Abstract
A variety of methods exist for inducible control of DNA transcription in yeast. These include the use of native yeast promoters or regulatory elements that are responsive to small molecules such as galactose, methionine, and copper, or engineered systems that allow regulation by orthogonal small molecules such as estrogen. While chemically regulated systems are easy to use and can yield high levels of protein expression, they often provide imprecise control over protein levels. Moreover, chemically regulated systems can affect many other proteins and pathways in yeast, activating signaling pathways or physiological responses. Here, we describe several methods for light mediated control of DNA transcription in vivo in yeast. We describe methodology for using a red light and phytochrome dependent system to induce transcription of genes under GAL1 promoter control, as well as blue light / cryptochrome dependent systems to control transcription of genes under GAL1 promoter or LexA operator control. Light is dose dependent, inexpensive to apply, easily delivered, and does not interfere with cellular pathways, and thus has significant advantages over chemical systems.
Keywords: photoreceptor, cryptochrome, phytochrome, optogenetics, two-hybrid, DNA transcription
1. Introduction
A number of inducible systems are available for controlling protein expression in yeast. These include promoters naturally responsive to molecules such as galactose, methionine, and copper, or engineered regulatory systems that respond to orthogonal molecules such as estrogen (Gal-ER-VP16) [1] or doxycycline (Tet-OFF) [2]. Using galactose inducible promoters, protein expression can be repressed in the presence of glucose and induced with addition of galactose [3]. Galactose induction is robust and rapid, with ~ 1000-fold expression levels achieved after approximately four hours [3, 4]. However, titration of galactose can give imprecise intermediate levels of protein [4], and as the system is repressed by glucose, the yeast sugar source must be changed from glucose to galactose unless mutant strains are used, resulting in large changes in yeast gene expression [5, 6]. CUP1 promoters allow for the control of protein expression by the addition of copper to the yeast media [7, 8], while the MET3 and MET25 promoters allow conditional expression in the absence of methionine [4, 9–11]. Reduction of protein levels with the MET25 promoter is straightforward with the addition of methionine, but reversal of protein expression requires removal of methionine from the media [4]. Using engineered regulatory systems such as Gal-ER-VP16, protein levels can be controlled by the addition of β-estradiol to the media [1]. The Tet-OFF system [2] and a related Tet-ON system [12] employ the VP16 activation domain of herpes simplex virus fused to the E. coli tetracycline repressor, and can be used to either induce (Tet-ON) or halt (Tet-OFF) gene transcription in response to doxycycline. While these systems have been proposed to have few adverse effects in yeast [13, 14] and can provide some degree of graded response, the induction and reversibility are still affected by the requirements for compound import and removal.
Recently, several groups have demonstrated use of light-responsive transcription factors that allow light-dependent control of DNA transcription and protein expression. Some of these consist of split transcription factors that are reconstituted by light-dependent protein-protein interactions as in yeast two-hybrid systems [15–17]. Others are single plasmid systems engineered to respond to light in bacteria [18] or mammalian cells [19]. Light-inducible transcription systems offer several advantages over chemical inducible systems. First, they are fast: delivery of light is immediate and does not require time for chemical uptake into cells. Second, light does not require manipulation of culture media, and can be easily automated. Third, light is minimally invasive. While high levels of constant blue light can induce cellular stress responses and toxicity [20], use of brief pulses of light to stimulate photoreceptor proteins can minimize this. Fourth, light is not affected by compound uptake or drug pump issues, which can confound studies that use chemical inducers. For example, using chemically-induced transcription systems to screen libraries of chemical compounds or yeast deletion strains could result in false-positives, where deletion strains or compounds that affect membrane transporters or drug efflux pumps result in changes in reporter protein expression levels that appear as hits. Finally, light induced systems allow exquisite dose-dependent control over protein levels, as light can be delivered for defined amounts of time and instantly removed.
