Abstract
DNA polymerase ζ (Pol ζ) plays a key role in DNA translesion synthesis (TLS) and mutagenesis in eukaryotes. Previously, a two-subunit Rev3–Rev7 complex had been identified as the minimal assembly required for catalytic activity in vitro. Herein, we show that Saccharomyces cerevisiae Pol ζ binds to the Pol31 and Pol32 subunits of Pol δ, forming a four-subunit Pol ζ4 complex (Rev3–Rev7–Pol31–Pol32). A [4Fe-4S] cluster in Rev3 is essential for the formation of Pol ζ4 and damage-induced mutagenesis. Pol32 is indispensible for complex formation, providing an explanation for the long-standing observation that pol32Δ strains are defective for mutagenesis. The Pol31 and Pol32 subunits are also required for proliferating cell nuclear antigen (PCNA)-dependent TLS by Pol ζ as Pol ζ2 lacks functional interactions with PCNA. Mutation of the C-terminal PCNA-interaction motif in Pol32 attenuates PCNA-dependent TLS in vitro and mutagenesis in vivo. Furthermore, a mutant form of PCNA, encoded by the mutagenesis-defective pol30-113 mutant, fails to stimulate Pol ζ4 activity, providing an explanation for the observed mutagenesis phenotype. A stable Pol ζ4 complex can be identified in all phases of the cell cycle suggesting that this complex is not regulated at the level of protein interactions between Rev3-Rev7 and Pol31-Pol32.
INTRODUCTION
DNA polymerase ζ (Pol ζ) is a B-family DNA polymerase participating in DNA translesion synthesis (TLS) and plays a predominant role in both spontaneous and damage-induced mutagenesis in all eukaryotes (1–3). Pol ζ bypasses a variety of DNA lesions and readily extends mismatched primer-template termini (4,5). Pol ζ was initially identified as a heterodimeric complex of the catalytic Rev3 subunit with the accessory Rev7 subunit that is also required for DNA polymerase activity (6). Mutations in REV3 or REV7 result in a severe decrease of induced mutagenesis. The rev3Δ and rev7Δ strains are also spontaneous antimutators, suggesting that Pol ζ acts to bypass naturally occurring damage or other structural blocks (7–9). Deficiency in the Rev3 catalytic subunit leads to embryonic lethality in mice (10). In humans, alterations in Pol ζ expression are associated with cancer, chromosome instability and cisplatin resistance (11).
All four eukaryotic B-family DNA polymerases, Pol α, δ, ε, and ζ, contain two conserved cysteine-rich metal-binding motifs, CysA and CysB, in the C-terminal domain (CTD) of their catalytic subunits [reviewed in (12,13)]. The four cysteine residues of CysA form a classical zinc ribbon motif. In the case of Pol δ, where the role of both CysA and CysB in metal binding has been studied most extensively, the four-cysteine motif of CysB coordinates a [4Fe-4S]2+ cluster (14). However, the other catalytic subunits have also been shown to bind [4Fe-4S] clusters. Indeed, expression of the CTD of Rev3 in Escherichia coli also indicated the presence of a [4Fe-4S] cluster in this domain (14). In Pol δ, the [Fe-S] cluster is required for stable binding of Pol3 to its second subunit Pol31 (14,15), which in turn binds to Pol32 (16–18). The CysB motif of the catalytic subunit of Pol α also coordinates interactions with its second subunit (19,20). Therefore, an arrangement analogous to that determined for Pol δ may also hold for Pol α and for Pol ε·
In contrast to the three replicative DNA polymerases, interactions between the Rev7 subunit of Pol ζ with the catalytic subunit Rev3 have been mapped to the N-terminal region of human Rev3 rather than its CTD (6,21). The possibility then exists that the [4Fe-4S]-containing CTD of Rev3 might provide interactions with other factors that function in mutagenesis. Indeed, two recent articles report on the interaction between Rev3 and Pol31. One interaction study was carried out in E. coli with the critical CTD of Rev3 (22), whereas the second study reported the purification of a four-subunit Pol ζ complex from yeast (23). Herein, we also report on the isolation and functional characterization of a four-subunit Pol ζ enzyme (Pol ζ4) and extend these previous studies by showing that the novel interactions with Pol31 and Pol32 are essential for proliferating cell nuclear antigen (PCNA)-mediated TLS. Mutation of the PCNA-binding domain (PIP) of Pol32 attenuates TLS, in accordance with a decrease in mutagenesis in the pol32-ΔPIP mutant (24). Furthermore, deletion of the non-essential POL32 gene results in a failure to form a complex of Pol31 with Rev3–Rev7, suggesting a logical explanation for the mutagenesis defect of pol32Δ mutants (16). Altogether our data suggest that the formation of Pol ζ4 complex is critical for the TLS function of Pol ζ in vitro and in vivo.
MATERIALS AND METHODS
Strains and plasmids
All yeast strains are listed in Supplementary Data. Plasmids are listed in Supplementary Table S1.
Enzymes
Saccharomyces cerevisiae Pol δ was expressed in yeast and purified as described previously (25). The replication protein A (RPA), replication factor C (RFC), PCNA and pcna-113 of Saccharomyces cerevisiae were expressed and purified from E. coli (26,27). Pol ζ4 (Rev3–Rev7–Pol31–Pol32), Pol ζ2 (Rev3–Rev7) and their mutant forms were produced in protease-defective strain FM113 or in pol32Δ derivative strain PY117, or in rev1Δ strain PY201, and purified as described previously with several modifications (28). The detailed protocol is described in Supplementary Data.
Yeast two-hybrid analysis
Indicator strain PJ69-4A was co-transformed with plasmids containing REV3-GAL4 DNA BD fusion genes (pBL816, pBL816A and pBL816B), and plasmids encoding for REV7 (pBL817), POL31 (pBL364) and POL32 (pBL391) fused to GAL4 activation domain (AD) or with empty vector pACT2. Transformants were grown on -His plates for 5 days to score protein–protein interactions as growth.
