Abstract
Clostridium histolyticum collagenase causes extensive degradation of collagen in connective tissue that results in gas gangrene. The C-terminal collagen-binding domain (CBD) of these enzymes is the minimal segment required to bind to a collagen fibril. CBD binds unidirectionally to the undertwisted C-terminus of triple helical collagen. Here, we examine whether CBD could also target undertwisted regions even in the middle of the triple helix. Collageneous peptides with an additional undertwisted region were synthesized by introducing a Gly → Ala substitution [(POG)xPOA(POG)y]3, where x + y = 9 and x > 3). 1H–15N heteronuclear single quantum coherence nuclear magnetic resonance (HSQC NMR) titration studies with 15N-labeled CBD demonstrated that the minicollagen binds to a 10 Å wide 25 Å long cleft. Six collagenous peptides each labeled with a nitroxide radical were then titrated with 15N-labeled CBD. CBD binds to either the Gly → Ala substitution site or to the C-terminus of each minicollagen. Small-angle X-ray scattering measurements revealed that CBD prefers to bind the Gly → Ala site to the C-terminus. The HSQC NMR spectra of 15N-labeled minicollagen and minicollagen with undertwisted regions were unaffected by the titration of unlabeled CBD. The results imply that CBD binds to the undertwisted region of the minicollagen but does not actively unwind the triple helix.
Keywords: collagen, binding, NMR, triple-helix, undertwisted collagen
Introduction
Histotoxic clostridia are invasive and cause extensive degradation of muscle and connective tissue. Clostridium histolyticum produces two collagenases (ColG and ColH). Although collagenases are harmful during infection, their ability to break down a wide variety of collagen types makes them beneficial as a treatment for excessive connective tissue build up. Recently, the Food and Drug Administration approved the use of a bacterial collagenase mixture of ColG and ColH for the nonsurgical treatment of Dupuytren's contracture.1 Dupuytren's contracture is characterized by excessive collagen deposition that appears as cords causing an extension deficit. The enzymes weaken the cord to allow regaining of the function of fingers. ColG and ColH are homologous but differ in segmental structure.2 In both enzymes, the C-terminal domains are responsible for collagen-binding function.3 The C-terminal collagen-binding domains (CBDs) can serve as an anchoring unit to deliver therapeutic drugs. Fusion proteins of growth factors and CBD stayed at the collagen rich tissues and remained active for more than 10 days.4–7 Without the CBD, growth factors themselves dispersed quickly in 24 h and required frequent doses to achieve equivalent effects.4–7
CBD only binds to collagen adopting a triple helix and does not bind to denatured collagen (gelatin).3 CBD binds to minicollagen in the presence of Ca2+.8,9 The X-ray crystal structures of CBD from ColG in its apo and Ca2+ bound form were determined at 1.0 and 1.7 Å resolutions, respectively. CBD consists of a β-sandwich fold with a dynamic helical linker at the N-terminus.8,10 Site-directed mutagenesis of the surface residues to Ala identified three probable binding modes for CBD-collagen interaction. Solution-mediated experiments using nuclear magnetic resonance (NMR) spectroscopy and SAXS revealed that CBD binds unidirectionally to the C-terminus of minicollagen.11 The C-terminal region in tropocollagen could be where the collagenolysis begins. The terminal end of the collagenous peptide is less structured as evident from its crystallographic B-factors12 and monomer like NMR chemical shifts.13,14 Furthermore, the C-terminal end of the collagen fibril is exposed as seen from fiber diffraction studies.15
It has been postulated that for collagenases to hydrolyze tissue collagen,1 the enzymes must anchor themselves onto an insoluble collagen fibril, which is a staggered array of tropocollagen, then2 isolate a single triple helical molecule from the bundle, and finally3 unwind the triple helix to expose a scissile peptide bond. Our earlier studies indicated that CBD positions the bacterial collagenase to the undertwisted and exposed C-terminus to initiate collagenolysis. Because tropocollagen is suggested to contain undertwisted regions in the middle of the molecule,16 we thought that it is also possible for CBD to target that region and initiate unwinding.