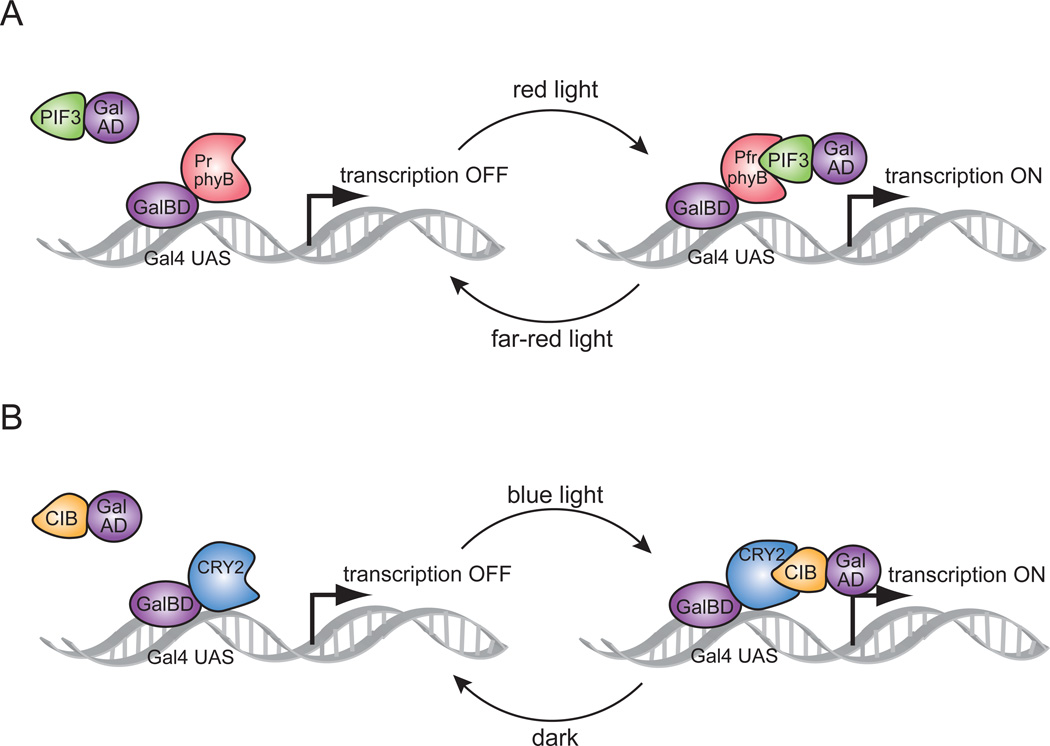
Here we describe the use of two different systems for regulation of DNA transcription by light. Both systems are based on a two-hybrid interaction [21] in which a light-mediated protein interaction brings together two halves (a binding domain and an activation domain) of a split transcription factor (Fig. 1). The first system (Fig. 1A) is based on an interaction between the Arabidopsis photoreceptor protein phytochrome B (PhyB) and an interacting partner protein, PIF3 [16]. PhyB can adopt two different conformations: a far-red absorbing ‘Pfr’ form after stimulation by red light, and a red absorbing ‘Pr’ form in the dark or after stimulation by far-red light. To adopt the Pfr form, PhyB also requires the binding of a bilin chromophore, which is not present in yeast but is added exogenously. PIF3 binds only to the Pfr form, and thus the PhyB/PIF3 interaction occurs upon illumination with red light in the presence of chromophore, and the complex is dissociated with the application of far-red light [22]. A second system that has been developed for light-induced transcriptional control is based on a blue light stimulated photoreceptor, cryptochrome 2 (CRY2) and its interacting partner, CIB1 [15] (Fig. 1B). The CRY2/CIB1 interaction is entirely genetically encoded and does not require addition of any exogenous cofactors. The binding naturally reverses within minutes in the dark, allowing rapid shutoff of transcription by placing samples in the dark. Here, we show use of both systems for light-dependent control of DNA transcription and protein expression in yeast, where we demonstrate regulation of reporter genes under GAL promoter control. In addition, we have developed an orthologous system allowing transcriptional induction of genes under LexA operator control using the CRY/CIB system, which can be used to regulate transcription in any strain.
Figure 1.
Schematic showing split transcription factor reconstitution using optical dimerizers. (A) Red light switches PhyB into the Pfr form, enabling binding to PIF3. The PhyB/PIF3 interaction allows reconstitution of a split Gal4 transcription factor and switches on DNA transcription. Far-red light reverts PhyB to the Pr form, which is unable to bind to PIF3 and halts transcription. (B) Blue light enables binding of CRY2 to CIB1, bringing together a split Gal4 transcription factor controlling DNA transcription. Dark reversion of CRY2 dissociates the interaction with CIB1 and halts Gal4-dependent transcription.
2. Methods
2.1 Strains and plasmids
Yeast strains used were PJ69-4a (MATa trp1-901 leu2-3,112 ura3-52 his3-200 Δgal4, Δgal80, LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) [23], W303-1A (MATa leu2-3,112, ura3-1 trp1-1 his3-11,15 ade2-1 can1-100).
PhyBNT-GalBD (amino acids 1–621 of Arabidopsis PhyB fused at the N-terminus of the Gal4 binding domain in plasmid D153) and GalAD-PIF3 (full length PIF3 fused at the C-terminus of the Gal4 activation domain) were provided by Peter Quail, and are described previously [16]. Gal4BD-CRY2PHR contains amino acids 1–498 of cryptochrome 2 in plasmid pDBTrp [24], Gal4AD-CIB1 contains full length CIB1 in pGADT7rec (Clontech), while Gal4AD-CIBN contains amino acids 1–170 of CIB1 in pGADT7rec, and have been described previously [15]. To generate pRH120, Gal4BD-CRY2PHR was amplified by PCR from the pDBTrp vector and homologous recombination in yeast was used to insert this fragment between the TEF promoter and CYC terminator of p414TEF digested with Spe I and Xho I. Next, a fragment containing the ADH promoter, Gal4AD-CIB1, and the ADH terminator was cloned via homologous recombination into p414TEF-Gal4BD-CRY2PHR that had been digested with KpnI.
Homologous recombination was used to replace the Gal4 activation domain in pGADT7rec with the VP16 activation domain, generating pRH-VP16-CIBN and pRH-VP16-CIB1. Similarly, homologous recombination was used to replace the Gal4 binding domain in pDBTrp with the LexA binding domain, generating pRH-LexA-CRY2PHR. To generate the YFP reporter, a destabilized YFP from pRB3 [25] was PCR amplified and cloned by ligation into pESC-Ura at Eco RI and Not I sites, enabling expression of the YFP construct with an C-terminal FLAG tag. A hyperstabilized Sic1-6xHis protein [26] was cloned into vector pSH18-34 (Invitrogen) cut with Spe I and Nar I using homologous recombination.