GST-pull down
Yeast cells transformed by plasmids encoding for GST-REV3, REV7, POL31 and POL32, all under control of the GAL1-10 promoter, were grown in 125 ml of selective medium containing 2% raffinose to O.D660 = 0.5. Protein expression was induced by 2% galactose, and cells were grown for another 8 hours. Cells were collected, resuspended in lysis buffer (50 mM Hepes (pH 7.4), 200 mM NaCl, 5% glycerol, 1 mM DTT, 0.1% Tween 20, 0.01% NP40, 10 μM pepstatin A, 10 μM leupeptin, 2.5 mM benzamidine, 0.5 mM PMSF) and lysed by vortexing with glass beads on ice. Cell lysates were clarified by centrifugation, and 0.8 ml of yeast extract containing 1 mg of protein was incubated with 40 μl of glutathione sepharose beads (GE Healthcare) for 1 h. Beads were washed six times with wash buffer (50 mM Hepes (pH 7.4), 800 mM NaCl, 5% glycerol, 1 mM DTT, 0.1% Tween 20, 0.01% NP40, 1 μM pepstatin A, 0.5 mM PMSF) and boiled for 2 min in 80 μl of 2× sodium dodecyl sulfate (SDS) sample buffer.
Cell cycle analysis and exposure to DNA-damaging agents
Cells containing GST-REV3 on plasmid pBL813 were grown in 125 ml of selective medium with 2% raffinose to O.D660 = 0.5 without galactose induction. They were arrested in G1 phase by α-factor (20 μg/ml for 2 h), in G2/M phase with nocodazole (15 μg/ml for 2 h) and in S phase by hydroxyurea (200 mM for 90 min). Then cells were treated with 4NQO (1 μg/ml) or methylmethane sulfonate (0.05%) for 30 min at 30°C. The cells from 200 μl of culture were fixed, stained with propidium iodide and DNA content was measured by flow cytometry. The remaining cultures were harvested, and extract preparation and GST-pull down were performed as described earlier.
Western blot and antibodies
Western blot analysis was performed to detect the presence of GST-Rev3, Rev7, Pol3, Pol31, Pol32 and Rev1 proteins in purified Pol ζ preparations and after pull-down experiments. To detect the Rev1, Rev3 and Rev7 proteins, rabbit polyclonal antisera were raised against purified yeast Rev1 and Pol ζ2. GST-Rev3 was detected with anti-GST antibody (ab9085, Abcam). Rabbit anti-Pol3, -Pol31 and -Pol32 antibodies were immunopurified. Detection was carried using alkaline phosphatase–conjugated secondary antibody (Sigma) and a BCIP/TNBT chromogenic substrate (Sigma).
DNA polymerase and translesion synthesis assays
Three different assays were used. (i) Measurement of basal DNA polymerase activity: This measures polymerase activity on activated calf thymus DNA, for 45 min at 30°C, as described (29). (ii) DNA replication assay on circular ssDNA: The assay on primed ssDNA (pSKII) was performed as described previously (24). The reactions containing 5 nM of 3 kb circular ssDNA, 500 nM RPA, 3 nM RFC and 10 nM of Pol ζ were incubated at 30°C for 50 min with increasing PCNA as shown in legends to figures. (iii) In vitro DNA translesion bypass assay: Sequences of the 107-nt template (with or without a model abasic site) and the primer are given in Supplementary Data. The standard 20 μl reaction contained 40 mM Tris–HCl, pH 7.8, 0.2 mg/ml bovine serum albumin, 8 mM Mg acetate, 120 mM NaCl, 100 μM each dNTPs, 0.5 mM ATP, 10 nM DNA, 15 nM RPA, 30 nM PCNA, 3 nM RFC and 10 nM Pol ζ. The DNA was preincubated with RPA, RFC and PCNA for 30 sec at 30°C, and the reaction was started by addition of Pol ζ and incubated at 30°C. Reactions were stopped with 15 mM ethylenediaminetetraacetic acid and 0.5% SDS and analyzed on a 12% polyacrylamide 7 M urea gel. Quantification was done by either phosphorimaging of the dried gel (32P) or fluorescence imaging on a Typhoon system.
Damage-induced mutagenesis assays
The rev3Δ strain BY4741 (rev3::KanMX4) contained empty vector or plasmid pBL811 (GST-REV3) or mutants of REV3 as shown in Table 1. Strains were grown for 2 days to saturation in selective minimal media. The cells were washed with sterile water and 2 × 107 cells plated on selective plates, with or without 80 µg/ml canavanine and either irradiated or not irradiated with 30 J/m2 of UV light. The plating efficiencies and the percent of UV survival were measured on plates without canavanine. Spontaneous frequencies to canavanine resistance were measured on unirradiated canavanine plates, and UV-induced frequencies to canavanine resistance were measured on irradiated canavanine plates. Colonies appearing after 3 days of growth at 30°C were counted. Frequencies of mutation to canavanine resistance were corrected for the UV survival percentage. The experiments were carried out on three independent cultures, and in duplicate, and the results are presented in Table 1.
Table 1.
Damage-induced mutagenesis efficiency of REV3 mutants
| REV3 | Spontaneous (10−6) | Survival (%) | Induced (10−6) |
|---|---|---|---|
| WT | 3.1 ± 0.2 | 56 ± 10 | 183 ± 30 |
| Δ | 2.5 ± 0.5 | 23 ± 3 | 1.5 ± 1 |
| cysA | 4.7 ± 2 | 58 ± 4 | 168 ± 10 |
| cysB | 2.0 ± 0.3 | 12 ± 4 | 6 ± 2 |
See ‘Materials and Methods’ section for details.
