Abstract
Red1, Hop1 and Mek1 are three yeast meiosis-specific chromosomal proteins that uphold the interhomolog (IH) bias of meiotic recombination. Mek1 is also an effector protein kinase in a checkpoint that responds to aberrant DNA and/or axis structure. The activation of Mek1 requires Red1-dependent Hop1-Thr(T)318 phosphorylation, which is mediated by Mec1 and Tel1, the yeast homologs of the mammalian DNA damage sensor kinases ATR and ATM. As the ectopic expression of Mek1-glutathione S-transferase (GST) was shown to promote IH recombination in the absence of Mec1/Tel1-dependent checkpoint function, it was proposed that Mek1 might play dual roles during meiosis by directly phosphorylating targets that are involved in the recombination checkpoint. Here, we report that Mek1 has a positive feedback activity in the stabilization of Mec1/Tel1-mediated Hop1-T318 phosphorylation against the dephosphorylation mediated by protein phosphatase 4. Our results also reveal that GST-Mek1 or Mek1-GST further increases Hop1-T318 phosphorylation. This positive feedback function of Mek1 is independent of Mek1’s kinase activity, but dependent on Mek1’s forkhead-associated (FHA) domain and its arginine 51 residue. Arginine 51 directly mediates the interaction of Mek1-FHA and phosphorylated Hop1-T318. We suggest that the Hop1–Mek1 interaction is similar to the Rad53-Dun1 signaling pathway, which is mediated through the interaction of phosphorylated Rad53 and Dun1-FHA.
INTRODUCTION
In most sexually reproducing organisms, programmed DNA double-strand breaks (DSBs) are necessary for generating crossovers (COs) between homologous chromosomes to ensure accurate segregation during the first meiotic division (1). In contrast, the recombination-mediated repair of DSBs uses the sister templates during mitosis (2). Thus, a unique aspect of meiotic recombination is the choice between three recombination templates (two homologs and one sister chromatid) to repair a DSB. In Saccharomyces cerevisiae, the formation of DSBs requires the evolutionarily conserved transesterase Spo11 and several accessory factors (1). These DSBs are resected by the Mre11–Rad50–Xrs2 (MRX) nuclease complex, Sae2 and Exo1 nuclease ensemble to generate 3′-single-strand DNA (ssDNA) tails that invade the intact DNA duplexes used in DNA repair (3–5). Overhangs of 3′-ssDNA are initially bound by replication protein A (RPA) and then replaced by RecA-like strand exchange proteins, Rad51 and Dmc1 (6). The resulting nucleoprotein filaments mediate the search for homology and catalyze DNA strand exchange to form joint molecule intermediates. Dmc1 and Rad51 have overlapping but also non-redundant functions in promoting recombination during meiosis (7). Eventually, the DSBs are repaired, resulting in COs, with the exchange of chromosome arms or non-COs (NCOs). Three meiosis-specific proteins, Mek1, Hop1 and Red1, act positively to promote interhomolog (IH) recombination (8–10) and negatively to slow the rate of intersister recombination (11). The faster rate of intersister repair is mediated by sister chromatin cohesion (Rec8), while Red1/Hop1/Mek1 counteracts this effect, thereby ensuring IH bias (12).
Mek1 is a protein kinase, whereas Red1 and Hop1 are structural components of the axial elements of the synaptonemal complex (SC). The SC is a zipper-like proteinaceous structure that mediates chromosome synapsis during the late prophase stage known as pachytene. The SC consists of a central region and two dense lateral elements. The lateral element constitutes the rod-like homolog axis, which is called an axial element prior to synapsis (13–17). The central region of the SC is made up of Zip1 (18). In response to DSBs, Red1 associates with SUMO polymeric chains (19) and the 9-1-1 complex (Ddc1–Mec3–Rad17) (20) to activate Mec1 and Tel1, the yeast homologs of the mammalian DNA damage sensor kinases ATM and ATR. These two kinases preferentially phosphorylate their substrates at serine (S) and threonine (T) residues that precede glutamine residues, so-called SQ/TQ motifs. Many known targets of the ATM/ATR family proteins contain the SQ/TQ cluster domains (SCDs). For example, Mec1 and Tel1 phosphorylate Hop1 at multiple SQ/TQ motifs within its N-terminal SCD (amino acid residues 298–319), in which the phosphorylation of T318 affects Hop1 activities most profoundly (21). Hop1-T318 phosphorylation is required for the activation of Mek1 and its recruitment to chromosomes (21), consistent with previous reports that Mek1 activation requires both Red1 and Hop1 (8,9). Mec1 and Tel1 also phosphorylate SQ and TQ sites in several other phosphoproteins during meiosis, including Sae2, histone H2A at S129 (i.e. γ-H2A), RPA, Rad53 and Zip1 (22–26). The Mec1-dependent phosphorylation of the Zip1 protein at S75 dynamically destabilizes homology-independent centromere pairing in response to DSBs (26). Protein phosphatase 4 (PP4) is responsible for the dephosphorylation of several Mec1/Tel1-dependent phosphoproteins during meiosis, as persistent hyperphosphorylation occurs in the absence of Pph3 and Psy2 (26). Pph3 and Psy2 are the catalytic subunit and the coactivator of PP4, respectively (27,28). Finally, the presence of aberrant DNA structures or incomplete synapsis activates a Mec1/Tel1-dependent recombination checkpoint that leads to the inhibition of Ndt80, a transcription factor required for the exit from pachytene (29,30).
