Abstract
Mitotic chromosome structure is pivotal to cell division but difficult to observe in fine detail using conventional methods. DNA catenation has been implicated in both sister chromatid cohesion and chromosome condensation, but has never been observed directly. We have used a lab-on-a-chip microfluidic device and fluorescence microscopy, coupled with a simple image analysis pipeline, to digest chromosomal proteins and examine the structure of the remaining DNA, which maintains the canonical ‘X’ shape. By directly staining DNA, we observe that DNA catenation between sister chromatids (separated by fluid flow) is composed of distinct fibres of DNA concentrated at the centromeres. Disrupting the catenation of the chromosomes with Topoisomerase IIα significantly alters overall chromosome shape, suggesting that DNA catenation must be simultaneously maintained for correct chromosome condensation, and destroyed to complete sister chromatid disjunction. In addition to demonstrating the value of microfluidics as a tool for examining chromosome structure, these results lend support to certain models of DNA catenation organization and regulation: in particular, we conclude from our observation of centromere-concentrated catenation that spindle forces could play a driving role in decatenation and that Topoisomerase IIα is differentially regulated at the centromeres, perhaps in conjunction with cohesin.
INTRODUCTION
Complete and equal transmission of DNA to daughter cells is crucial during mitosis. Within each chromosome, two dimensions of organization are at play: condensation along the axes ensures the entire chromatid, end-to-end, is kept together, while the tight association of sister chromatids until anaphase, termed sister chromatid cohesion (SCC), ensures that each daughter cell receives only one copy (1). Two mechanisms are known to play a role in SCC: DNA catenation, which physically interlocks (catenates) DNA across the sister chromatids (2); and protein linkages through the cohesin complex, which physically tether the sister chromatids to one another (3,4).
Both linkages must be resolved to complete mitosis: cohesin is specifically cleaved by separase at anaphase onset (5); topoisomerase II (Topo II) in yeast and its human analogue (topoisomerase IIα, Topo IIα) is required to complete DNA decatenation.
While Topo II can both introduce and remove DNA catenation (6), the enzyme strongly biases its actions towards decatenation through ATP hydrolysis (7) and by acting on severely bent DNA (8), which has been shown by mathematical models to be a key feature of knotted and catenated DNA (9,10). This mechanism for preferential DNA decatenation is supported by the crystal structure of yeast Topo II, in which bound DNA is bent by 150° (11). Although this preference towards decatenation is able to prevent Topo II from extensively intertwining newly replicated DNA, it alone is not sufficient to ensure the complete decatenation necessary for sister chromatid separation (6). Through stochastic action alone, a few DNA intertwines would necessarily remain (7) and these would need to be completely removed to complete mitosis (6). Complete decatenation of sister chromatids by Topo II is therefore driven by external factors.
Recent experiments have demonstrated a complex choreography between condensin, cohesin and DNA catenation: electrophoresis of 5 kb-long yeast centromeric plasmids have been used to show that positive supercoiling generated by condensin and induced by kinetochore attachment can drive Topo II to fully decatenate DNA before anaphase (12), though the use of longer plasmids (26 kb) and linear ‘minichromosomes’ (42 kb) indicated that centromeric catenation was still present at metaphase and maintained by cohesin (13). This work reconciled yeast models with previous work involving immunolabeling of fixed mammalian cells, which showed that DNA catenation was present at metaphase and into anaphase (14–16) and could only be resolved after cohesin cleavage (17). Whether the persistence of catenation is due to direct blocking of Topo II activity by cohesin or a consequence of the physical proximity of the chromatids promoted by cohesin and stochastic catenation–decatenation by Topo II has not yet been determined (18).