RESULTS
The [4Fe-4S] cluster is required for the interaction of Rev3 with the Pol31 subunit of Pol δ
The CTD of Pol3 shows strong sequence homology with that of Rev3, particularly in a region C-terminal of the CysB motif (Figure 1A), suggesting the possibility of an interaction between Rev3 and the Pol31–Pol32 subunits of Pol δ. To test this, we performed a yeast two-hybrid analysis using full-length Rev3 as bait (Figure 1B). We co-expressed REV3, fused to the GAL4 DNA BD, together with either REV7, as positive control, or with POL31 or POL32 fused to the GAL4 AD, or empty vector. Significant interaction signals were obtained between Rev3 and Rev7 and between Rev3 and Pol31. No interaction between Rev3 and Pol32 was detected by this assay. Importantly, double mutations from cysteine to serine in the CysB motif (rev3-CC1449,1473SS), which ligands the [4Fe-4S] cluster, abrogated the Rev3-Pol31 interactions without affecting the Rev3–Rev7 signal. In contrast, double mutations from cysteine to serine in the CysA motif (rev3-CC1401,1417SS), did not significantly decrease the Rev3–Pol31 signal (Figure 1B). These data suggest that Pol31 binds to Rev3 through the CysB region, and an intact iron–sulfur cluster is required for interaction. This is the same binding specificity as observed between Pol3 and Pol31 (14).
Figure 1.
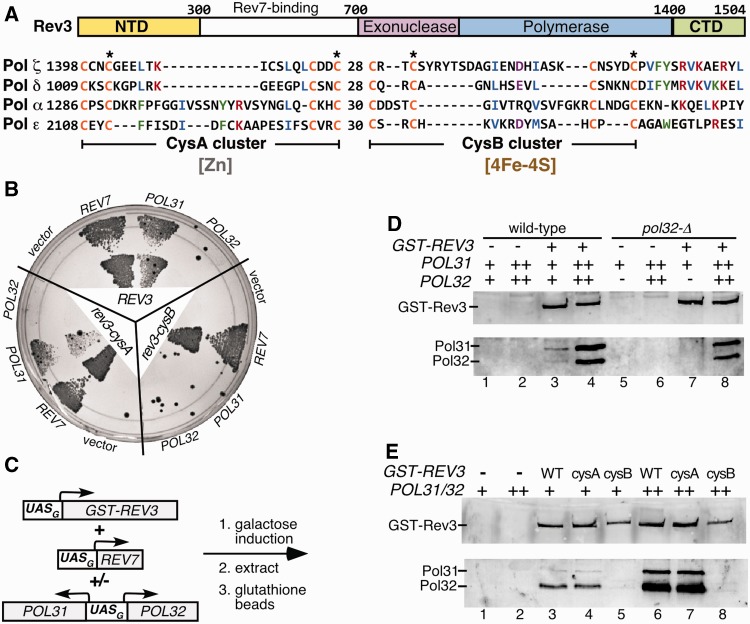
Interaction of Pol ζ catalytic subunit Rev3 with Pol31 and Pol32. (A) Domain organization of S. cerevisiae Rev3 and alignment of the CTDs of B-family DNA polymerases. The second and fourth residues of each cysteine-rich cluster were mutated in REV3 to create the CysA (CC1401,1417SS or CC1401,1417AA) and CysB (CC1449,1473SS) mutants. (B) Yeast two-hybrid analysis. REV3, rev3-cysA or rev3-cysB was fused to the GAL4 DNA-binding domain. REV7, POL31 or POL32 was fused to the GAL4 AD; empty vector pACT2 was the negative control. Analysis was in two-hybrid indicator strain PJ69-4A. Cells were grown on His-selective medium. (C) Scheme for overexpression of GST-REV3, REV7, POL31 and POL32, and affinity pull down of complexes. (D) Pull down of Pol31 and Pol32 with GST-Rev3. GST-Rev3-Rev7 complex was overexpressed alone or together with Pol31–Pol32 subunits in either wild-type or Δpol32 yeast. Cell extracts were incubated with glutathione sepharose beads and washed extensively. GST-Rev3 and Pol31 and Pol32 were detected by western analysis. -, gene deleted; +, native level; ++, overexpression. (E) Analysis of the interaction between Pol31–Pol32 and GST–Rev3 mutants by GST-pull down. Details are as in (D).
We next analyzed these interactions by pull-down experiments using GST-Rev3 trapping. We overexpressed GST-REV3 and REV7 and assayed for Rev3–Rev7–associated factors by glutathione chromatography (Figure 1C). Significant levels of Pol31 and Pol32 were detected, when compared with controls (Figure 1D, lane 3 vs. 1 and 2). When POL31 and POL32 were also overexpressed, a strong interaction signal was detected (lane 4). However, when the same experiment was carried out in a pol32Δ strain, Pol31 was undetectable after affinity co-purification (lane 7 vs. lane 3). This defect was rescued by providing back overexpressed POL32 (lane 8). These data strongly suggest the existence of a four-subunit Rev3–Rev7–Pol31–Pol32 complex called Pol ζ4. Importantly, unlike Pol δ, in which a Pol3–Pol31 complex is a stable assembly (30), Pol32 is required to stabilize the interactions between Rev3 and Pol31. These important differences in polymerase complex stabilities between Pol δ and Pol ζ explain why pol32Δ mutants are viable, but defective for mutagenesis (16).
In agreement with the yeast two-hybrid experiments, we found that Pol31 and Pol32 fail to bind the CysB mutant of GST-Rev3, independent of overexpression (Figure 1E, lanes 5 and 8). In contrast, the CysA mutant of GST-Rev3 pulled down Pol31–Pol32 with the same efficiency as wild-type (compare lane 3 with 4 and 6 with 7).
Rev3-cysB mutant is defective for mutagenesis
Our model suggests that the four-subunit form of Pol ζ is involved in mutagenesis and predicts that mutations disrupting this complex result in a defect in mutagenesis. We measured UV damage-induced mutagenesis in the CysA and CysB mutants of REV3, using a forward mutation assay to canavanine resistance (Table 1). Mutations in the CysB motif that are predicted to disrupt iron–sulfur cluster binding disrupt Rev3–Pol31 interactions (Figure 1B and E), which are almost completely defective for damage-induced mutagenesis, although the observed residual signal is higher than that of a rev3Δ mutant. However, double cys->ser, or double cys->ala mutations in the CysA motif that should disrupt metal binding to the zinc-ribbon motif show no damage-induced mutagenesis phenotype. Our genetic analysis of the CysA and CysB mutants is in complete agreement with a similar analysis reported recently by Baranovskiy et al. (22).