Mek1 phosphorylates multiple targets in meiosis, including Rad54 at T132 (31), and histone H3 at T11 (32). Rad54 is a dsDNA-dependent ATPase that is required for Rad51 recombinase activity (33). The mek1Δ mutant generates very few viable spores. However, a rad54T132A allele that substitutes T132 with alanine (A) displayed no apparent defect in spore viability. Phosphorylated Rad54 might act synergistically with the meiosis-specific protein Hed1 to suppress the recombinase activity of Rad51 (31). Alternatively, Rad54 phosphorylation may have little effect on preventing intersister recombination or another protein Tid1/Rdh54 can substitute Rad54 and use Dmc1 for IH recombination (34,35). The role of H3-T11 phosphorylation is still unclear. It was also suggested that Mek1 might directly phosphorylate targets that are involved in establishing IH bias (10). Mek1 activation can be artificially maintained through glutathione S-transferase (GST)-mediated dimerization. Mek1-GST and GST-Mek1 can rescue the spore viability of mek1Δ but not hop1Δ or red1Δ (8,36). Intriguingly, the ectopic expression of Mek1-GST in wild-type cells results in increased IH bias, but the extraneous IH events are preferentially repaired as NCOs rather than COs. In contrast, Mek1-GST promotes both IH bias and CO formation in the absence of RAD17- and PCH2-dependent checkpoint functions (36). Rad17 is a component of the 9-1-1 checkpoint complex (20). Pch2 is a widely conserved, putative ATPase associated with cellular activities (AAA)-type ATPase with important roles in pachytene checkpoint, normal SC assembly, and the distribution of DSBs and CO events (37–48). Recently, Pch2 was shown to physically and functionally interact with Xrs2 (49) and Tel1 (Wang,T.-F., unpublished data). Xrs2 is a member of the MRX complex, which acts at the sites of unprocessed DSBs. It has been suggested that the Pch2–Xrs2 interaction might enable Pch2 to remodel chromosome structures adjacent to DSB sites to promote access by Tel1 and Mec1 kinases for Hop1 phosphorylation (49). As Mek1-GST is involved in promoting IH bias in the absence of RAD17- and PCH2-dependent checkpoint functions, Mek1 was proposed to phosphorylate proteins that are directly involved in the recombination checkpoint (36). In this report, we further explored the roles of Mek1, GST-Mek1 and Mek1-GST in regulating the recombination checkpoint. Our data support an alternative model in which Mek1 provides positive feedback in the stabilization of Mec1/Tel1-mediated Hop1-T318 phosphorylation.
MATERIALS AND METHODS
Antisera
Guinea pig antisera against phosphorylated Zip1-S75 were raised using a synthetic phosphopeptide (C65KKLITSMSLS[P]QRNHGYS82) as an antigen. Rabbit antisera against phosphorylated Rad54-T132 and phosphorylated Hop1-T318 were raised using the synthetic phosphopeptides (T126LRRSFT[P]VPIKGYV139 and S311QASIQPT[P]QFVSNN324) as antigens, respectively. The antisera were pre-cleared by affinity chromatography using the corresponding non-phosphorylated peptides coupled to agarose beads. Phosphopeptide synthesis and animal immunization were conducted by LTK BioLaboratories, Taiwan. Anti-phospho-Histone 2A (H2A)-S129 antiserum (50) was from Millipore, Billerica, MA, USA. Anti-GST antibody was from Genescript, NJ, USA. Anti-YFP (or anti-GFP) antibody was from Clontech, CA, USA.
Yeast strains, sporulation and western blot analysis
All meiotic experiments were performed using diploid cells from the SK1 strain background. Western blot analyses were performed as previously described (19,51). Spore viability was determined by tetrad dissection.
Inhibition of the analog-sensitive GST-Mek1-as mutant
4-Amino-1-tert-butyl-3(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1-NM-PP1) (52) was provided by Cellular Genomics (New Haven, CT, USA). Five micromolars of 1-NM-PP1 was added into sporulating cell culture to inhibit GST-Mek1-as kinase activity.
Quantitative western blot analyses
Yeast total protein extracts were prepared by trichloroacetic acid precipitation as previously described (51). Western blots were developed using the ChemiLucent ECL Detection System (Millipore) and imaged by exposure to X-ray film. The relative intensities of protein bands of interest from X-ray films were obtained with a Biospectrum 600 imaging system (UVP, Upland, CA, USA) containing an OptiCam 600 camera (Canon, Japan). For quantification, protein bands were plotted on a bar graph using VisionWorksLS Image Acquisition and Analysis Software (UVP). The intensities of phosphorylated Hop1-T318 proteins (72–95 kDa) and of the loading control (Hsp104) were acquired from X-ray films by subtracting an equally sized background, respectively. For normalization, intensity of phosphorylated Hop1-T318 protein was divided by that of the loading control at the indicated time point. To reduce the quantitative variations resulting from different X-ray film exposure time, the protein samples with the strongest phosphorylated Hop1-T318 signals were collected from different cultures and simultaneously compared in a separate western blot.
RESULTS
Validation of antisera against phosphorylated Hop1-T318, Zip1-S75 and Rad54-T132 proteins
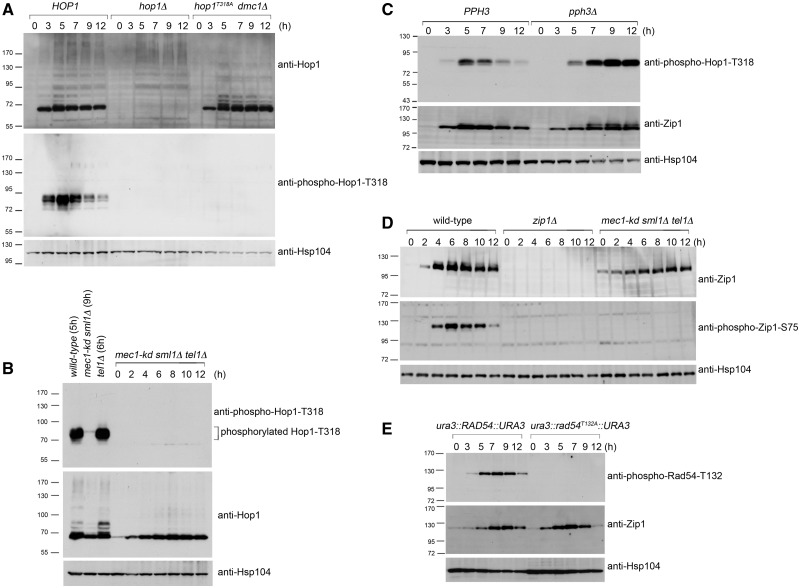
Phosphorylated peptides were synthesized and used to generate antisera specific to phosphorylated Hop1-T318, Zip1-S75 and Rad54-T132 proteins (see ‘Materials and Methods’ section). Synchronous meiosis was induced in wild-type and mutant strains. Following transfer to sporulation medium, meiotic cells were harvested at the indicated time points, and total cell lysates were prepared (51). Western blot analyses were performed to validate these antisera. As shown in Figure 1A (middle panel), at least two bands (∼85 kDa) were detected by anti-phospho-Hop1-T318 antibodies in the wild-type lysate, but not in the lysates from strains carrying mutant alleles hop1Δ or hop1T318A dmc1Δ. The hop1T318A variant encodes a mutant protein in which the T318 residue of Hop1 has been mutated to alanine (A); this strain produces no viable spores (Table 1) (21). The dmc1Δ mutation results in the accumulation of resected DSBs (or ssDNAs) (53) and phosphorylated Hop1 (21). T318 is not the sole phosphorylation site in Hop1 (21), as shifted bands are still detected by anti-Hop1 antisera in wild type and hop1T318A dmc1Δ (Figure 1A). Next, we showed that the extent of Hop1-T318 phosphorylation is comparable between wild type and tel1Δ but reduced in mec1-kd (=kinase dead) sml1Δ (Figure 1B; sml1Δ suppresses the lethality conferred by mec1Δ or mec1-kd mutations). Phosphorylated Hop1-T318 is not detected with anti-phospho-Hop1-T318 antisera in the mec1-kd sml1Δ tel1Δ triple mutant. Furthermore, shifted Hop1 band is hardly recognized by anti-Hop1 antisera in the mec1-kd sml1Δ tel1Δ triple mutant (Figure 1B).
