Abstract
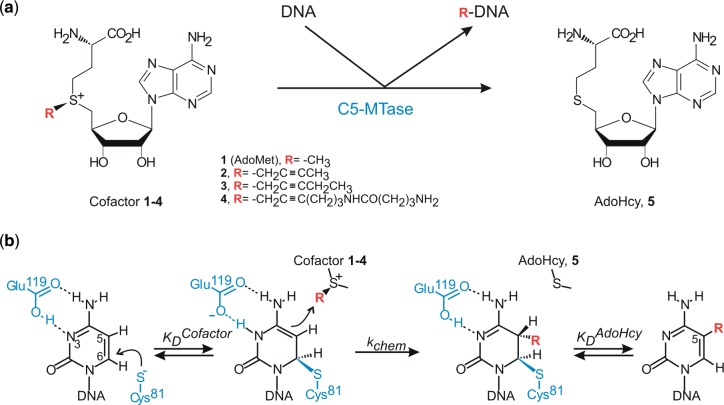
DNA methyltransferases catalyse the transfer of a methyl group from the ubiquitous cofactor S-adenosyl-L-methionine (AdoMet) onto specific target sites on DNA and play important roles in organisms from bacteria to humans. AdoMet analogs with extended propargylic side chains have been chemically produced for methyltransferase-directed transfer of activated groups (mTAG) onto DNA, although the efficiency of reactions with synthetic analogs remained low. We performed steric engineering of the cofactor pocket in a model DNA cytosine-5 methyltransferase (C5-MTase), M.HhaI, by systematic replacement of three non-essential positions, located in two conserved sequence motifs and in a variable region, with smaller residues. We found that double and triple replacements lead to a substantial improvement of the transalkylation activity, which manifests itself in a mild increase of cofactor binding affinity and a larger increase of the rate of alkyl transfer. These effects are accompanied with reduction of both the stability of the product DNA–M.HhaI–AdoHcy complex and the rate of methylation, permitting competitive mTAG labeling in the presence of AdoMet. Analogous replacements of two conserved residues in M.HpaII and M2.Eco31I also resulted in improved transalkylation activity attesting a general applicability of the homology-guided engineering to the C5-MTase family and expanding the repertoire of sequence-specific tools for covalent in vitro and ex vivo labeling of DNA.
INTRODUCTION
The most abundant covalent modification of DNA is cytosine methylation which serves to expand the information content of the genome in organisms from bacteria to humans. In higher vertebrates, it is part of an intricate epigenetic regulatory network (1,2). DNA methylation patterns are brought about by DNA methyltransferases (MTases)—a class of enzymes that transfer a methyl group from the ubiquitous cofactor S-adenosyl-L-methionine (AdoMet) onto predefined target sites on DNA (Figure 1). Three classes of DNA MTases are known which modify the exocyclic amino group of adenine, the exocyclic amino group of cytosine or the carbon at the 5 position of cytosine (C5-MTases), respectively. DNA C5-MTases are found both in prokaryotes and eukaryotes and share a common mechanism of catalysis which involves flipping of the target cytosine from the DNA helix to the active site followed by its covalent activation (3). The enzymatic catalysis involves transient formation of a Michael adduct between the catalytic cysteine and C6 of the cytosine, permitting a direct SN2 transfer of the methyl group onto C5. These enzymes also share a set of conserved sequence motifs (I–X) and a large intervening variable region, which fold to make the catalytic and target recognition domains, respectively (4). The bacterial HhaI C5-MTase (M.HhaI), which recognizes the sequence GCGC and methylates the inner cytosine (underlined), serves as a structural and mechanistic paradigm for this class of enzymes (5).
Figure 1.
Alkylation reactions catalysed by the AdoMet-dependent methyltransferases. (a) General reaction scheme for enzymatic transfer of methyl or extended groups from natural cofactor AdoMet (1) or its analogs (2–4) onto DNA. (b) Catalytic mechanism of C5-alkylation of cytosine catalysed by DNA cytosine-C5 methyltransferases. Residue numbers correspond to those of M.HhaI.
Most, if not all, bacterial and archaeal DNA MTases exhibit clearly defined sequence and base specificity. To expand the practical utility of this highly specific enzymatic reaction, two major classes of synthetic AdoMet analogs have been developed (6). One such design is based on exploiting highly reactive aziridine (7) or N-mustard (8) chemistries, which lead to coupling of a whole cofactor molecule to the target DNA (named Sequence-specific Methyltransferase-Induced Labeling, SMILing). Subsequently, cofactors with activated sulfonium-bound side chains have been produced, which permit targeted transfer of these linear side chains alone (named methyltransferase-directed Transfer of Activated Groups, mTAG) (9,10). Both chemistries have been used to demonstrate the potential utility of this approach for a number of DNA manipulations such as nanoparticle design (11), in-cell DNA tracking (12) or optical genome mapping (13,14). In the mTAG series, cofactors with extended propargylic side chains were chemically synthesized (Figure 1) and examined in enzymatic reactions with all three types of DNA methyltransferases (9,10). However, the efficiency of transalkylations observed with the wild-type C5-MTases proved insufficiently high for routine applications.
