SUMMARY
Circadian rhythms govern a large array of metabolic and physiological functions. The central clock protein CLOCK has HAT properties. It directs acetylation of histone H3 and of its dimerization partner BMAL1 at Lys537, an event essential for circadian function. We show that the HDAC activity of the NAD+-dependent SIRT1 enzyme is regulated in a circadian manner, correlating with rhythmic acetylation of BMAL1 and H3 Lys9/Lys14 at circadian promoters. SIRT1 associates with CLOCK and is recruited to the CLOCK:BMAL1 chromatin complex at circadian promoters. Genetic ablation of the Sirt1 gene or pharmacological inhibition of SIRT1 activity lead to disturbances in the circadian cycle and in the acetylation of H3 and BMAL1. Finally, using liver-specific SIRT1 mutant mice we show that SIRT1 contributes to circadian control in vivo. We propose that SIRT1 functions as an enzymatic rheostat of circadian function, transducing signals originated by cellular metabolites to the circadian clock.
INTRODUCTION
The epigenetic basis of many developmental, physiological, and metabolic processes is manifest. Epigenetic mechanisms control gene expression by potentially reversible changes in DNA methylation and chromatin structure. The remodeling of chromatin is largely elicited by enzyme-catalyzed posttranslational modifications of the core histone N-terminal tails (Kouzarides, 2007; Li et al., 2007a; Peterson and Laniel, 2004; Strahl and Allis, 2000). These include acetylation, poly(ADP-ribosylation), ubiquitination, methylation, and phosphorylation and represent critical regulatory events of a large array of nuclear responses. Unique combinations of these modifications, for which the “histone code” hypothesis has been formulated (Strahl and Allis, 2000), induce conformational changes of chromatin, rendering it permissive to transcription, silencing, DNA replication, and repair (Cheung et al., 2000a; Kouzarides, 2007; Kurdistani and Grunstein, 2003; Li et al., 2007a; Strahl and Allis, 2000).
Histone acetylation is recognized as one of the most prominent epigenetic marks leading to activation of gene expression (Strahl and Allis, 2000). Acetylation of the 3-amino groups of specific lysine residues in the N termini of core histones is generally associated with transcription activity, as it is thought to induce an open chromatin conformation that allows the transcription machinery access to promoters (Cheung et al., 2000a; Li et al., 2007a; Struhl, 1998). Indeed, acetylation of lysines in histones neutralizes the positive electric charge between the negatively charged DNA backbone and tips the balance toward relaxing chromatin. Deacetylation, on the other hand, would shift the balance back to condensing chromatin and silencing gene expression. The enzymes that elicit these critical transitions are histone acetyltransferases (HAT) and histone deacetylases (HDAC). HDAC-mediated deacetylation of histones correlates with gene silencing (Grunstein, 1997; Struhl, 1998; Wade and Wolffe, 1997; Workman and Kingston, 1998). HDACs have also been implicated in the reversible acetylation of nonhistone proteins, including p53 (Luo et al., 2001; Vaziri et al., 2001), Hsp90 (Kovacs et al., 2005), MyoD (Mal et al., 2001), and E2F1 (Martinez-Balbas et al., 2000). Mammalian HDACs have been classified into four classes based on their structure and regulation (Yang and Seto, 2008). There are seven mammalian enzymes constituting class III; these are homologs of yeast Sir2 (silencing information regulator) and are known as SIRT1 to SIRT7. These proteins are structurally distinct from the other HDACs and have the property of dynamically sensing cellular energy metabolism (Bordone and Guarente, 2005). Indeed, unlike other HDACs, SIRT proteins catalyze a unique reaction that requires the coenzyme NAD+ (nicotinamide adenine dinucleotide). In this reaction, nicotinamide (NAM) is liberated from NAD+ and the acetyl group of the substrate is transferred to cleaved NAD+, generating the metabolite O-acetyl-ADP ribose (Sauve et al., 2006). Due to the NAD+ dependency, SIRTs are thought to constitute one of the functional links between metabolic activity and genome stability and, finally, aging (Bishop and Guarente, 2007).
In yeast, the Sir2 complex mediates transcriptional silencing at telomeres and regulates the pace of aging (Chopra and Mishra, 2005; Oberdoerffer and Sinclair, 2007). Because of the NAD+ requirement for Sir2 deacetylase activity, it is evident that silencing is likely coupled to the metabolic cycle of cells. In C. elegans, one of the Sir2 orthologs, Sir2.1, has been shown to prevent aging (Tissenbaum and Guarente, 2001).
SIRT1, the mammalian ortholog of Sir2, is a nuclear protein that occupies a privileged position in the cell and governs critical metabolic and physiological processes. SIRT1 helps cells to be more resistant to oxidative or radiation-induced stress (Brunet et al., 2004; Luo et al., 2001), promotes mobilization of fat from white adipose tissues, an event that contributes to extending the life span (Picard et al., 2004), and mediates the metabolism of energy sources in metabolically active tissues (Lagouge et al., 2006; Rodgers et al., 2005). At the level of chromatin, SIRT1 enzymatic activity preferentially targets histone H3 at Lys9 and Lys14 and histone H4 at Lys16 (Imai et al., 2000). In addition, a number of nonhistone proteins, including p53 (Luo et al., 2001; Vaziri et al., 2001), FOXO3 (Brunet et al., 2004; Motta et al., 2004), PGC-1a (Nemoto et al., 2005; Rodgers et al., 2005), and LXR (Li et al., 2007a), are regulated by SIRT1-mediated deacetylation, stressing the pivotal function that this regulator plays in cellular control and responses.
A remarkable array of metabolic and physiological processes display daily oscillations (Panda et al., 2002; Storch et al., 2002; Ueda et al., 2002), and an intimate interplay exists between circadian clocks and metabolic rhythms in all organisms (Wijnen and Young, 2006). The discovery that a core element of the circadian clock machinery, the protein CLOCK, is an enzyme with HAT activity (Doi et al., 2006) revealed the crucial role that chromatin remodeling plays in the circadian regulation of gene expression (Hardin and Yu, 2006; Nakahata et al., 2007). More recently, the finding that CLOCK specifically also acetylates nonhistone targets, such as its own partner BMAL1, suggested that it may control a number of physiological cellular functions (Hirayama et al., 2007). The intrinsic nature of SIRT1 as a NAD+-dependent HDAC prompted us to explore the possibility that SIRT1 could participate in circadian control by regulating the HAT function of CLOCK. This would uncover a unique example of control of gene expression by metabolites (Ladurner, 2006).