Figure 1.
Validation of antisera against phosphorylated Hop1-T318 (A–C), Zip1-S75 (D) and Rad54-T132 (E). Western blot time course analyses of meiotic cells were performed as described previously (19,51). Wild-type and mutant strains at different sporulation time points are indicated. Anti-Hop1, anti-Zip1, anti-phospho-Hop1-T318, anti-phospho-Rad54-T132 and anti-phospho-Zip1-S75 antibodies were used to detect the corresponding proteins. Antibody against Hsp104 was used as a loading control. Molecular weights (in kiloDaltons) are indicated to the left of the blots.
Table 1.
Genotypes and spore viabilitya
| Strain | Genotype | % Viable spores | Numbers of spores |
|---|---|---|---|
| WHY3285 | ho::hisG/″, leu2::hisG/″, HIS4::LEU2-(BamHI)/his4-X::LEU2-(BamHI)–URA3 | 98 | 204 |
| WHY2947 | hop1Δ/″ | 0 | 144 |
| WHY8853b | hop1Δ/″, ura3::hop1T318A::URA3/″, dmc1Δ/″ | n.d.c | n.d. |
| WHY7706 | tel1Δ/″ | 85 | 240 |
| WHY9047b | mec1-kd/″, sml1Δ/″ | 28 | 216 |
| WHY9754 | mec1-kd/″, sml1Δ/″, tel1Δ/″ | 0 | 216 |
| WHY9174 | pph3Δ/″ | n.d. | n.d. |
| WHY10228 | mek1Δ/″, PMEK1-mek1-as-V5 (2 µ) | 96 | 216 |
| WHY9384 | mek1Δ/″, mock vector (2 µ) | 3 | 212 |
| WHY9978 | mek1Δ/″, PMEK1-MEK1 (2 µ) | 94 | 208 |
| WHY9385 | mek1Δ/″, PMEK1-GST-mek1-as (2 µ) | 95 | 212 |
| WHY10134 | mek1Δ/″, PMEK1-GST-mek1R51A-as (2 µ) | 4 | 212 |
| WHY9386 | hop1Δ/″, ura3::hop1T318A::URA3/″, mock vector (2 µ) | <1 | 108 |
| WHY9387 | hop1Δ/″, ura3::hop1T318A::URA3/″, PMEK1-GST-mek1-as (2 µ) | <1 | 96 |
| WHY9755d | rad54Δ/″, ura3:: RAD54::URA3/″ | 90 | 212 |
| WHY9746d | rad54Δ/″, ura3:: rad54T132A::URA3/″ | 92 | 212 |
| WHY2766 | rad51Δ/″, dmc1Δ/″ | n.d. | n.d. |
| WHY9954 | rad51Δ/″, dmc1Δ/″, mek1Δ/″ | n.d. | n.d. |
| WHY9083 | pch2Δ/″, | 99 | 144 |
| WHY9382e | pch2Δ/″, rad17Δ/″, MEK1-GST/″ | 52 | 144 |
| WHY9383e | pch2Δ/″, rad17Δ/″ | <1 | 128 |
| WHY10175 | mek1Δ/″, PMEK1-gst(nd)-mek1-as (2 µ) | 80 | 216 |
| WHY10281 | RAD54-YFP/RAD54-YFP | 89 | 216 |
| WHY10315 | RAD54-YFP/RAD54-YFP, mek1Δ/″ | 4 | 208 |
aAll strains are SK1 MATa/MATα diploids. Tetrads with four spores were dissected and analyzed following sporulation in liquid medium at 30°C. bWHY8853 and WHY9047 were gifts from Rita Cha (21). cn.d. (not determined). dWHY9755 and WHY9746 were gifts from Nancy Hollingsworth (31). eWHY9382 and WHY9383 were gifts from Sean Burgess (36).
The dephosphorylation of Hop1-T318 is mediated by PP4 in meiosis, and the persistent hyperphosphorylation of Hop1-T318 was observed in pph3Δ (Figure 1C). Pph3 is the catalytic subunit of PP4. The pph3Δ mutant also accumulated higher levels of phosphorylated Zip1; shifted bands were detected with anti-Zip1 antibody (Figure 1C). The anti-phospho-Zip1-S75 antibodies were able to detect phosphorylated Zip1-S75 in the wild type but not in the zip1Δ or the mec1-kd sml1Δ tel1Δ triple mutant (Figure 1D). These results confirmed that Mec1 and Tel1 mediate Hop1 and Zip1 phosphorylation (21,26) and that PP4 is responsible for Hop1 and Zip1 dephosphorylation (26). Finally, the anti-phospho-Rad54-T132 antibody specifically recognizes phosphorylated Rad54-T132, as suggested by the absolute dependence of a positive signal in western blotting on the phosphorylation of T132. This band is absent in a strain carrying the rad54T132A mutation despite normal entry into meiosis, as indicated by timely appearance of SC protein Zip1 (Figure 1E).
