Abstract
In the past decade since the discovery of NAD-dependent deacetylase activity of the SIR2 (silent information regulator 2) family, now called “sirtuins,” many exciting connections between protein deacetylation and energy metabolism have been revealed. The importance of sirtuins have been firmly established in the regulation of many fundamental biological responses to a variety of nutritional and environmental stimuli. Sirtuins have also emerged as critical regulators for aging and longevity in model organisms. Their absolute requirement of NAD has revived an enthusiasm in the study of mammalian NAD biosynthesis. Furthermore, sirtuin-targeted pharmaceutical and nutriceutical interventions against age-associated diseases are now on the horizon. This review will summarize the recent progress in sirtuin research, particularly in the field of mammalian sirtuin biology, and reevaluate the connection between sirtuins, metabolism, and age-associated diseases, such as type 2 diabetes, to set a milepost for the next 10 years of sirtuin research.
Introduction
We have arrived at the 10th anniversary of this discovery of NAD-dependent SIR2 family deacetylases. The sir2 gene was identified as mar1, a gene affecting yeast mating ability, in 1979 1. Over the past decade, more than 1,500 papers have been published on the SIR2 family, now called “sirtuins,” while there were less than 100 before this period. We believe this is a good opportunity to assess what has been done and what still needs to be achieved in this exciting field of sirtuin biology. Therefore, in this review, we will summarize the recent progress in sirtuin research, particularly in the field of mammalian sirtuin biology, and reevaluate the connection between sirtuins, metabolism, and age-associated diseases.
Chronology of sirtuin research over the past 10 years
A brief history
In 1998, Tsang and Escalante-Semerena reported that a protein called CobB in Salmonella typhimurium can substitute for the function of CobT, a protein that transfers phosphoribose from nicotinic acid mononucleotide to dimethylbenzimidazole in the cobalamin biosynthesis pathway 2, 3. Intriguingly, CobB has signature motifs of the evolutionarily conserved SIR2 family 4, which turned to be the first important clue suggesting that SIR2 proteins might catalyze a related pyridine nucleotide transfer reaction. Following their paper, in 1999, Roy Frye showed that SIR2 proteins from Escherichia coli and humans were able to transfer 32P from [32P]NAD to bovine serum albumin, albeit weakly 5. Subsequent to this study, Danesh Moazed’s group demonstrated that SIR2 was indeed able to transfer ADP-ribose from NAD to histones, leading them to the conclusion that the observed ADP-ribosyltransferase activity of SIR2 was involved in gene silencing in yeast 6.
Meanwhile, we noticed that peptides of the amino-terminal tails of histone H3 or H4 could accept 32P from [32P]NAD in the reactions mediated by recombinant yeast SIR2 or mammalian SIRT1 proteins, but only if the peptides were acetylated 7. Like Frye’s findings, however, this transfer reaction was extremely weak even using the acetylated peptides as substrates. Believing the low concentrations of labeled NAD may be below the Km of the enzyme, we attempted to significantly increase the NAD concentration in the SIR2 enzymatic reaction (using unlabeled NAD), which necessitated analyzing the reaction products directly by mass spectrometry. Using this assay in October 1999, we found that the relative molecular weight of the reaction product was not larger but actually smaller than that of the acetylated peptide exactly by 42 Da. Moreover, both the yeast and mammalian sirtuin proteins robustly converted the substrate peptides to their smaller products. This serendipitous experiment was the first demonstration that the major enzymatic activity of SIR2 is NAD-dependent deacetylase 8. Importantly, yeast and mammalian SIR2 proteins were able to specifically deacetylate lysine 16 of H4 in an NAD-dependent manner, strongly suggesting that the NAD-dependent deacetylase activity plays a critical role in establishing silenced chromatin structures in vivo 8. Sirtuins require NAD, but not NADH, NADP, and NADPH, for their enzymatic activity, immediately suggesting that sirtuins might function as sensors of the cellular energy status represented by NAD. Within months of the paper on these findings, reports appeared showing that SIR2 and HST2, a yeast SIR2 homolog, catalyze both NAD-nicotinamide exchange reaction and NAD-dependent deacetylation 9, 10.
Major advances and questions in sirtuin research
The above findings set the stage for sirtuin biology, and in the past 10 years, significant progress has been made in this quickly evolving field. First, the catalytic mechanism of NAD-dependent deacetylation has been extensively studied. Importantly, the deacetylation reaction has been demonstrated to be tightly coupled with the cleavage of NAD into nicotinamide and ADP-ribose and the formation of a previously unidentified compound, O-acetyl-ADP-ribose 11, 12. Crystal structures of yeast, mammalian, and archaeobacterial sirtuins have been determined, and the structure-based catalytic mechanism of this reaction have been proposed 13–21.
In mammals, there are seven sirtuin family members, named SIRT1 through SIRT7 (Table 1). Numerous target proteins have been identified for sirtuins, particularly for the mammalian SIR2 ortholog SIRT1, and it has been established that NAD-dependent deacetylation of those target factors by sirtuins plays a critical role in the regulation of fundamental biological responses to nutritional and environmental stimuli in each subcellular compartment 22–26. SIR2 and its orthologs have also emerged as critical regulators for aging and longevity in model organisms, such as yeast, worms, and flies 27–32. In certain genetic backgrounds, they mediate anti-aging and life span-extending effects of caloric restriction, a dietary regimen low in calories without malnutrition that delays aging and extends life span in a wide variety of organisms 30, 33–36.