DNA catenation has also been implicated in chromosome condensation: knockdown of the condensin complex only delays condensation (19), whereas Topo II is essential (20). In vitro micromanipulation experiments—in which the ends of chromosomes are aspirated into pipette tips and subjected to axial stretching forces to measure chromosome elasticity—have shown that while nuclease treatment completely ‘dissolves’ chromosomes, protease treatment only relaxes them (21,22). These results suggest that DNA forms the ‘backbone’ of the chromosome to which proteins are attached, and not vice versa. In subsequent experiments, Topo IIα was shown to relax mitotic chromosomes, indicating that DNA catenation has a structural role in chromosome architecture. The changes in chromosome structure, however, could not be observed visually (23).
One of the biggest challenges in elucidating the extent, structure and function of DNA catenation is the fact that DNA catenation itself ‘has never been observed directly within mitotic chromosomes,’ as Farcas et al. have noted (13). Microscopy allows for obtaining visual images of DNA, but direct staining of DNA typically produces high background from dense chromatid bodies and observing fine structure is difficult (14). Indirect methods have included the use of fluorescent histone fusion proteins in live cells (24) and antibody labelling of DNA-associated proteins in fixed cells (14,16) that were shown to be co-local to DNA by counterstaining with antibodies to bromodeoxyuridine, which had been incorporated into the DNA (15).
In this article, we demonstrate the use of a microfluidic lab-on-a-chip device and integrated image-processing pipeline capable of providing observation of DNA catenation and its effect on chromosome structure. Fluidic systems have long been used to handle metaphase chromosomes (25) and for more complex manipulations, such as on-chip cell lysis and chromosome extraction (26) and isolation for single-chromosome sequencing (27). In order to combine the benefits of microscopy, biochemistry and micromanipulation techniques, we have used a microfluidic device (28) with a central reaction chamber that allows for real-time observation, temperature control, reagent exchange and gentle fluidic manipulation in order to tease apart chromosomes and examine their structure in more detail while chromosomes remain free and unfixed in solution.
MATERIALS AND METHODS
Solutions and reagents
Chromosomes were isolated from mammalian cell culture (Jurkat cell line, DSMZ, Germany) based on a modified polyamine isolation protocol (25,29). YOYO-1 DNA intercalating dye was used without further purification (Life Technologies, Naerum, Denmark). Chemicals and buffers were purchased from Sigma Aldrich (Copenhagen, Denmark) and any water used was purchased nuclease-free unless otherwise noted. Buffer solutions for chip-handling work were prepared on the day of use from degassed 0.5x TBE buffer. A working buffer solution for chip handling (BwT) was composed of 3% 2-Mercaptoethanol (BME), 0.5% Triton-X and 0.5x degassed Tris-Borate EDTA. For chromosome pre-treatment, fresh solutions (described below) were prepared and care was taken to avoid inadvertent nuclease contamination. Stock 1 M DTT was diluted to 10 mM working concentration. Human topoisomerase IIα solution (Topo IIα), dexrazoxane and etoposide were purchased from Sigma Aldrich (UK). Topo IIα was used as supplied and the inhibitors were dissolved in DMSO at 30 mg/ml prior to use.
Lab-on-a-chip device
The fluidic chip was fabricated in silicon as previously described (28); the chip contained a chromosome inlet channel, leading to a central reaction chamber, and a chromosome outlet channel. Reagents could be flushed around this chamber from separate inlets and allowed to diffuse inwards to the chromosome without dislodging it. The surface of the interior channels of the chip was coated with a bovine serum albumin solution (BwT+BSA: 0.1 mg/ml BSA in BwT) prior to chromosome loading. The entire chip was mounted in a custom holder on a Nikon TE-2000 microscope that contained wells for loading solutions and air pressure was used to drive fluid flow from these wells. A cartridge heater (Omega Engineering, UK) set at 37°C was used to maintain temperature during digestion.
Chromosome pre-treatment, staining and digestions
To observe native chromosomes, staining was performed by supplementing chromosome suspensions from cell culture with YOYO-1 dye to 1 µM and incubating in the dark for 1 h at room temperature, followed by 1 h at 50°C. Chromosomes were diluted in BwT+BSA to ∼105 chromosomes per ml, then loaded onto the microfluidic chip and positioned in the central reaction chamber by fluid flow. ProK (0.1 mg/ml in BwT+BSA buffer supplemented with YOYO-1 to 1 µM) was continuously flushed around the chamber from where it diffused into the reaction chamber and digested chromosomal proteins.