Here we report that the HDAC activity of SIRT1 is regulated in a circadian manner in cultured cells and in the liver. SIRT1 physically associates with CLOCK and contributes to the acetylated state of CLOCK targets, such as Lys9/Lys14 in the tail of histone H3 and Lys537 in the BMAL1 protein. CLOCK, BMAL1, and SIRT1 colocalize in a chromatin-associated regulatory complex at promoters of clock-controlled genes. Pharmacological inhibition of SIRT1 activity by NAM and the drug splitomicin causes a loss in stringency of circadian gene expression, an effect equally observed in mouse embryo fibroblasts (MEFs) derived from Sirt1 null mice. Importantly, this effect is paralleled by a significant reduction in the oscillation of H3 and BMAL1 acetylation. Finally, using tissue-specific mutant mice, in which the Sirt1 gene is mutated uniquely in the liver, we demonstrate that SIRT1 contributes to circadian regulation in vivo. We propose that SIRT1 functions as an enzymatic rheostat of CLOCK function, thereby transducing signals originated by cellular metabolites to the circadian machinery.
RESULTS
SIRT1 Deacetylase Activity Is Circadian
The CLOCK protein is one of the few core circadian regulators whose levels do not oscillate in most settings (Lee et al., 2001). Thus, we predicted that its HAT activity would oscillate in a circadian manner, thereby explaining the physiological remodeling of chromatin (Doi et al., 2006). An alternative scenario implicates a regulated HDAC, whose activity may function as rheostat of the HAT’s function of CLOCK.
To assess whether SIRT1 may be regulated in a circadian manner we prepared RNA and protein extracts from serum-stimulated cultured MEFs and from mouse liver at various Zeitgeber times (ZT). In both cases, the transcript and protein levels of SIRT1 remained constant, as determined using two anti-SIRT1-specific antibodies (Figure 1A; see also later, Figure 7B) and reverse transcription (RT)-PCR (Figures 1B and 1C, where dbp is shown as a control from the same RNA preparations). We have also determined the levels of SIRT1 in nuclear fractions prepared in various manners. Again SIRT1 protein levels showed either modest or no oscillation (Figure S1 available online). Thus, we turned to determining whether SIRT1 deacetylase activity may oscillate.
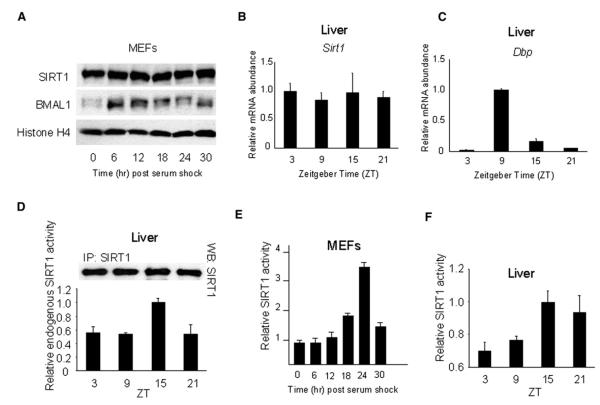
Figure 1. SIRT1 Deacetylase Activity Is Circadian in Serum-Shocked MEFs and in the Liver.
(A) Endogenous SIRT1 and BMAL1 expression in MEFs after serum shock were determined by western blot.
(B and C) Sirt1 (B) and Dbp (C) genes expression in liver was analyzed by quantitative PCR. The highest expression time point was set to 1.
(D) Endogenous SIRT1 proteins from livers of entrained mice at indicated time points were immunoprecipitated and subjected to deacetylation assays. The value at ZT15 was set to 1. Top panel shows the amount of immunoprecipitated SIRT1.
(E) Cell extracts prepared from serum-shocked MEFs at indicated time points were complemented with recombinant SIRT1 protein and acetylated p53 peptide as substrate and subjected to deacetylation assay. Value at time 0 was set to 1.
(F) Liver extracts prepared from entrained mice were complemented with recombinant SIRT1 protein and acetylated p53 peptide as substrate and subjected to deacetylation assay. Values at ZT15 were set to 1. All data presented are the means ± standard errors of the mean (SEM) of three independent samples.
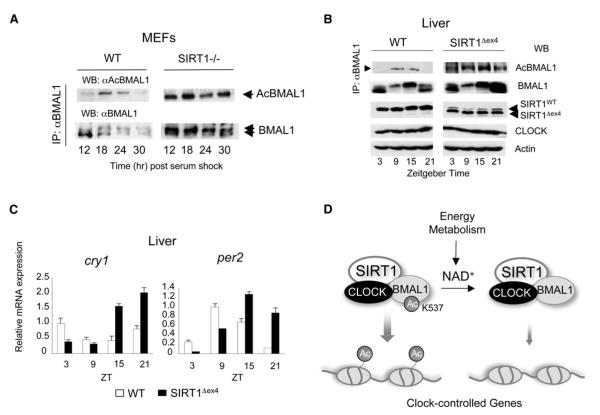
Figure 7. Circadian Dysfunction and BMAL1 Upregulated Acetylation in Liver-Specific SIRT1-Deficient Mice.
(A) BMAL1 Lys537 acetylation profile in WT or SIRT1-deficient MEFs was investigated. Cell extracts prepared from indicated time points were immunoprecipitated with BMAL1 antibody and acetylation of BMAL1 was detected by probing with the acetylated BMAL1 antibody.
(B) BMAL1 Lys537 acetylation profile in WT and SIRT1Dex4 mice was investigated. Liver extracts prepared from indicated time points were immunoprecipitated by BMAL1 antibody and acetylation of BMAL1 was detected by using the acetylated BMAL1 antibody. Acetylation of BMAL1 in the SIRT1-Dex4 mutant mice is non-rhythmic and elevated compared to WT mice. Overall levels of BMAL1 are also higher in the SIRT1-Dex4 mutant mice. The pattern of BMAL1 phosphorylation is also altered (see also Figure S5). The lower SIRT1 band corresponds to the SIRT1-Dex4 deletion. Levels of the CLOCK protein and actin were used as control.