2.2 Isolation of phycocyanobilin (PCB) from Spirulina
The basis for the conformational switch in PhyB is a bond isomerization in the phytochromobilin (PϕB), the chromophore that binds phytochrome in plants. While yeast do not make phytochromobilin, a functional homolog, phycocyanobilin (PCB), can be extracted from Spirulina by methanolysis [27] and added to the yeast medium, where it is taken up into cells and assembles to form holo-PhyB [16]. The methanolysis protocol requires a 500 ml round bottom flask, a reflux condenser, a water bath and hot plate, a ring stand with two clamps, two 500 ml vacuum filtration flasks, and a 250 ml separatory funnel. Also needed is a dark room outfitted with a green safety light (for example a green LED light) and a centrifuge. While it is advisable to cover the methanolysis apparatus in aluminum foil to prevent light penetration and subsequent photobleaching of the chromophore, brief ambient light exposure when placing the methanolysis fractions into the centrifuge did not harm the chromophore.
Protocol
50 g of Spirulina powder (Seltzer Ingredients) was resuspended in 1.5 L water in a 2 L Erlenmeyer flask, which had been wrapped with foil to protect it from light. After stirring for 10 min, the solution was centrifuged for one hour (8000 RPM, 4°C). The supernatant was decanted into a 2 L flask and 15 g of TCA was added. After stirring for 1 hour in the dark at 4°C, the solution was centrifuged again (8000 RPM, 4°C, 10 min) and the supernatant discarded. The remaining pellets were resuspended in 25 ml of MeOH and vigorously vortexed. During this process the pellets were kept on ice. After resuspension, additional MeOH was added to a final volume of 250 ml and samples were again centrifuged (8000 RPM, 4°C, 10 minutes). The supernatant was discarded. The MeOH wash step was repeated once more, and then the pellets were stored overnight, wrapped in foil, at −20°C.
The next day, in a dark room under green safelight, the six pellets were resuspended and pooled in a final volume of 400 ml MeOH. A methanolysis apparatus consisting of a 75 °C water bath, a condensing coil cooled with tap water, a ring stand and a 500 ml round bottom flask was used to boil the methanolic solution for 8 hours. After boiling, the solution was transferred to two centrifuge bottles and spun at 8000 RPM for 20 min at 4 °C. The resulting supernatant was then decanted into a foil-wrapped centrifuge bottle and stored at −20°C. The pellet was then resuspended in MeOH (400 ml) and boiled again overnight at 75°C. The resulting solution was again centrifuged and the supernatant decanted and stored at −20°C.
Next, two vacuum flasks were set up, one stoppered and foiled to hold the PCB-MeOH solution and connected to the second flask with plastic tubing via its sidearm. The second flask was connected to a house vacuum line via its sidearm and was also partially submerged in a dry ice/EtOH bath. The collection of MeOH (350 ml) took approximately 16 hours. The concentrated PCB/MeOH solution (50 ml) was transferred to a round bottom flask, foiled, stoppered, and stored at −20°C. The second methanolysis fraction was then concentrated under vacuum and stored at −20°C.
The next day, 50 ml of the concentrated PCB solution was combined with 50 ml chloroform and 100 ml H20 in a 250 ml separatory funnel and shaken. The chloroform phase (bottom) was collected in an Erlenmeyer flask. Two additional 50 ml chloroform extractions were performed on the same PCB solution (resulting in 150 ml of extract). This process was repeated for the concentrated second methanolysis fraction as well. The combined extracts were transferred to a 500 ml round bottom flask (wrapped securely in foil) and the chloroform was evaporated with a stream of house nitrogen. The dried PCB was resuspended in 5 ml of DMSO. The concentration of PCB was calculated by measuring the absorption at 680 nm in 95/5 MeOH:HCl and using the formula C(mM) = (A680/37.9)*dilution factor.
2.4 Light induction systems using split Gal4
To induce expression of proteins using the PhyB/PIF3 split Gal4 system, we used a strain of yeast (PJ69-4a) with deletions in the Gal4 and Gal80 genes, as the Gal80 protein binds to the Gal4 activation domain and represses Gal4 transcriptional activation. As a reporter, we used a destabilized YFP-FLAG construct that was cloned downstream of a Gal4 upstream activating sequence (UAS). Yeast containing PhyBNT-GalBD, GalAD-PIF3 and the GalUAS-YFP reporter were grown overnight in SC –Trp/-Leu/-Ura media and diluted to a density of OD600 = 0.2. PCB was added at a final concentration of 2 µM under a green safety light, and the culture was wrapped in foil and grown an additional 4 hours at 30°C to log phase. To induce transcription and protein expression, cultures were exposed to a pulse of red light (660 nm) every 30 min – 1 hr using an automated timer. To shut off induction, cultures were exposed to a single pulse of far-red light (730 nm).
To induce protein expression using the CRY2/CIB1 split Gal4 system, we also used PJ69-4a yeast. Yeast containing Gal4BD-CRY2 and Gal4AD-CIB1 and the Gal4 UAS-YFP reporter were grown to log phase, then exposed to blue light. In most of the experiments outlined here, we delivered pulses of blue light generated by an Arduino-controlled LED light source (461 nm), to provide greater light intensity and quantifiable control over the amount of light applied. However, for many studies, such precise control is not needed, and light can be delivered using constant illumination from a LED light bulb, as was used in the initial two-hybrid studies carried out in Kennedy et al. 2010 [15]. Full spectrum white light can also be used as a light source for the CRY/CIB system, as no light wavelength has been found that reverses this system. Thus, activation of the CRY/CIB system can be achieved by incubating yeast cultures in room light. However, many applications that are of interest to the experimentalist require greater light intensity and control over the light source.
Upon stimulation of CRY2 with a pulse of blue light, the protein converts to a conformation that is able to interact with CIB1, but only remains in this stimulated conformation for several minutes. Over a time course of approximately 10–12 minutes (τ = 5.5 min), the CRY2-CIBN interaction completely (> 90%) dissociates. Thus, a single pulse of light will promote on average 5 minutes of activity, and pulsing cultures with brief (1 s) flashes of blue light every 2–3 minutes promotes near constant association, while minimizing light illumination that can activate stress responses.
