in the developing mouse embryo (28). Considering the association of long ncRNA with SUMOylated HP1 in mouse cells (27), it remains to be determined if SUMOylation of HP1 and strand-specific Major satellite transcripts are also critical for HP1 accumulation and formation of the chromocentre in the early embryo.
Pericentric heterochromatin transcription during differentiation, cellular stress and disease
Pericentric heterochromatin transcription dynamics has been linked to cell proliferation and differentiation, or cellular stress. Large, 1.8 kb Major satellite transcripts have been detected in the developing mouse embryo as well as in adult liver and testis tissue. These transcripts were down-regulated in the presence of the differentiation agent retinoic acid, suggesting a link between Major satellite transcription and cell proliferation (29). However, the detection of Major satellite RNA in the senescent cardiac muscle of aging mice (30) suggests that the relationship between pericentric transcription and proliferative capacity is not straightforward.
Pericentric transcription in human cells have also been linked to the cellular stress response. Relatively less is known of pericentric transcription in human cells, and this is mostly due the homogeneity of α-satellite DNA in pericentric and centromere core regions in human cells. This makes it difficult to discern the origin of α-satellite transcripts—unlike in mouse centromeres, in which Minor and Major satellite repeats distinguish the centromere core and pericentric regions, respectively. However, within the human pericentric regions, there are other repeat DNAs, such as SINEs, LINEs (31) and Satellite III (predominantly found at the pericentric regions of chromosomes 9, 13–15, 21, 22 and Y) (32)). RNAPII-mediated Satellite III transcription has been linked to the heat-shock stress response (33,34), and has been shown to be required for the targeting of splicing factors to nuclear stress granules (35). More recently, Satellite III RNA expression has been implicated in cellular stress responses to heavy metal exposure, oxidative and hyper-osmotic stress (36). Altogether, these studies indicate that pericentric RNA transcription maybe differentially activated in response to stress conditions.
The accumulation of RNA transcripts may reflect the derepression of heterochromatic regions under diseased or stressed conditions. A recent study showed that a higher abundance of α-satellite and Satellite II transcripts was found in pancreatic and epithelial cancer cells (37). In addition, α-satellite expression was also found to be elevated in Breast Cancer Susceptibility Gene 1 (BRCA1)-deficient cancer cells. These BRCA1-deficient cancer cells show defective heterochromatin formation, as indicated by the severe reduction in levels of HP1 and ubiquitylation of histone H2A (H2Aub) at the satellite repeats. Importantly, the ectopic α-satellite expression in normal cells has been shown to cause genomic instability and an impaired spindle checkpoint, similar to BRCA1 deficiency (38). It will be exciting in the future to investigate if overexpression of pericentric sequences is merely a consequence of global heterochromatin derepression, or of etiological significance in cancer formation.
TRANSCRIPTION AT THE CENTROMERE CORE DOMAIN
In comparison to the relatively well-characterized transcription at the pericentric regions, the centromere core transcription is more enigmatic. However, evidence is accumulating that the centromere core domain is also transcribed, though much less is known of the dynamics or function of this transcriptional activity.
Centromere core transcription in the two model yeasts
Although much of the RNAi-linked pericentric heterochromatin data were originally elucidated in S. pombe, only recently have both model yeasts (S. pombe and the budding yeast S. cerevisiae) been utilized to examine the transcriptional status of the centromere core domain.
In S. cerevisiae, Cbf1, a transcription factor (TF) involved in methionine biosynthesis (39), binds the core centromere DNA as a bona fide constitutive centromere protein (40). Recently, it was shown that Cbf1 is required for the production of centromeric transcripts. Furthermore, two novel centromere-binding proteins and TFs, Ste12 and Dig1 regulate Cbf1 activity. Loss of Cbf1, Ste12 or Dig1 resulted in chromosomal instability. Interestingly, this chromosomal instability phenotype of Cbf1 mutants could be rescued by driving centromere transcription from an artificial promoter, indicating that transcription at the centromere is vital to centromere function in S. cerevisiae (41).
In S. pombe, the insertion of a reporter gene within the CENP-A containing central core domain (cnt and imr DNA) resulted in variable levels of reporter gene repression (42). This silencing activity correlated with the increase in CENP-A incorporation at the centromere core domain, which suggested that CENP-A nucleosomes have a negative impact on transcription at the yeast centromere core domain (43). In contrast, the binding of a GATA-type zinc finger TF was necessary for the continued association of CENP-A at the centromere in S. pombe (44). Indeed, ncRNA transcripts have been detected emanating from the CENP-A domain of S. pombe centromere. These transcripts were generated by RNAPII, but were subjected to rapid turnover rates by exosome subunits (45). The function of these centromeric transcripts, and the significance of their rapid degradation is currently unclear.