(C) Altered circadian expression of Cry1 and Per2 clock genes in the livers of SIRT1Dex4 mice. Error bars represent SEM.
(D) Scheme of the NAD+-dependent regulation exerted by SIRT1 on the circadian clock machinery. SIRT1 interacts with CLOCK and thereby establishes a functional and molecular link between energy metabolism and circadian physiology.
We found that the endogenous liver SIRT1 obtained by immunoprecipitation from entrained mice, although constant in levels, displays circadian HDAC activity that peaks at ZT15 (Figure 1D), a time that remarkably parallels the minimal transcriptional levels of various clock-controlled genes in the liver (such as dbp,Figure 1C).
To confirm that equal levels of SIRT1 could still generate circadian HDAC function, we complemented cellular extracts with equal amounts of recombinant SIRT1 protein and acetylated p53 peptide as SIRT1 substrate (Luo et al., 2001; Vaziri et al., 2001). This assay likely reflects the intracellular relative concentrations of NAD+ and NAM, or of yet undefined circadian metabolites, whose ratio determines SIRT1 activity (Imai et al., 2000; Luo et al., 2001). Extracts from wild-type (WT) MEFs were prepared every 6 hr post-serum shock (Figure 1E), and liver extracts were prepared at four different ZT from entrained mice (Figure 1F). Also, under these conditions we found that SIRT1 deacetylase activity is rhythmic, peaking 24 hr post-serum shock in MEFs (Figure 1E) and at ZT15 in the liver (Figure 1F). Importantly, the peak of SIRT1 deacetylase activity is consistent with the cyclic acetylation of histone H3 at promoters of clock-controlled genes; at 24 hr, this acetylation is at its lowest levels (see later,Figure 4).
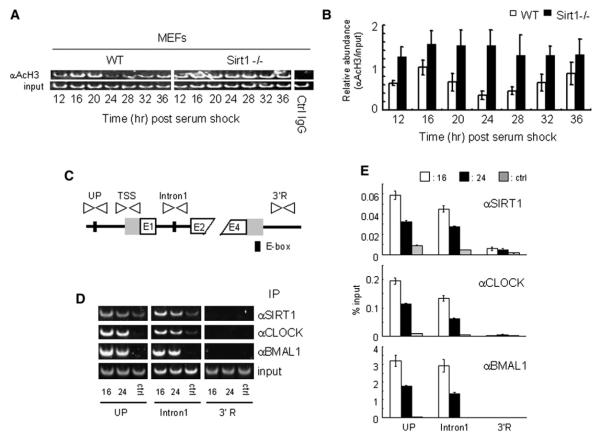
Figure 4. SIRT1 Is Recruited to the E-box and Regulates Circadian Histone Acetylation on the Dbp Gene.
(A and B) Histone H3 (Lys9/Lys14) acetylation at Dbp TSS is hyperacetylated in SIRT1-deficient MEFs. Crosslinked cell extracts were isolated from WT or SIRT1-deficient MEFs at indicated time points after serum shock, subjected to ChIP assay with anti-acetyl histone H3 (Lys9/Lys14) and control IgG, and analyzed by semiquantitative PCR (A) or quantitative PCR (B) with TSS primers. Control IgG was used as a control for immunoprecipitation. The value at time 16 in WT MEF was set to 1.
(C) Schematic diagram of the mouse Dbp promoter and primers used for ChIP assay.
(D) Representative results of the ChIP assay analyzed by semiquantitative RT-PCR. Dual crosslinked cell extracts were isolated from MEF after 16 or 24 hr serum shock and subjected to ChIP assay with anti-SIRT1, anti-CLOCK, anti-BMAL1, or no antibody (ctrl). No antibody and 3′R primers were used as controls for immunoprecipitation and PCR, respectively.
(E) Quantification of ChIP by quantitative PCR. Quantitative PCR was performed on the same samples as described in (D). All data presented are the means ± SEM of three independent samples.
SIRT1 Contributes to the Stringency of Circadian Gene Expression
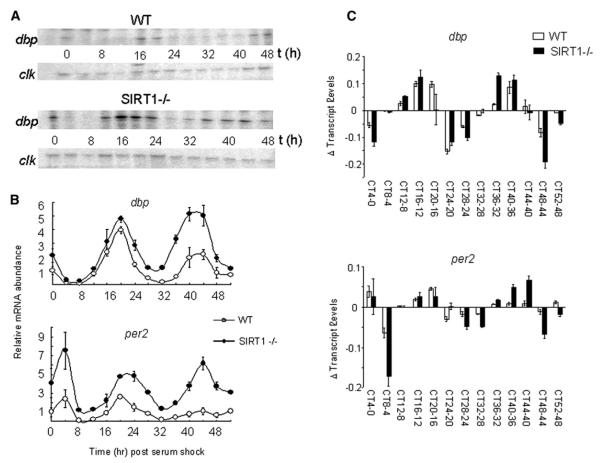
The finding that SIRT1 activity is regulated in a circadian manner prompted us to investigate its role in clock gene expression and chromatin remodeling. MEFs generated from WT and Sirt1 null mice were serum-shocked, RNA was prepared at various times, and quantitative RNase protection assay was used to monitor dbp circadian gene transcription (Figure 2A). The analysis reveals that genetic ablation of SIRT1 causes changes in circadian gene expression, including an overall increase in the transcription levels and a broadening of the oscillation cycles, with earlier onsets of increasing transcription and later decreases. Importantly, the expression of a nonoscillating gene (clock) is not affected (Figure 2A). These observations are consistent with SIRT1 having a role in controlling the stringency of circadian gene expression and being involved in the oscillatory silencing that periodically follows a transcriptional peak. These results were confirmed by quantitative RT-PCR and reproduced also on the Per2 gene (Figure 2B). Finally, a point-by-point circadian analysis of the differential transcription of both dbp and per2 between WT and Sirt1−/− MEFs confirms that the lack of Sirt1 induces a significantly higher transcriptional efficacy at specific times that normally precede and follow each circadian expression peak (Figure 2C).
Figure 2. SIRT1 Regulates Circadian mRNA Expression of dbp and per2 Genes.