Immunoblotting and ONPG assays
For immunoblotting, samples were pelleted and frozen at −80°C. Lysis was induced by adding 50 µl of 2% SDS to the frozen pellet, vortexing 7 min at full speed, boiling 3 min, then adding 100 µl of 2X LSB + BME and boiling 1 additional min. Samples were then spun at 14,000 × g for 1 min before gel electrophoresis. Proteins were run on a SDSPAGE gel and transferred to nitrocellulose membranes. Membranes were probed with an anti-Flag antibody (Sigma) and an IRDye 800CW goat anti-rabbit IgG secondary antibody (Li-COR). Alternatively, cultures were pelleted and lysed with Y-PER (Thermo Scientific) and assayed for β-galactosidase activity as described previously [15].
2.6 Construction of a programmable LED light source
Light control of the PhyB/PIF3 and CRY2/CIB1 systems can be achieved through a variety of light sources. PhyB is responsive to illumination in the red and far-red wavelengths (660 nm and 730 nm), while CRY2 is responsive to blue light (460 nm, range 390–530 nm). The CRY2/CIB1 interaction requires fairly intense blue light (> 1 mW power) to achieve high levels of transcriptional activation [28], but exposure to constant blue light of a very high intensity can activate cell stress responses [20]. To provide precise control of light pulse duration and frequency and reduce the possibility of toxicity, we generated a modular programmable LED array (Supplementary Fig. 1). The modular design of the device allows use with LED arrays of different wavelengths, allowing activation of blue or red light systems as needed. The control module consists of an Arduino Uno microcontroller and a LED driver (BuckPuck DC Driver, LuxeonStar, 3021-DI-700) that interfaces with the LED arrays through MOLEX connectors. The microcontroller is connected by a USB connector to a computer, where it can be programmed then unplugged from the computer for use. The LED modules house 4-by-3 arrays of LEDs. For the blue LED array, we used LuxeonStar LXML-PR01-500 (448 nm) LEDs. The device exhibited linear calibration between expected and observed pulse frequency (R2 = 1.00) and expected and observed pulse duration (R2= 1.00). The root mean square error (RMSE) of pulse frequency and duration were 6.16 *10−5 Hz and 0.51 s, respectively, for the prototype. The device constructed was capable of generating light pulses of frequencies between 1*10−4 Hz and 1 Hz and durations between 1 s and 5 min.
The user interface of the device is Arduino software that accepts a pulse duration of seconds or minutes, a pulse interval of seconds or minutes, and a run-time. The Arduino software can be downloaded from the Arduino website (www.arduino.cc), and the source code (provided in Supplementary Materials) can be copied into the text area. The parameters are modified in the source code section entitled “Enter experiment values below:”. To set the number of hours or minutes the experiment will run, change “ExprTimeHrs” or “ExprTimeMins” values to the specified hours or minutes, respectively (otherwise leave “0”). To set the pulse duration, change “PulseDurMins” or “PulseDurSecs” to the duration of minutes or seconds, respectively (otherwise, leave as “0”). To set the pulse frequency, change “PulseFreqMins” or “PulseFreqSecs” to the frequency of the pulse in minutes or seconds, respectively. For example, to modify the pulse duration from 1 to 2 seconds, change ‘float PulseDurSecs = 1’ to ‘float PulseDurSecs = 2’. Once the parameters have been set, click the upload button to load the parameters into the device. Once uploaded, the device can be disconnected from the computer.
In the absence of the above pulse device, user controlled LED arrays or filtered light sources can also be used. With PhyB, which remains in a photostimulated state for much longer than CRY2, a red LED array connected to a standard 24-hour programmable outlet timer can be used to deliver a brief pulse of red light once per hour.
3. Results and Discussion
3.1 Establishment of PhyB/PIF3 activation conditions
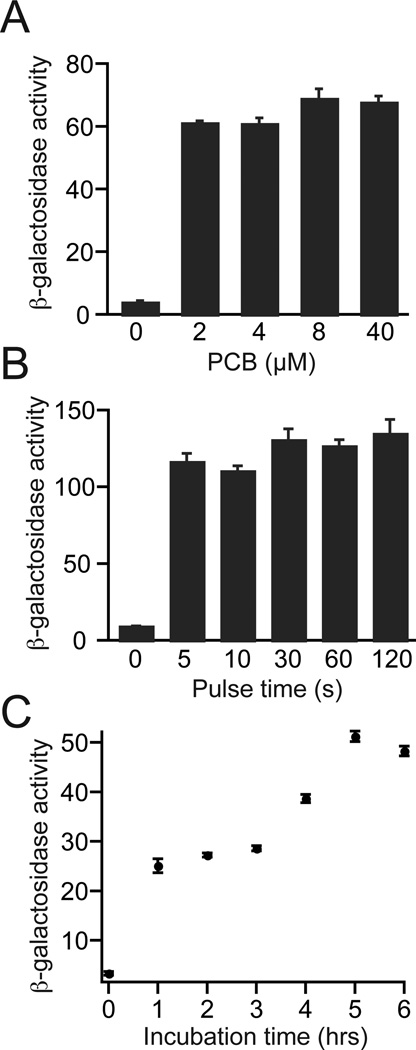
In previous work, a PhyB/PIF3 reconstituted Gal4 transcription factor was found to show strong light-dependent induction of protein expression in yeast [16]. Using PhyB-GalBD and GalAD-PIF3 constructs, we carried out several experiments to determine the optimal conditions for use of this system. To determine the minimal amount of PCB needed, we examined concentrations ranging from 2 µM to 40 µM. Using a preincubation time of 4 hours to allow entry of the compound and conjugation with PhyB, we found that 2 µM PCB was sufficient for maximal activation (Fig. 2A). We also investigated the minimum duration of red light pulse required for PhyB activation in cultures (Fig. 2B). Light was flashed for durations ranging from 5 s to 2 min, and we observed that a 5 s pulse of red light (2.8 mW/cm2) gave similar activation levels as a 2 min light application. Finally, we investigated the minimum incubation time required for the PCB to be taken up into yeast and incorporated into holo-PhyB (Fig. 2C). Using 2 µM PCB, we tested incubation times of 1–6 hours. The optimal incubation time appeared to be 5 hours, as beyond this amount of time a slight decrease in reporter levels occurred. However, a 4 hour incubation period gave ~75% of maximal activity, and so we used this incubation interval for further experiments. One hour of incubation with PCB gave ~ 50% of maximal levels, which is likely sufficient for most experiments. Use of higher concentrations of PCB may also be more effective for shorter incubation times (based on results from Fig. 3B, below).