RNA Polymerase III-mediated chromatin barrier within S. pombe centromeres
Schizosaccharomyces pombe centromeres also have distinct, non-RNAPII transcriptional activity within its core domain. Sequencing of S. pombe centromeres indicated that all three S. pombe centromeres contain clusters of tRNA genes (46,47), which demarcate the pericentric heterochromatin chromatin (48) from the centromere core domain. Despite their proximity to the repressive pericentric heterochromatin, the tRNA genes were actively transcribed by RNA Polymerase III (RNAPIII), (49). Deletion of the tRNA clusters resulted in expansion of heterochromatin into the CENP-A chromatin—indicating that the tRNA genes functioned as chromatin barriers at the centromere. This barrier activity was independent of tRNA gene orientation, but reliant on the DNA sequence required for RNAPIII complex formation (49,50). However, it is currently unknown whether transcription, or simply the formation of the RNAPIII complex is sufficient for the chromatin barrier activity of the tRNA genes.
To date, no equivalent RNAPIII complex binding at mammalian centromeres has been reported. The retrotransposon SINE, which is abundant throughout mammalian genomes, as well as human pericentric regions, is transcribed by RNAPIII. SINE transcription has been implicated in the establishment of boundary elements and chromatin insulators in organization of chromatin domains elsewhere in the genome (51,52). RNAPIII driven transcription of SINE, or of yet unidentified DNA elements at mammalian centromeres could function to insulate centromere chromatin from bulk chromatin.
Lessons from transcription within variant and neo-centromeres
There are instances where fully functional ectopic centromeres (called neocentromeres) arise at non-repetitive regions. Clinical neocentromeres forming within gene-poor regions initially supported the view that centromere function was incompatible with transcription (for review, see (53)). Furthermore, analysis of three independent human neocentromeres at the 13q32 genomic region showed no overlap of CENP-A chromatin within coding sequences (54). Non-tandem-repetitive DNA-based centromeres have also been found to occur in some model organisms. In chicken, centromeres of chromosomes 5, 27 and Z were devoid of tandem-repetitive sequences, and were proposed to be evolutionary new centromeres (ENCs) (55). Interestingly, analyses of Equus and Orangutan genomes indicated that evolutionary centromere repositioning events had given rise to neocentromeres at gene-free genomic sites (56,57).
In Candida albicans, deletion of endogenous centromeres led to neocentromere formation at intergenic regions, where they exerted repressive effects on nearby genes. Neocentromere formation on a marker gene led to its repression. When induced for marker gene expression, the neocentromere shifted away from the expressed loci, only to shift back again when restored onto non-selective media (58). This not only illustrated the plasticity of the neocentromere domain, but also suggested that in C. albicans, centromere core transcription is not favourable.
In S. pombe, conditional deletion of the endogenous centromere also led to neocentromere formation, but mostly at the subtelomeric regions. The CENP-A regions were devoid of repeat DNA, but overlapped with several open reading frames. Interestingly, these open reading frames were low-expression genes that are normally upregulated upon nitrogen starvation. However, these gene clusters remained lowly expressed post-neocentromere formation, even upon nitrogen media withdrawal. This indicated that neocentromere formation could not tolerate high rates of transcription (59).
Despite the above evidence that CENP-A chromatin is incompatible with transcriptional activity, it is intriguing that CENP-A chromatin could assemble over non-alphoid sequences, including that of an expressed marker gene during the construction of alphoid-based human artificial chromosomes (HACs), (60). Furthermore, coding sequences were found embedded within repetitive sequences of native rice centromeres, and it has been demonstrated that these genes are actively transcribed in most plant tissues (61–63). This is in contrast to most canonical centromeres, which are filled with repetitive DNA and do not contain any genes. A human neocentromere, the 10q25 Mardel(10) neocentromere, is unusual in that the transcriptional competency within the neocentromeric region remains intact, despite the fact that genes are embedded within CENP-A domains (64). However, it is important to note that none of the genes associated with 10q25 Mardel(10) neocentromere CENP-A domains are expressed at high levels.