(A) dbp and clock mRNA expression levels in wild-type (WT) and Sirt1 null (Sirt1−/−) MEFs after serum shock were examined by RNase protection assay. Representative results are shown and experiments were done using four independent samples.
(B) dbp and per2 mRNA expression profiles in WT and Sirt1−/− MEFs after serum shock were analyzed by quantitative PCR. Time 0 value in WT MEF for each gene was set to 1. All data presented are the means ± SEM of three independent samples.
(C) Relative increment/decrement of dbp and per2 gene expression. Plus values on vertical lines mean increment and minus values on vertical lines indicate decrement.
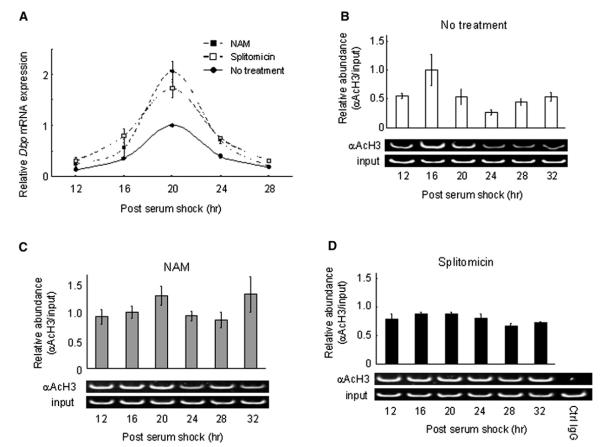
The effect of Sirt1 genetic ablation on circadian gene expression was confirmed with pharmacological treatments using SIRT1 inhibitors (splitomicin and NAM) (Figure 3A), which had no effect on the Sirt1−/− MEFs (not shown). Analysis of dbp expression levels in WT MEFs treated with either inhibitors revealed a pattern highly similar to the one obtained with the Sirt1 null cells, basically displaying a significantly broader phase and higher amplitude in the oscillation (Figure 3A).
Figure 3. Circadian Histone H3 (Lys9/Lys14) Acetylation at dbp TSS Is Dependent on SIRT1 Activity.
(A) Dbp expression levels in MEFs either treated with SIRT1 inhibitors (10 mM NAM or 120 mM splitomicin) or not were analyzed by quantitative PCR. The value at time 20 in MEFs without treatment was set to 1.
(B–D) Crosslinked cell extracts were isolated from MEF without treatment (B) or with SIRT1 inhibitors, 10 mM NAM (C), or 120 mM splitomicin (D) at indicated time points after serum shock, subjected to ChIP assay, and analyzed by quantitative PCR with dbp TSS primers (See Figure 4C). Results of semiquantitative PCR are also shown at the bottom of each experiment. The value at time 16 in MEFs without treatment was set to 1. All data presented are the means ± SEM of three independent samples.
SIRT1 Controls Circadian Histone Acetylation
The finding that CLOCK is a HAT revealed that chromatin remodeling is intimately connected to circadian physiology (Doi et al., 2006; Grimaldi et al., 2007). The remarkable effect of SIRT1 on clock gene expression (Figures 2 and 3) suggested that this effect may be mediated by changes in histone acetylation at specific sites. We analyzed the acetylation at Lys9 and Lys14 of histone H3 because we had previously found these to be preferential sites of CLOCK’s HAT activity (Doi et al., 2006), and also because SIRT1 deacetylase function was described to be targeted to these lysines (Imai et al., 2000). We performed chromatin immunoprecipitation (ChIP) assays using WT MEFs stimulated by a serum shock. Acetylation of H3 at the Dbp transcription start site (TSS) follows a circadian profile (Figure 3B and Ripperger and Schibler, 2006), presumably due to the concerted action of CLOCK and of an HDAC. Pharmacological treatment with the SIRT1 inhibitors (splitomicin and NAM) induced a loss in the circadian oscillation of acetylation, generating a noncyclic, high level of Lys9/Lys14 acetylation (Figures 3C and 3D).
Next, we extended this analysis to the MEFs from the Sirt1−/− mice. Again, histone H3 displays a robust cyclic acetylation at the Dbp promoter in WT MEFs, while genetic ablation of Sirt1 results in a constitutive, high acetylation at Lys9/Lys14 (Figures 4A and 4B). Thus, SIRT1 plays a critical role in maintaining a controlled rhythmicity in histone acetylation, thereby contributing to circadian chromatin remodeling.
SIRT1 Is in a Chromatin Complex with CLOCK:BMAL1 on the Dbp Promoter
Based on the pattern of histone acetylation associated with circadian genes (Figures 3 and 4A), it is conceivable that CLOCK and SIRT1 converge in a coordinate manner to the same regulatory regions. Thus, we decided to test whether SIRT1 is recruited to E-box elements present in the regulatory region of CLOCK: BMAL1-controlled genes. To do so, we performed a dual crosslinking ChIP assay and analyzed two E-box elements within the Dbp gene. As predicted (Ripperger and Schibler, 2006), CLOCK and BMAL1 are recruited to E-box elements in the Dbp gene in a time-dependent manner (Figure 4D). Importantly, we found that SIRT1 is jointly recruited to the same E-box elements in the Dbp gene. Furthermore, the presence of SIRT1 is temporally regulated and parallels the recruitment of the CLOCK:BMAL1 dimer (Figures 4D and 4E). Since SIRT1 is not a DNA-binding protein, we favor a scenario in which SIRT1 recruiting to a circadian promoter is mediated by the CLOCK:BMAL1 dimer. These results indicate that CLOCK:BMAL1 and SIRT1 coexist in a chromatin regulatory complex that operates on circadian promoters.