Table 1.
Mammalian sirtuins
| Enzymatic activity |
Homologues | Subcellular localization |
Function | |
|---|---|---|---|---|
| SIRT1 | Deacetylase | Sir2p (S. cerevisiae) Hst1p (S. cerevisiae) SIR-2.1(C. elegans) dSIR2 (D. melanogaster) |
Nuclear, Cytoplasmic | Glucose production (liver) Fatty acid oxidation (liver) Cholesterol regulation (liver) Fatty acid mobilization (WAT) Adipokine regulation (WAT) Fatty acid oxidation (skeletal muscle) Insulin secretion (pancreatic β cells) Neuroprotection (brain) Regulation of cellular differentiation Stress resistance&apoptosis control Mediator for caloric restriction |
| SIRT2 | Deacetylase | Hst2p (S. cerevisiae) SIRT2 (D. melanogaster) |
Cytoplasmic, Nuclear | Tublin deacetylation Cell cycle control |
| SIRT3 | Deacetylase | Mitochondrial | Mitochondrial protein deacetylation Acetate metabolism regulation ATP production |
|
| SIRT4 | ADP-ribosyltransferase | SIR-2.2 (C. elegans) SIR-2.3 (C. elegans) SIRT4 (D. melanogaster) |
Mitochondrial | Amino acid-stimulated insulin secretion (pancreatic β cells) |
| SIRT5 | Deacetylase | Mitochondrial | Urea cycle regulation (liver) | |
| SIRT6 | ADP-ribosyltransferase Deacetylase |
SIR-2.4 (C. elegans) SIRT6 (D. melanogaster) |
Nuclear | Base excision repair Telomeric chromatin structure NF-κB regulation |
| SIRT7 | Deacetylase | SIRT7 (D. melanogaster) | Nucleolar | Pol I transcription |
These studies also potentiated screening for small molecule sirtuin activators. These screens initially identified a plant polyphenolic compoud resveratrol, but more potent activators were indentified later 28, 32, 37–42. These compounds have so far provided a hope of new therapeutic interventions against age-associated complications, such as type 2 diabetes and Alzheimer’s disease 43, 44.
Lastly, the biological significance of NAD biosynthetic pathways for the regulation of sirtuin enzymatic activity has been brought front and center 33, 45–53. In particular, nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme in a major NAD biosynthetic pathway from nicotinamide in mammals, has recently been shown to play important roles in a variety of biological events related to sirtuin biology, metabolism, cancer, and immune response 54, 55. With all these great advances, there are currently two major questions in the field of sirtuin biology: 1) Do mammalian sirtuins play critical roles in the pathogenesis of major age-associated diseases, such as type 2 diabetes, neurodegenerative diseases, osteoporosis, and others? 2) Do mammalian sirtuins, particularly SIRT1, indeed function as a key regulator for aging and longevity in mammals? In the next two sections, we will address these questions and summarize our current knowledge on the diverse functions of mammalian sirtuins.
Mammalian sirtuins and age-associated metabolic complications
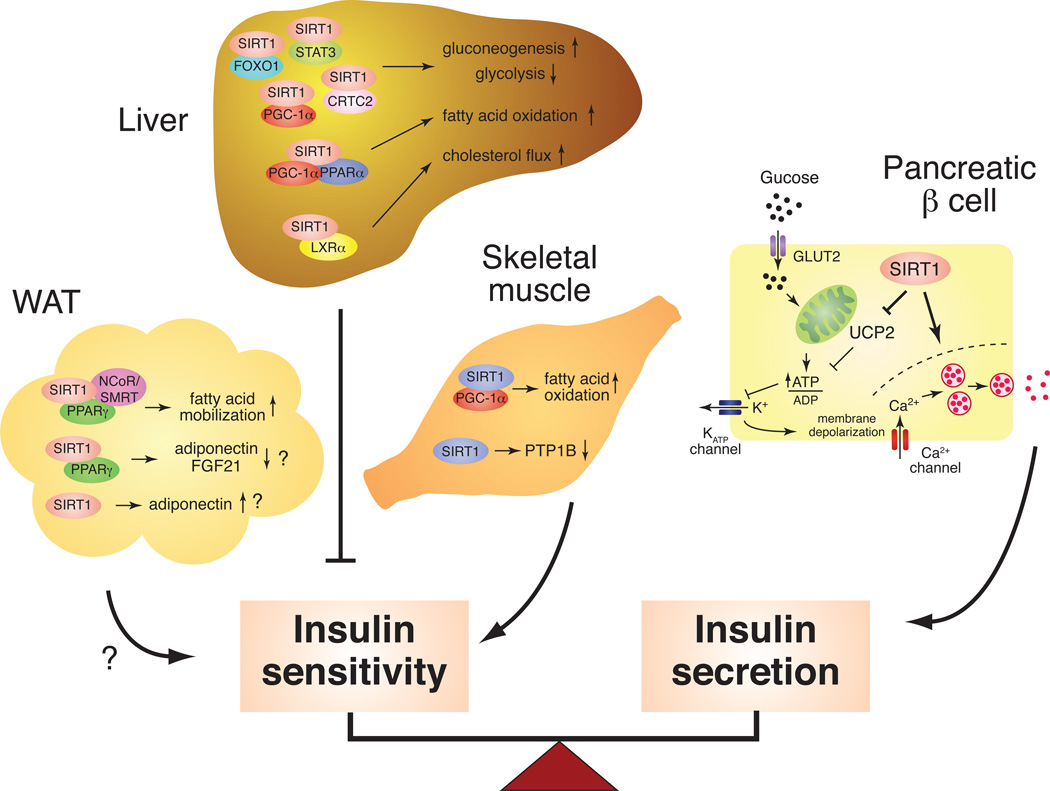
It is not yet clear whether mammalian sirtuins determine longevity, but they have been implicating in retarding aging, as defined by degenerative processes leading to diseases. For example, increasing lines of evidence have firmly established that mammalian sirtuins, particularly SIRT1, regulate metabolic responses to changes in nutritional availability in multiple tissues 23, 24, 26, implying that mammalian sirtuins play essential roles in the pathogenesis of age-associated metabolic diseases. In the pathogenesis of type 2 diabetes, a delicate balance between insulin sensitivity and secretion is compromised by both environmental and genetic factors 56–58, resulting in the development of insulin resistance and β cell dysfunction. SIRT1 contributes to both aspects of type 2 diabetes pathogenesis in liver, skeletal muscle, adipose tissue, and pancreatic β cells (Figure 1).