Centromere RNA as an integral component of centromere chromatin and mitotic kinetochore
Early electron microscopy studies indicated the presence of RNA at the kinetochore of animal cells (65), but the sequence and origin of the RNA was unknown. Recently, we showed that RNAPII and its TFs localize to the mitotic kinetochore of mammalian cells. Importantly, this kinetochore-bound RNAPII complex is engaged in active transcription and is required for kinetochore function (66). Although it is not known whether this mitotic transcriptional activity traverses the CENP-A domains, it indicated that the centromere core region is amenable to transcription, and most interestingly, it is transcribed during mitosis (see below for further discussion), a purportedly transcriptionally silent phase of the cell cycle—further highlighting the uniqueness of the centromere chromatin. Other studies have also shown the association of centromere core-derived transcripts and centromere core proteins—raising interesting questions about the function of centromere core transcriptional activity and its resultant ncRNA. For example, in maize, both forward and reverse strands of centromeric CentC satellite repeat and CRM retrotransposon transcripts have been found tightly associated with CENH3 (maize CENP-A). These transcripts did not resemble siRNAs, they were single stranded, and much larger in size—ranging 40–200 nt (67).
In mammals, centromere core transcripts have also been detected, however, their size and dynamics appear variable (Table 1 and detailed below). In tammar wallaby cells, a novel class of small RNAs, 34–42 nt Centromere Repeat-Associated Small-Interacting RNAs (crasi-RNAs) derived from the marsupial-specific KERV1 retrotransposon has been detected (68). In mouse cell lines, the centromere core Minor satellite transcripts were found to be tightly associated with centromere core chromatin (69,70). These Minor satellite transcripts were large, ranging from 2 kb to 4 kb. In non-cycling, differentiated or stressed cells, a 120 nt RNA population is also found (69).
In human cells, single-stranded centromeric α-satellite transcripts, along with centromere core proteins CENP-C and INCENP, have been detected in the nucleolus in interphase cells (71). Besides transcription of endogenous alphoid-based centromeres, transcripts derived from a full-length LINE-1 (L1) retrotransposon found within the Mardel(10) 10q25 neocentromere the CENP-A domain was found to bind CENP-A chromatin. Specific depletion of the Mardel(10) L1 transcripts resulted in the loss of CENP-A binding and mitotic stability of the neocentromeric marker chromosome (72). The close association of these centromeric transcripts, in particular those derived from retrotransponsons, with CENP-A chromatin, suggests that centromere core derived, ncRNA function as critical epigenetic determinants of centromere chromatin identity.
Centromere RNA also associates with centromere proteins other than CENP-A. In mouse cells, Minor satellite RNA is associated with the M-phase-specific Chromosomal Passenger Complex (CPC; consisting of Aurora Kinase B (AUKB), INCENP, Survivin) and has been implicated in regulating AUKB activity (70). Depletion of centromere core transcripts led to reduced AUKB kinase activity (70), which may result in failure of detection and correction of erroneous microtubule–kinetochore interactions, and eventual loss of mitotic checkpoint integrity. Similarly, in human cells, the localization of CENP-C, INCENP and Survivin at the mitotic kinetochore was also dependent on the presence of single-stranded α-satellite RNA. CENP-C, a DNA-binding protein was shown to have RNA-binding capacity (71). An in vitro study showed that the DNA binding of CENP-C is enhanced and stabilized by the presence of long single-stranded RNA (ssRNA) (73). Our recent study also provided further evidence of centromere transcripts as key components for the assembly of the mitotic kinetochore. Inhibition of RNAPII activity, and consequent depletion of α-satellite RNA in mitotic cells induced chromosome missegregation as a result of reduced CENP-C binding at lagging chromosomes (66). This suggests that RNAPII-generated mitotic α-satellite transcripts may function to stabilize CENP-C binding at the kinetochore (66). Considering centromeric transcripts associate with SUMOylated HP1 for pericentric targeting (27), and that CENP-C has been shown to genetically interact with the SUMO pathway (74), it would be interesting to see if CENP-C SUMOylation or other post-translation modifications could regulate CENP-C/RNA association.
In consideration of the evidence above, we propose that centromere transcripts are important for mitotic kinetochore function during mitosis. The evidence of centromere RNA association with CENP-C and non-enzymatic partners of the CPC (70,71,73) suggests that centromere core RNA could act as a molecular scaffold in the recruitment and organization of key centromere proteins (Figure 2A). An analogous example is telomerase RNA, which tethers proteins to the ends of the chromosomes (75). The centromere RNA discussed above could function to recruit centromere/kinetochore proteins for maturation of the kinetochore, and to stabilize the overall kinetochore structure during the G2/M-phase. Thus, depletion of centromere transcripts could potentially affect the structural integrity of the kinetochore, which in turn could further erode the spatial regulation of CPC, including AUKB and/or other mitotic checkpoint signalling protein activities at the kinetochore–microtubule interface (76).