Purification and characterization of two forms of Pol ζ: Pol ζ2 and Pol ζ4
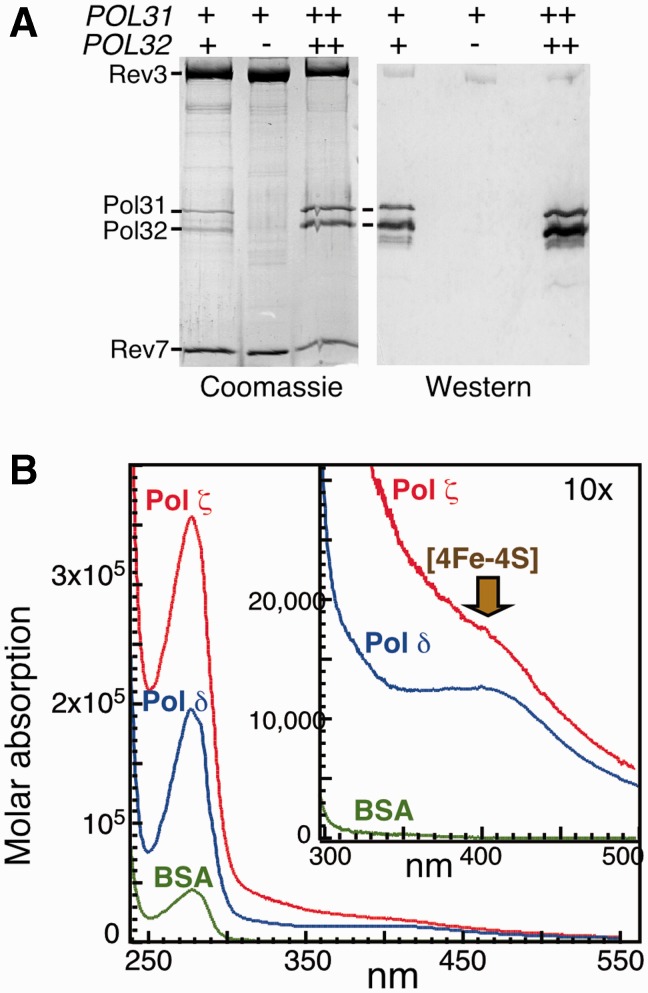
To obtain a Pol ζ4 complex containing an intact [4Fe-4S] cluster, we overexpressed all four genes from galactose-inducible promoters (Figure 1C) and modified the purification protocol of Pol ζ that was described previously (28). Overexpression was carried out in a rev1Δ strain to eliminate trace contamination of the purified preparation with Rev1 (see below). The modified procedure made use of two affinity purification tags, an N-terminal GST tag on Rev3 and an N-terminal His7 tag on Pol32. First, the extract, after an ammonium sulfate precipitation step, was subjected to glutathione-affinity chromatography. The resistance of the Pol ζ4 complex to ammonium sulfate precipitation indicates that the interaction between Rev3–Rev7 and Pol31–Pol32 is very strong and specific. This procedure yielded a preparation that was slightly substoichiometric for Pol31–Pol32 (∼80–90% in three purifications). Next, after cleavage of the GST-tag by rhinoviral 3C protease, the complex was further purified by Ni-chelate affinity chromatography with ∼100 % stoichiometry (Figure 2A). The Pol32-His7 tag did not influence the activity of the Pol ζ4 complex (data not shown).
Figure 2.
Purification of Pol ζ2 and Pol ζ4. (A) Subunit composition of substoichiometric Pol ζ4, Pol ζ2 and stoichiometric Pol ζ4 complex. Pol ζ preparations were analyzed by Coomassie blue staining following SDS-PAGE and by western analysis, probed with a mixture of Pol31 and Pol32 antibodies, as indicated. POL31 and POL32 were expressed at endogenous levels (+), overexpressed (++) or absent from cells (-). (B) UV-VIS spectra of Pol ζ4, Pol δ and bovine serum albumin. Spectra were collected at ∼0.3 to 1 mg/ml protein and recalculated to molar absorptions. Absorption maximum due to the presence of [4Fe-4S] cluster in proteins is indicated.
In agreement with the yeast two-hybrid analysis and pull-down experiments, Pol31 and Pol32 were present in affinity-purified preparations of Pol ζ with mutations in the CysA cluster (Rev3-CC1401,1407SS or Rev3-CC1401,1407AA) but not in the purified preparation of Pol ζ sample with mutations in the CysB cluster (Rev3-CC1449S,1473SS) (Supplementary Figure S1A). The Pol ζ2 complex was purified from a pol32Δ strain, and in agreement with the pull-down data in Figure 1D, this two-subunit complex lacks any detectable level of Pol31 by Coomassie staining after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and by western analysis (Figure 2A).
Overexpression of REV3 and REV7 in wild-type yeast without concomitant overexpression of POL31 and POL32 yielded affinity-purified preparations that were severely substoichiometrical for Pol31 and Pol32, with abundances ranging from 3 to 15% (Figure 2A). We had previously noted that different Pol ζ preparations were quite variable in activity, but because of the extreme difficulty in purifying the enzyme and the very low yields, it had not been feasible to investigate those issues further at that time (28). We now think that the variations in activity were due to the variable presence of low levels of Pol31–Pol32 that escaped detection. With improved expression and purification methodologies and increased yields, we re-investigated the protein composition of our purified preparations. First, because Rev1 is known to interact with Pol ζ through Rev7 (31), we probed Pol ζ preparations for the presence of Rev1 by western analysis. Both Pol ζ2 and Pol ζ4 complexes, as well as all preparations of Pol ζ mutants, contain similar levels of Rev1 (∼1–2% compared with Rev3, Supplementary Figure S1B). Therefore, we have purified Pol ζ2 and Pol ζ4 from a rev1Δ mutant strain without loss of complex stability, indicating that Rev1 is not required for the formation of the Pol ζ4 complex (data not shown). Second, because Pol31 interacts with the catalytic subunit of Pol δ, we investigated the possibility of the presence of Pol3 by western analysis. However, none of the Pol ζ preparations contained Pol3 at detectable levels (detection limit is ∼0.1%), suggesting that Pol31 binds either Pol3 or Rev3, but not both catalytic subunits (Supplementary Figure S1C). Therefore, we conclude that our current forms of Pol ζ4 and Pol ζ2 contain the expected subunits without contamination by other proteins that may function in TLS and mutagenesis.