Figure 1.
The tissue-specific metabolic functions of SIRT1 in the regulation of insulin sensitivity and secretion. SIRT1 plays a critical role in maintaining a delicate balance between insulin sensitivity and secretion in major metabolic tissues, such as liver, skeletal muscle, white adipose tissue (WAT), and pancreatic β cells. In the liver, SIRT1 regulates glucose production through PGC-1α, FOXO1, CRTC2, and STAT3 and appears to repress insulin sensitivity. SIRT1 also regulates LXRα and PPARα for cholesterol and fatty acid metabolism. In skeletal muscle, SIRT1 improves insulin sensitivity by increasing fatty acid oxidation through PGC-1α and repressing the expression of PTB1B. In WAT, SIRT1 promotes fatty acid mobilization by repressing PPARγ function and also regulates the production/secretion of adiponectin and FGF21 through FOXO1 and/or PPARγ. In pancreatic β cells, SIRT1 promotes glucose-stimulated insulin secretion and likely contributes to β cell adaptation in response to insulin resistance.
SIRT1 in liver
SIRT1 regulates glucose production, fatty acid oxidation, and cholesterol flux in the liver. In response to fasting, SIRT1 enhances and represses gluconeogenesis and glycolysis, respectively, by interacting with and deacetylating peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), FOXO1, CRTC2 (a.k.a. TORC2), and STAT3, in an NAD-dependent manner 59–63. SIRT1 also promotes fatty acid oxidation through PGC-1α and peroxisome proliferator-activated receptor α (PPARα), a nuclear receptor that controls physiological responses to fasting 64. Furthermore, SIRT1 regulates cholesterol flux by deacetylating and activating LXRα, a critical nuclear receptor that controls cholesterol and lipid homeostasis 65. As a total sum of these responses, SIRT1 appears to contribute to the negative regulation of insulin sensitivity in the liver because adenovirus-mediated hepatic SIRT1 knockdown results in improved glucose and insulin tolerance in mice, while hepatic SIRT1 overexpression causes moderate glucose intolerance (Figure 1) 60. This notion is also consistent with a report that knockdown of SIRT1 in the liver decreases fasting plasma glucose and increases hepatic insulin sensitivity in a type 2 diabetes rat model 61. High fat diet (HFD)-fed liver-specific SIRT1 knockout (LKO) mice also show improved glucose tolerance and lower levels of blood glucose and insulin 66. On the other hand, it is reported that HFD-fed SIRT1 LKO mice also develop hepatic steatosis, hepatic inflammation, and endoplasmic reticulum stress 64. It is possible that differences in the cholesterol and fat content between the various HFDs exert subtly different effects on liver metabolism, and further investigation will be necessary to clarify the hepatic function of SIRT1 in diet-induced diabetes models.
SIRT1 in skeletal muscle
SIRT1 induces mitochondrial fatty acid oxidation in response to nutrient deprivation through deacetylation of PGC-1α67. Given that intramyocellular fatty acid metabolism and the expression of PGC-1α and mitochondrial OXPHOS genes are reduced in skeletal muscle of insulin-resistant or type 2 diabetes patients 68–70, SIRT1 might contribute in this way to the improvement of insulin sensitivity in skeletal muscle (Figure 1). Consistent with this notion, SIRT1 improves insulin sensitivity through the transcriptional repression of the protein tyrosine phosphatase 1B (PTP1B) gene in skeletal myotube cells 71. PTP1B is a key insulin receptor phosphatase, and PTP1Bdeficient mice have been shown to be more insulin-sensitive and more resistant to dietinduced obesity compared to controls 72. Although these reported effects of SIRT1 should be validated in vivo, these findings indicate that SIRT1 might play an important role in improving insulin sensitivity in skeletal muscle.