Earlier studies by Toyoshima et al.,17 using gold-labeled CBD, demonstrated that CBD binds to collagen fibril that do not exhibit a periodicity of 67 nm. A lack of periodicity implied that CBD could be binding to more sites than just the C-terminus of tropocollagen. Bacterial collagenase differs from vertebral collagenase [matrix metalloproteinase (MMP)] in sequence, tertiary structure, and substrate specificity. Unlike MMPs, bacterial collagenase is much less sequence specific. MMPs may target the specific sequence, because the region is less ordered thereby avoiding the energetically costly task of unwinding a triple-helical collagen.18–21 Whereas Nagase et al.22 have demonstrated that MMP could actively unwind tropocollagen. These studies of evolutionarily unrelated MMPs provided ideas for our study.
A single Gly → Ala substitution in the middle of the collageneous peptide sequence induces local changes to the helical twist, and the peptide adopts a triple-helical conformation with a helical relaxation.23 This single mutation in [(POG)10]3 decreases the stability of the triple helix (Tm = 29°C, Supporting Information Table 1). The missense mutation in procollagen influences both the molecular nanomechanics as well as the interaction between molecules to cause connective tissue diseases such as osteogenesis imperfecta, Ehlers–Danlos syndrome, and hypochondrogenesis.24 CBD uses a saddle-like surface to interact with minicollagen such as [(POG)10]3.11 Whether CBD uses the known binding surface to interact with minicollagen with undertwisted regions or not was demonstrated by monitoring the main-chain H–N environment in 1H–15N heteronuclear single quantum coherence (HSQC) spectra. Resonances of all nonprolyl residues of CBD except K896, G925, D926, K937, L958, R967, I968, and Y970 have been unambiguously assigned using various multidimensional NMR experiments.25 The peptides were further modified to accommodate a paramagnetic-spin label at either the N- or C-terminus. One peptide was modified to accommodate the spin label at the middle of the peptide. Because the paramagnetic relaxation enhancement is a distance-dependent phenomenon, the residues on 15N-labeled CBD closest to the spin label can be identified.11 To obtain the three-dimensional shape of CBD-collagen complex in solution, SAXS measurements were used.
The possibility of active unwinding of the triple helix by CBD was examined using unlabeled CBD and 15N-labeled minicollagen molecules. The earlier studies described two distinct structural environments of 15N-labeled Gly in 1H–15N HSQC, that is, monomeric and trimeric.26 Liu et al.13 monitored the folding kinetics of 15N-enriched Gly in minicollagen by measuring the loss of trimer intensity as a function of temperature using HSQC NMR spectroscopy. The effect of unlabeled CBD on both 15N-labeled minicollagen and 15N-labeled minicollagen with Gly → Ala mutation was monitored using 1H–15N HSQC NMR titration.
Results and Discussion
Conformation and stability of collagenous peptides with Gly → Ala substitution
The conformational stability of the peptides was verified using CD (Supporting Information Figs. 2 and 3). Thermal denaturation experiments revealed that melting temperatures (Tm) of all the minicollagens were greater than ∼28°C and adopted stable triple-helical conformation between 4 and 20°C. The conformation in solution was also verified by small-angle X-ray scattering (SAXS). For collagen that possesses a helical symmetry, the pair-distance distribution function P(r) emerges as a “saw-tooth” pattern.11 Each minicollagen exhibited the “saw-tooth” profile unique to a molecule with translational symmetry (Supporting Information Fig. 5). The SAXS-derived structure is a rod-shaped molecule with ∼60 Å in length and ∼10 Å in diameter (Supporting Information Fig. 6). 1H–15N HSQC NMR spectra of the minicollagen molecule selectively labeled with 15N-Gly either at near the N- or C-termini also confirmed the triple-helical structure [Fig. 6(A–D)]. The spectra were indistinguishable from each other and also from published spectra of minicollagen.13,14,27,28 Throughout the manuscript, the minicollagen with the Gly → Ala mutation is referred to as a minicollagen with “undertwisted regions.” Analysis for local changes in helical twist revealed that the triple-helix is “undertwisted” at the POA region followed by an “overtwisting” at the POG region.16
Figure 6.