Figure 2.
Establishment of parameters for use of PhyB/PIF3 split Gal4 system. PJ69-4a yeast cultures expressing PhyBNT-GalBD and GalAD-PIF3 were diluted to OD600 = 0.2 and grown for indicated times in the dark with indicated amounts of PCB before red light exposure (2.8 mW/cm2, 660 nm). (A) PCB titration. Yeast were grown 4 hours in the dark with indicated amounts of PCB, then illuminated with red light (5 s pulse). Cultures were incubated in the dark for an additional hour, then analyzed for β-galactosidase reporter activity. (B) Light pulse titration. Yeast cultures were grown for 4 hours in the dark with 2 µM PCB, then exposed to red light for indicated amounts of time. Samples were returned to the dark for 3 hours, then analyzed for β-galactosidase reporter activity. (C) Yeast were grown for indicated times in the dark before exposing to red light (5 sec pulse) then returned to the dark for 1 hour before analysis of β-galactosidase reporter activity.
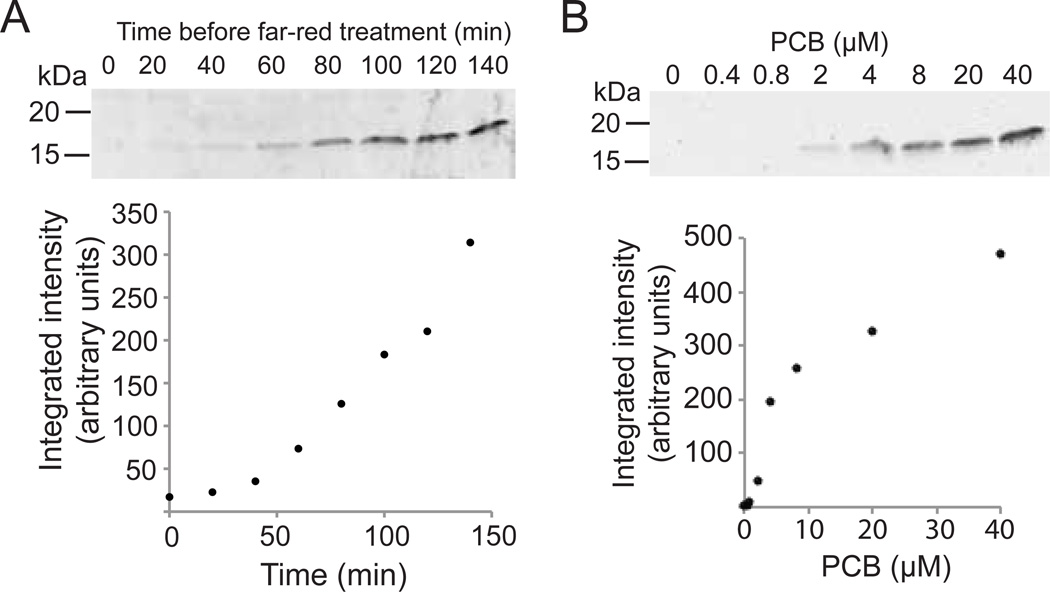
Figure 3.
Induction of protein expression using PhyB/PIF3 split Gal4 system. (A) PJ69-4a yeast cultures expressing PhyBNT-Gal4BD, Gal4AD-PIF3, and the GalUAS-YFP reporter were diluted to OD600 = 0.2 and grown for 4 hrs in the dark with 2 µM PCB then exposed to a pulse of red light (5 s, 2.8 mW/cm2, 660 nm). Cultures were returned to the dark, then flashed with far-red light (5 s, 6.9 mW/cm2) at indicated times to halt transcription. Samples were then returned to the dark, and all samples were harvested simultaneously 140 minutes after the initial red light exposure. The graph below show integrated intensities of immunoblot bands (arbitrary units). (B) Yeast cultures as in (A) were incubated with indicated amounts of PCB at OD600 = 0.2 and grown for 6 hours, treated with a pulse of red light (5 s, 2.8 mW/cm2) each hour. All samples were harvested simultaneously at the end of the 6-hour incubation period.