Mek1 promotes not only Rad54-T132 phosphorylation but also Hop1-T318 phosphorylation
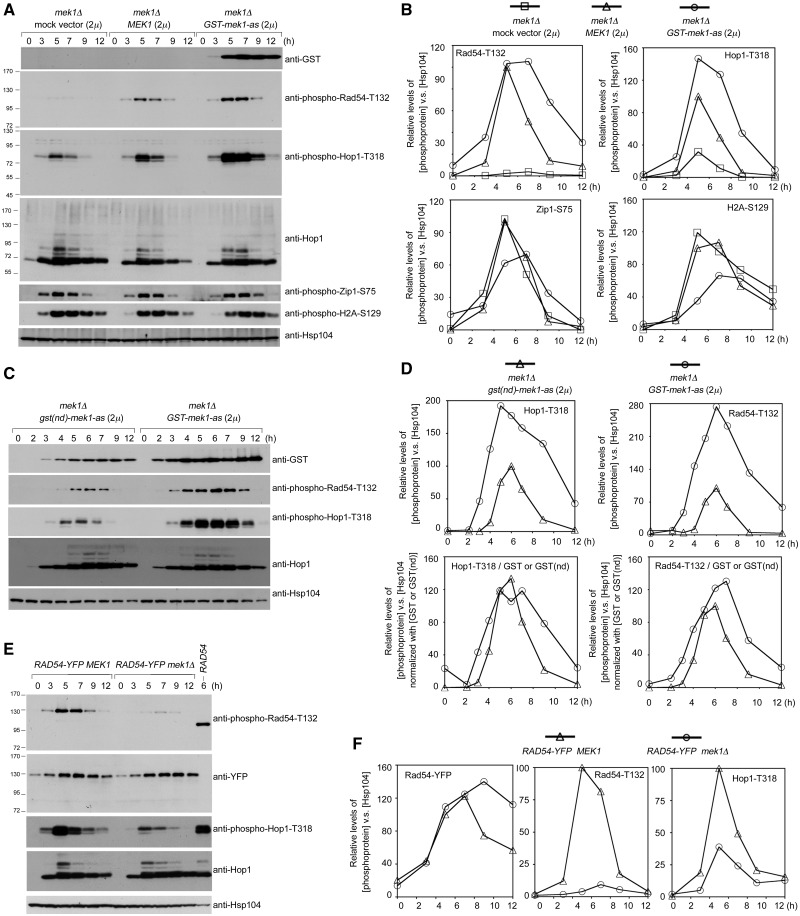
Mek1 mediates Rad54-T132 phosphorylation during meiosis (31). Phosphorylated Rad54-T132 was detected with anti-phospho-Rad54-T132 antisera in mek1Δ diploids transformed with either a PMEK1-MEK1 or PMEK1-GST-mek1-as expression vector, in which the expression of Mek1 or GST-Mek1-as, respectively, is under the control of the MEK1 gene promoter (PMEK1). GST-mek1-as, an analog-sensitive mutant, has the advantage that it can be specifically inhibited by the addition of the purine analog 1-NM-PP1, whereas wild-type GST-Mek1 is unaffected by inhibitor (9,31). Quantitative western blot analyses (see ‘Materials and Methods’ section) showed that Rad54-T132 phosphorylation was negligible in a mek1Δ strain carrying a mock 2 µ vector. Higher steady-state levels of phosphorylated Rad54-T132 were induced by GST-Mek1-as (in the absence of 1-NM-PP1) than by Mek1 in mek1Δ (Figure 2A and B). Thus, in accordance with the semi-dominance of GST-Mek1 and Mek1-GST, semi-dominance can also be assumed for GST-mek1-as (8,36).
Figure 2.
Mek1 and GST-Mek1-as enhance Rad54-T132 phosphorylation and Hop1-T318 phosphorylation. The mek1Δ diploid cells were transformed with an empty 2 µ vector or a vector carrying MEK1 (A), GST-mek1-as (A and C) or gst(nd)-mek1-as (C). The MEK1 and mek1Δ diploid cells express Rad54-YFP proteins (E). The expression levels of various phosphoproteins were visualized by western blot analysis. (B, D and F) Quantitation of western blot results in (A, C and E) (see ‘Materials and Methods’ section). The relative levels of different phosphoproteins versus Hsp104 (loading control) at each time point are shown.
Mek1 is a downstream effector of phosphorylated Hop1-T318 (21). Surprisingly, we observed that Mek1 and GST-Mek1-as enhanced Hop1-T318 phosphorylation but not Zip1-S75 phosphorylation or H2A-S129 phosphorylation in mek1Δ. Overexpression of GST-Mek1-as in mek1Δ induced persistent and higher steady-state levels of Hop1-T318 phosphorylation, whereas overexpression of Mek1 in mek1Δ induced a transient increase of Hop1-T318 phosphorylation (Figure 2A and B). In contrast, the steady-state levels of Zip1-S75 phosphorylation and H2A-S129 phosphorylation were slightly lower in PMEK1-GST-mek1-as mek1Δ than in PMEK1-MEK1 mek1Δ or in mek1Δ (Figure 2A and B). As all these three phosphoproteins are mediated by the Mec1 and Tel1 kinases, it is unlikely that the positive feedback function of GST-Mek1-as or Mek1 on Hop1-T318 phosphorylation is due to increased Mec1 and Tel1 kinase activities.
Since we were unable to generate or acquire antisera that could specifically recognize both phosphorylated and non-phosphorylated Rad54 in yeast total cell lysates, we next constructed yeast strains expressing yellow fluorescent protein (YFP) tagged Rad54 proteins (i.e. Rad54-YFP) (54) to determine whether mek1Δ mutation would affect steady-state levels of Rad54 as detected by an anti-YFP antibody (Figure 2E). Similar to the wild-type strain, the RAD54-YFP strain generates 89% viable spores. The mek1Δ RAD54-YFP mutant produces 4% viable spores (Table 1). The results of quantitative western blot analyses confirm that these two strains display similar levels of Rad54-YFP proteins during meiosis (Figure 2E and F).
GST-mediated Mek1 dimerization resulted in enhanced Hop1-T318 phosphorylation
GST-Mek1-as is better than Mek1 in enhancing Hop1-T318 and Rad54-T132 phosphorylation. GST artificially enhances dimerization of wild-type Mek1, although the role of dimerization in promoting Hop1-T318 phosphorylation is not yet clear. This possibility that GST-enhanced dimerization was responsible for the increased phosphorylation of Hop1-T318 and Rad54-T132 was addressed by constructing a non-dimerizing gst(nd)-mek1-as allele (8). The gst(nd)-mek1-as strain, similar to GST-mek1-as (Table 1), mek1-GST and mek1-gst(nd) (36), produces a large number of viable spores. Quantitative western blot analyses revealed that GST-Mek1-as appeared ∼1 h earlier than GST(nd)-Mek1-as. Moreover, the expression levels of GST-Mek1-as were greater than those of GST(nd)-Mek1-as. We also observed that the GST-mek1-as strain resulted in higher levels of Hop1-T318 phosphorylation and Rad54-T132 phosphorylation compared with the gst(nd)-mek1-as strain (Figure 2C and D). As the relative ratios of phosphorylated Rad54-T132 or phosphorylated Hop1-T318 versus GST-Mek1-as were similar to those of phosphorylated Rad54-T132 or phosphorylated Hop1-T318 versus GST(nd)-Mek1-as, we inferred that GST-mediated dimerization was unlikely directly responsible for the enhancement of Hop1-T318 and Rad54-T132 phosphorylation. Rather, GST-Mek1-as is apparently more stable than GST(nd)-Mek1-as.