In this work, we performed steric engineering of the cofactor pocket in the HhaI DNA cytosine-5 methyltransferase by systematic replacement of three non-essential positions with shorter residues in conserved sequence motifs IV and X and in the variable region. We found that double and triple replacements confer substantial improvements of the transalkylation activity and a reduction of the methyltransferase activity in M.HhaI and, to a smaller degree, in related M2.Eco31I and M.HpaII C5-MTases recognizing the CCGG and GGTCTC target sites, respectively. Binding and kinetic studies of M.HhaI mutants showed that these replacements substantially enhance the rate of alkyl transfer and reduce the enzyme affinity toward the natural cofactor AdoMet and its product AdoHcy. Importantly, the engineered MTases can efficiently utilize such synthetic analogs in the presence of AdoMet, and the modified DNA is not degraded by methylation-dependent restriction systems in bacteria. These findings open new ways for targeted covalent deposition of reporter groups onto DNA for a variety of ex vivo and in vivo applications.
MATERIALS AND METHODS
Recombinant DNA WT and mutant MTases M.HhaI were produced as previously described (15). Recombinant DNA MTases M2.Eco31I and M.HpaII were expressed as HisTaged proteins and purified according manufacturer recommendations (Supplementary Data). HPLC-purified oligodeoxyribonucleotides were obtained from MWG (Germany). McrBC endonuclease was purchased from New England Biolabs.
Synthesis of AdoMet analogs
Chemical synthesis and purification of AdoMet analogs were performed as previously described by direct chemoselective S-alkylation of S-adenosyl-L-homocysteine (9). Cofactor 2 was obtained as a diasteromeric mixture of R,S- and S,S-isomers starting from butyn-2-ol-1 (16). Cofactor 3 was synthesized starting from pentyn-2-ol-1 and purified to an over 85% chiral purity of the enzymatically active S,S-isomer (17). For analog 4, the alkylation was performed with N-BOC-protected 6-(4-aminobutanamido)hex-2-yn-1-yl 4-nitrobenzenesulfonate, which was obtained in three steps from 5-chloropentyne-1 following previously described procedures (10), and the S,S-diastereomer was isolated by reversed-phase HPLC.
DNA protection assays
DNA protection assays were performed essentially as previously described (9). Briefly, 2-fold serial dilutions of DNA MTases were incubated with phage lambda (M.HhaI and M.HpaII, 0.6–1.2 µg per reaction) or pUC19 (M2.Eco31I, 1.0 µg) DNA in a corresponding reaction buffer containing 300 µM cofactors at 37°C for 1–4 h. MTase reaction buffers were as follows: M.HhaI: 50 mM Tris–HCl, 15 mM NaCl, 0.5 mM EDTA, 2 mM 2-mercaptoethanol, 0.2 mg/ml BSA, pH 7.4; M2.Eco31I: 20 mM MOPS, 20 mM Tris–HCl; 20 mM CAPS, 15 mM NaCl, 0.5 mM EDTA, 0.2 mM 2-mercaptoethanol, 0.2 mg/ml BSA, pH 8.0; M.HpaII: 20 mM MES, 20 mM HEPES, 30 mM NaCl, 0.5 mM EDTA, 0.2 mM 2-mercaptoethanol, 0.2 mg/ml BSA, pH 7.0. After incubation the reactions were stopped by heating to 80°C for 10 min and the modified DNA was challenged with 10–12 u of corresponding restriction endonuclease for 1 h at 37°C. M.HhaI modification was assessed with R.Hin6I, M2.Eco31I—with R.Eco31I and R.PvuII and M.HpaII—with R.HpaII. Samples were analysed by agarose gel (1–1.5%) electrophoresis. Full protection of substrate DNA from endonuclease cleavage in reaction in which the MTase is present in an N-fold dilution relative to its target sites indicates that the enzyme carried out at least N turnovers; the turnover rate was calculated by dividing the number of turnovers by reaction time t (kobs ≥ N/t)
Single-turnover kinetics
Reactions were performed using 596 bp DNA fragment with a single M.HhaI target site as a substrate. The reaction components in syringes A (200–1000 nM M.HhaI plus 100 or 200 nM DNA) and B (50 µM cofactor) were mixed rapidly in M.HhaI reaction buffer at 37°C, and after a specified period the reaction was quenched with 6 M guanidinium chloride in a Rapid-Quench-Flow instrument RQF-3 (KinTek). Modified DNA was precipitated with propanol-2, dissolved in 30 µl of water and treated with 10 units of R.Hin6I. DNA fragments were analysed by 1.5% agarose gel electrophoresis and visualized by scanning with 473 nm laser after staining with ethidium bromide (EtBr). Bands intensity was quantified using Multi gauge v3.0 software. Normalized progress curves were fitted to single exponential equations using GraFit 5 (18).
Analysis of cofactor binding
0.2 µM M.HhaI was preincubated with 0.3 µM duplex oligonucleotide I (Supplementary Table S1) and then titrated by incremental addition of AdoHcy, AdoMet or its analogs in M.HhaI reaction buffer with no BSA. Tryptophan fluorescence quenching upon cofactor binding (19) was measured at 25°C on PerkinElmer LS50B spectrofluorimeter at an excitation wavelength of 290 nm (slit width 5 nm) and emission wavelength of 350 nm (slit width 7.5 nm). Titration data (Supplementary Figure S3) were analysed with the equilibrium solver routine of GraFit 5 (18).
Chromatographic analysis of DNA composition
Analyses were performed essentially as described previously (9). M.HhaI (12.5 μM), oligonucleotide duplex I (Supplementary Table S1) and AdoMet or its analogs (300 μM) in M.HhaI reaction buffer were incubated at 37°C for 4 h. Samples were then digested with Nuclease P1 (1.5 u) and calf intestine alkaline phosphatase (30 u) at 42°C for 4 h. Digested DNA samples were loaded onto a reversed-phase HPLC column (Discovery HS C18, 3 μm, 75 × 2.1 mm, Supelco) and eluted for 3 min with 20 mM ammonium formate (pH 3.5), then by a linear gradient of methanol to 20% in 15 min, followed by a linear gradient from 20% to 80% methanol in 2 min and 80% methanol for 5 min at a flow of 0.3 ml/min and at 30°C. Post-column equal co-flow of 96% methanol, 4% formic acid and 1 mM sodium formate was used for the MS detection of modified nucleosides and its derivatives in the 50–500 m/z range.