SIRT1 in white adipose tissue
SIRT1 triggers lipolysis and promotes free fatty acid mobilization in response to fasting by repressing PPARγ, a nuclear receptor that promotes adipogenesis 73. SIRT1 also regulates the production and/or the secretion of insulin sensitizing factors, such as adiponectin and FGF21, through the regulation of FOXO1 and PPARγ 74–76. Nonetheless, further investigation with in vivo models will be necessary to determine the actual effect of SIRT1 in adipose tissue on systemic insulin sensitivity (Figure 1).
SIRT1 in pancreatic β cells
SIRT1 positively regulates glucose-stimulated insulin secretion (GSIS) in part by repressing the expression of uncoupling protein 2 (Ucp2), a mitochondrial inner membrane proton pore that uncouples respiration from ATP production, and increasing cellular ATP levels 77, 78. Indeed, pancreatic β cell-specific SIRT1-overexpressing (BESTO) transgenic mice show enhanced insulin secretion and improved glucose tolerance in response to glucose 78. Furthermore, BESTO mice are still able to maintain significantly improved glucose tolerance with enhanced GSIS compared to controls under a HFD condition 79. Consistent with this observation, SIRT1 also plays an important role in protecting pancreatic β cells from metabolic stress- and cytokine-induced β cell death by deacetylating FOXO1 and the p65 subunit of NF-κB, respectively 80, 81. Therefore, SIRT1 plays an important role in protecting pancreatic β cells from their dysfunction caused by increasing peripheral insulin resistance (Figure 1).
SIRT1 and type 2 diabetes
Does SIRT1 promote or prevent type 2 diabetes at a systemic level? Accumulating bodies of evidence have so far suggested that SIRT1 functions to provide overall protection against type 2 diabetes. SIRT1-overexpressing transgenic mice using large genomic fragments that contain the entire Sirt1 gene locus show significant protection from the adverse effects of HFD or normal aging on metabolism, including hepatic inflammation and impaired insulin sensitivity 74, 82. Additionally, resveratrol and new SIRT1-activating non-polyphenolic compounds are able to improve glucose homeostasis and insulin sensitivity in diet-induced and genetic type 2 diabetes animal models 37–40, 42. In humans, it has recently been reported that certain genetic variations of the SIRT1 gene influence survival of subjects with type 2 diabetes in interaction with dietary niacin and smoking and risk of obesity in Dutch populations 83, 84. Taken together, all these findings strongly suggest that SIRT1 can protect animals and possibly humans from type 2 diabetes.
Other mammalian sirtuins and metabolic complications
The connection between other mammalian sirtuin members and age-associated metabolic complications is still unclear. Mitochondrial sirtuins SIRT3-5 might be the next targets for intensive studies on their potential connection to metabolic diseases. SIRT3 deacetylates and activates the mitochondrial enzyme acetyl-CoA synthetase 2 (AceCS2) 85, 86. It has recently been reported that AceCS2-deficient mice exhibit a significant defect in acetate oxidation necessary for the generation of ATP and heat under low-glucose or ketogenic conditions 87. Consistent with this finding, SIRT3-deficient mice also show a significant reduction in basal ATP levels in multiple tissues 88. On the other hand, SIRT4 negatively regulates amino acid-stimulated insulin secretion by ADP-ribosylating and inhibiting glutamate dehydrogenase in pancreatic β cells 89. Finally, SIRT5 deacetylates and activates CPS1, the first and committed step in the urea cycle for ammonia detoxification 49. This activity is particularly important when diets are limiting in energy, forcing the use of amino acids for energy and the generation of excess ammonia via their catabolism. Therefore, it will be of great interest to examine metabolic responses of SIRT3- and SIRT4-deficient mice to HFD conditions, which might provide critical insights into their connection to the pathogenesis of type 2 diabetes.