The protein kinase activity of Mek1 is not required for enhanced Hop1-T318 phosphorylation
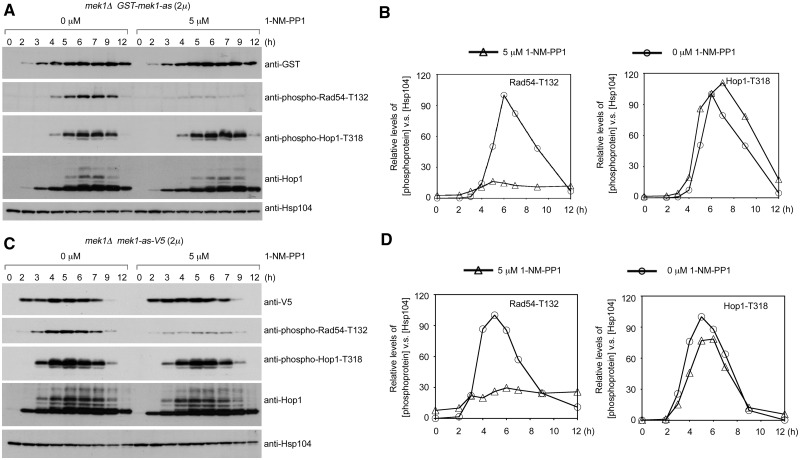
Next, we confirmed that the protein kinase activities of GST-Mek1-as and Mek1-as are not required for the enhancement of Hop1-T318 phosphorylation. To monitor the Mek1-as protein levels, the mek1Δ mutant was transformed with a PMEK1-mek1-as-V5 expression vector. The mek1-as-V5 allele encodes a V5-tagged Mek1-as protein. An addition of 5 µM 1-NM-PP1 to mek1Δ PMEK1-GST-mek1-as (Figure 3A and B) and mek1Δ PMEK1-mek1-as-V5 (Figure 3C and D) meiotic cells greatly diminished Rad54-T132 phosphorylation (31). In contrast, Hop1-T318 phosphorylation in mek1Δ PMEK1-GST-mek1-as (Figure 3A and B) and mek1Δ PMEK1-mek1-as-V5 (Figure 3C and D) was not affected or was only slightly affected by 1-NM-PP1, respectively. Thus, GST-Mek1-as and Mek1-as may not phosphorylate proteins that are directly involved in the Mec1/Tel1 recombination checkpoint.
Figure 3.
Mek1 kinase activity is not required for the enhancement of Hop1-T318 phosphorylation. The mek1Δ GST-mek1-as (2 µ) (A) and mek1Δ mek1-as-V5 (2 µ) (C) diploid cells underwent sporulation in SPM with 0 or 5 µM 1-NM-PP1. The expression levels of various phosphoproteins were visualized by western blot analysis. (B, D) Quantitation of western blot results in (A) and (C).
Mek1 promotes Hop1-T318 phosphorylation in the presence of unrepaired DSBs
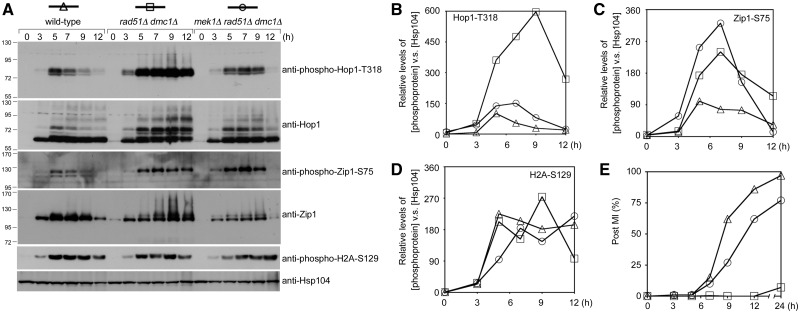
DSBs are more rapidly repaired through intersister recombination in the absence of Mek1 kinase activity (11). Mec1/Tel1-mediated Hop1 phosphorylation is DSB dependent (8,21). Accordingly, the lower Hop1-T318 phosphorylation levels detected in mek1Δ might simply be a consequence of faster DSB repair. This hypothesis was further examined here by comparing Hop1 and Hop1-T318 phosphorylation in rad51Δ dmc1Δ and mek1Δ rad51Δ dmc1Δ. The rad51Δ dmc1Δ double mutant persistently accumulates resected DSBs (or ssDNA) and produces markedly fewer CO products than the rad51Δ or dmc1Δ single mutants (53). Contrary to the hypothesis, we observed that the persistent hyperphosphorylation of Hop1-T318 and Hop1 was more profound in the rad51Δ dmc1Δ double mutant than in the mek1Δ rad51Δ dmc1Δ triple mutant (Figure 4A and B). This demonstrates that the loss of Hop1-T318 phosphorylation in the absence of Mek1 is not due to precocious DSB repair.
Figure 4.
Mek1 promotes Hop1-T318 phosphorylation in the presence of unrepaired DSBs. (A–D) Quantitative western blot results of various phosphoproteins in rad51Δ dmc1Δ and rad51Δ dmc1Δ mek1Δ diploid cells. (E) Timing of nuclear division (MI). Cells that had completed MI were identified by determining the number of cells with two to four DAPI-staining nuclei.
To further explore the effect of Mek1 on the overall Mec1 and Tel1 kinase activities in rad51Δ dmc1Δ, we examined Zip1-S75 phosphorylation and H2A-S129 phosphorylation by quantitative western blot analyses with anti-phospho-Zip1-S75 and anti-phospho-H2A-S129 antisera. Zip1-S75 phosphorylation was higher in mek1Δ rad51Δ dmc1Δ and lower in the wild-type strain than in rad51Δ dmc1Δ (Figure 4A and C). In contrast, similar levels of H2A-S129 phosphorylation were detected in these three strains (Figure 4A and D). The mek1Δ mutation is known to alleviate the prophase arrest or delay conferred by dmc1Δ and/or rad51Δ mutation (55). We found that the mek1Δ dmc1Δ rad51Δ triple mutant completed the first meiotic nuclear divisions (MI) faster than the dmc1Δ rad51Δ double mutant (Figure 4E). Thus, because the steady-state levels of Zip1 proteins were lower in the mek1Δ dmc1Δ rad51Δ triple mutant than in wild type or the dmc1Δ rad51Δ double mutant (Figure 4A, third bottom panel), we inferred that Zip1 proteins might be degraded faster in the mek1Δ dmc1Δ rad51Δ triple mutant. These results indicate that Mek1 does not affect overall Mec1 and Tel1 kinase activities. Mek1 might use a novel mechanism to stabilize Mec1/Tel1-mediated Hop1-T318 phosphorylation.