Direct Interaction of SIRT1 and CLOCK
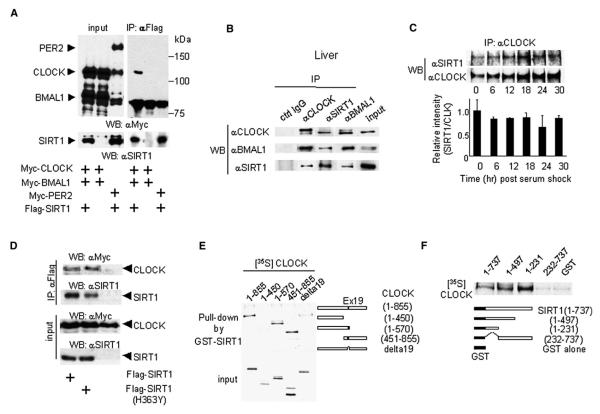
The coordinated recruiting of the CLOCK:BMAL1 dimer and SIRT1 to circadian gene promoters suggested that these regulators may physically interact. To test this possibility we coexpressed SIRT1 with CLOCK in cultured cells. In coimmunoprecipitation experiments we reveal that SIRT1 interacts with CLOCK but not with PER2 (Figure 5A). Next, we wished to establish whether native, endogenous cellular SIRT1 interacts with CLOCK. Native SIRT1 can be coimmunoprecipitated with both CLOCK and BMAL1 in liver extracts, indicating that it interacts with the CLOCK:BMAL1 complex (Figure 5B). It is unclear why BMAL1 in transfected cells does not seem to coimmunoprecipitate with the SIRT1-CLOCK complex (Figure 5A), but the results on native proteins (Figure 5B) suggest the requirement of some specific physiological conditions.
Figure 5. Interaction of SIRT1 with CLOCK.
(A) JEG3 cells were cotransfected with a series of expression vectors as described. Flag-tagged SIRT1 proteins were immunoprecipitated by FLAG antibody, and abundance of coimmunoprecipitated proteins was determined by western blotting using anti-Myc antibody (top right panel) and anti-SIRT1 antibody (bottom right panel). Left panels show the immunoblotting results of total cell lysates as an input.
(B and C) SIRT1 and CLOCK interaction in vivo. Endogenous CLOCK:BMAL1 and SIRT1 interactions in the mice liver (B) and in cultured MEFs after serum shock
(C) were determined by coimmunoprecipitation assays.
(D) The deacetylase-deficient (H363Y) mutant SIRT1 interacts with CLOCK as well as WT SIRT1.
(E) CLOCK (aa 450–570) is required for the interaction with SIRT1. In vitro-translated [35S]-labeled truncated CLOCK proteins were pulled down by GST-SIRT1.
(F) SIRT1 N terminus (aa 1–231) is required for CLOCK interaction. In vitro-translated [35S]-labeled full-length CLOCK proteins were pulled down by truncated GST-SIRT1.
We also followed the SIRT1-CLOCK interaction during the circadian cycle (Figure 5C). To do so, we coimmunoprecipitated SIRT1 from MEFs by using anti-CLOCK-specific antibody at various times after serum shock. While it would appear that the association undergoes some mild variations, after quantification of three different experiments we concluded that the SIRT1-CLOCK interaction is mostly stable during the circadian cycle (Figure 5C). We have also found that the CLOCK-SIRT1 interaction is not significantly modulated by agents known or likely to influence SIRT1 function, including NAD+, pyruvate, resveratrol, splitomicin, desferroxamine, and glucose (Figure S2).
Finally, the CLOCK-SIRT1 interaction does not appear to require the HDAC function. We used a SIRT1 mutant with a single amino acid substitution that diminishes the deacetylase activity (SIRT1-(H363Y); Vaziri et al., 2001). The mutated protein interacts with CLOCK with efficacy equivalent to that of normal SIRT1 (Figure 5D).
To identify the protein regions involved in SIRT1-CLOCK association, we performed GST pulldown assays (Figures 5E and 5F). We found that the central region of CLOCK (aa 450–570) is necessary for interaction with SIRT1. Interestingly, this region contains the serine/threonine-rich domain, which we predicted to be involved in regulated protein interactions (Doi et al., 2006), and exon 19, the domain originally found to be essential for CLOCK function (Antoch et al., 1997). In SIRT1, the N-terminal region (aa 1–231) is necessary and sufficient for eliciting efficient interaction with CLOCK (Figure 5F). This information is of interest because the same SIRT1 domain is involved in the interaction with other regulatory proteins. Specifically, it has been recently found to mediate the interaction with the histone methyltransferase SUV39H1 (Vaquero et al., 2007), a regulatory event that results in increased levels of the H3K9me3 modification and thereby control of heterochromatin formation.
BMAL1 Acetylation at Lys537 Is Regulated by SIRT1
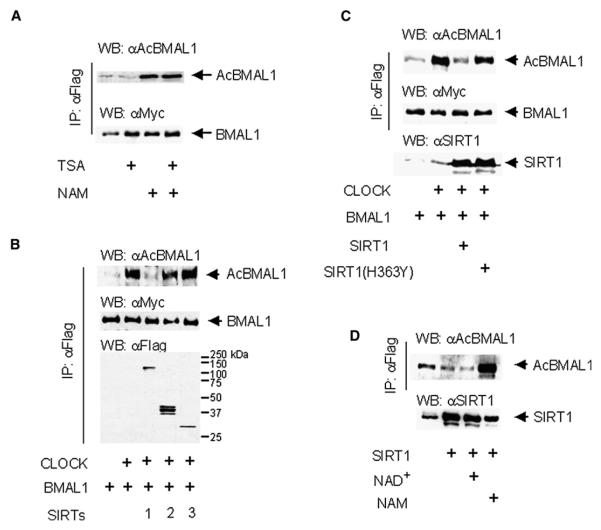
Recently we have reported that BMAL1 is rhythmically acetylated by CLOCK and that this event is essential for control of circadian function (Hirayama et al., 2007). We have generated an antibody that specifically recognizes acetylated BMAL1 at Lys537 (Figure S3). Because of the interplay between CLOCK and SIRT1, we suspected that the deacetylase that could regulate the dynamic levels of BMAL1 acetylation could be SIRT1. To identify which class of HDAC is responsible for deacetylation of BMAL1, we treated cultured cells expressing Myc-CLOCK and Flag-Myc-BMAL1 with class I and II inhibitor, trichostatin A(TSA), and/or class III inhibitor, NAM, for 6 hr and 16 hr, respectively. Acetylation of BMAL1 at Lys537 is significantly increased by NAM treatment but not by TSA treatment (Figure 6A).
Figure 6. SIRT1 Regulates BMAL1 Lys537 Acetylation.