Transformation of Escherichia coli with alkylated DNA
2–3 µg of pUC19 plasmid DNA (reaction volume 20–30 µl, 1 µM HhaI sites) was modified with 300 µM AdoMet, cofactors 2 or 4 in the presence of 0.5 µM M.HhaI (variant Q82A/Y254S/N304A). Modified DNA was extracted twice with an equal volume of Roti®-Phenol/C/I mix, 3 times with equal volume of chloroform, precipitated with propanol-2 and dissolved to a final concentration of 0.4 µg/µl in water.
Aliquots (0.8 µg) of modified pUC19 were digested with R.Hin6I or McrBC according to manufacturer recommendations. Samples were analysed by 1.5% agarose gel electrophoresis and EtBr staining. For the transformation experiments, equal aliquots of modified pUC19 DNA (1 ng/µl) were used to transform competent E. coli JM109 mcrB+ and mcrB− cells prepared according to methodology described in (20). The Mcr restriction rate in vivo is determined as the ratio of ampicillin resistant colonies obtained after growing these strains for 16 h at 37°C on the LB agar medium supplemented with ampicillin (0.1 mg/ml).
M2.Eco31I-directed labeling of lambda DNA
Reaction solution containing 72 nM M2.Eco31I (variant N127A/Q233A) and 1.8 nM phage lambda DNA in reaction buffer (50 mM Tris–HCl pH 7.4, 2.5 mM EDTA, 0.2 mg/ml BSA, 20 mM NaCl, 0.4 mM 2-mercaptoethanol) with 40 μM cofactor 4 was incubated at 37°C for 1 h. Afterwards samples were mixed with an equal amount of water and extracted with Roti®-Phenol (pH 8.0), twice with Roti®-Phenol/C/I and three times with chloroform and DNA was precipitated with propanol-2. Pellets were washed with ice cold 75% ethanol and dried. Amino-modified DNA was dissolved in water to a final concentration of 0.2 μg/μl and treated with fluorescein-NHS (125 μM) in 0.15 M sodium bicarbonate (pH 8.3) for 1 h at room temperature. Approximately 1.5 μg of labeled DNA was fragmented with R.Eco91I and analysed by 0.8% agarose gel electrophoresis. Gels were scanned with a Fuji FLA-5100 imaging system using a 473 nm laser and an LPB filter set for fluorescein emission.
RESULTS
Steric engineering of M.HhaI for improved transalkylations
Previously we found that the reactivity of M.HhaI with extended AdoMet analogs was increased by replacing Gln82 to alanine. The observed rates were still lower as compared to those achieved with the natural cofactor (9), although in theory the SN2 alkylation rates with propargyl and allyl compounds should be similar or higher than the methylation rates (21). Directed evolution of M.HhaI for altered sequence specificity produced variants in which Tyr254 is replaced with serine, and which also showed an enhanced transalkylation activity with certain extended cofactor analogs (17). To guide our engineering effort, we built a structure-based model of M.HhaI–DNA–(cofactor 2) complex (Figure 2A and B). The model structure showed that Gln82, Tyr254 and Asn304 residues are in close proximity and thus might sterically interfere with the extended transferable side chains. We therefore selected these positions for steric engineering of the cofactor pocket. Gln82 and Asn304 were replaced with an Ala; due to extensive solvent exposure and close interactions with bound DNA Tyr254 was replaced with a small polar residue (Ser).
Figure 2.
Engineering the transalkylation reactions in the HhaI methyltransferase. Models of cofactor 2 bound in the active site of the WT (a) and an engineered variant (b) of M.HhaI (Q82A/Y254S/N304A) with DNA. Modeling was based on M.HhaI-DNA-AdoMet X-ray structure (PDB code 6mht). Red arrows point along the trajectory from attacking the C5 atom of the target cytosine residue to the transferable carbon in the cofactor. Pictures are made with Swiss-PdbViewer program (3.7 sp5 version) (22). (c) AdoMet and its analogs activity with WT M.HhaI and its variants. Turnover rates were estimated using a DNA protection assay.
Initially, the three single mutants were analysed by comparing their activity with AdoMet and three of its extended analogs using a DNA protection assay (Figure 2C). This semi-quantitative end point assay (Supplementary Figure S1) estimates the number of enzymatic turnovers based on the number of modified target sites in substrate DNA (9). The estimated turnover rate for the WT enzyme with AdoMet (2.1 min−1) was in good agreement with published kcat = 1–2 min−1 observed on a variety of DNA substrates (19), but no activity was observed with the extended cofactor analogs. All single-mutant enzymes demonstrated an increased activity toward the AdoMet analog 2. The Gln82 substitution resulted in a 4-fold increased enzymatic activity with cofactor 2 and an ∼16-fold decreased activity with AdoMet. A barely detectable activity was observed with cofactors 3 and 4 respectively. In contrast, the Y254S mutation caused little effect on the methylation activity, but significantly increased catalysis of the chain transfer from cofactors 2 and 3. The most dramatic activity enhancement came from the N304A mutation which showed an 8-fold and 4-fold faster transfer of the four-carbon and five-carbon chains from analog 2 and 3 as compared to the methyl group. However, none of the single-mutant variants showed detectable reactivity with the most extended cofactor 4. This observation suggests that although the shortening of the 304 residue creates adequate space in the active site for the transfer of moderately extended groups, it fails to accommodate the transfer of the larger functionalized group.