Expression in E. coli of the entire CTD of Rev3, containing both CysA and CysB motifs, yielded a yellow-brown preparation that after reduction by dithionite was converted into an electron spin resonance (EPR) active form with the spin signal of that of a [4Fe-4S]1+ cluster (14). This suggests that, like Pol3, Rev3 has a [4Fe-4S]2+ cluster. Indeed, similar to Pol δ, purified Pol ζ4 has a UV-spectral signature that is indicative of the presence of an iron–sulfur cluster (Figure 2B). Unfortunately, we were unable to obtain sufficiently high quantities of the CysB mutant form to query whether the iron–sulfur cluster was eliminated in the mutant, but on the basis of the strong sequence homology between Pol3 and Rev3 CTD, we accept this as a likely explanation.
Pol31 and Pol32 are essential for functional interactions between PCNA and Pol ζ
The presence of Rev7 is required for DNA polymerase activity of Rev3 (6). We measured basal DNA polymerase activity of Pol ζ preparations on activated DNA in the absence of PCNA. The presence of the Pol31 and Pol32 subunits in Pol ζ4 enhanced the activity 5- to 10-fold compared with the Pol ζ2 preparations, which were either obtained by purification from a pol32Δ strain or by mutation of the CysB motif in REV3 (Supplementary Figure S2A).
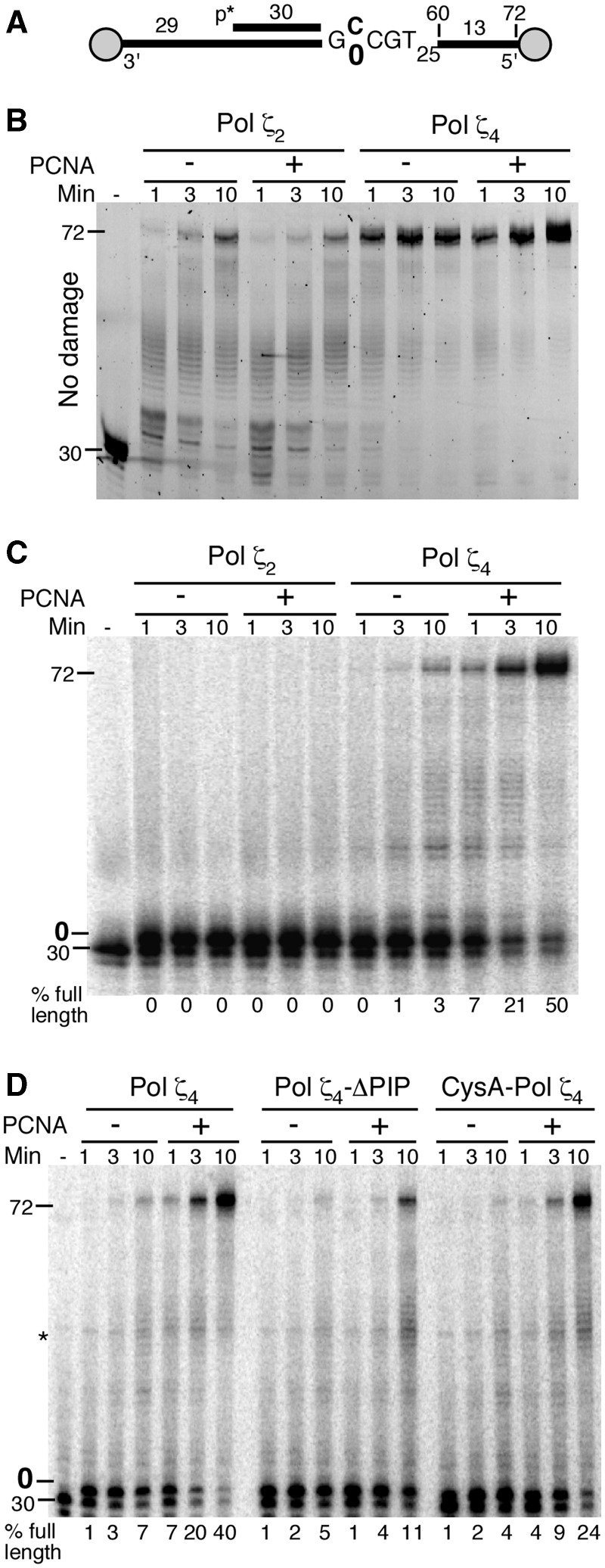
To determine the role of PCNA in TLS by Pol ζ, we used an oligonucleotide-based replication system with defined template damage. The substrate is incubated with RPA to coat the ssDNA regions, and PCNA is loaded by RFC and ATP. To prevent sliding of PCNA off the DNA, biotin-streptavidin bumpers are added to the 5′- and 3′-termini of the template (Figure 3A). We first assayed the replication by Pol ζ2 on an undamaged template-primer. Pol ζ2 activity on this template was much less efficient compared with the Pol ζ4 complex (Figure 3B). In addition, the presence of PCNA had no detectable effect on DNA replication by Pol ζ2. Because of the robust activity of Pol ζ4 on this DNA substrate, PCNA stimulation could not be detected under these conditions. However, PCNA stimulation of Pol ζ4 on undamaged DNA was readily detected using primed single-stranded plasmid DNA substrates (Supplementary Figure S2C).
Figure 3.