(A) Class III HDAC inhibitor enhances BMAL1 Lys537 acetylation. JEG3 cells transfected with Myc-CLOCK and Flag-Myc-BMAL1 were treated with HDAC I and II inhibitor, TSA (1 mM), for 6 hr and HDAC III inhibitor, NAM (10 mM), for 16 hr before harvest. Immunoprecipitated BMAL1 proteins by FLAG antibody were subjected to SDS-PAGE and probed with acetylated BMAL1 or Myc antibodies.
(B) SIRT1 deacetylates acetylated Lys537 in BMAL1. JEG3 cells transfected with Myc-CLOCK and Flag-Myc-BMAL1 were cotransfected with SIRT1-Flag, SIRT2-Flag, or SIRT3-Flag. Immunoprecipitated BMAL1 proteins by FLAG antibody were probed with acetylated BMAL1 or Myc antibodies. Immunoprecipitated SIRT proteins by FLAG antibody were probed with FLAG M2 antibody.
(C) Acetylated BMAL1 is not deacetylated by deacetylase-deficient mutant SIRT1. JEG3 cells transfected with Myc-CLOCK and Flag-Myc-BMAL1 were cotransfected with WT or mutant (H363Y) SIRT1-HA. Immunoprecipitated BMAL1 proteins by FLAG antibody were probed with acetylated BMAL1 or Myc antibodies. SIRT1 protein amount was detected by SIRT1 antibody.
(D) Deacetylation of BMAL1 by SIRT1 is NAD+ dependent manner. JEG3 cells transfected with Myc-CLOCK and Flag-Myc-BMAL1 and SIRT1-HA were treated with 1 mM NAD+ or 10 mM NAM for 16 hr before harvest. Immunoprecipitated BMAL1 proteins by FLAG antibody were probed with acetylated BMAL1 antibody. SIRT1 protein amount was detected by SIRT1 antibody.
We then coexpressed BMAL1 with CLOCK and confirmed that the anti-AcBMAL1 antibody readily recognizes CLOCK-induced acetylation at Lys537 (Figure 6B). In the same assay, coexpression of SIRT1, but not SIRT2 and SIRT3, induced specific deacetylation of BMAL1 (Figure 6B), indicating that SIRT1 specifically deacetylates BMAL1. We also confirmed that SIRT1 readily deacetylates BMAL1 in an in vitro deacetylation assay (Figure S4). Importantly, the SIRT1(H363Y) enzymatically deficient mutant did not affect the acetylation state of BMAL1 (Figure 6C). Furthermore, BMAL1 deacetylation by SIRT1 is responsive to NAD+ and significantly attenuated by NAM (Figure 6D), suggesting that the acetylation of BMAL1 is an event regulated by cellular metabolism.
SIRT1 Contributes to Circadian Control In Vivo
To determine the effect of SIRT1 on the cyclic acetylation of BMAL1 we first compared MEFs from WT mice and Sirt1−/− animals. Upon serum shock, acetylation at Lys537 is cyclic in WT cells, whereas it is sustained and mostly constant in Sirt1−/− MEFs (Figure 7A). Interestingly, lack of acetylation appears to also influence BMAL1 phosphorylation levels (Figure 7A). As phosphorylation has been linked to BMAL1 stability (Cardone et al., 2005; Kondratov et al., 2003), it is interesting to observe that BMAL1 appears indeed to be expressed at higher levels in the absence of SIRT1. These results suggest that SIRT1-controlled acetylation could constitute a critical regulatory step in the control of BMAL1 protein stability.
Next we sought to demonstrate the role of SIRT1 in vivo. To do so, we used tissue-specific Sirt1−/− mice in which the loxed gene was selectively deleted by albumin promoter-driven Cre recombinase in the liver. The original mutant mice have the unique deletion of exon 4 of the Sirt1 gene, which encodes the conserved SIRT1 catalytic domain (Cheng et al., 2003). Livers were collected from mice entrained at different times of the circadian cycle and used to analyze BMAL1 acetylation and gene expression levels. Paralleling the results obtained with the Sirt1−/− MEFs (Figure 7A), BMAL1 acetylation is significantly increased and only mildly rhythmic in the livers from the mutant mice (Figure 7B). BMAL1 phosphorylation is also slightly increased with respect to WT mice, although not to the extent observed in the Sirt1−/− MEFs (see also Figure S5). Finally, expression of the circadian genes Cry1 and Per2 is also significantly altered (Figure 7C), reminiscent of the results obtained with the Sirt1−/− MEFs (Figure 2).
DISCUSSION
A large array of metabolic processes follows the rhythmicity of the circadian cycle. The presence of molecular links that reveal functional wiring between the clock machinery and metabolic pathways has been invoked (Rutter et al., 2002; Schibler and Sassone-Corsi, 2002), and much compelling evidence has accumulated (Turek et al., 2005; Wijnen and Young, 2006). We have proposed that the HAT function of CLOCK may be controlled by changing cell energy levels or, conversely, could regulate them (Doi et al., 2006; Grimaldi et al., 2007). The finding that a core element of the clock machinery directly elicits histone modifications underscored the link between circadian physiology and chromatin remodeling. These notions suggested that NAD(H)-dependent energy pathways in the cell could influence the HAT function of CLOCK:BMAL1. We reasoned that CLOCK-mediated acetylation, and thereby transcriptional activation, could be counterbalanced by transcriptional silencing induced by NAD+-dependent HDACs (Imai et al., 2000; Landry et al., 2000). Intriguingly Sir2, a NAD+-dependent HDAC, had been functionally linked to Sas2 (Kimura et al., 2002; Suka et al., 2002), a protein of the MYST family of HATs to which CLOCK belongs (Doi et al., 2006; Nakahata et al., 2007).