3.2 Dose-dependent induction of transcription using PhyB/PIF3
To examine how protein expression varies with different light doses, we reconstituted the Phy/PIF-controlled split Gal4 activator (using PhyB-GalBD and GalAD-PIF3) by pulsing with red light for different amounts of time, followed by a pulse of far-red light at the end of the time period to dissociate the transcription factor (Fig 3A). For a reporter, we used a destabilized YFP that has a half life of ~ 1 hour [25]. After inducing protein expression with a single 5 s pulse of red light, cultures were incubated in the dark for times from 0–140 min, then exposed to a pulse of far-red light (6.9 mW/cm2). The yeast cultures were then harvested simultaneously 3 hours after the initial light pulse. Using this approach, protein levels correlated linearly with light dosage (Fig. 3A, bottom).
The PhyB/PIF3 interaction requires the presence of PCB in the yeast media. While we saw no reporter expression in the absence of PCB, in many experiments, such as shown in Figure 2, we observed a small amount of reporter expression in the dark in the presence of PCB. We thus examined whether PCB itself, an inexpensive, orthogonal chemical, could be used for dose-dependent induction of protein expression in yeast with less light-independent background. As the PhyB/PIF3 interaction is dissociated with far-red light, we reasoned that this system would provide tight control over protein levels as expression could not only be induced with PCB, but also turned off by exposure to far-red light. To examine this, we subjected yeast to flashes of red light (5 s flash each hour), but added a different amount of PCB chromophore to each sample (Fig 3B). Cultures were harvested after a 6 hr incubation period. Using this approach, we observed that protein production depended on levels of PCB added to the media (Fig. 3B, bottom). Thus, PCB concentration, in addition to light, can be used to produce defined quantities of protein.
3.3 Control of protein expression using CRY2/CIB1
In previous work, the plant proteins CRY2 and CIB1 were used to induce transcription and protein expression with light [15]. One large advantage of light-regulated systems is the potential to rapidly shut off DNA transcription by removal of light. Often, cycloheximide is used to rapidly halt DNA transcription, but this chemical is not specific and affects all gene transcription. Alternatively, glucose can be used to halt protein expression from a galactose-regulated promoter, but changing the carbon source has widespread effects on yeast cell physiology and gene expression.
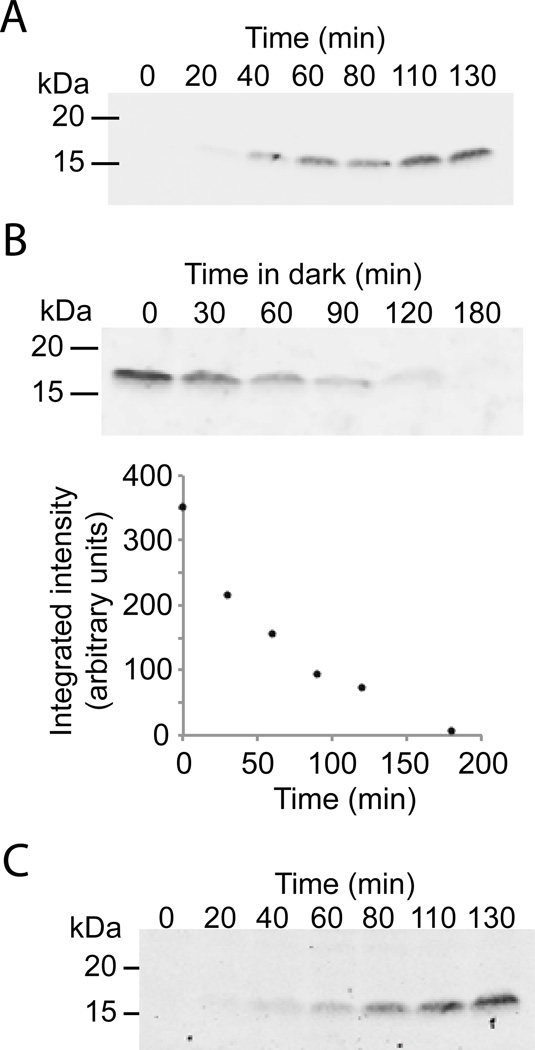
To explore transcriptional shutoff with the CRY2/CIB1 split Gal4 system, we used the destabilized YFP reporter that has a short half-life of ~1 hour. Based upon previous studies [15], we expected that the CRY/CIB reconstituted transcription factor would dissociate and turn off within 10–12 min, enabling rapid termination of transcription in the dark. We first illuminated yeast with pulses of blue light, inducing accumulation of the YFP reporter (Fig. 4A). After illuminating the yeast culture with pulses of blue light for 5 hours, we placed the samples in the dark and removed an aliquot every 30 minutes to track levels of the short-lived reporter protein (Fig 4B). We observed rapid degradation of the reporter protein, supporting that DNA transcription and subsequent protein expression stops quickly after the culture is returned to the dark (Fig 4B). Thus, using a single system, we are able to regulate protein production in rapid fashion.
Figure 4.
Control of protein expression using CRY2/CIB1 split Gal4 system. (A) PJ69-4a cultures expressing Gal4BD-CRY2, Gal4AD-CIB1, and the GalUAS-YFP reporter were inoculated at OD600 = 0.2 and grown for 3 hours in the dark. Blue light flashes were then applied (2 s, every 4 minutes, 2.7 mW/cm2), and samples were taken at indicated times for immunoblot analysis. (B) Samples as in (A) were grown for 5 hours with blue light flashes (2 s, every 4 minutes, 2.7 mW/cm2). After 5 hours, the samples were placed in the dark for indicated amounts of time, then harvested for immunoblot analysis (top). The graph below shows the integrated intensity of blot bands (arbitrary units). (C) PJ69-4a containing pRH120 (which expresses Gal4BD-CRY2PHR and Gal4ADCIB1 from a single plasmid) and a GalUAS-YFP reporter were treated as in (A).