To further optimize M.HhaI for the transalkylation reactions, all combinations of double and triple mutants involving the above three residues were constructed and examined. Surprisingly, none of the double (Q82A/N304A or Y254S/N304A) or triple (Q82A/Y254S/N304A) mutant enzymes gave a significant enhancement in transferring the alkyne groups from cofactor 2 or 3 compared to the N304A variant (Figure 2C). However, the transfer of the largest group is accelerated in the Y254S/N304A and Q82A/Y254S/N304A variants, which both show a similarly high activity with the cofactor analog 4. Moreover, the Q82A mutants displayed a small, if any, enhancement of the transalkylation rate, but led to consistent reduction of the methylation rate (Figure 2C). Altogether, both double mutants involving N304A and the triple mutant accepts the largest cofactor 4 in par with AdoMet; the achieved turnover rates of 0.13–0.25 min−1 (Supplementary Table S2) permit complete derivatization of DNA in <15 min, which makes the reaction suitable for routine laboratory applications.
The sequence specificity of the engineered M.HhaI variants in the presence of cofactor analogs is indirectly attested by the DNA protection experiments, in which complete protection of HhaI sites in bacteriophage DNA was observed. To verify that the protein engineering does not alter the specificity of base modification, an oligonucleotide duplex was modified by the triple mutant Q82A/Y254S/N304A and the model cofactor analogs and its nucleoside composition was determined using reversed-phase HPLC. In addition to the four major nucleosides, novel compounds with longer retention times were observed and identified as corresponding alkylated 2′-deoxycytidines using in-line ESI-MS analysis (Supplementary Figure S2).
Mechanistic effects of the Y254S/N304A mutations
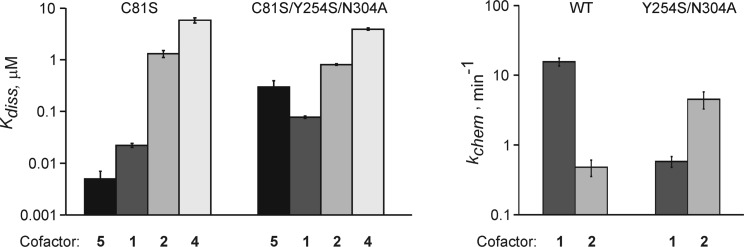
To assess whether the cofactor binding or the catalytic power is a major factor for the enhanced alkylation activity, the best performing and least altered variant (Y254S/N304A) was subjected to further biochemical studies. First, we determined the binding affinity of the cofactors (KDcofactor) in the ternary pre-catalytic complex M.HhaI–DNA–cofactor. Catalytic turnover of the ternary complex during the cofactor binding measurements was abolished by replacing the catalytic Cys81 residue with a serine (23) in both the wild-type enzyme and in the engineered variant. The cofactor binding experiments (Supplementary Figure S3) exploited fluorescence changes of a unique tryptophan (Trp41) (19). Surprisingly, we found that binding of the analogs 2 and 4 to the engineered variant of M.HhaI (C81S/Y254S/N304A) was barely improved (<2-fold) as compared to the catalytic C81S mutant (Figure 3, left). On the other hand, we found a strong negative effect on AdoMet (4-fold) and AdoHcy (60-fold) binding.
Figure 3.
Effects of steric engineering on cofactor binding and catalysis by M.HhaI. Left, binding affinity (KDcofactor) of cofactors 1–5 in the ternary pre-catalytic complex M.HhaI-DNA-cofactor was determined for a catalytic (C81S) mutant and a corresponding engineered variant (C81S/Y254S/N304A); right, single-turnover reaction rates (kchem) of the WT and engineered (Y254S/N304A) M.HhaI with cofactors 1 and 2 as indicated.
Furthermore, we determined the single-turnover reaction rates (kchem) of the WT and Y254S/N304A using rapid-quench flow technique (Supplementary Figure S4). Remarkably, under these saturating conditions ([E]>[DNA], [cofactor]>>KDcofactor), the mutant enzyme displayed a 24-fold enhanced transfer of the butynyl groups from cofactor 2 as compared to the WT enzyme, whereas the rate of methyltransfer was decreased by a similar margin (27-fold) (Figure 3, right). Altogether, our analyses indicate that the described mutations lead to changes in the catalytic transfer rather than cofactor binding. In addition, a weaker retention of the reaction product AdoHcy in the engineered enzyme also contributes to the overall catalytic efficiency in vitro since it leads to a faster dissociation of product complex MTase-modified DNA-AdoHcy (19) and reduced non-productive binding of AdoHcy to the binary M.HhaI–DNA complex (Figure 1B).