To pre-treat chromosomes with Topo IIα, unstained chromosomes were diluted in Topo II reaction buffer with a final concentration of 50 mM Tris at pH 8, 10 mM MgCl2, 120 mM KCl and 0.5 mM DTT. When used, inhibitors were added to a final concentration of 2.6 mg/ml. The reactions were then incubated for 45 min at room temperature, after which the reaction was supplemented with ATP to 0.5 mM and with 0.5 µl Topo IIα solution. The reaction was mixed by gently pipetting up and down, incubated for 30 min at 37°C, and then heated to 65°C to inactivate the Topo IIα enzyme. Chromosomes were then stained, loaded and digested as described above.
Image acquisition and processing pipeline
Imaging was conducted on a custom Nikon TE-2000 inverted fluorescence microscope using metal halide lamp illumination and appropriate excitation and emission filters for YOYO-1 dye (B2E/C filter, Nikon Instruments, Amstelveen, Netherlands). Image acquisition was conducted using MetaMorph 7 (Molecular Devices, Sunnyvale, CA, USA), which controlled a Cascade II:512 back-illuminated EMCCD camera (Photometrics, Tuscon, AZ, USA), and an electromechanical illumination shutter (Prior Scientific, Jena, Germany). Time-lapse images of each chromosome digestion were acquired using a macro which sampled a set of 5 frames every 30 s and averaged each set in Matlab (Mathworks, Natick, MA, USA) to reduce image jitter and real-time images of DNA catenation were frame-averaged to produce clearer static images for publication. ImageJ and Fiji were used to apply automatic thresholding and extract feature length and roundness during timelapse reactions. Roundness was calculated as 4π(area/perimeter2). The full macro used for analysis is presented in Supplementary Methods.
RESULTS AND DISCUSSION
Observation of DNA catenation between sister chromatids
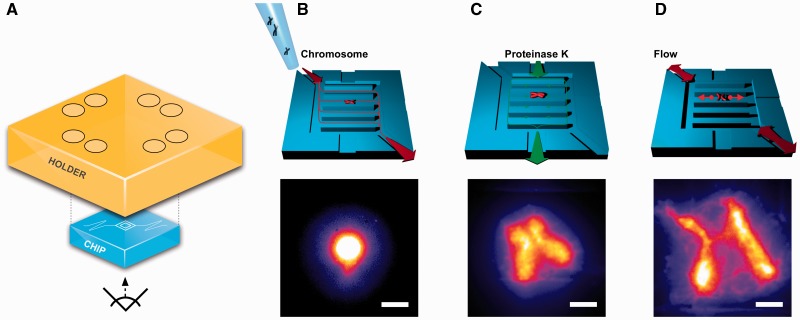
After mounting the microfluidic chip on its holder on an inverted epifluorescence microscope (Figure 1A) and passivating the surface with BSA, a single chromosome was positioned within the central microfluidic reaction chamber using fluid flow. Chromosomes were stained with a DNA intercalating dye prior to loading onto the microfluidic chip and appeared as bright, dense masses of fluorescent DNA floating freely in solution (Figure 1B). To digest chromosomal proteins, proteinase K (proK) was continuously flushed around the chamber and allowed to diffuse inwards so that the chromosome could be digested without being dislodged (Figure 1C). Chromosomes rapidly expanded during digestion, as shown in Supplementary Movie S1: after filling the height of the reaction chamber, they spread out horizontally—analogous to inflating a balloon in a thin rectangular box—but did not adhere to the ‘ceiling’ and ‘floor’ of the microfluidic chip and could be moved with fluid flow. Digestion was complete within 20 min and chromosomes possessed a canonical ‘X’ shape, with close association of sister chromatids at the centromere.
Figure 1.