Figure 2.
(A) Centromere RNA has been shown to associate with CENP-C and stabilize its DNA-binding ability. The localization of CENP-C has been shown to be dependent on the presence of ssRNA at the mitotic kinetochore (71). Centromere RNA has also been shown to associate with CPC proteins, Survivin, INCENP and to mediate the kinase activity of another CPC proteins, AUKB (70). This suggests that centromere RNA could act as a molecular scaffold at the mitotic kinetochore to recruit and organize kinetochore proteins at the centromere. (B) The act of transcription could also have an important function. The histone chaperone and chromatin remodeller, FACT complex and CHD1, has been shown to be important for CENP-A loading (90). The nucleosome destabilization activity of FACT could function to promote RNAPII transcription through the compact CENP-A chromatin, while RNAPII transcription could drive further chromatin remodelling at the centromere domain. In particular transcription could promote histone acetylation. A peak of histone acetylation has been reported to occur during mitosis (92). RNAPII transcription could recruit HAT complexes at the mitotic kinetochore to generate an acetylated chromatin environment, which has been shown to be favourable for CENP-A loading.
Low-level RNAPII transcription is vital for centromere function
Extended-chromatin fibre experiments have shown that human and Drosophila centromeres consist of clusters of CENP-A nucleosomes interspersed with canonical H3 chromatin (77), which were enriched with active chromatin marks H3 dimethylated lysine 4 (H3K4me2) and hyper-methylated K36, (78,79), but lacked the repressive H3K9me normally associated with pericentric heterochromatin (80,81) (Figure 1). Recently, it has been shown that the replication-independent histone variant H3.3 is loaded together with H3 onto centromere core chromatin during S-phase (82) (Figure 1). H3.3 is commonly associated with transcriptional competency—it is enriched at transcribed regions and it promotes post-translational modifications associated with active chromatin. However, it is not known whether this population of H3.3 at the centromere is enriched for the active modifications reported in earlier studies.
Recent studies have utilized HACs and centromere-targeted chromatin modifiers to manipulate the epigenetic state of centromere chromatin. Both targeting of transcriptional activators and silencers to HAC centromeres, to induce a more open or closed chromatin configuration, resulted in the loss of kinetochore function and increased chromosome missegregation, suggesting that centromere function required a dynamic balance between permissive and repressive chromatin (83). It is however interesting that the induction of a more closed configuration and the accumulation of H3K9me3 at the centromere caused a more severe phenotype, with higher rates of HAC loss due to rapid loss of centromere proteins CENP-A, CENP-B and CENP-C (83). The importance of non-repressive chromatin state at the centromere was also demonstrated by a recent work that showed that depletion of H3K4me2 at a HAC centromere rapidly induced loss of RNAPII transcription at the underlying DNA, and loss of CENP-A loading due to failure in HJURP (CENP-A chaperone) recruitment (80). On the other hand, a completely open chromatin configuration permissive for high level of active transcription was incompatible with centromere function as well (83), indicating that centromere chromatin state must be carefully modulated to maintain centromere identity.
Other than the maintenance of histone modifications at the centromere core domain, the level of transcriptional activity also appears to be important to centromere function. In a recent study, the artificial seeding of H3-acetylated chromatin at a HAC kinetochore caused no deleterious effects. However, when the same hyperacetylated modification was coupled with elevated transcription rates, the HAC kinetochore was rapidly inactivated due to defective CENP-A chromatin assembly (84). This, in combination with the data of neocentromeres forming at lowly expressed regions (59,64), suggest that only low levels of transcriptional activity is compatible with centromere function. High level of transcription has been associated with nucleosome eviction (85,86), and this rapid turnover could cause untimely erasure of histone modifications and loss of centromeric histone CENP-A at the centromere core domain. This need for low transcriptional activity is reflected by the lack of euchromatic marks such as H3K4-trimethylation and histone hyperacetylation at the centromere core chromatin (80,81). Thus, meticulous regulation of the level of transcriptional activity within the centromere core is likely to be vital for centromere chromatin maintenance.