Enzymatic transalkylation of DNA in the presence of AdoMet
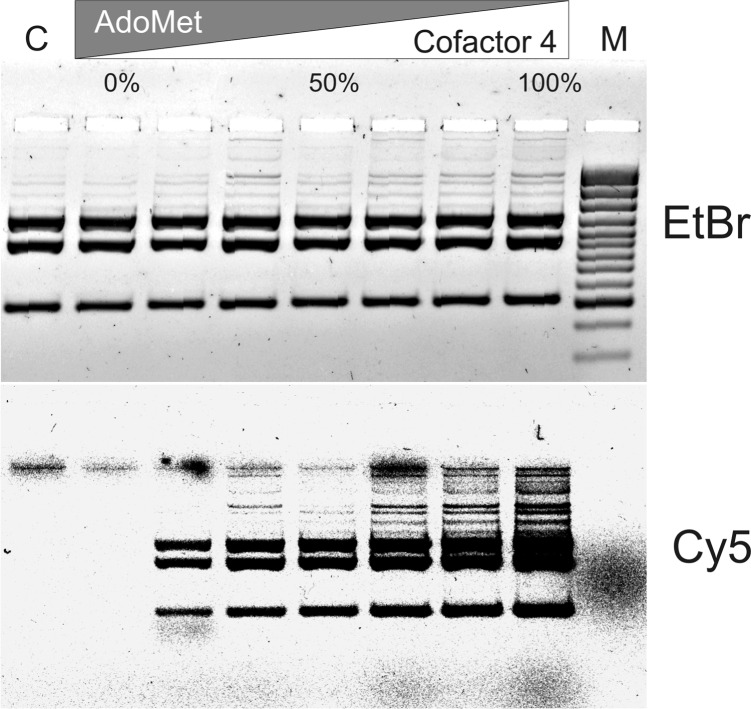
Taking together the enhanced catalytic power of the engineered M.HhaI variants in alkyltransfer reactions (especially with cofactors 2 and 3) and their reduced affinity toward the natural cofactor one might expect a shifted or reversed cofactor preference during reaction. Efficient aminoalkylation reactions in the presence of endogenous AdoMet would potentially permit in situ or even in vivo derivatization and subsequent labeling of DNA. Our DNA protection experiments indicated that the turnover rates of the triple (Q82A/Y254S/N304A) mutant were similar with AdoMet or the Ado-11-amine cofactor 4 (Figure 2C), however the cofactor preference of the enzyme in direct competition between the two cofactors remained unclear. As a proof of principle, enzymatic modification of pUC19 plasmid DNA in the presence of varied molar ratios of AdoMet to its analog was performed. Methylation and alkylation of DNA was probed using combination of the R.Hin6I and R.BspLI endonucleases (Supplementary Figure S5). Moreover, incorporation of the extended chain with a reactive amino group was assessed by labeling the modified DNA with Cy5-NHS (Figure 4). Visualization of Cy5-labeled DNA showed that Q82A/Y254S/N304A variant is capable of efficiently alkylating DNA in presence of physiologically relevant concentrations of AdoMet (8.5–300 µM) (24). The label is incorporated even in presence of 5-fold excess of AdoMet (Figure 4). This result opens the possibility for sequence-specific labeling of DNA in vivo or in the cell in presence of endogenous AdoMet.
Figure 4.
mTAG aminoalkylation and labeling of DNA in the presence of AdoMet. pUC19 plasmid was modified with M.HhaI (Q82A/Y254S/N304A mutant) and a mixture of AdoMet and cofactor 4 supplied in different molar fractions as indicated (total cofactor concentration was 50 µM). Modified DNA was labeled with Cy5-NHS, fragmented with R.BseSI and separated by agarose gel electrophoresis. Bulk DNA was visualized after staining with EtBr. Visualization was performed using 473 nm (EtBr) and 635 nm (Cy5) lasers. C is control sample incubated without cofactor; M, DNA size marker GeneRuler™ 100 bp Plus DNA Ladder.
Restriction of C5-alkylated DNA in bacterial cells
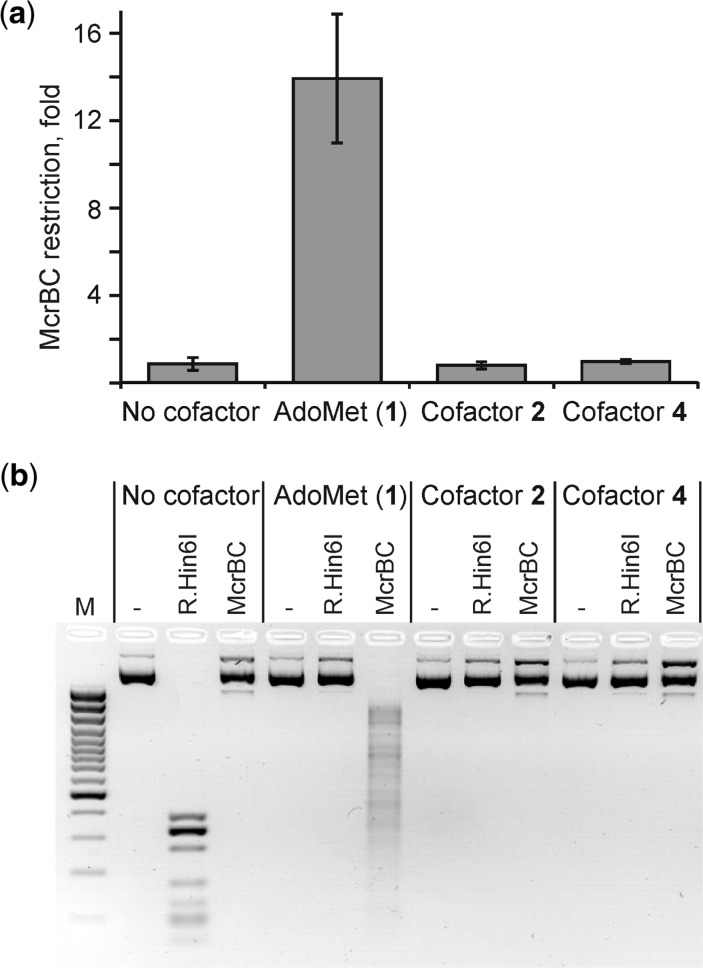
The presence of natural cytosine modifications in DNA such as 5mC, 4mC or 5hmC is thoroughly surveyed by dedicated DNA restriction systems in bacteria. For example, the McrBC system recognizes the above modified cytosine residues in certain sequence contexts and destroys modified DNA by endonucleolytic cleavage, whereby restricting entry of many types of methylated and hydroxymethylated plasmids into E. coli cells (25). For this reason, plasmid DNA premethylated by M.HhaI (Figure 5) and many other DNA MTases is strongly restricted by the McrBC system both in vitro and in vivo. However, the sensitivity of the system to DNA artificially modified with larger groups has not been studied before. Experiments presented in Figure 5, bottom panel, show that the McrBC endonuclease fails to attack the DNA modified using AdoMet analogs 2 or 4.The McrBC system renders a 14-fold restriction of the M.HhaI-methylated plasmid, however equal transformation efficiency was observed in experiments involving alkylated plasmid DNAs in either mcrB− or mcrB+ E. coli strain (Figure 5, top panel). These observations clearly indicate that the McrBC restriction system is inert to mTAG-alkylated DNA in vitro and in vivo. This observation is consistent with recent McrBC-DNA co-crystal structures, which show tight binding of the flipped out 5-hydroxymethylcytosine (26), however residues carrying larger groups seem unlikely to be accommodated in the binding pocket.