Direct observation of DNA catenation between sister chromatids. (A) Experimental setup: a custom fluidic chip is mounted in a holder on an inverted fluorescence microscope. (B) Native, unfixed human mitotic chromosomes are loaded onto the chip and a single chromosome is positioned in the central reaction chamber. (C) Chromosomal proteins are digested; chromosomes expand and exhibit a canonical ‘X’ shape, as shown in Supplementary Movie S1. Back-and-forth fluid flow (D) separates sister chromatids, revealing the DNA catenating the sister chromatids, which is concentrated at the centromere. Scale bars, 10 µm.
Fluidic manipulation was then used to pry the sister chromatids apart to examine the structure of the DNA tethering them. After stopping proK flow, gentle back-and-forth fluid flow was introduced in the reaction chamber. As a result, the two chromatid bodies moved apart, permitting visualization of DNA catenation that persisted between the chromatids without being obstructed by the bright fluorescence of the main mass of DNA (Figure 1D). This DNA catenation between the sister chromatids was concentrated at the centromeres, though some was present along the arms, particularly at their ends.
Our observation of centromere-concentrated DNA catenation has implications for what forces might drive the decatenation of sister chromatids to ensure correct separation and complete mitosis. On its own, Topo II will both catenate and decatenate DNA, so some external ‘driver’ must impose directionality on Topo II to fully remove the DNA catenation. The most obvious driving force during mitosis is the spindle force that pulls the sister chromatids apart. Recent discussions in the literature, however, have assumed that DNA catenation is spread evenly along the length of the chromosome (13,18) and it has therefore been argued that the spindle force cannot be an efficient driver of decatenation since its force would be high at the centromeres and would diminish along the length of the chromatid arms (18). In contrast, our observation of centromere-concentrated catenation would suggest that it is premature to rule out spindle forces as a factor in driving the decatenation of sister chromatid DNA.
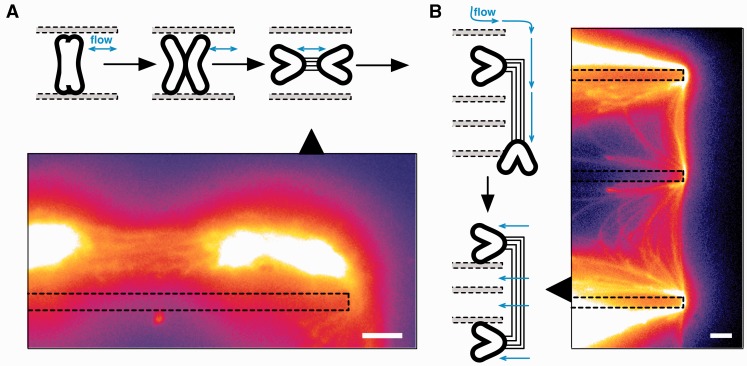
We then sought to examine the structure of the catenating DNA in more detail. Continued fluid flow allowed further separation of the sister chromatids (Figure 2 and Supplementary Movie S2): no longer constrained by chromosomal proteins, the arms of each chromatid folded outwards, leaving the centromeric regions at the centre of the mass of DNA. The fluid flow forced the bodies of the chromatids apart, pulling apart the centromere and giving a direct view of the DNA fibres catenating the two sister chromatids at the centromere (Figure 2A). The presence of fibres was confirmed by trapping the chromatids on the edges of the reaction chamber to stretch them further apart (Figure 2B). Observation under fluid flow (Supplementary Movie S3) showed the dynamics of the catenating DNA, which was organized as discrete fibres.
Figure 2.
DNA catenation at centromeres is composed of discrete fibres. Sister chromatids are further separated with fluid flow (schematic) and the structure of the DNA catenation between the chromatids becomes visible, (A). The handling process is shown in Supplementary Movie S2 and a 3D projection of the fibres is shown in Supplementary Movie S3. The DNA catenation is robust and continues to tether the chromatids even after entrapping the chromatids on edges within the microfluidic reaction chamber (B, dotted lines). Scale bars, 10 µm.