PCNA-mediated translesion activity of Pol ζ2 and Pol ζ4. (A) A schematic diagram of the oligonucleotide substrate. The template is a 102-mer with streptavidin-biotin blocks at the 5′ and 3′ ends. The template at the +2 position is either a C (in (B)) or an abasic site, indicated as a ‘0’ (in (C) and (D)). The 72-mer products represent full extension of the 30-mer primer to the end of the template. PCNA (30 nM) was added where indicated in (B–D). See Materials and Methods for details. (B) Time course of reactions of Pol ζ2 and Pol ζ4 on undamaged template DNA. (C) Time course of translesion synthesis by Pol ζ2 and Pol ζ4 on an abasic site (0) template. (D) Stimulation by PCNA of the DNA polymerase activity of Pol ζ4, Pol ζ4-ΔPIP and Pol ζ4-CysA on template DNA with an abasic site. Asterisk indicates an impurity in the radiolabeled primer.
To study the role of PCNA in DNA damage TLS, we used the oligonucleotide assay system with a model abasic site at the +2 position after the primer terminus. We observed that Pol ζ2 readily extended the primer by one nucleotide but did not insert a nucleotide opposite the abasic site, and PCNA did not enhance this activity (Figure 3C). In contrast, the Pol ζ4 complex bypassed the abasic site damage even in the absence of PCNA. Remarkably, a dramatic stimulation of damage bypass synthesis was detected in the presence of PCNA. These data indicate that formation of the Pol ζ4 complex is essential for both efficient damage bypass in the absence of PCNA and stimulation of Pol ζ-mediated TLS in the presence of PCNA. Therefore, we conclude that functional interactions between Pol ζ and PCNA require its Pol31 and Pol32 subunits. However, ubiquitination of PCNA did not significantly enhance TLS by Pol ζ4 (Supplementary Figure S2B). This is consistent with a model in which ubiquitination of PCNA exerts its TLS-promoting activity through Rev1 (32).
The observation that interactions with Pol31–Pol32 enhanced the PCNA-dependent activity of Pol ζ raised the possibility that the PCNA-binding motif is localized in the Pol31 or Pol32 subunit. Previously, we have identified a C-terminal PCNA-binding motif in Pol32 (24). Deletion of this motif only marginally affected processive DNA replication by Pol δ; however, the pol32-ΔPIP mutant showed a substantial reduction in the efficiency of damage-induced mutagenesis, particularly at higher loads of DNA damage. We purified a mutant Pol ζ4 containing a truncated form of Pol32 that lacks its PCNA-binding motif (Pol ζ4-ΔPIP). Although the basal activities of Pol ζ4 and Pol ζ4-ΔPIP were comparable, PCNA stimulation of the mutant complex was substantially reduced (Figure 3D and Supplementary Figure S2A and S2C). We conclude that the PCNA BD of Pol32 contributes to the functional interaction between Pol ζ4 and PCNA.
DNA replication by Pol δ requires an intact CysA motif, as CysA mutants are severely compromised for PCNA-dependent replication (14). In contrast, the CysA mutant form of Pol ζ4 demonstrated close to wild-type basal DNA polymerase activity (Supplementary Figure S2A). Although its TLS activity was slightly reduced (∼60% of wild-type), PCNA stimulated this TLS activity to the same degree as it did wild-type Pol ζ4 (Figure 3D). This lack of a strong in vitro phenotype is consistent with the lack of a damage-induced mutagenesis phenotype of the same CysA mutations in yeast (Table 1).
The pol30-113 mutant of PCNA shows severe defects in damage-induced mutagenesis, without affecting the efficiency of a proper DNA damage response through PCNA ubiquitination at Lys164 (27,33). Pol30-113 has mutations at Glu113 and Leu151 near the monomer–monomer interface of PCNA. Previously, we showed that this mutant form of PCNA was defective for PCNA-mediated TLS in vitro (27). With our increased knowledge of the assembly state of Pol ζ, we assume that the previous preparations of Pol ζ contained low levels of Pol31–Pol32 that drove the observed PCNA stimulation. Indeed, the stoichiometrical Pol ζ4 complex was unable to perform processive replication with pcna-113 (Supplementary Figure S2C).
The Pol ζ4 complex is stable throughout the cell cycle
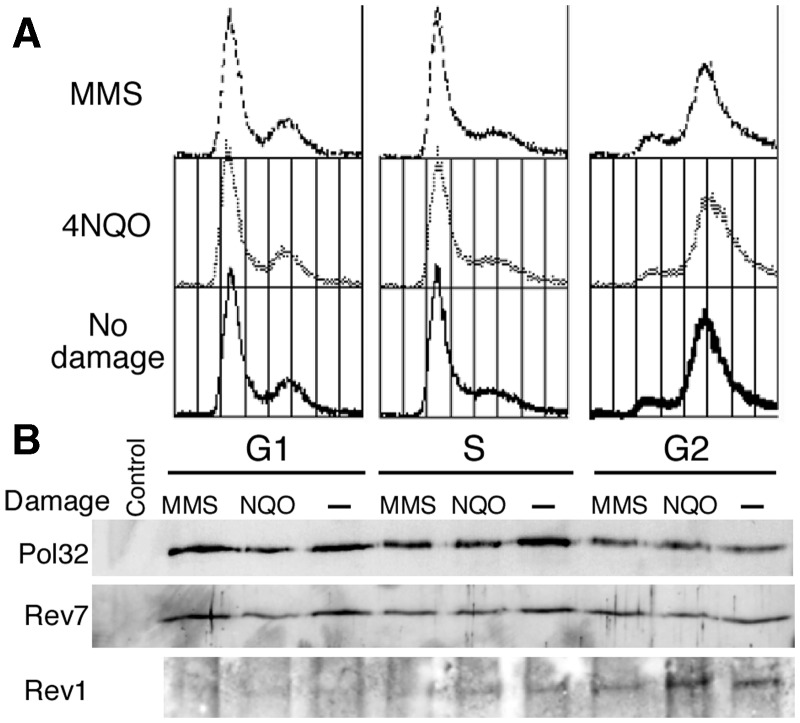
To test whether the formation of the Pol ζ4 complex is subject to either cell cycle or DNA damage control, we prepared synchronized cell populations and determined co-purification of Pol31 and Pol32 with GST-Rev3 on gluthathione sepharose beads. For this experiment, we used the GST-REV3 expression plasmid, however, omitted galactose induction to maintain Rev3 at low levels. Under the same growth conditions, this construct fully complemented the mutagenesis defect of a rev3Δ mutant (data not shown). POL31 and POL32 were not overexpressed in these experiments. Cells were arrested in G1 phase with α-factor, in S phase with hydroxyurea and in G2/M phase with nocodazole. About 80–95% of cells were arrested in the appropriate phase of the cell cycle in our experiments (Figure 4A). Synchronized cells were also treated with MMS or 4NQO to induce the DNA damage response. After affinity purification on glutathione beads, the presence of Rev7, Pol32 and Rev1 was monitored by western analysis (Figure 4B). The data indicate that Pol ζ can exist as a four-subunit complex in all phases of the cell cycle. Furthermore, treatment with DNA-damaging agents did not alter the formation or stability of the complex. Interestingly, Rev1 association with Pol ζ is highest in G2 phase. This study addressed the question whether the four-subunit complex, or its stability, is regulated at the level of posttranstional modification, and we found it is not, but we cannot exclude the possibility of cell cycle–specific transcriptional regulation of Rev3.