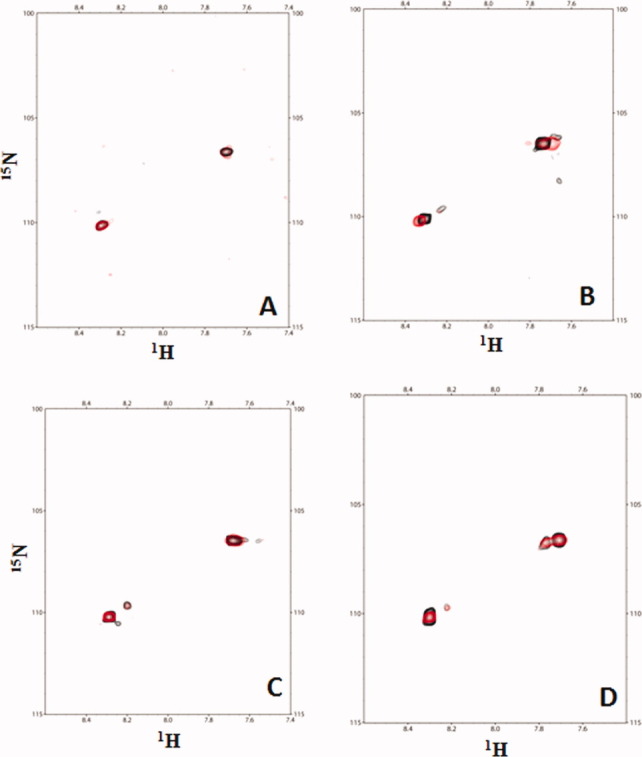
Titrations of various 15N-minicollagen with CBD demonstrate that CBD does not induce further untwisting. A: Overlay of the 1H–15N HSQC spectrum of [POGPO-15N-G-(POG)8]3 (black) with the 1H–15N HSQC spectrum of the [POGPO-15N-G-(POG)8]3:CBD complex (red) at 1:1 molar ratio shows little difference. B: Overlay of the 1H–15N HSQC spectrum of [(POG)8PO-15N-G-POG]3 (black) with the 1H–15N HSQ spectrum of the [(POG)8PO-15N-G-POG]3:CBD complex (red) at 1:1 molar ratio shows little difference. C: Overlay of the 1H–15N HSQC spectrum of [POGPO-15N-G-(POG)2-POA(POG)5]3 (black) with the 1H–15N HSQC spectrum of the [POGPO-15N-G-(POG)2POA(POG)5]3:CBD complex (red) at 1:1 molar ratio shows little difference. D: Overlay of the 1H–15N HSQC spectrum of [(POG)4POAPO-15N-G-POG]3 (black) with the 1H–15N HSQC spectrum of the [(POG)4POAPO-15N-G-POG]3:CBD complex (red) at 1:1 molar ratio shows little difference.