Hop1-T318 phosphorylation has a critical role in enhancing Mek1-mediated Hop1 phosphorylation
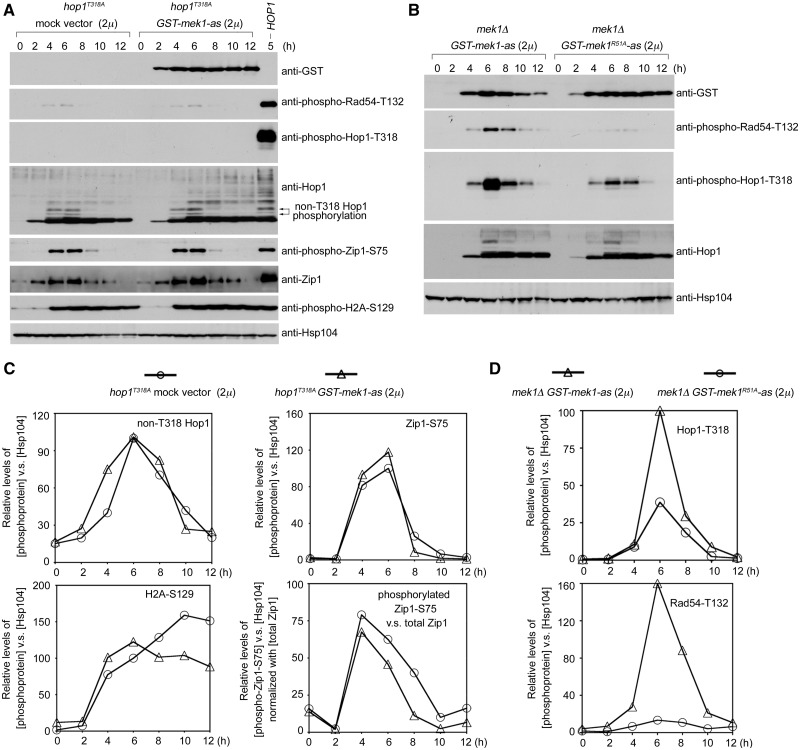
Hop1-T318 phosphorylation is required for both the activation and the chromosomal recruitment of Mek1 (21). To determine whether Hop1-T318 phosphorylation is also a prerequisite for the positive feedback function of Mek1, GST-mek1-as (2 µ) was introduced into the hop1T318A strain. In hop1T318A, the GST-Mek1-as protein failed to rescue the levels of phosphorylated Rad54-T132 (Figure 5A) or spore viability (Table 1). No phosphorylated Hop1-T318 was detected in either strain (Figure 5A). GST-Mek1-as did not increase overall ‘non-T318’ Hop1 phosphorylation or H2A-S129 phosphorylation, as indicated by similar band shifts in hop1T318A strains carrying GST-mek1-as (2 µ) or a mock vector. Although GST-Mek1-as increased Zip1-S75 phosphorylation ∼1.2-fold, the relative ratios of phosphorylated Zip1-S75 versus total Zip1 were similar in the presence or absence of GST-Mek1-as (Figure 5C). These results indicate that GST-Mek1-as unlikely affects overall Mec1 and Tel1 kinase activities and that Hop1-T318 phosphorylation is required for the positive feedback function of Mek1 in enhancing Hop1 phosphorylation.
Figure 5.
Hop1-T318 phosphorylation and arginine (R) 51 of Mek1 are required for the enhancement of Hop1-phosphorylation by Mek1. (A) hop1T318A diploids were transformed with a mock 2 µ vector or a vector carrying GST-MEK1-as. (B) mek1Δ diploids were transformed with GST-mek1-as or GST-mek1R51A-as, respectively. Western blot analyses of various phosphoproteins and proteins by the corresponding antibodies. (C and D) Quantitation of western blot results in (A) and (B).
Arginine 51 of Mek1 is required for the enhancement of Hop1-T318 phosphorylation
Several lines of evidence suggest that Hop1 and Mek1 may act similar to the Rad53-Dun1 signaling pathway. Dun1 and Rad53 are part of the DNA damage response during S. cerevisiae vegetative growth. Mec1 and Tel1 first activate Rad53, which in turn activates Dun1 (56,57). These two effector kinases, similar to Mek1 (9), are characterized by the forkhead-associated (FHA) domains that are essential for their activation in response to DNA damage. Rad53 contains two SCDs (SCD1 and SCD2) with high concentrations of Mec1/Tel1 phosphorylated sites. Two threonine residues, T5 and T8, are phosphorylated in the Rad53-SCD1 (1MENI5T[p]QP8T[p]QQSTQA15TQRFLI21E), and the diphosphorylated Rad53-SCD1 is bound by the Dun1-FHA domain (amino acid residues 19–159) to promote the phosphorylation-dependent activation of Dun1 (58,59). Two evolutionarily conserved positively charged residues (arginine 60 and lysine 100) in the Dun1-FHA domain have been shown to mediate the interaction between Dun1-FHA and diphosphorylated Rad53-SCD1 peptide (60). A structure-based sequence alignment revealed that arginine (R) 51 in Mek1-FHA is analogous to R60 in Dun1-FHA. A mutation of R51 to alanine (A) in the Mek1-FHA domain was shown to create a null allele (mek1R51A) (9). Notably, Rad53-SCD1 (5T[p]QP8T[p]QQST12) and Hop1-SCD (315IQP318T[p]QFVS322) share four identical amino acid residues (QPTQ) and a phosphorylated threonine. Accordingly, Mek1-R51 and Hop1-T318 phosphorylation may act together to mediate the interaction between Mek1-FHA and the phosphorylated Hop1-SCD. Thus, we inferred that R51 of Mek1 might stabilize Hop1-T318 phosphorylation. Therefore, we examined Rad54-T132 phosphorylation and Hop1-T318 phosphorylation in mek1Δ diploids transformed with PMEK1-GST-mek1-as or PMEK1-GST-mek1R51A-as. Overexpression of GST-Mek1-as or GST-Mek1R51A-as did not significantly alter total Hop1 protein levels (Figure 5B, second bottom panel). We found that, unlike GST-Mek1-as, GST-Mek1R51A-as promoted neither Hop1-T318 and Rad54-T132 phosphorylation (Figure 5B) nor spore viability (Table 1). These results support the model in which Mek1-FHA binds to phosphorylated Hop1-T318 and then stabilizes phosphorylated Hop1 against PP4-mediated dephosphorylation. Subsequently, Mek1 or GST-Mek1 is activated to phosphorylate Rad54-T132.