Mammalian sirtuins and their connection to the regulation of aging and longevity
SIRT1 and physiological aspects of aging
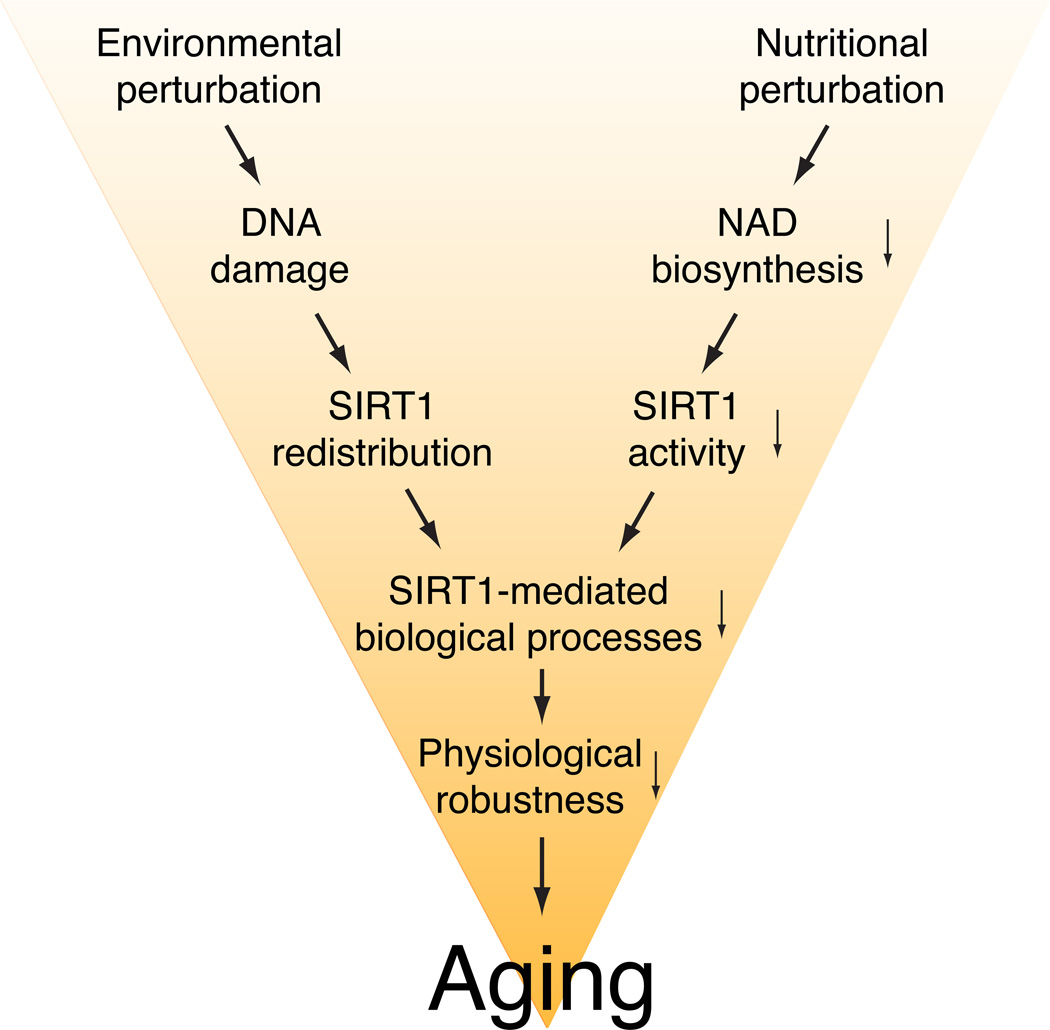
Recent studies have provided significant support for the importance of SIRT1 in the induction of age-associated physiological changes (Figure 2). Most recently, it has been demonstrated that oxidative stress-induced redistribution of SIRT1 causes the desilencing of SIRT1-bound target genes in mouse embryonic stem (ES) cells 90. This damage-induced SIRT1 redistribution requires DNA damage signaling through a mammalian PI3-kinase ATM and histone H2AX. Importantly, these SIRT1-bound target genes that are derepressed by oxidative stress in mouse ES cells are also derepressed in aged mouse brain, and SIRT1 overexpression is able to suppress these age-associated changes, suggesting that the damage-induced SIRT1 redistribution might trigger certain age-associated physiological changes in mouse brain (Figure 2) 90, 91.
Figure 2.
The role of SIRT1 in the induction of age-associated physiological changes. A variety of environmental and nutritional perturbations likely induce DNA damage and a reduction in NAD biosynthesis over time, resulting in the redistribution of SIRT1 and the reduction in SIRT1 activity in many different tissues. These events might affect SIRT1- mediated biological processes in tissues, causing the reduction in physiological robustness and aging.
Another line of evidence is that SIRT1 activity is significantly reduced in aged β cells, likely due to a significant decrease in NAMPT-mediated systemic NAD biosynthesis over age 54, 55, resulting in the reduction in GSIS in aged β cells 79. Indeed, a progressive age-associated decline in β cell function has been suggested to be one of the major contributing factors to the pathogenesis of type 2 diabetes 92–94. Interestingly, administration of nicotinamide mononucleotide (NMN), a reaction product of NAMPT and a key NAD intermediate, restores higher GSIS in both aged wild-type and BESTO females 79. These findings suggest that the age-associated reduction in SIRT1 activity due to NAD insufficiency plays a critical role in inducing the age-associated decrease in GSIS in pancreatic β cells (Figure 2) 95, 96.
SIRT1 and caloric restriction (CR)
CR is well known to retard aging and extend life span in many organisms, and recent studies have strongly suggested that SIRT1 mediates adaptive responses to CR in mammals. For example, whereas wild-type CR mice show a significant increase in physical activity compared to ad libitum-fed controls, SIRT1-deficient mice do not exhibit such an increase 97. The CR-induced enhancement of mitochondrial biogenesis by nitric oxide-induced up-regulation of SIRT1 is also blunted in endothelial nitric oxide synthase (eNOS)-deficient mice 98. Furthermore, it has recently been reported that SIRT1-deficient mice are metabolically inefficient and unable to adapt to CR normally 99. SIRT1-overexpressing transgenic (SIRT1-KI) mice in which the SIRT1 cDNA is knocked into the β-actin locus display phenotypes that mimic some of the physiological changes in response to CR, including decreased blood insulin and glucose levels, improved glucose tolerance, reduced fat mass and circulating free fatty acid and leptin levels, reduced total blood cholesterol levels, enhanced oxygen consumption, improved activity in rotarod tests, and delayed reproductive timing. Two other independent lines of SIRT1 whole-body transgenic mice also show significant protection from the adverse effects of HFD or normal aging on metabolism, which CR has also been reported to convey 74, 82. Given that CR delays the onset of and decreases the incidence of age-associated pathophysiological changes, these findings support the notion that SIRT1 counteracts the detrimental effects of aging in mammals.
Does SIRT1 promote mammalian longevity?