Figure 4.
Stability of Pol ζ4 during the cell cycle. (A) Fluorescence-activated cell sorting analysis of the DNA content of cells. Cells expressing low levels of GST-REV3 and REV7, and POL31 and POL32 at native levels, were arrested in G1, S, or G2 phase, followed by treatment with MMS or 4NQO. (B) Extracts were prepared from arrested cells, and Pol32, Rev7 and Rev1 were detected by western analysis after GST-Rev3 pull down with glutathione sepharose beads. Control: Western analysis of extracts made from cells lacking GST-Rev3 and subjected to glutathione affinity purification.
DISCUSSION
Pol ζ is a low-fidelity, B-family DNA polymerase and the sixth eukaryotic DNA polymerase to be described (6). The original article described a form of Pol ζ that was overexpressed in yeast, and all subsequent studies, including those from our laboratory, used forms that were also purified from yeast overexpression systems (5, 28,30). Therefore, it is likely that these forms contained low, variable levels of Pol31 and Pol32 in the preparations. Our previous observations that TLS by Pol ζ is stimulated by PCNA likely originated from the use of preparations that contained such low levels of Pol31–Pol32, which we now know varied from 3 to 15% over the years. Coupled with the fact that Pol ζ2 has much lower basal polymerase activity than Pol ζ4 (Supplementary Figure S2A and Figure S3), the latter species would have contributed more to the observed activity than considerations of abundance suggest. This also explains the variability in activity of different Pol ζ preparations that we remarked on several years ago (28).
Previously, we have shown that the catalytic subunits of the yeast B-family DNA polymerases contain an [4Fe-4S]2+ cluster, coordinated by the CysB motif in their CTDs, and we and others have suggested that all B-family polymerases are similarly organized (14,20). However, a comparison between the architecture of Pol δ and Pol ζ reveals some interesting differences that may underlie their different functions in the cell. Both Pol3 and Rev3 bind Pol31 through their CysB motif as mutations in this motif abrogate binding, while mutations in the CysA motif do not. However, Pol3 forms a stable complex with Pol31 alone (34), but Rev3 does not (Figure1D). As a result, pol32Δ mutants are viable, but they are defective for damage-induced mutagenesis (16,35). Furthermore, transformation studies with plasmids containing specific DNA damage show that pol32Δ is defective for the bypass of abasic site damage similar to rev3Δ, but proficient for the bypass of thymine dimers, which is Pol η dependent (36). This is consistent with Pol32 functioning as an integral part of the Pol ζ complex.
CysA mutations in POL3 are lethal, most likely because the mutant form of Pol δ is severely defective for PCNA-mediated processive replication (14). However, the analogous mutations in the CysA motif of REV3 show no defect in mutagenesis [Table 1, (22)] nor is the mutant polymerase defective for PCNA-mediated processive replication (Figure 3D). Functional interactions of Pol δ with PCNA is imparted by multiple potential PCNA-binding motifs in the various subunits of Pol δ (14,24,37–40). In Pol ζ4, PCNA interacts through the consensus PIP box in the extreme C-terminus of Pol32 as deletion of this motif reduces TLS in vitro (Figure 3D). This POL32 mutant also has a reduced efficiency in damage-induced mutagenesis (24). The residual PCNA stimulation observed in vitro and mutagenesis in vivo suggests that Pol ζ4 contains additional PCNA interaction motif(s). The striking difference in CysA phenotype between Pol δ and Pol ζ4 suggests a different positioning of the PCNA clamp in relation to this motif in these enzymes. Consistent with this, mutations in PCNA differentially affect its interactions with Pol δ versus Pol ζ. A pcna-113 mutant functions as a processivity clamp for Pol δ, although its activity is somewhat reduced (27); however it is defective with Pol ζ4 (Supplementary Figure S2C). This provides a rational explanation for the mutagenesis defect in this mutant.
The formation and stability of the Pol ζ4 complex was unaffected by the cell cycle or by exposure to DNA-damaged agents (Figure 4). This result suggests that Pol ζ-mediated mutagenesis can occur throughout the cell cycle. However, other factors, for example, Rev1 and PCNA, show cell cycle and/or DNA damage control, and overall pathway control is likely mediated through those factors. Ubiquitination of PCNA is a key switch in this pathway, and both damage-induced mutagenesis as well as spontaneous mutagenesis in response to replisome dysfunction is dependent on ubiquitination of PCNA (27,41,42). The Rev1 protein, considered to be the scaffold onto which the mutasome assembles through binding of ubiquitinated PCNA on one hand and Pol ζ on the other hand, is most highly expressed in G2 phase (43). Indeed, it has been shown that PCNA ubiquitination and mutagenesis can be restricted to the G2 phase of the cell cycle (44,45). We found that Rev1 association with Pol ζ4 is also highest during G2 phase (Figure 4). Therefore we suggest that the regulation of Pol ζ4-dependent mutagenesis is likely mediated by the formation of multisubunit complexes of higher order, for example with Rev1 and ubiquitinated PCNA, but not through the assembly of the Pol ζ4 complex. Finally, the cell cycle kinase CDC7/DBF4 promotes the efficiency of UV mutagenesis (46).