Overall, we observe a relatively large amount of DNA catenating the sister chromatids. Although past work in yeast with short model DNAs showed that there was little DNA catenation at metaphase (30), immunofluorescence data from mammalian cells were contradictory: DNA catenation was shown to be present at metaphase and diminish as cells progress through anaphase (14,15) as a result of Topo II action after cohesin cleavage (17). Recent work has reconciled these results with yeast models by using longer yeast minichromosomes and demonstrating that longer DNAs have greater amounts of DNA catenation present at metaphase (31). Though we cannot exclude the possibility that the large amount of DNA we observed is being ‘pulled along’ by a smaller number of actual DNA catenanes, our observation of a mass of DNA catenation—as opposed to a few single fibres—is in line with these recent results that imply larger natural chromosomes will have substantial amounts of DNA catenation at metaphase (32).In addition to the amount of DNA we observe, its organization has implications for the current debate over how Topo II is regulated at the centromere. While Topo II must decatenate DNA to complete mitosis, centromeric cohesin prevents premature Topo II-mediated DNA decatenation at the centromere prior to anaphase. Two models have been proposed for the mechanism of Topo II inhibition by cohesin (13): the first model proposes that the presence of cohesin directly blocks or inhibits the activity of Topo II, which is then prevented from decatenating sister chromatids. The second model proposes that Topo II remains fully active, but that cohesin serves to keep the sister chromatids in close proximity at the centromere. This proximity would then facilitate the action of Topo II, which would randomly catenate (and decatenate) the sisters while they are nearby. The result of this stochastic action of Topo II would be a mass of stochastically inter-catenated DNA and we would expect the structures to rapidly become entangled in vivo and to observe a ‘snapshot’ of this disordered catenation after proK digestion.
It is noteworthy, therefore, that the DNA catenates are maintained as discrete fibres at the centromere, despite being in close proximity to each other; our observations are consistent with the first model in which Topo II is not active at the centromere in vivo. This model would therefore support the hypothesis that cohesin can actively block Topo II activity—or at least that Topo II is co-regulated with cohesin maintenance at the centromere.
DNA catenation maintains overall shape and structure of metaphase chromosomes
After examining the structure of DNA catenation at the centromeres of metaphase chromosomes, we explored how DNA catenation affected their overall structure. We disrupted DNA catenation in vitro with Topo IIα before loading them into the microfluidic reaction chamber, digesting away chromosomal proteins, and quantitating differences in chromosome shape and size using an image-processing routine.
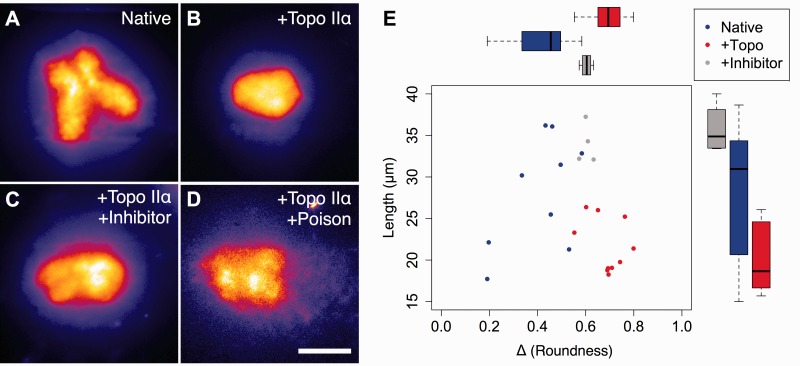
While native chromosomes maintained a distinct ‘X’ shape after proK digestion (Figure 3A), Topo IIα-treated chromosomes exhibited a significant alteration in structure: chromatids no longer had distinct axes, distal arm separation was abolished, and the overall shape of the chromosome was more globular (Figure 3B). This alteration in morphology is the result of the stochastic decatenation (and catenation) action of Topo II during in vitro treatment: disordering the catenation along the arms of the chromatids removed topological constraints that provided the axial definition of the arms, while some catenation was introduced between the sister chromatids, bringing the arms together.