1H–15N HSQC NMR titration-CBD targeting the undertwisted site in minicollagen
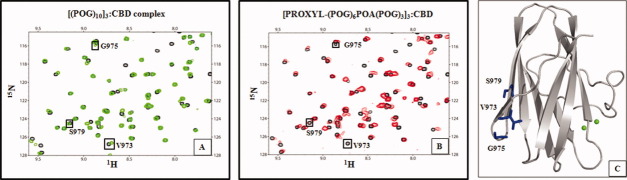
The collagenous peptide [(POG)6POA(POG)3]3 that has Ala in the 21st position from the N-terminus was synthesized. This peptide was further modified to accommodate a paramagnetic spin label at the N-terminus (Supporting Information Fig. 1). 1H–15N HSQC NMR titrations were performed with [PROXYL-(POG)6POA(POG)3]3 and 15N-labeled CBD at molar ratios ranging from 0.02:1 to 1.5:1. As demonstrated earlier,11 a total of 11 residues on the collagen-binding interface (S928, W956, G971, K995, Y996, L924, T957, Q972, D974, L991, and V993) either disappeared from the HSQC spectrum or exhibited significant chemical shift perturbation from their original position during the course of the titration. The paramagnetic PROXYL group on the N-terminus of the collagenous peptides can cause a distance-dependent line broadening of the NMR signals of CBD.11 In addition to the 11 residues, three more residues, V973, G975, and S979, exhibited appreciable line broadening, and these residues eventually disappeared from the 1H–15N HSQC spectrum [Fig. 1(A,B)]. When the paramagnetic nitroxide functionality in [PROXYL-(POG)6POA(POG)3]3:CBD complex was reduced to diamagnetic hydroxylamine by the addition of ascorbic acid, those three residues reappeared in the 1H–15N HSQC spectrum (data not shown). The disappearance of these three residues was consistent with the titration of [PROXYL-G(POG)7]3 (C-terminus is at 22nd position from the PROXYL) in our earlier publication.11 The comparison of the titration results demonstrates that CBD is targeting the Gly → Ala-substituted site. If CBD had only bound to the C-terminus of [PROXYL-(POG)6POA(POG)3]3 (C-terminus is at 30th position from the N-terminal PROXYL), we would expect to observe the disappearance of only one residue (V973), as in the published titration of [PROXYL-G(POG)7(PRG)]3.11 The disappearance of the residues (V973, G975, and S979) located at distal side from the Ca2+-binding site [Fig. 1(C)] confirmed that CBD binds unidirectionally. The collagen-binding surface in CBD is the 10 Å wide and 25 Å long cleft.11 The width of the binding cleft in CBD matches the diameter of the triple helix and its length of [(POG)3]3. NMR results imply that CBD is binding to the undertwisted [(POG)2POA]3 region of the minicollagen.
Figure 1.
Titration of 15N-CBD with a spin-labeled minicollagen demonstrates that CBD binds unidirectionally to a minicollagen. A: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no differences in chemical shifts of V973, G975, and S979. B: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [PROXYL-(POG)6POA(POG)3]3:CBD complex (red) at 1:1 molar ratio shows differences in chemical shifts of V973, G975, and S979. C: Three residues are found distal from Ca2+ ions indicated as green spheres.
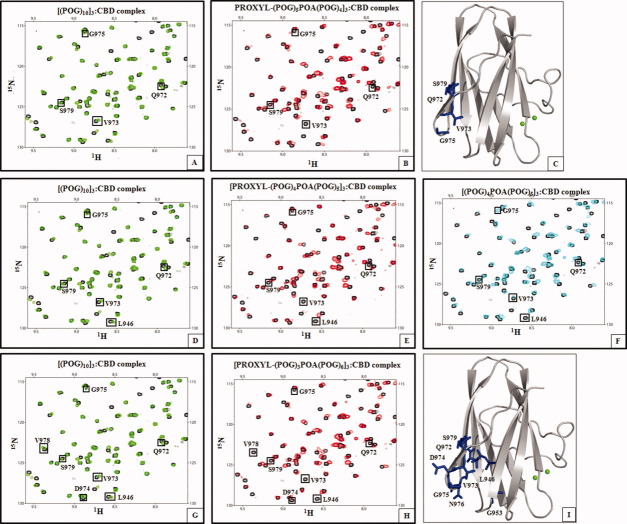
As paramagnetic relaxation enhancement is a distance-dependent phenomenon, Gly → Ala substitution made closer to the N-terminal PROXYL group should result in the disappearance of additional residues on CBD. PROXYL-containing collagenous peptides, [PROXYL-(POG)5POA(POG)4]3 (Ala at 18th position from the N-terminal PROXYL), [PROXYL-(POG)4POA(POG)5]3 (Ala at the 15th position from the PROXYL), and [PROXYL-(POG)3POA(POG)6]3 (Ala at the 12th position from the PROXYL) were synthesized (Supporting Information Fig. 1). Then, the line-broadening effects at residues were examined as the previous titrations. The shorter the distance between Gly → Ala substitution site and the N-terminal PROXYL, more residues in CBD disappeared (Fig. 2 and Table 1). The magnitude of intensity change for four amide resonances (Q972, G975, S979, and L946) was also a function of the distance (Fig. 3). The shorter the distance between Ala site and the N-terminal PROXYL, the greater the intensity loss was for the amide resonances. The NMR results are consistent with CBD binding to the [(POG)2POA]3 region in each of the four minicollagen. The binding constants obtained from all the NMR titrations were <100 μM indicating a moderate binding affinity between CBD and the minicollagen.