Based on these findings, one could expect that increasing SIRT1 dosage or activity throughout a body might extend life span in mammals. Unfortunately, life span results of currently existing SIRT1 transgenic mice have not yet been reported. However, an extra caution might be necessary because the roles of SIRT1 in each organ and tissue are significantly diverged so that beneficial effects of enhanced SIRT1 activity on life span in some organs/tissues might be balanced out with its unfavorable effects in others. Thus, SIRT1 whole-body transgenic mice might not show a significant extension of life span. Indeed, it has been reported that long-term treatment of regular chow-fed mice with resveratrol fails to extend life span, even though it recapitulates transcriptional profiles induced by CR in some organs/tissues100.
It is possible that SIRT1 dosage or activity is down-regulated in certain tissues by CR, as has been reported in the liver 66. Moreover, recent findings indicated that SIRT1 levels could be both up- and down-regulated locally within the same tissues, such as brain 101. Therefore, the idea that global activation of SIRT1 could extend the life span might be too simplistic. It will be of great importance to examine the effects of SIRT1 enhancement on pathophysiological aging of tissues and their propensity for diseases as well as longevity.
Sirtuin-targeted anti-aging interventions
Sirtuin activator compounds (STACs)
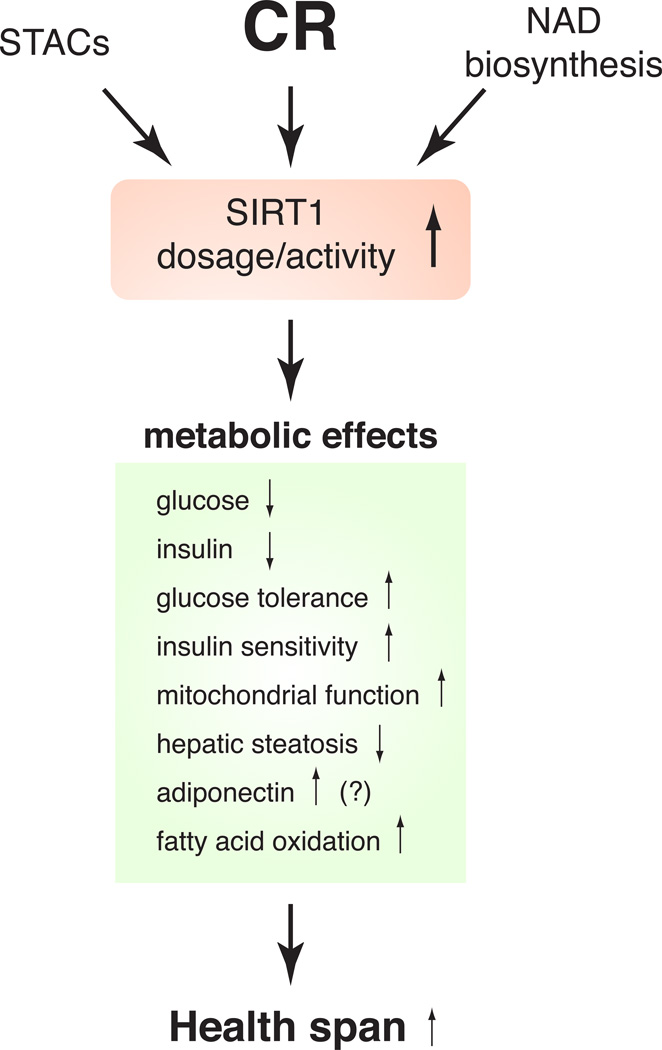
For the past decade, the developments summarized above have raised a broad interest in sirtuin-targeted anti-aging interventions with a hope to mimic beneficial effects of CR (Figure 3)44, 102, 103. The first case of the concept was the finding that a group of polyphenolic compounds, such as resveratrol, fisetin, and butein, was able to activate SIR2 and its orthologs in vitro and extend the life spans of yeast, worms, and flies 28, 32. Following these findings, it has been reported that different doses of resveratrol counteract detrimental effects of HFD on metabolism and other physiological parameters and increase survival 37, 39. Resveratrol also induces gene expression profiles similar to those induced by every-other-day feeding, which is known to convey physiological effects similar to those in CR, particularly in liver and skeletal muscle 100. Furthermore, new non-polyphenolic STACs have recently been shown to improve glucose homeostasis and insulin sensitivity in diet-induced and genetically obese rodent models, implicating a novel therapeutic intervention for the treatment of type 2 diabetes 40. In particular, SRT1720, one of those new STACs, has been further examined and shown to mediate beneficial effects on energy metabolism in obese, insulin-resistant mice 38, 41, 42. These findings indicate that STACs might function as potential CR mimetics, at least in part, through the activation of SIRT1 and be effective to treat age-associated metabolic complications, such as type 2 diabetes (Figure 3). However, whether STACs directly activate SIRT1 in vivo is still controversial. Recent studies have shown that AMP-activated protein kinase (AMPK) plays a critical role in mediating the effect of resveratrol 104, 105. Concern has also been raised by studies on the substrate specificity of SIRT1-activating effects in vitro 106–109. Therefore, careful assessments will be necessary to clarify the mechanism of action of STACs.
Figure 3.
SIRT1 as a molecular target for pharmaceutical and nutriceutical anti-aging interventions. Small molecule SIRT1-activating compounds (STACs) and the enhancement of NAD biosynthesis could mimic metabolic responses to caloric restriction (CR) through SIRT1 activation, leading to a possible extension of health span in mammals.