Figure 3.
Disruption of DNA catenation alters the overall shape and structure of metaphase chromosomes. Native chromosomes (A) after proteinase K digestion on-chip exhibit typical morphology with compact axes and well-separated arms. Topo IIα-pretreated chromosomes (B) lose their axial definition and appear more round. A catalytic inhibitor, dexrazoxane, partially inhibits Topo IIα and results in an intermediate morphology, (C). In contrast, the presence of a poisonous inhibitor, etoposide, causes the Topo IIα to act as an endonuclease. No intact chromosomes are observed and any large DNA-containing objects (D) disintegrate upon proteinase K digestion. Morphometry analysis (E) of microscopy data quantitates the effects shown in (A–C) in terms of the chromosomal roundness and length and shows distinct effects of DNA catenation on gross chromosome structure. Scale bar, 20 µm.
We then modulated Topo IIα activity in vitro using two different types of specific inhibitors. In order to catenate and decatenate DNA, Topo IIα must generate a double-strand break (DSB) in one DNA molecule, weave the second one through the cut, and then re-ligate the first molecule. ‘Standard’ catalytic Topo IIα inhibitors prevent the generation of the initial DSB, while ‘poisonous’ inhibitors stabilize the DSB-Topo IIα intermediate and prevent DNA re-ligation, effectively rendering Topo IIα an endonuclease (33). Chromosomes were pre-treated with Topo IIα that was partially inhibited by a standard inhibitor, dexrazoxane (ICRF-187), and separately with Topo IIα in the presence of a poisonous inhibitor, etoposide (VP-16).
Chromosomes pre-treated with partially inhibited Topo IIα in the presence of dexrazoxane had a shape in between native and Topo IIα-treated chromosomes: chromatid axes were visible and slightly elongated, but the ends of the arms were not well separated, with the chromosome appearing more as an ‘H’ than an ‘X’ shape and overall slightly round in shape (Figure 3C). In contrast, Topo IIα treatment in the presence of etoposide resulted in few intact chromosomes. Those that appeared partially intact were subjected to on-chip proK digestion to remove chromosomal proteins (Figure 3D), but quickly released many small DNA fragments and disintegrated, consistent with the conversion of Topo IIα to nuclease-like activity.
In order to quantitate the differences we observed in chromosome shape after disrupting DNA catenation, we developed an automated image-processing pipeline: greyscale images were converted to binary black-and-white images using automatic thresholding, any internal dark ‘holes’ missed by the thresholding were filled, and morphometry analysis was used to determine chromosome length and roundness. This pipeline was applied to timelapse images of chromosome digestions for native, Topo IIα-treated, and Topo IIα+dexrazoxane-treated chromosomes. Chromosomes that had been pre-treated with Topo IIα+etoposide disintegrated and left no intact chromosomes to analyse.
After proK digestion, native chromosomes (n = 9) had an average length of 28 µm, whereas Topo IIα-treated chromosomes (n = 10) were shorter by 6 µm, with an average length of 22 µm. Topo IIα-treated chromosomes also had a greater degree of roundness than native chromosomes: 0.69 versus 0.41. The intermediate morphology of chromosomes pre-treated with partially inhibited Topo IIα (n = 4) was also apparent: chromatid axes were slightly elongated, with an average length of 34 µm as a result of their arms remaining associated, and an intermediate roundness of 0.60. Plotting roundness versus length for each chromosome after proK digestion (Figure 3E) revealed separate populations for each type, with catenation-disrupted chromosomes having a distinct, altered morphology as compared to native chromosomes.
Overall, these results highlight the importance of DNA catenation in higher-level chromosome structure. In particular, catenation plays a role within each individual chromatid, maintaining the definition of the arms while the chromosomes are condensed; this role for DNA catenation is in addition to its function at the centromere, serving to maintain SCC. While there is general agreement that chromosome compaction—whether by topological changes in DNA or protein linkages—leads to decatenation between chromosomes and sister chromatids outside of the centromeres (34), results from small model DNAs in yeast have led to a model proposing supercoiling as being the primary method of DNA compaction (35). This mechanism has yet to be reconciled with studies of larger natural chromosomes.