Our results indicate that SIRT1 could function as a molecular rheostat of CLOCK-mediated HAT function, by modulating the timing of histone lysine acetylation (Figure 7D). SIRT1 also modulates the circadian machinery by controlling the acetylation levels of BMAL1 (Figure 6), a core circadian element whose CLOCK-induced acetylation is important for circadian physiology (Hirayama et al., 2007). BMAL1 is acetylated at a key, conserved lysine at position Lys537. We have shown that Lys537 acetylation increases the efficacy of the repressor CRY to silence CLOCK: BMAL1-mediated transcription, an event essential to obtaining proper circadian oscillations (Hirayama et al., 2007). Importantly, the oscillatory acetylation patterns of H3 and BMAL1 differ in their timing: BMAL1 acetylation is sustained at a circadian time when H3 acetylation is at minimal levels (at 24 hr post-serum shock; compare Figures 4A and 7A). This difference nicely fits the scenario of a dual role for CLOCK-mediated acetylation, implicated both in transcriptional activation of circadian promoters (acetylation of H3; Doi et al., 2006) and in their subsequent down-regulation following acetylation of BMAL1- and CRY-mediated repression (Hirayama et al., 2007). These findings raise the fascinating possibility that CLOCK and SIRT1 enzymatic activities may be temporally regulated by additional posttranslational modifications. Interestingly, H3 Lys14 acetylation was shown to be significantly modulated by the phosphorylation of the nearby Ser10 residue (Cheung et al., 2000b; Lo et al., 2000). Of relevance to circadian control, phosphorylation at Ser10 has been linked to light-induced activation of clock gene expression in the supra-chiasmatic nucleus (SCN) (Crosio et al., 2000).
Another important question relates to whether SIRT1 would operate on other nonhistone circadian targets. CLOCK was found to interact with some nuclear receptors, including RARa and RXRa (McNamara et al., 2001). Since periodic availability of nuclear hormones has been implicated in the resetting of peripheral clocks (McNamara et al., 2001; Yin et al., 2007), and since SIRT1 has been found to control a number of nuclear receptors (see for example Li et al., 2007b), it is reasonable to speculate that the CLOCK-SIRT1 interaction described in this study represents a key event in the processes of fat and energy metabolism. In this respect, it is worth noting that PGC-1, a transcriptional coactivator that regulates energy metabolism and that acts in combination with SIRT1 (Nemoto et al., 2005; Rodgers et al., 2005), is rhythmically expressed in the liver and skeletal muscle and is required for cell-autonomous clock function (Liu et al., 2007; Sonoda et al., 2007). Thus, the CLOCK-SIRT1 interplay seems to occupy a privileged position in the control of gene expression by metabolites.
CLOCK and SIRT1 appear to be associated at all times of the circadian cycle (Figure 5), suggesting that they would not only coordinately contribute to the dynamic oscillation of histone acetylation but also regulate a number of nonhistonic targets.
The identification of additional molecular elements within the CLOCK:BMAL1/SIRT1 complex will define its functional features, leading to the unraveling of intracellular regulatory pathways yet poorly understood. In this respect, a fascinating connection is apparent between circadian metabolism, aging, and cancer. DNA damage accumulates with age and defects in DNA repair may lead to phenotypes reminiscent of premature aging (Lombard et al., 2005; Saunders and Verdin, 2007). The conceptual and functional link existing between the circadian clock and the cell cycle (Hunt and Sassone-Corsi, 2007; Chen and McKnight, 2007) has been extended to implicate the circadian machinery in the DNA-damage response (Collis and Boulton, 2007). The circadian genes per1 and tim have been shown to play an important role in DNA-damage control (Gery et al., 2006; Unsal-Kaçmaz et al., 2005), and phase resetting of the mammalian circadian clock is readily obtained by DNA-damaging agents (Oklejewicz et al., 2008). Finally, the role of SIRT1 in the aging process (Oberdoerffer and Sinclair, 2007; Bishop and Guarente, 2007) is intriguingly paralleled by recent observations of early aging and age-related pathologies observed in BMAL1-deficient mice (Kondratov et al., 2006). The far-reaching implications of our findings are thereby multiple, including the identification of novel strategies for the study of diabetes, obesity, and aging.
EXPERIMENTAL PROCEDURES
Animals
Male BALB/c mice and liver-specific Sirt1−/− mice were housed under 12 hr light/12 hr dark (LD) cycles over 2 weeks. All protocols using animals were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine.
Plasmids
FLAG-tagged and Myc-tagged plasmids have been described (Doi et al., 2006; Travnickova-Bendova et al., 2002). Full and truncated mouse Clock ORFs were amplified by PCR and cloned in pG4MpolyII vector. Mouse Clock(D19) was amplified by PCR from c/c MEFs. Mouse SIRT1 ORF was sub-cloned into pcDNA3 with a FLAG epitope sequence at the 5′ end. hSIRT1-Flag/pcDNA3.1, hSIRT2-Flag /pcDNA3.1, and hSIRT3-Flag /pcDNA3.1 were kind gifts of E. Verdin. Flag-hSIRT1/pECE, Flag-hSIRT1(H363Y)/pECE, hSIRT1-HA/pECE, and hSIRT1(H363Y)-HA/pECE were kind gifts of A. Brunet.
Antibodies
Antibodies against acetyl-histone H3, histone H4, and SIRT1 were from Millipore, antibodies against CLOCK and rabbit IgG from Santa Cruz Biotechnology, and antibodies against Flag (M2) and b-actin from Sigma. Antibodies against BMAL1 and Myc were described (Cardone et al., 2005). Polyclonal acetyl-lysine 537 BMAL1antibody was generated by immunizing rabbits with KLH-conjugates of the peptide NH2-ASSPGG[acetyl-K]KILN-(mouse BMAL1).
Cell Culture
MEFs were generated from WT or homozygous Sirt1−/− sibling mice and cultured in DMEM (4.5 g/l glucose) supplemented with 7.5% newborn bovine serum, 2.5% FBS, and antibiotics. JEG3 cells were grown in Basal Medium Eagle supplemented with 10% FBS and antibiotics.
Preparation of Cell Extracts and Nuclear Extracts from Cultured Cell Lines
Cells were washed twice with phosphate-buffered saline (PBS) and lysed in RIPA buffer (50 mM Tris/HCL [pH 8.0], 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% NP40, 13 protease inhibitor cocktail [Roche], 1 mM DTT, 1 mM trichostatin A [TSA], 10 mM NAM, 10 mM NaF, 1 mM PMSF). For nuclear extracts (Andrews and Faller, 1991), after washing cells with cold PBS, cells were lysed with hypotonic buffer (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 13 protease inhibitor cocktail [Roche], 1 mM DTT, 1 mM TSA, 10 mM NAM, 10 mM NaF, 1 mM PMSF). Following a brief centrifugation, pellet was resuspended in hypertonic buffer (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl 1.5 mM MgCl2, 0.2 mM EDTA, 13 protease inhibitor cocktail [Roche], 1 mM DTT, 1 mM TSA, 10 mM NAM, 10 mM NaF, 1 mM PMSF). Supernatants were recovered as nuclear extracts.