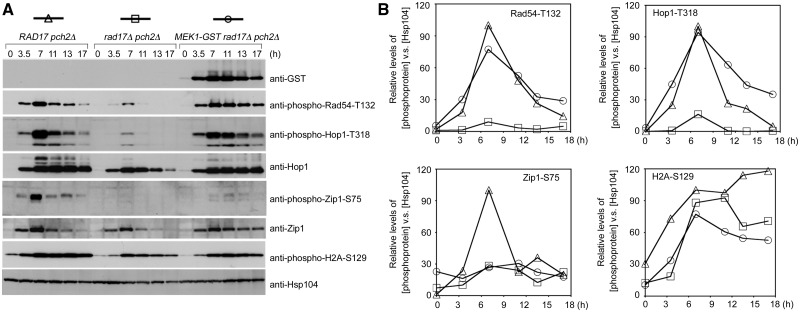
GST-Mek1 specifically enhances Hop1-T318 phosphorylation in pch2Δ rad17Δ
Previously, it was reported that the ectopic expression of Mek1-GST results in increased IH bias and CO formation, as well as a delayed MI phenotype, in the pch2Δ rad17Δ mutant (36). Rad17 is a component of the 9-1-1 checkpoint complex that promotes Mec1 kinase activity. Pch2 is a putative AAA-ATPase with important roles in pachytene checkpoint, normal SC assembly, and the distribution of DSBs and CO events (37–48). Compared with pch2Δ, pch2Δ rad17Δ exhibited lower levels of phosphorylated Rad54-T132, Hop1-T318, Zip1-S75 and H2A-S129 (Figure 6). Additionally, it has been shown that pch2Δ rad17Δ exhibits faster MI progression and lower spore viability than pch2Δ (36). We further observed markedly higher levels of Hop1-T318 phosphorylation and Rad54-T132 phosphorylation in MEK1-GST pch2Δ rad17Δ than in pch2Δ rad17Δ. Therefore, the MEK1-GST pch2Δ rad17Δ triple mutant is more similar to the pch2Δ single mutant than to the rad17Δ pch2Δ double mutant in terms of higher spore viability and delayed MI phenotypes (36). Consistently, the steady-state levels of phosphorylated Hop1 in MEK1-GST pch2Δ rad17Δ are similar to or slightly lower than those in pch2Δ rad17Δ, as revealed by anti-Hop1-T318 antibody and by the shifted bands detected with anti-Hop1 antibody (Figure 6). Due to a delayed MI phenotype (36), the levels of phosphorylated Rad54-T312 and Hop1-T318 persisted longer in the MEK1-GST pch2Δ rad17Δ triple mutant (Figure 6A). Notably, MEK1-GST did not significantly increase the maximum steady-state levels of H2A-S129 phosphorylation or Zip1-S75 phosphorylation in pch2Δ rad17Δ (Figure 4). We conclude that MEK1-GST did not increase the overall Mec1 and Tel1 kinase activities in pch2Δ rad17Δ, and that the Mek1-GST-dependent stabilization of Hop1-T318 phosphorylation is responsible for the partial rescue of the mek1-GST pch2Δ rad17Δ triple mutant.
Figure 6.
Mek1-GST specifically stabilizes Hop1-T318 phosphorylation in a pch2Δ rad17Δ mutant. Hsp104 was used as a loading control.
DISCUSSION
The current manuscript provides new insights into the roles of the chromosome axis-associated kinase Mek1 in regulating IH recombination and checkpoint responses during yeast meiosis. Our results suggest that R51 of Mek1-FHA is critical for recognizing phosphorylated Hop1-T318 to activate Mek1 and that this interaction also stabilizes Hop1-T318 phosphorylation against PP4-dependent dephosphorylation during meiosis. The positive feedback function of Mek1 apparently does not affect Mec1 and Tel1 kinase activities on other substrates (such as Zip1-S75, H2A-S129 or non-T318 Hop1 sites). Our results also indicate that the GST-mediated dimerization may increase the protein stability of the Mek1 protein and thereby indirectly enhance Hop1-T318 phosphorylation. These findings can account for the semi-dominant phenotypes of GST-Mek1 and Mek1-GST (8,36), which rescue the spore viability of mek1Δ in the presence of Red1 and Hop1.
The stabilization of Hop1-T318 phosphorylation by GST-Mek1 also explains why the ectopic expression of Mek1-GST in pch2Δ rad17Δ promotes increased IH recombination and recombination checkpoint responses (36). The pch2Δ rad17Δ double mutant progresses more rapidly though MI than does the pch2Δ single mutant (40), and MEK1-GST promotes a delayed MI phenotype in pch2Δ rad17Δ (36). Our results indicate that the delayed MI phenotype of the pch2Δ single mutant and the MEK1-GST pch2Δ rad17Δ triple mutants are compatible with sustained elevated levels of phosphorylated Hop1-T318 and phosphorylated Rad54-T132. In contrast, less phosphorylated Hop1-T318 and phosphorylated Rad54-T132 proteins were detected in the pch2Δ rad17Δ double mutant, which progresses more rapidly through MI (Figure 6). Thus, the sustained elevation of Mek1 kinase activity apparently accounts for the delay in MI progression. It will be interesting to identify the downstream target(s) of Mek1 that are directly responsible for inhibiting cell cycle progression during meiotic prophase.
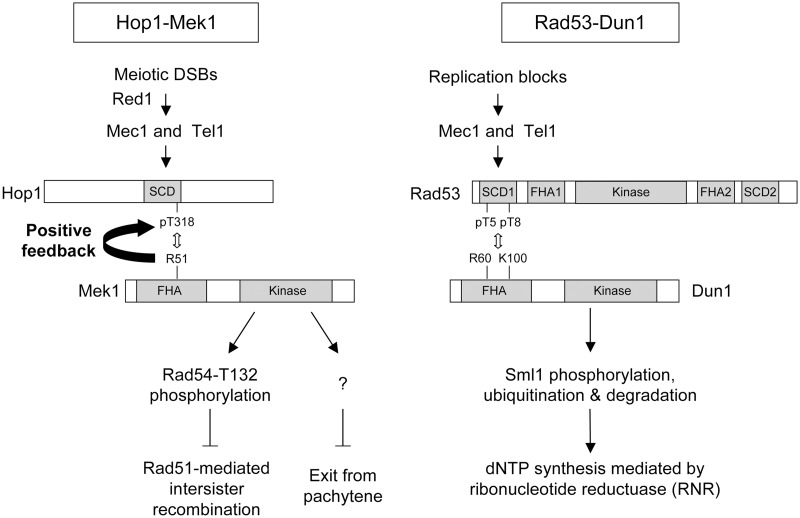
Our findings support a model in which Mek1 is a meiotic paralog of Dun1 in mediating cellular responses to DNA damage (Figure 7). These two checkpoint effector kinases apparently share similar upstream and downstream signaling mechanisms. The Dun1 signaling cascade involves the Mec1/Tel1-dependent phosphorylation of Rad53-SCD1. The phosphorylated Rad53-SCD1 (at T5 and T8) specifically recognizes Dun1-FHA, leading to the recruitment and activation of Dun1 to sites of DNA damage during DNA replication (56,59,60). One important function of Dun1 is the regulation of cellular dNTP levels (57,61–63). On the other hand, the Mek1 signaling cascade involves the Mec1/Tel1-dependent phosphorylation of the Hop1-SCD (21,64). The phosphorylated Hop1-SCD (T318) specifically recognizes Mek1-FHA, leading to the recruitment and activation of Mek1 at the sites of developmentally programmed DSBs during meiotic prophase. The best-known target of Mek1 is Rad54, and Rad54 phosphorylation prevents Rad54 from binding to and activating Rad51 in vitro (31). Further studies will be conducted to examine whether phosphorylation-dependent inactivation (or degradation) is a general mechanism by which Mek1 regulates its downstream targets at DSB sites and chromosome axes.