In addition to our observations, others have observed that DNA catenation maintains chromosome rigidity (23) and that Topo II inhibition in vivo at metaphase causes chromosomes to visibly decondense (24). It is possible that further studies using larger model DNAs may reveal that catenation within each chromatid is more extensive than previously thought, similar to recent results regarding inter-chromatid DNA catenation (13). It also remains unclear whether DNA catenation is simply a side effect of the actions of other compacting forces such as condensin, or whether it has distinct role in compaction on its own, in addition to any role in tethering sister chromatids.
This dual role for DNA catenation would appear to be a fundamental contradiction since catenation must be removed in order to disjoin sister chromatids and complete mitosis, while simultaneously maintained to preserve tight chromatid packing and the structure of the arms. How can both of these processes be carried out? One possible answer is that condensin blocks Topo II activity within the arms, either directly or by inducing changes in the DNA (e.g. conformation) that disfavour Topo II binding or decatenation. This mechanism would be somewhat surprising since the presence of condensin does not block Topo II activity earlier during chromosome compaction. Another possibility is that Topo II is differentially regulated at the centromere and the arms, similar to how cohesin is preserved at the centromeres by the localized actions of protein phosphatase 2A (PP2A)—which, perhaps not coincidentally, associates with Topo IIα during interphase (36) and regulates the chromosomal association of condensin II (37). Such differential regulation would be consistent with our observations of fibrous centromeric DNA catenation, as we have discussed above. It remains to be determined whether the process of decatenation that takes place to disjoin the chromatids arises from a ‘wave’ of supercoiling and decatenation (12), or as a result of spindle forces (13). Our observations, however, suggest that any process that induces DNA decatenation in chromosomes at anaphase would necessarily be localized to the centromeres to ensure full disjunction of sister chromatids while maintaining intra-chromatid compaction until two separate daughter cells are formed as mitosis ends.
CONCLUSION
In this article, we have combined microfluidics with simple image analysis in order to explore chromosome structure and provide a direct observation of DNA catenation and its organization. We are able to observe key roles for DNA catenation, maintaining chromatid compaction and promoting SCC, and advance the discussion involving current models for the mechanism of sister chromatid decatenation and the regulation of Topo II. In addition, the method presented here will extend the existing toolkit for studying the molecular organization of chromosomes beyond conventional methods involving electrophoresis of plasmids, indirect cell labelling, and micropipette manipulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods and Supplementary Movies 1–3.
FUNDING
European Union Seventh Framework Programme (FP7/2007-2013), [201418] (READNA); Danish Council for Strategic Research [10-092322/DSF] (Polynano); Wellcome Trust [075491/Z/04/B, 090532/Z/09/Z]; Rhodes Trust [Rhodes Scholarship to D.L.V.B.]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jacob M. Lange and Winnie E. Svendsen at DTU for providing the metaphase chromosomes used in this study, and Timothy D. Craggs at Oxford for comments on this manuscript.