RNAPII transcription requires the action of chromatin modifiers and histone chaperons, which allow the RNAPII access to the DNA template and to reassemble chromatin in the wake of the receding polymerase (87). At the centromere core, the low-level transcriptional activity could be important to recruit the remodelling complexes required to modify centromere chromatin. Specifically, RNAPII transcription at the centromere could facilitate CENP-A loading. Interestingly, Facilitates Chromatin Transcription (FACT) complex, a factor required for RNAPII transcription through chromatinized DNA templates, (88) co-purified with CENP-A nucleosomes (89) and has been shown to be required for the incorporation of CENP-A in both chicken and human cells (90). Like RNAPII, FACT subunit was also been found to be present at the core centromere during mitosis (66,90). During mitosis, the nucleosome destabilization activity of FACT may function to promote RNAPII transcriptional activity through condensed CENP-A chromatin domain such as the centromere core. Additionally, the RNAPII transcriptional activity could help to maintain the CENP-A domain in an ‘open’ chromatin state. Indeed, although the endogenous centromere chromatin is predominantly maintained in a hypoacetylated state (91), an ‘open’ chromatin state enriched with acetylated H3K9 has been shown to be favourable for CENP-A loading (91,92). Interestingly, a histone acetyltransferase (HAT) complex has been detected at active mitotic kinetochores (93), while more recently, a transient peak of centromeric H3K9 acetylation has been detected during mitosis (92), prior to CENP-A loading in telophase and early G1 (94). The RNAPII transcription and associated chromatin remodelling activities could have an important role in promoting histone acetylation to generate a chromatin environment favourable for CENP-A loading; however, this hypothesis awaits confirmation (Figure 2B).
Centromere RNA and CENP-C in the regulation of centromere core chromatin
In addition to what is already known about the interaction of CENP-C and centromere RNA, there is new evidence that indicate that CENP-C also interacts with other chromatin modifying proteins to regulate centromere core chromatin (Figure 3). CENP-C has been shown to recruit DNA Methyltransferase 3A-B (DNMT3A-B) to direct DNA methylation at the centromere. CENP-C depletion led to loss of DNA methyltransferase 3B (DNMT3B) recruitment and DNA methylation, accompanied by loss of H3K4me2, H3K9me3 and HP1 binding, and increased centromere transcription (95). Interestingly, independent studies have shown CENP-C interacts with Mis18 complex proteins, Mis18α and M18BP1 (96,97), which control the histone acetylation profile at the centromere core domain (91). Mis18α, through its interaction with DNMT3A-B could control DNA methylation and the histone modification profile at centromere core chromatin (96), whereas CENP-C interaction with M18BP1 was important to promote the recruitment of HJURP for CENP-A loading (97).
Figure 3.
RNAPII localizes to the metaphase kinetochore during mitosis. Transcription of the underlying DNA generates non-coding transcripts that stabilizes the binding of CENP-C at the mitotic kinetochore. The centromere RNA/CENP-C complex recruits DNMT3B complex at the metaphase, where it persists until telophase. The centromeric enrichment of DNMT3 complex presumably leads to methylation of centromeric DNA. As cells undergo transition into telophase, the Mis18 complex (M18BP1, Mis18α and Mis18β) is recruited to the centromere. M18BP1 is recruited by CENP-C (97), while it has been shown that Mis18α-DNMT3B directly interact (96). The CENP-C/DNMT3/Mis18 complex may facilitate the recruitment of other histone modifiers to further remodel the centromeric chromatin. As cells enter G1, the continued presence of Mis18 complex at the centromere recruits the CENP-A chaperone HJURP, which facilitates CENP-A loading to ensure centromere chromatin inheritance.
Considering the role of RNA in stabilization of CENP-C at the mitotic kinetochore (66,71), centromere RNA could also stabilize the localization of CENP-C/DNMT3A-B/Mis18 complex at the kinetochore. Although we have cannot rule out that RNAPII transcription at the centromere occurs during other parts of the cell cycle, RNAPII localization at the mitotic kinetochore is most apparent from metaphase through to anaphase. Similarly, DNMT3A/B recruitment is most prominent from metaphase through to telophase (95), whereas the Mis18 complex first localizes to the mitotic kinetochore during late anaphase (91). The stabilization of the CENP-C/DNMT3A-B/Mis18 complex by centromere RNA could facilitate targeting of HJURP (through M18BP1) to initiate CENP-A loading (Figure 2). Thus, we propose that centromere RNA is required as a vital component of the stepwise remodelling of centromere chromatin required for centromere chromatin inheritance (Figure 3).