Figure 7.
Mek1 is a meiotic paralog of Dun1 in mediating cellular responses to DNA damage.
FUNDING
Funding for open access charge: The National Science Council and Academia Sinica, Taiwan (to T.F.W.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Sean Burgess (University of California, Davis), Rita Cha (MRC, London), Valentin Börner (Cleveland State University), Nancy Hollingsworth (Stony Brook University) and Douglas Bishop (University of Chicago) for the yeast strains used in this study; Chung Wang (IMB, Academia Sinica) for providing anti-Hsp104 antibodies and AndreAna Peña (IMB, Academia Sinica) for English editing.
REFERENCES
- 1.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 2.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodgson A, Terentyev Y, Johnson RA, Bishop-Bailey A, Angevin T, Croucher A, Goldman AS. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair. 2011;10:138–148. doi: 10.1016/j.dnarep.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 8.Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terentyev Y, Johnson R, Neale MJ, Khisroon M, Bishop-Bailey A, Goldman AS. Evidence that MEK1 positively promotes interhomologue double-strand break repair. Nucleic Acids Res. 2010;38:4349–4360. doi: 10.1093/nar/gkq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc. Natl Acad. Sci. USA. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockmill B, Roeder GS. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 15.Hollingsworth NM, Ponte L. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell. Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 18.Sym M, Engebrecht JA, Roeder GS. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 19.Lin FM, Lai YJ, Shen HJ, Cheng YH, Wang TF. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010;29:586–596. doi: 10.1038/emboj.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichinger CS, Jentsch S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc. Natl Acad. Sci. USA. 2010;107:11370–11375. doi: 10.1073/pnas.1004248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Carballo JA, Cha RS. Meiotic roles of Mec1, a budding yeast homolog of mammalian ATR/ATM. Chromosome Res. 2007;15:539–550. doi: 10.1007/s10577-007-1145-y. [DOI] [PubMed] [Google Scholar]
- 23.Cartagena-Lirola H, Guerini I, Viscardi V, Lucchini G, Longhese MP. Budding yeast Sae2 is an in vivo target of the Mec1 and Tel1 checkpoint kinases during meiosis. Cell Cycle. 2006;5:1549–1559. doi: 10.4161/cc.5.14.2916. [DOI] [PubMed] [Google Scholar]
- 24.Cartagena-Lirola H, Guerini I, Manfrini N, Lucchini G, Longhese MP. Role of the Saccharomyces cerevisiae Rad53 checkpoint kinase in signaling double-strand breaks during the meiotic cell cycle. Mol. Cell. Biol. 2008;28:4480–4493. doi: 10.1128/MCB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair. 2009;8:1127–1138. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Falk JE, Chan AC, Hoffmann E, Hochwagen A. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev. Cell. 2010;19:599–611. doi: 10.1016/j.devcel.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 28.O’Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates JR, 3rd, Aparicio OM, Romesberg FE. Pph3–Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl Acad. Sci. USA. 2007;104:9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 30.Tung KS, Hong EJ, Roeder GS. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl Acad. Sci. USA. 2000;97:12187–12192. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev. 2010;24:1772–1786. doi: 10.1101/gad.1954910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl Acad. Sci. USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu HY, Ho HC, Burgess SM. Mek1 kinase governs outcomes of meiotic recombination and the checkpoint response. Curr. Biol. 2010;20:1707–1716. doi: 10.1016/j.cub.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 38.San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- 40.Wu HY, Burgess SM. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 2006;16:2473–2479. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3:e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl Acad. Sci. USA. 2008;105:3327–3332. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi N, Barot A, Jamison C, Borner GV. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joyce EF, McKim KS. Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics. 2009;181:39–51. doi: 10.1534/genetics.108.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanders S, Alani E. The pch2Δ mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 2009;5:e1000571. doi: 10.1371/journal.pgen.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 2010;6:e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vader G, Blitzblau H, Tame M, Falk J, Curtin L, Hochwagen A. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477:115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho HC, Burgess SM. Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genet. 2011;7:e1002351. doi: 10.1371/journal.pgen.1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 2003;4:678–684. doi: 10.1038/sj.embor.embor871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai YJ, Lin FM, Chuang MJ, Shen HJ, Wang TF. Genetic requirements and meiotic function of phosphorylation of the yeast axial element protein red1. Mol. Cell. Biol. 2011;31:912–923. doi: 10.1128/MCB.00895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bishop AC, Shokat KM. Acquisition of inhibitor-sensitive protein kinases through protein design. Pharmacol. Ther. 1999;82:337–346. doi: 10.1016/s0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 53.Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- 54.Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell. Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez Y, Zhou Z, Huang M, Kemp BE, Elledge SJ. Analysis of budding yeast kinases controlled by DNA damage. Methods Enzymol. 1997;283:398–410. doi: 10.1016/s0076-6879(97)83033-9. [DOI] [PubMed] [Google Scholar]
- 57.Chen SH, Smolka MB, Zhou H. Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:986–995. doi: 10.1074/jbc.M609322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SJ, Schwartz MF, Duong JK, Stern DF. Rad53 phosphorylation site clusters are important for Rad53 regulation and signaling. Mol. Cell. Biol. 2003;23:6300–6314. doi: 10.1128/MCB.23.17.6300-6314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H, Yuan C, Hammet A, Mahajan A, Chen ES, Wu MR, Su MI, Heierhorst J, Tsai MD. Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade. Mol. Cell. 2008;30:767–778. doi: 10.1016/j.molcel.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 63.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl Acad. Sci. USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]