NAD biosynthesis and sirtuins
Accumulating bodies of evidence have demonstrated that NAMPT-mediated NAD biosynthesis plays an important role in the regulation of SIRT1 activity in a number of cell types, such as pancreatic β cells 79, vascular smooth muscle cells 47, 110, skeletal myoblasts 46, cardiac myocytes 48, 111, and others 50, 51, 112, 113. Most recently, SIRT1 has been connected to the circadian transcriptional regulation mediated by key circadian transcription factors CLOCK and BMAL1 114, 115. Furthermore, the CLOCK:BMAL1 complex produces the circadian oscillation of NAMPT and NAD levels in vivo, thus comprising a novel circadian clock feedback loop involving NAMPT/NAD and SIRT1/CLOCK:BMAL1 50, 51, 116. Because the core molecular clock machinery is one of the most powerful modifiers of metabolism 117, 118, it is conceivable that enhancing NAD biosynthesis at a systemic level by administering key NAD intermediates, such as NMN, might be able to activate SIRT1 and convey beneficial effects on metabolism and other physiological processes (Figure 3). Indeed, this idea has been demonstrated at least in Nampt-heterozygous mice and aged BESTO mice, in both of which NMN significantly improves GSIS and glucose tolerance 79. The mitochondrial sirtuins SIRT3-5 can also be activated by increasing NAMPT-mediated NAD biosynthesis 49, 119. Therefore, it will be of great importance to examine whether intermediates in NAD biosynthesis can be used as an effective anti-aging nutriceutical intervention to ameliorate age-associated metabolic complications and thus improve the quality of life 120.
Concluding remarks
The past 10 years have been exciting and innovative for sirtuin-related research. Sirtuins have created a new arena where we can further investigate an intimate connection between NAD biology, metabolism, and aging. Many novel NAD-dependent biological processes have been revealed, and our knowledge on the dynamic interaction between protein acetylation and energy metabolism has been revolutionized. In this respect, it is striking that acetyl-CoA is both the product of carbohydrate and fat metabolism and the substrate for protein acetylation. Yet several important questions remain: What are the functions of other mammalian sirtuin family members in each subcellular compartment? How is the connection between sirtuins and NAD biosynthesis regulated in each subcellular compartment and through the whole body? Are there any other known or even unknown NAD intermediates and metabolites that are involved in the regulation of sirtuin activities? Can we really develop an effective sirtuin-targeted intervention to improve the quality of life in our aging society? Those questions will hopefully be addressed in the next 10 years, and we may be able to enjoy our lives with anti-aging pharmaceutical and nutriceutical interventions that target sirtuins.
Acknowledgments
We apologize to those whose work is not cited due to space limitations. We thank members in the Imai lab and the Guarente lab for critical discussions and suggestions. S.I. is supported by grants from the National Institute on Aging (AG024150), Ellison Medical Foundation, and Longer Life Foundation. L.G. is supported by the Glenn Foundation and grants from NIH and the American Asthma Foundation. Both S.I. and L.G. serve as scientific advisory board members for Sirtris pharmaceuticals, a GSK company.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klar AJS, et al. MAR1-A regulator of the HMa and HMα loci in Saccharomyces cerevisiae. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 3.Trzebiatowski JR, Escalante-Semerena JC. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J. Biol. Chem. 1997;272:17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- 4.Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 5.Frye RA. Characterization of five human cDNAs with homology to yeast SIR2 gene: Sir2-like proteins (Sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 6.Tanny JC, et al. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 7.Imai S, et al. Sir2: An NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harbor Symp. Quant. Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 8.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 9.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner KG, et al. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avalos JL, et al. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Avalos JL, et al. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 15.Finnin MS, et al. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 16.Hawse WF, et al. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson MD, Denu JM. Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 18.Jin L, et al. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J. Biol. Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauve AA, et al. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 20.Schuetz A, et al. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007;15:377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhao K, et al. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 22.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 23.Dali-Youcef N, et al. Sirtuins: the 'magnificent seven', function, metabolism and longevity. Ann. Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 24.Imai S, Guarente L. Sirtuins: A universal link between NAD, metabolism, and aging. In: Guarente L, editor. The Molecular Biology of Aging. Cold Spring Habor Laboratory Press; 2007. pp. 39–72. [Google Scholar]
- 25.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Astrom SU, et al. The Drosophila melanogaster sir2+ gene Is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 29.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 32.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 33.Anderson RM, et al. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S-J, et al. Life span extension by calorie restriction in S. cerevisiae requires NAD and SIR2. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 35.Lin S-J, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Lagouge M, et al. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith JJ, et al. Small molecule activators of SIRT1 replicate signaling pathways triggered by calorie restriction in vivo. BMC Syst. Biol. 2009;3:31. doi: 10.1186/1752-0509-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki Y, et al. Treatment with SRT1720, a SIRT1 Activator, Ameliorates Fatty Liver with Reduced Expression of Lipogenic Enzymes in MSG Mice. Am. J. Physiol. Endocrinol. Metab. 2009 doi: 10.1152/ajpendo.90997.2008. Epub on Sept 1. [DOI] [PubMed] [Google Scholar]
- 43.Lamming DW, et al. Small molecules that regulate lifespan: evidence for xenohormesis. Mol. Microbiol. 2004;53:1003–1009. doi: 10.1111/j.1365-2958.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- 44.Westphal CH, et al. A therapeutic role for sirtuins in diseases of aging? Trends Biochem. Sci. 2007;32:555–560. doi: 10.1016/j.tibs.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Anderson R, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 46.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho C, et al. SIRT1 markedly extends replicative lifespan if NAD(+) salvage is enhanced. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.08.031. Epub on Aug 28. [DOI] [PubMed] [Google Scholar]