REFERENCES
- 1.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 2.Murray AW, Szostak JW. Chromosome segregation in mitosis and meiosis. Ann. Rev. Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 3.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 4.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Ann. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 5.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 6.Hardy CD, Crisona NJ, Stone MD, Cozzarelli NR. Disentangling DNA during replication: a tale of two strands. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2004;359:39–47. doi: 10.1098/rstb.2003.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 8.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnier Y, Weber C, Flammini A, Stasiak A. Local selection rules that can determine specific pathways of DNA unknotting by type II DNA topoisomerases. Nucleic Acids Res. 2007;35:5223–5231. doi: 10.1093/nar/gkm532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 11.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 12.Baxter J, Sen N, Martínez VL, De Carandini MEM, Schvartzman JB, Diffley JFX, Aragón L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–1332. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 13.Farcas A-M, Uluocak P, Helmhart W, Nasmyth K. Cohesin's concatenation of sister DNAs maintains their intertwining. Mol. Cell. 2011;44:97–107. doi: 10.1016/j.molcel.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 15.Chan K-L, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LH-C, Mayer B, Stemmann O, Nigg EA. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 2010;123:806–813. doi: 10.1242/jcs.058255. [DOI] [PubMed] [Google Scholar]
- 18.Aragón L. A double lock on sister chromatids by cohesin. Mol. Cell. 2011;44:5–6. doi: 10.1016/j.molcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Hirano T. Condensins: organizing and segregating the genome. Curr. Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 21.Pope LH, Xiong C, Marko JF. Proteolysis of mitotic chromosomes induces gradual and anisotropic decondensation correlated with a reduction of elastic modulus and structural sensitivity to rarely cutting restriction enzymes. Mol. Biol. Cell. 2006;17:104–113. doi: 10.1091/mbc.E05-04-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marko JF. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–497. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura R, Pope LH, Christensen MO, Sun M, Terekhova K, Boege F, Mielke C, Andersen AH, Marko JF. Mitotic chromosomes are constrained by topoisomerase II-sensitive DNA entanglements. J. Cell Biol. 2010;188:653–663. doi: 10.1083/jcb.200910085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young BD, Ferguson-Smith MA, Sillar R, Boyd E. High-resolution analysis of human peripheral lymphocyte chromosomes by flow cytometry. Proc. Natl Acad. Sci. USA. 1981;78:7727–7731. doi: 10.1073/pnas.78.12.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prinz C, Tegenfeldt JO, Austin RH, Cox EC, Sturm JC. Bacterial chromosome extraction and isolation. Lab Chip. 2002;2:207–212. doi: 10.1039/b208010a. [DOI] [PubMed] [Google Scholar]
- 27.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen KH, Marie R, Lange JM, Svendsen WE, Kristensen A, Mir KU. A device for extraction, manipulation and stretching of DNA from single human chromosomes. Lab Chip. 2011;11:1431–1433. doi: 10.1039/c0lc00603c. [DOI] [PubMed] [Google Scholar]
- 29.Cram LS, Bell CS, Fawcett JJ. Chromosome sorting and genomics. Methods Cell Sci. 2002;24:27–35. doi: 10.1023/a:1024108923475. [DOI] [PubMed] [Google Scholar]
- 30.Koshland D, Hartwell LH. The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science. 1987;238:1713–1716. doi: 10.1126/science.3317838. [DOI] [PubMed] [Google Scholar]
- 31.Farcas A-M, Uluocak P, Helmhart W, Nasmyth K. Cohesin's concatenation of sister DNAs maintains their intertwining. Mol. Cell. 2011;44:97–107. doi: 10.1016/j.molcel.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aragón L. A double lock on sister chromatids by cohesin. Mol. Cell. 2011;44:5–6. doi: 10.1016/j.molcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Jensen LH, Liang H, Shoemaker R, Grauslund M, Sehested M, Hasinoff BB. A three-dimensional quantitative structure-activity relationship study of the inhibition of the ATPase activity and the strand passing catalytic activity of topoisomerase IIalpha by substituted purine analogs. Mol. Pharmacol. 2006;70:1503–1513. doi: 10.1124/mol.106.026856. [DOI] [PubMed] [Google Scholar]
- 34.Cuylen S, Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Baxter J, Aragón L. A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet. 2012;28:110–117. doi: 10.1016/j.tig.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Escargueil AE, Larsen AK. Mitosis-specific MPM-2 phosphorylation of DNA topoisomerase IIalpha is regulated directly by protein phosphatase 2A. Biochem. J. 2007;403:235–242. doi: 10.1042/BJ20061460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takemoto A, Maeshima K, Ikehara T, Yamaguchi K, Murayama A, Imamura S, Imamoto N, Yokoyama S, Hirano T, Watanabe Y, et al. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat. Struct. Mol. Biol. 2009;16:1302–1308. doi: 10.1038/nsmb.1708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.