Transcription or RNA?
Despite the heterogeneity of the resultant transcripts, transcription of the pericentric heterochromatin and centromere core domains function to direct distinct, vital chromatin modifying events for the definition of centromere chromatin identity. Pericentric heterochromatin ncRNA, be it siRNAs in S. pombe, or long ssRNA in mammalian cells, are inextricably linked to HP1 dynamics, whereas centromere core-derived transcripts is tightly linked to CENP-A chromatin and mitotic kinetochore assembly. It is not impossible to imagine a scenario in which both transcription and the ncRNA function synergistically—transcription through the centromere core domain could drive chromatin remodelling with its accessory TFs, whereas the RNA functions to stabilize important centromere proteins and chromatin modifiers to ensure centromere chromatin inheritance (Figure 3).
FUNDING
Funding for open access charge: National Health and Medical Research Council (NHMRC) of Australia [AP1031872].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
L.H.W. received a Career Development Award from the NHMRC.
REFERENCES
- 1.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 3.Ketting RF. The many faces of RNAi. Dev. cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 5.Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E, et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 2009;28:3832–3844. doi: 10.1038/emboj.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 8.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 12.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 14.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 15.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 16.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 21.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 22.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Gene. Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 28.Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- 30.Gaubatz JW, Cutler RG. Mouse satellite DNA is transcribed in senescent cardiac muscle. J. Biol. Chem. 1990;265:17753–17758. [PubMed] [Google Scholar]
- 31.Prades C, Laurent AM, Puechberty J, Yurov Y, Roizes G. SINE and LINE within human centromeres. J. Mol. Evol. 1996;42:37–43. doi: 10.1007/BF00163209. [DOI] [PubMed] [Google Scholar]
- 32.Tagarro I, Fernandez-Peralta AM, Gonzalez-Aguilera JJ. Chromosomal localization of human satellites 2 and 3 by a FISH method using oligonucleotides as probes. Hum. Genet. 1994;93:383–388. doi: 10.1007/BF00201662. [DOI] [PubMed] [Google Scholar]
- 33.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc'h C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metz A, Soret J, Vourc'h C, Tazi J, Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 2004;117:4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- 36.Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell KF, Baker RE. Possible cross-regulation of phosphate and sulfate metabolism in Saccharomyces cerevisiae. Genetics. 1992;132:63–73. doi: 10.1093/genetics/132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemmerich P, Stoyan T, Wieland G, Koch M, Lechner J, Diekmann S. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc. Natl Acad. Sci. USA. 2000;97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 2011;21:1695–1703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 43.Castillo AG, Mellone BG, Partridge JF, Richardson W, Hamilton GL, Allshire RC, Pidoux AL. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007;3:e121. doi: 10.1371/journal.pgen.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 45.Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011;286:23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J. Mol. Biol. 1991;218:13–17. doi: 10.1016/0022-2836(91)90867-6. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn RM, Clarke L, Carbon J. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc. Natl Acad. Sci. USA. 1991;88:1306–1310. doi: 10.1073/pnas.88.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Gene. Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 49.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 50.Scott KC, White CV, Willard HF. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE. 2007;2:e1099. doi: 10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roman AC, Gonzalez-Rico FJ, Molto E, Hernando H, Neto A, Vicente-Garcia C, Ballestar E, Gomez-Skarmeta JL, Vavrova-Anderson J, White RJ, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 53.Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- 55.Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 60.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl Acad. Sci. USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 62.Yan H, Ito H, Nobuta K, Ouyang S, Jin W, Tian S, Lu C, Venu RC, Wang GL, Green PJ, et al. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell. 2006;18:2123–2133. doi: 10.1105/tpc.106.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH. Transcription within a functional human centromere. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 65.Rieder CL. Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J. Cell Biol. 1979;80:1–9. doi: 10.1083/jcb.80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan FL, Marshall OJ, Saffery R, Won Kim B, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl Acad. Sci. USA. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl Acad. Sci. USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O'Neill MJ, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 69.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl Acad. Sci. USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genetics. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukagawa T, Regnier V, Ikemura T. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 2001;29:3796–3803. doi: 10.1093/nar/29.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Welburn JP, Vleugel M, Liu D, Yates JR, III, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 79.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 80.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 87.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol. Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 89.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, III, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 90.Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 92.Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 94.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum. Mol. Genet. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, et al. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 97.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]