- 48.Hsu CP, et al. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ. Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa T, et al. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramsey KM, et al. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Revollo JR, et al. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 53.Revollo JR, et al. Nampt/PBEF/visfatin regulates insulin secretion in β cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garten A, et al. Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol. Metab. 2009;20:130–138. doi: 10.1016/j.tem.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imai S. Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 2009;15:20–28. doi: 10.2174/138161209787185814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in β-cell function. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- 57.Kahn SE, et al. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 58.Weir GC, Bonner-Weir S. Five stages of evolving beta-cell dysfunction during progression to diabetes. Diabetes. 2004;53(Suppl 3):S16–S21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 59.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 60.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erion DM, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc. Natl. Acad. Sci. USA. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nie Y, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 66.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petersen KF, et al. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. EnglJMed. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun C, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 72.Elchebly M, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 73.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/EBPalpha transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, et al. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Mol. Cell. Biol. 2008;28:188–200. doi: 10.1128/MCB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moynihan KA, et al. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 79.Ramsey KM, et al. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in β cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kitamura YI, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 81.Lee JH, et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009;58:344–351. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfluger PT, et al. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zillikens MC, et al. SIRT1 genetic variation is related to body mass index and risk of obesity. Diabetes. 2009 doi: 10.2337/db09-0536. Epub on Sept 11, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zillikens MC, et al. SIRT1 genetic variation and mortality in type 2 diabetes: interaction with smoking and dietary niacin. Free Radic. Biol. Med. 2009;46:836–841. doi: 10.1016/j.freeradbiomed.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 85.Hallows WC, et al. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwer B, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sakakibara I, et al. Fasting-induced hypothermia and reduced energy production in mice lacking acetyl-CoA synthetase 2. Cell Metab. 2009;9:191–202. doi: 10.1016/j.cmet.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 88.Ahn BH, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 90.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell. Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- 92.Basu R, et al. Mechanisms of the age-associated deterioration in glucose tolerance: contribution of alterations in insulin secretion, action, and clearance. Diabetes. 2003;52:1738–1748. doi: 10.2337/diabetes.52.7.1738. [DOI] [PubMed] [Google Scholar]
- 93.Iozzo P, et al. Independent influence of age on basal insulin secretion in nondiabetic humans. European Group for the Study of Insulin Resistance. J. Clin. Endocrinol. Metab. 1999;84:863–868. doi: 10.1210/jcem.84.3.5542. [DOI] [PubMed] [Google Scholar]
- 94.Muzumdar R, et al. Decrease in glucose-stimulated insulin secretion with aging is independent of insulin action. Diabetes. 2004;53:441–446. doi: 10.2337/diabetes.53.2.441. [DOI] [PubMed] [Google Scholar]
- 95.Imai S. The NAD World: a new systemic regulatory network for metabolism and aging - Sirt1, systemic NAD biosynthesis, and their importance. Cell. Biochem. Biophys. 2009;53:65–74. doi: 10.1007/s12013-008-9041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Imai S, Kiess W. Therapeutic potential of SIRT1 and NAMPT-mediated NAD biosynthesis in type 2 diabetes. Front. Biosci. 2009;14:2983–2995. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen D, et al. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 98.Nisoli E, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 99.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pearson KJ, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen D, et al. The role of calorie restriction and SIRT1 in prion-mediated neurodegeneration. Exp. Gerontol. 2008;43:1086–1093. doi: 10.1016/j.exger.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 103.Milne JC, Denu JM. The Sirtuin family: therapeutic targets to treat diseases of aging. Curr. Opin. Chem. Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 104.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. USA. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Um JH, et al. AMPK-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2009 Epub on Nov 23, 2009. [Google Scholar]
- 106.Borra MT, et al. Mechanism of human SIRT1 activation by resveratrol. J Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 107.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 108.Pacholec M. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2005 doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beher D, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 110.van der Veer E, et al. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ. Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- 111.Alcendor RR, et al. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ. Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 112.Skokowa J, et al. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat. Med. 2009;15:151–158. doi: 10.1038/nm.1913. [DOI] [PubMed] [Google Scholar]
- 113.Zhang T, et al. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J. Biol. Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 115.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Imai S. "Clocks" in the NAD World: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim. Biophys. Acta. 2009 doi: 10.1016/j.bbapap.2009.10.024. Epub on Nov 6, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Green CB, et al. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ramsey KM, et al. The clockwork of metabolism. Annu. Rev. Nutr. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 119.Yang H, et al. Nutrient-sensitive mitochondrial NAD(+) levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Imai S. A possibility of nutriceuticals as an anti-aging intervention: Activation of sirtuins by promoting mammalian NAD biosynthesis. Pharmacol. Res. 2010 doi: 10.1016/j.phrs.2010.01.006. Epub on Jan 18. [DOI] [PMC free article] [PubMed] [Google Scholar]