The initial CRY/CIB system contains the split Gal4 transcription factor on two different plasmids. To increase the utility of the system, we generated a new plasmid, pRH120, containing both Gal4BD-CRY2PHR and Gal4AD-CIB1 on the same construct, which has a p414TEF backbone. This single construct behaved similar to the two-plasmid system, and can be used in similar ways to activate transcription from a GAL1 promoter (Fig 4C).
3.4 An orthogonal LexA-VP16 transcription factor controlled by CRY2/CIB1
While the previously developed CRY2/CIB1 controlled split Gal4 system worked well in a Gal4 / Gal80 deleted strain [15], we were interested in the ability to use this system in other strains. Thus, we first replaced the Gal4 activation domain with the herpes simplex virus VP16 activation domain. VP16 has a very strong transcriptional activation domain that is orthogonal to yeast [29]. We evaluated the efficiency of different CRY2 and CIB1 truncations in the context of the GalBD/VP16 pair using a β-galactosidase assay, testing yeast containing GalBD-CRY2 or GalBD-CRY2PHR and the VP16AD fusions VP16-CIB1 or VP16-CIBN (Fig 5A). We found that both GalBD-CRY2PHR and GalBD-CRY2 coupled well with VP16-CIB1 for light-inducible protein expression. As seen in prior work [15], the CRY2PHR/CIB1 pair gave the strongest protein induction but also resulted in some background light-independent activity. We tested the ability of the GalBD-CRY2PHR/VP16-CIB1 pair to induce expression of the destabilized YFP reporter (Fig. 5B). Using this reporter, we observed induction of protein expression in blue light and very little background expression in the dark (Fig. 5B).
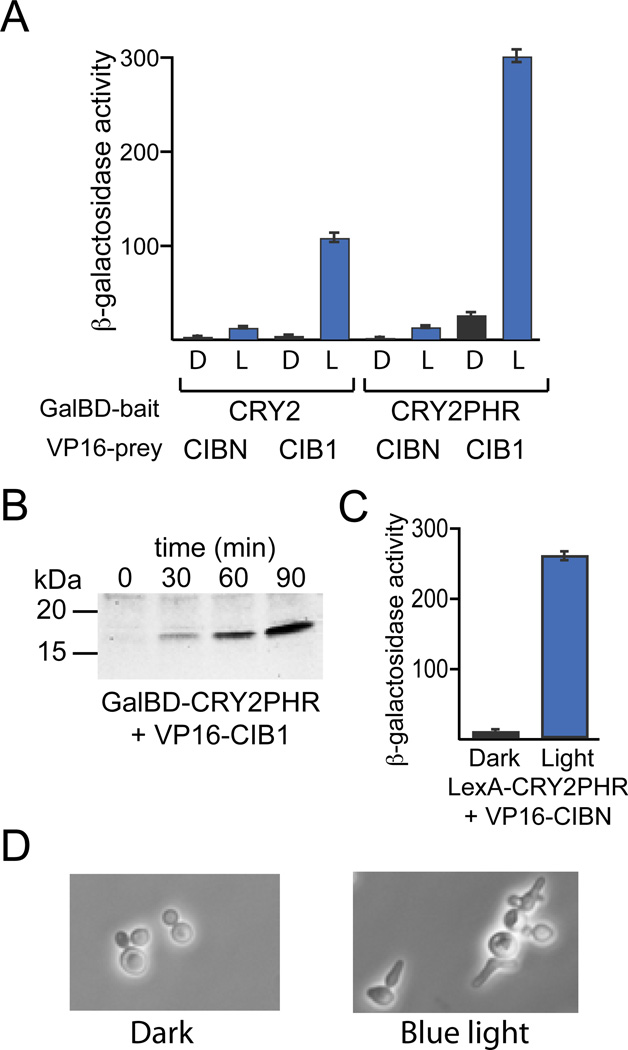
Figure 5.
Orthogonal light-regulated LexA-VP16 transcription system. (A) PJ69-4a yeast expressing GalBD-CRY2 or GalBD-CRY2PHR and VP16-CIB1 or VP16-CIBN were tested for activation of a β-galactosidase reporter after a 5 hr incubation in the dark (‘D’) or exposure to blue light pulses (‘L’) (1 s, every 4 min, 1.9 mW/cm2). (B) PJ69-4a yeast expressing GalBD-CRY2PHR, VP16-CIB1, and a GalUAS-YFP reporter were exposed for indicated times to blue light pulses (1 s, every 4 min, 1.7 mW/cm2). (C) W303-1A yeast expressing LexA-CRY2PHR, VP16-CIBN, and a pSH18-34 reporter (containing 8 LexA operators driving expression of LacZ) were incubated in the dark or exposed to blue light pulses (2 s, every 4 min, 1.9 mW/cm2) for 3 hours, then extracts were assayed for β-galactosidase activity. (D) W303-1A yeast expressing LexA-CRY2PHR, VP16-CIBN, and a LexA-8xop- SIC10p construct, containing a hyperstable Sic1 protein, were kept in the dark (left) or exposed to blue light pulses (2 s, every 4 min, 2.7 mW/cm2) for 4 hours (right).
To explore the use of a completely orthogonal transcription factor, we replaced the Gal4BD of the GalBD-CRY2PHR construct with a DNA binding domain from the E. coli LexA protein (Brent and Ptashne 1985), generating LexA-CRY2PHR. LexA-CRY2PHR and VP16-CIBN were coexpressed in strain W303-1A, along with the pSH18-34 reporter, which contains 8 LexA operators driving expression of LacZ. Use of LexA-CRY2PHR / VP16-CIBN resulted in very little background protein expression in the dark and robust activation in the light, activating transcription 32-fold over dark levels (Fig. 5C).