ChIP Assays
Conventional ChIP assay was used for histones from MEFs (Yamamoto et al., 2004). For nonhistone proteins, dual crosslinking ChIP assay (Nowak et al., 2005) was used with slight modifications. After serum shock, cells were washed three times with room temperature PBS, then PBS/1 mM MgCl2 was added. Disuccinimidyl Glutalate (DSG) was added to a final concentration of 2 mM for crosslinking. After 45 min at room temperature, formaldehyde was added to a final concentration of 1%(v/v) and cells incubated for 15 min. After dual crosslinking, glycine was added to a final concentration of 0.1 M and incubated for 10 min to quench formaldehyde out. After harvesting, cells were lysed in 500 ml ice-cold cell lysis buffer (50 mM Tris/HCl [pH 8.0], 85 mM KCl, 0.5% NP40, 1 mM PMSF, 13 protease inhibitor cocktail [Roche]) for 10 min on ice. Nuclei were precipitated by centrifugation (3000 g for 5 min), resuspended in 600 ml ice-cold RIPA buffer (50 mM Tris/HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM PMSF, 13 protease inhibitor cocktail [Roche]), and incubated on ice for 30 min. Sonication was done to obtain DNA fragments 100–600 bp in length.
Quantitative Real-Time RT-PCR
Each quantitative real-time RT-PCR was performed using the Chromo4 real time detection system (BIO-RAD). The PCR primers for mDbp mRNA, mPer2 mRNA, mCry1 mRNA, 18S rRNA, Dbp UP, Dbp E-box, Dbp 3′R, and mSIRT1 mRNA were described (Ripperger and Schibler, 2006; Rodgers et al., 2005; Yamamoto et al., 2004). PCR primers for Dbp TSS were designed using Real-Time PCR Primer Design (https://www.genscript.com/ssl-bin/app/primer), and the sequences are available upon request. For a 20 ml PCR, 50 ng of cDNA template was mixed with the primers to final concentrations of 200 nM and 10 ml of iQ SYBR Green Supermix (BIO-RAD), respectively. The reaction was first incubated at 95°C for 3 min, followed by 40 cycles at 95°C for 30 s and 60°C for 1 min.
RNase Protection Assays
RNA extractions were done using TRIzol (GIBCO BRL). RNase protection assays (RPAs) were performed as described (Pando et al., 2002). The riboprobes were generated using an in vitro transcription kit (Promega). Data were quantified using a phosphorimager.
Recombinant Proteins, [35S] Labeling, and GST Pulldown Assay
GST-fused recombinant proteins were expressed in E. coli BL21. Recombinant proteins were lysed by CelLytic B Cell Lysis Reagent (Sigma) according to the manufacturer’s protocol and purified by glutathione Sepharose 4B (Amersham). 35S-methionine-labeled proteins were made in vitro using the TNT-T7 quick-coupled transcription-translation system (Promega). Twenty microliters of in vitro-translated 35S-methionine-labeled proteins and 1 mg of GST-mSIRT1 or GST on glutathione Sepharose were added in a 1 ml binding buffer (50 mM Tris/HCl [pH 8.0], 150 mM NaCl, 1% NP-40), incubated over-night at 4°C. After washing sepharose with binding buffer three times, proteins were analyzed on SDS-PAGE.
SIRT1 Deacetylation Assay
SIRT1 deacetylase activity was determined using a SIRT1 Fluorimetric Activity Assay/Drug Discovery Kit (AK-555; BIOMOL International) following the manufacturer’s protocol. Extracts from serum-stimulated MEFs and liver from entrained mice lysed by RIPA buffer were used for measuring SIRT1 deacetylase activity. Complementation assays were performed by adding recombinant E. coli-generated SIRT1, and they included 1 U/reaction of SIRT1 protein and 25 mM of substrate (acetylated p53) in a 50 ml final volume. Endogenous SIRT1 from liver was obtained by immunoprecipitation and then incubated in deacetylase buffer with the substrate and 0.1 mM NAD+ for 1 hr at 37°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Alt, A. Brunet, S. Masubuchi, D. Gauthier, S. Katada, S.B. Curto, E. Verdin, S. Dilag, and all members of the Sassone-Corsi laboratory for help, reagents, and discussions. This work was supported by grants of the Cancer Research Coordinating Committee of the University of California and of the NIH (R01-GM081634-01) to P. S.-C.
Footnotes
SUPPLEMENTAL DATA Supplemental Data include five figures and can be found with this article online at http://www.cell.com/cgi/content/full/134/2/329/DC1/.
REFERENCES
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat. Rev. Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cardone L, Hirayama J, Giordano F, Tamaru T, Palvimo JJ, Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cell cycle to the circadian and metabolic cycles. Cell Cycle. 2007;6:2906–2912. doi: 10.4161/cc.6.23.5041. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000a;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000b;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Chopra VS, Mishra RK. To SIR with Polycomb: linking silencing mechanisms. Bioessays. 2005;27:119–121. doi: 10.1002/bies.20191. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Boulton SJ. Emerging links between the biological clock and the DNA damage response. Chromosoma. 2007;116:331–339. doi: 10.1007/s00412-007-0108-6. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Nakahata Y, Sahar S, Kaluzova M, Gauthier D, Pham K, Patel N, Hirayama J, Sassone-Corsi P. Chromatin remodeling and circadian control: Master regulator CLOCK is an enzyme. Cold Spring Harb. Symp. Quant. Biol. 2007;72:105–112. doi: 10.1101/sqb.2007.72.049. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Yu W. Circadian transcription: passing the HAT to CLOCK. Cell. 2006;125:424–426. doi: 10.1016/j.cell.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbatcheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related patologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Ladurner AG. Rheostat control of gene expression by metabolites. Mol. Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007b;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr. Opin. Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GTJ. Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- Saunders LR, Verdin E. Sirtuins: critical regulators at the cross-roads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Ching LW, Nofsinger RR, Evans RM. PGC-1 beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Unsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol. Cell. Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wade PA, Wolffe AP. Histone acetyltransferases in control. Curr. Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.