Figure 5.
Sensitivity of mTAG-alkylated DNA to the McrBC endonuclease in vitro and in vivo. (a) The McrBC restriction rate in vivo is determined as the ratio of ampicillin resistant colonies obtained in mcrB- versus mcrB+ strains after transformation with pUC19 plasmid DNA modified in the presence of cofactor AdoMet or its analogs 2 and 4. (b) mTAG-modified pUC19 DNA as above was treated with the McrBC endonuclease or R.Hin6I (control for complete modification) in vitro and analysed by agarose gel electrophoresis. ‘-’, no endonuclease added; M, DNA size marker GeneRuler™ 100 bp Plus DNA Ladder.
Homology-based engineering of M.HpaII and M2.Eco31I
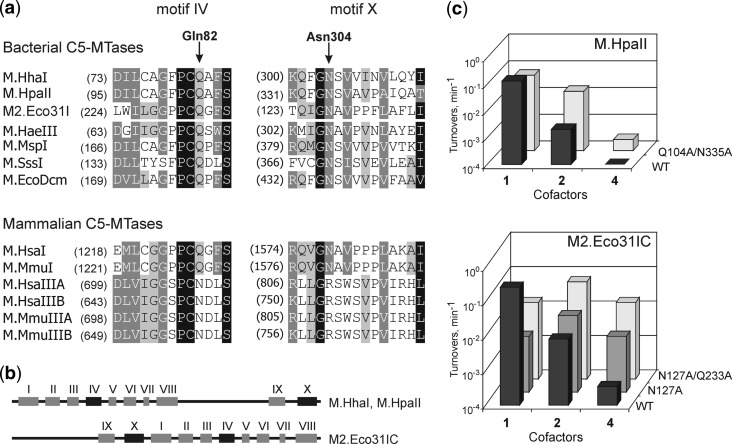
The structural conservation of C5-MTases and successful engineering of M.HhaI suggested that other orthologs can be similarly engineered based on sequence alignment even in the absence of crystal structures (Figure 6). We first turned to the HpaII C5-MTase (M.HpaII), which recognizes CCGG targets in DNA (27) and which thus can potentially be useful to study mammalian DNA modification at CpG sites. As expected, the WT enzyme showed low or no activity with AdoMet analogs 2 and 4, respectively. Replacement of the Q104 and N335 residues (correspond to Q82 and N304 in M.HhaI) to alanines resulted in an enzyme with a significantly enhanced activity toward these analogs (Figure 6, Supplementary Figure S1B and Supplementary Table S3).
Figure 6.
Homology-based engineering of C5-MTases in conserved sequence motifs IV and X. (a) Amino acid sequence alignment of regions corresponding to IV and X conserved motifs of prokaryotic and eukaryotic cytosine-C5 MTases. Arrows indicate Gln82 and Asn304 positions of M.HhaI in IV and X conserved motifs. Four degrees of conservation in descending order: black background with white text, dark gray background with white text, gray background with black text and white background with black text. Amino acid conservation of the Gln82 position (HhaI) in DNA C5-MTases is depicted, based on an alignment of ∼530 DNA C5-MTase sequences from the Pfam database (http:www.sanger.ac.uk/Software/Pfam/) (b) Permutation of conserved motifs in the M.HhaI, M.HpaII and M2.Eco31I DNA methyltransferases. (c) Enzymatic transalkylation of DNA by engineered variants of M.HpaII and M2.Eco31I. Turnover rates (shown in logarithmic scale) were estimated using the DNA protection assay.
Another studied example was the bacterial C5-MTase Eco31IC (M2.Eco31I) which recognizes an asymmetric hexanucleotide sequence GGTCTC in DNA and attaches methyl group to the inner cytosine residue on one strand of the target site (28). Therefore this MTase could be used for strand- and sequence-specific labeling of larger DNA molecules. Initial testing of the wild-type enzyme showed that it possesses low activity toward cofactor 2 and negligible activity with cofactor 4. The single mutation N127A (corresponds to N304A in M.HhaI) resulted in a significant increase of the alkyltransferase activity with cofactors 2 and 4. However the N127A/Q233A variant was capable of catalysing extended group transfer from AdoMet analog 4 with a 50-fold higher efficiency as compared to the WT MTase (Figure 6, Supplementary Figure S1C and Supplementary Table S3). Bacteriophage lambda contains only two Eco31I target sites, and is thus well suited to demonstrate a highly specific fluorescent labeling of a large natural DNA molecule (Supplementary Figure S6).