To test the ability of the LexA/VP16 light-inducible system to induce an experimental phenotype in yeast, we used the above constructs to induce transcription of a hyperstable mutant of Sic1, a yeast cell cycle protein [26]. Degradation of the protein Sic1 is required for yeast to transition from G1 to S phase in the cell cycle. Previous work had shown that mutation of specific residues in Sic1 (SIC10p) prevents its ubiquitination and degradation, resulting in abnormal accumulation of Sic1 and G1/S arrest [26]. We generated a LexA-8op-hyperstable Sic1 and expressed this construct in W303-1A yeast containing the LexA-CRY2PHR and VP16-CIBN light-activated transcription system (Fig. 5D). Yeast grew normally in the dark, but showed dramatic phenotype changes after only 4 hours of exposure to blue light pulses. The light-treated yeast showed a multi-budded phenotype similar to that previously seen with the same mutant induced for 3 hours using a galactose inducible promoter (pGAL1-SIC10p) [30]. In contrast to the galactose-induced mutant phenotype, induction using this system was accomplished without any manipulation of the yeast media.
4. Summary
Using plant photoreceptor protein interactions coupled to split transcription factor binding and activation domains based on two-hybrid systems, we can achieve precise light-dependent control over protein levels in yeast. These optically controlled transcriptional systems will be of interest to researchers requiring precise control over protein expression, or who wish to avoid chemicals and use a completely exogenous inducer. Here, we show that we can control protein production with light in any yeast background strain, a critical advance for the practical application of this technology. In addition, as GAL-regulated promoters are commonly used for activation of transcription in flies, zebrafish, and even mouse, these constructs can be adapted to allow regulation of transcription in different organisms.
Supplementary Material
Supplementary Material 1. Source code for controlling pulse length and frequency using Arduino software.
Supplementary Figure 1. Diagram of Blue LED control circuit. The LEDs are arranged such that 4 LEDs are in each series branch. 3 LED branches are connected in parallel.
Acknowledgements
We thank Steve Haase (Duke University) for the hyperstable Sic1 mutant protein, Ryan Baugh (Duke University) for the destabilized YFP reporter, and Peter Quail (UC Berkeley) for the PhyB and PIF3 two hybrid constructs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Louvion JF, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131(1):129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 2.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronicke V, Graulich W, Mumberg D, Muller R, Funk M. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol. 1997;283:313–322. doi: 10.1016/s0076-6879(97)83025-x. [DOI] [PubMed] [Google Scholar]
- 5.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 6.Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci U S A. 1997;94(24):13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butt TR, Sternberg EJ, Gorman JA, Clark P, Hamer D, Rosenberg M, Crooke ST. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci U S A. 1984;81(11):3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labbe S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 9.Cherest H, Kerjan P, Surdin-Kerjan Y. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5' non-coding region to that of MET25. Mol Gen Genet. 1987;210(2):307–313. doi: 10.1007/BF00325699. [DOI] [PubMed] [Google Scholar]
- 10.Kerjan P, Cherest H, Surdin-Kerjan Y. Nucleotide sequence of the Saccharomyces cerevisiae MET25 gene. Nucleic Acids Res. 1986;14(20):7861–7871. doi: 10.1093/nar/14.20.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sangsoda S, Cherest H, Surdin-Kerjan Y. The expression of the MET25 gene of Saccharomyces cerevisiae is regulated transcriptionally. Mol Gen Genet. 1985;200(3):407–414. doi: 10.1007/BF00425724. [DOI] [PubMed] [Google Scholar]
- 12.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268(5218):1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 13.McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti AA, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22(22):4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart JA, Hayes A, Wardleworth L, Zhang N, Oliver SG. Doxycycline, the drug used to control the tet-regulatable promoter system, has no effect on global gene expression in Saccharomyces cerevisiae. Yeast. 2005;22(7):565–569. doi: 10.1002/yea.1225. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7(12):973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20(10):1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 17.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of proteinprotein interactions in live cells using light. Nat Biotechnol. 2009;27(10):941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 18.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. J Mol Biol. 2012;416(4):534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9(3):266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 20.Bodvard K, Wrangborg D, Tapani S, Logg K, Sliwa P, Blomberg A, Kvarnstrom M, Kall M. Continuous light exposure causes cumulative stress that affects the localization oscillation dynamics of the transcription factor Msn2p. Biochim Biophys Acta. 2011;1813(2):358–366. doi: 10.1016/j.bbamcr.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 22.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400(6746):781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 23.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker CL, Peteya LA, Pittman AM, Zhong J. A genetic test for yeast two-hybrid bait competency using RanBPM. Genetics. 2009;182(4):1377–1379. doi: 10.1534/genetics.109.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324(5923):92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 26.Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278(5337):455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 27.Arciero DM, Bryant DA, Glazer AN. In vitro attachment of bilins to apophycocyanin. I. Specific covalent adduct formation at cysteinyl residues involved in phycocyanobilin binding in C-phycocyanin. J Biol Chem. 1988;263(34):18343–18349. [PubMed] [Google Scholar]
- 28.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322(5907):1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 30.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117(7):899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1. Source code for controlling pulse length and frequency using Arduino software.
Supplementary Figure 1. Diagram of Blue LED control circuit. The LEDs are arranged such that 4 LEDs are in each series branch. 3 LED branches are connected in parallel.