As stated before, two other groups have recently reported that Rev3 interacts with Pol31 and Pol32. The article by Baranovskiy et al. reported the co-purification from E. coli of the CTD of human Rev3 together with human Pol31 and Pol32 (22). Although this approach did not permit functional polymerase studies, it allowed these authors to probe the relevance of the CysA and CysB motifs for complex formation. In agreement with our results in Figure 1E, CysB mutations, but not CysA mutations, abrogated complex formation. Similarly, their genetic studies of the CysA and B motifs in yeast yielded analogous results to ours (Table 1). The second article by Johnson et al. reported the isolation of a Pol ζ4 complex from a yeast overexpression system and is in accord with ours when all four genes are overexpressed (23). However, our conclusion that Pol32 is required for stable complex formation between Rev3 and Pol31 is at variance with their study. These authors reported the purification of a three-subunit Rev3-Rev7-Pol31 complex from a strain that overexpressed just the REV3, REV7, and POL31 genes, and based on this purification concluded that Pol32 was not required for complex formation. However, this three-subunit preparation was purified from a wild-type yeast strain rather than a pol32Δ strain and was highly non-stoichiometric containing predominantly the Pol31 polypeptide, to which the purification tag was fused. Given the low levels of Rev3 in this preparation, and the close migration of Pol31 and Pol32 by SDS-PAGE, low levels of Pol32 may have escaped detection. Unfortunately, a more sensitive western analysis with Pol32 antibodies was not used as a detection method in this study. We think that these are important considerations, because our study indicates that Pol32 is absolutely required for complex formation and thereby provides a logical explanation for the long-standing observation that pol32Δ strains are defective for damage-induced mutagenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Text, Supplementary Table 1, Supplementary Figures 1 and 2 and Supplementary Reference [47].
FUNDING
Funding for open access charge: National Institutes of Health [GM032431 to P.B.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Carrie Stith and Bonnie Yoder for strain and plasmid construction and for help with protein purification, Sandeep Kumar for help with fluorescence-activated cell sorting analysis, and Sara Binz for advice during the course of this study.
REFERENCES
- 1.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence CW, Maher VM. Eukaryotic mutagenesis and translesion replication dependent on DNA polymerase zeta and Rev1 protein. Biochem. Soc. Trans. 2001;29:187–191. doi: 10.1042/0300-5127:0290187. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc. Natl Acad. Sci. USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo D, Wu X, Rajpal DK, Taylor JS, Wang Z. Translesion synthesis by yeast DNA polymerase zeta from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 2001;29:2875–2883. doi: 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol. Cell. Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 7.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 9.Larimer FW, Perry JR, Hardigree AA. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J. Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3 gene causes embryonic lethality. Curr. Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Trang J, Okuda T, Howell SB. DNA polymerase zeta accounts for the reduced cytotoxicity and enhanced mutagenicity of cisplatin in human colon carcinoma cells that have lost DNA mismatch repair. Clin. Cancer Res. 2006;12:563–568. doi: 10.1158/1078-0432.CCR-05-1380. [DOI] [PubMed] [Google Scholar]
- 12.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem. Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacNeill SA, Moreno S, Reynolds N, Nurse P, Fantes PA. The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase delta, binds to Pol3 and Cdc27. EMBO J. 1996;15:4613–4628. [PMC free article] [PubMed] [Google Scholar]
- 16.Gerik KJ, Li X, Pautz A, Burgers PM. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA. The C-terminal zinc finger of the catalytic subunit of DNA polymerase delta is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 2004;32:3005–3016. doi: 10.1093/nar/gkh623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle. 2008;7:3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno T, Yamagishi K, Miyazawa H, Hanaoka F. Molecular architecture of the mouse DNA polymerase alpha-primase complex. Mol. Cell. Biol. 1999;19:7886–7896. doi: 10.1128/mcb.19.11.7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L. 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 2009;28:1978–1987. doi: 10.1038/emboj.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 2010;285:12299–12307. doi: 10.1074/jbc.M109.092403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl Acad. Sci. USA. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 25.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 27.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg P, Stith CM, Majka J, Burgers PM. Proliferating Cell Nuclear Antigen Promotes Translesion Synthesis by DNA Polymerase zeta. J Biol Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 29.Bauer GA, Heller HM, Burgers PMJ. DNA polymerase III from Saccharomyces cerevisiae. I. Purification and characterization. J. Biol. Chem. 1988;263:917–924. [PubMed] [Google Scholar]
- 30.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 31.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 32.Amin NS, Holm C. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics. 1996;144:479–493. doi: 10.1093/genetics/144.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakumo Y. The property of DNA polymerase zeta: REV7 is a putative protein involved in translesion DNA synthesis and cell cycle control. Mutat. Res. 2002;510:37–44. doi: 10.1016/s0027-5107(02)00250-6. [DOI] [PubMed] [Google Scholar]
- 34.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 35.Huang ME, Rio AG, Galibert MD, Galibert F. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002;160:1409–1422. doi: 10.1093/genetics/160.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 Protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou JQ, He H, Tan CK, Downey KM, So AG. The small subunit is required for functional interaction of DNA polymerase delta with the proliferating cell nuclear antigen. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J. Biol. Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds N, Warbrick E, Fantes PA, MacNeill SA. Essential interaction between the fission yeast DNA polymerase delta subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21(Cip1)-like PCNA binding motif. EMBO J. 2000;19:1108–1118. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S. PCNA binding domains in all three subunits of yeast DNA polymerase delta modulate its function in DNA replication. Proc. Natl Acad. Sci. USA. 2011;108:17927–17932. doi: 10.1073/pnas.1109981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 42.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 43.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl Acad. Sci. USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 46.Pessoa-Brandao L, Sclafani RA. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 2004;167:1597–1610. doi: 10.1534/genetics.103.021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freudenthal BD, Gakhar L, Ramaswamy S, Washington MT. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol. 2010;17:479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.