These examples clearly show that a high degree of structural conservation of the catalytic domain of C5-MTases permits successful engineering of the methyltransferase reaction for sequence-specific labeling of DNA.
DISCUSSION
Structure-guided engineering of the cofactor pocket in M.HhaI produced four variants that show faster transfer of extended linear groups containing four or five carbon units (cofactor analogs 2 and 3) as compared to the natural AdoMet cofactor (Figure 2C). The transfer of the butyne group from cofactor 2 can occur several-fold faster (10 min−1) than the catalytic rate of WT M.HhaI with AdoMet (1.3 min−1) (19) and approaches the rate of methyltransfer observed with WT M.HhaI under single-turnover conditions (15–30 min−1) (Figure 3, and (19)). This indicates that the HhaI methyltransferase has been actually converted into a sequence-specific DNA alkyltransferase and that a full catalytic power has been attained in the engineered C5-transferase reaction. Further extension of the transferable side chain to 12 linear units (cofactor 4) leads to somewhat lower but still useful turnover rates of 0.3 min−1.
In contrast to our expectations, a major contributor to the efficiency of the transalkylation reactions in the engineered MTase turned out to be enhanced catalytic transfer rather than improved cofactor binding. In other words, the extended cofactors can bind to the WT and to the mutant variants of M.HhaI with similar affinity, however, the structure of the ternary complex in the engineered proteins is more favorable for catalysis. Inspection of the available crystal structures indicates that a wider solvent channel in the cofactor pocket permits a higher conformational flexibility of the extended cofactor, and rotation of the cofactor side chain into the newly created space may position the transferable carbon atom into a more favorable pre-catalytic conformation in which one of the two hydrogen atoms assumes a less obstructive position for an in-line attack of the C5 nucleophile (Supplementary Figure S7).
The improvement of the catalytic rate with the extended cofactors was accompanied with a substantial reduction of AdoHcy (60-fold) and AdoMet (4-fold) binding and a strong decrease of the rate of methylation (30-fold). Among the three mutated residues, Gln82 proved to be the most important for the methyl group transfer since all of the M.HhaI variants containing the Q82A mutation displayed a significant reduction of the methylation rate but no change of the transalkylation rates (Figure 2C). This residue is located in the mobile catalytic loop and thus the Q82A mutation reduces the number of intramolecular contacts to the core of the protein (N304A and Y254S would be expected to have similar but smaller effects), which in turn may destabilize the closed conformation of the mobile catalytic loop in the enzyme leading to a reduction of the apparent methylation rate. The reason for the strongly impaired binding of AdoHcy as compared to AdoMet and the other extended cofactors is not fully clear. One might expect that, in general, cofactors with smaller surface areas would be less efficient in bridging the expanded space between the catalytic loop and the rest of the protein, which is critical for to maintaining the closed catalytic conformer. On the other hand, a higher conformational flexibility of the thioether moiety in AdoHcy (as compared to tri-substituted sulfonium centers) may be another destabilizing factor in the context of fewer restraining contacts from adjacent protein residues. As discussed above, the weaker retention of AdoHcy contributes to the overall catalytic efficiency of the engineered enzyme due a faster dissociation of product complex AdoHcy-MTase-modified DNA and reduced inhibition potency of AdoHcy (Figure 1B).
Since two of the three mutations lie within highly conserved sequence motifs (IV and X), corresponding mutations in other DNA C5-MTases were expected to improve the acceptance of the extended AdoMet analogs. Although the magnitudes of the observed effects estimated in DNA protection experiments appeared quite distinct, both M.HpaII and M2.Eco31I showed improved reactivity toward AdoMet analogs attesting a general utility of homology-based engineering of the cofactor pocket in C5-MTases. For M2.Eco31I, which is an example of sequence permutation in C5-MTases (Figure 6B), the cofactor preference of the double mutant was very similar to that of M.HhaI (cofactor 2 > cofactors 1, 4), whereas the engineered M.HpaII still showed a profile characteristic of a WT MTase (cofactor 1 > cofactor 2 > cofactor 4). Nonetheless, taking into account a common structural organization of the AdoMet binding pocket (3,6,29) and recent examples from protein MTases (30,31), successful engineering of other classes of DNA and RNA MTases can be well anticipated.
The MTase-directed derivatizations of DNA occur in a highly specific manner, with full retention of sequence-, base- and atomic specificity whereby a linear moiety is attached on a DNA molecule. Such labeling offers an important advantage over endonuclease-polymerase-based techniques (32) since it does not rely on replacing natural nucleotides with modified ones in adjacent regions and thus preserves all pre-existing modifications in the DNA, which could be further subjected to investigation. Examples of practical application of sequence-specific mTAG labeling for single-molecule genotyping (13) and epigenome profiling (Kriukiene et al., submitted elsewhere) have recently been provided. Moreover, we demonstrate that (i) mTAG labeling can be performed in the presence of competing AdoMet (Figure 4) and that (ii) mTAG-modified DNA is immune to modification-dependent restriction systems such as McrBC of E. coli (Figure 5), which open ways to studying artificially modified DNA in bacterial cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–7, Supplementary Methods and Supplementary Reference [33].
FUNDING
Lithuanian State Science and Studies Foundation [P-03/2007]; National Institutes of Health [HG004535, HG005758]. Funding for open access charge: FP7-REGPOT-2009-1 program [245721, MoBiLi].
Conflict of interest statement. G.L. and S.K. are inventors on related patents.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr E. Merkienė, Z. Staševskij and Dr G. Vilkaitis for pHH5.3, pETHH2111 and pACAR1 plasmids, respectively, Dr A. Lubys for a kind gift of pUC19Eco31IRM plasmid and E. coli JM109 strains, and Prof. E. Weinhold for insightful discussions.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Doerfler W. The almost-forgotten fifth nucleotide in DNA: an introduction. Curr. Top. Microbiol. Immunol. 2006;301:3–18. doi: 10.1007/3-540-31390-7_1. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin: Landes Bioscience; 2009. [Google Scholar]
- 6.Klimašauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Pljevaljcic G, Schmidt F, Weinhold E. Sequence-specific methyltransferase-induced labeling of DNA (SMILing DNA) Chembiochem. 2004;5:265–269. doi: 10.1002/cbic.200300739. [DOI] [PubMed] [Google Scholar]
- 8.Comstock LR, Rajski SR. Conversion of DNA methyltransferases into azidonucleosidyl transferases via synthetic cofactors. Nucleic Acids Res. 2005;33:1644–1652. doi: 10.1093/nar/gki306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat. Chem. Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 10.Lukinavičius G, Lapienė V, Staševskij Z, Dalhoff C, Weinhold E, Klimašauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- 11.Braun G, Diechtierow M, Wilkinson S, Schmidt F, Huben M, Weinhold E, Reich NO. Enzyme-directed positioning of nanoparticles on large DNA templates. Bioconjug. Chem. 2008;19:476–479. doi: 10.1021/bc700275h. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt FH, Huben M, Gider B, Renault F, Teulade-Fichou MP, Weinhold E. Sequence-specific Methyltransferase-Induced Labelling (SMILing) of plasmid DNA for studying cell transfection. Bioorg. Med. Chem. 2008;16:40–48. doi: 10.1016/j.bmc.2007.04.054. [DOI] [PubMed] [Google Scholar]
- 13.Neely RK, Dedecker P, Hotta J-I, Urbanavičiūtė G, Klimašauskas S, Hofkens J. DNA fluorocode: a single molecule, optical map of DNA with nanometre resolution. Chem. Sci. 2010;1:453–460. [Google Scholar]
- 14.Kim S, Gottfried A, Lin RR, Dertinger T, Kim AS, Chung S, Colyer RA, Weinhold E, Weiss S, Ebenstein Y. Enzymatically incorporated genomic tags for optical mapping of DNA-binding proteins. Angew. Chem. Int. Ed. Engl. 2012;51:3578–3581. doi: 10.1002/anie.201107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daujotytė D, Vilkaitis G, Manelytė L, Skalicky J, Szyperski T, Klimašauskas S. Solubility engineering of the HhaI methyltransferase. Protein Eng. 2003;16:295–301. doi: 10.1093/proeng/gzg034. [DOI] [PubMed] [Google Scholar]
- 16.Dalhoff C, Lukinavičius G, Klimašauskas S, Weinhold E. Synthesis of S-adenosyl-L-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat. Protoc. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- 17.Gerasimaitė R, Vilkaitis G, Klimašauskas S. A directed evolution design of a GCG-specific DNA hemimethylase. Nucleic Acids Res. 2009;37:7332–7341. doi: 10.1093/nar/gkp772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leatherbarrow RJ. 3.0 edn. Staines, UK: Erithacus Sortware Ltd; 1992. [Google Scholar]
- 19.Merkienė E, Klimašauskas S. Probing a rate-limiting step by mutational perturbation of AdoMet binding in the HhaI methyltransferase. Nucleic Acids Res. 2005;33:307–315. doi: 10.1093/nar/gki175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. 3rd edn. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Dorwald FZ. Side Reactions in Organic Synthesis. Weinheim: WILEY-VCH; 2005. pp. 59–142. [Google Scholar]
- 22.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 23.Mi S, Roberts RJ. The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res. 1993;21:2459–2464. doi: 10.1093/nar/21.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clin. Chem. 2000;46:265–272. [PubMed] [Google Scholar]
- 25.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 26.Sukackaite R, Grazulis S, Tamulaitis G, Siksnys V. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 2012;40:7552–7562. doi: 10.1093/nar/gks332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Card CO, Wilson GG, Weule K, Hasapes J, Kiss A, Roberts RJ. Cloning and characterization of the HpaII methylase gene. Nucleic Acids Res. 1990;18:1377–1383. doi: 10.1093/nar/18.6.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitinaitė J, Manelienė Z, Menkevičius S, Klimašauskas S, Butkus V, Janulaitis AA. Alw26I, Eco31I and Esp3I – type IIs methyltransferases modifying cytosine and adenine in complementary strands of the target DNA. Nucleic Acids Res. 1992;20:4981–4985. doi: 10.1093/nar/20.19.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R, Zheng W, Yu H, Deng H, Luo M. Labeling substrates of protein arginine methyltransferase with engineered enzymes and matched S-adenosyl-L-methionine analogues. J. Am. Chem. Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling. ACS Chem. Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao M, Phong A, Ha C, Chan T, Cai D, Leung L, Wan E, Kistler A, DeRisi J, Selvin P, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2006;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holz B, Klimašauskas S, Serva S, Weinhold E. 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res. 1998;26:1076–1083. doi: 10.1093/nar/26.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.