Figure 2.
Titrations of 15N-CBD with various spin-labeled minicollagen molecules identified that CBD binds to [(POG)2POA]3 region in a minicollagen. A: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the spectrum of the [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no difference in chemical shifts of Q972, V973, G975, and S979. B: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the spectrum of the [PROXYL-(POG)5POA(POG)4]3:CBD complex (red) at 1:1 molar ratio shows differences in chemical shifts of Q972, V973, G975, and S979. C: The four residues shifted are found distal from Ca2+-binding site. D: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the spectrum of the [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no difference in chemical shifts of L946, Q972, V973, G975, and S979. E: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the spectrum of the [PROXYL-(POG)4POA(POG)5]3:CBD complex (red) at 1:1 molar ratio shows differences in chemical shifts of L946, Q972, V973, G975, and S979. F: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the spectrum of the [(POG)4POA(POG)5]3:CBD complex (cyan) at 1:1 molar ratio shows no difference in chemical shifts of L946, Q972, V973, G975, and S979. G: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no difference in chemical shifts of L946, G953, Q972, V973, D974, G975, N976, V978, and S979. H: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [PROXYL-(POG)3POA(POG)6]3:CBD complex (red) at 1:1 molar ratios shows differences in chemical shifts of L946, G953, Q972, V973, D974, G975, N976, V978, and S979. I: Nine residues shifted due to the spin label are found distal from Ca2+-binding site.
Table I.
15N-CBD Residues Experiencing Intermolecular Paramagnetic Nuclear Spin Relaxation Effects From Spin-Labeled Minicollagen
| Peptides titrated | Ala position | Residues disappeared due to PROXYL |
|---|---|---|
| Control | ||
| [(POG)10]3a | None | |
| [(POG)4POA(POG)5]3 | 15 | None |
| [(POG)4POA(POG)5C-carbamidomethyl]3 | 15 | None |
| [(POG)3PCG(POG)4]3 | None | |
| PROXYL at N-terminus | ||
| [PROXYL-(POG)6POA(POG)3]3 | 21 | V973, G975, S979 |
| [PROXYL-(POG)5POA(POG)4]3 | 18 | Q972, V973, G975, S979 |
| [PROXYL-(POG)4POA(POG)5]3 | 15 | L946, Q972, V973, G975, S979 |
| [PROXYL-(POG)3POA(POG)6]3 | 12 | L946, G953, Q972, V973, D974, G975, N976, V978, S979 |
| [PROXYL-G(POG)7PRG]3a | V973 | |
| [PROXYL-G(POG)7]3a | V973, G975, S979 | |
| PROXYL at C-terminus | ||
| [(POG)4POA(POG)5C-PROXYL]3 | 15 | S906, R929, S997, G998 |
| [GPRG(POG)7C-PROXYL]3a | S906, R929, S997, G998 | |
| PROXYL in the middle | ||
| [11PROXYL-(POG)3PCG(POG)4]3 | S906, R929, S997, G998 (at 0.2:1. molar ratio) | |
| Also Q972,V973, G975, S979 (at 0.3:1 molar ratio) | ||
The Gly→Ala substitution position of mini-collagen is indicated.
Results from previously published peptides are shown for comparison.11
Figure 3.
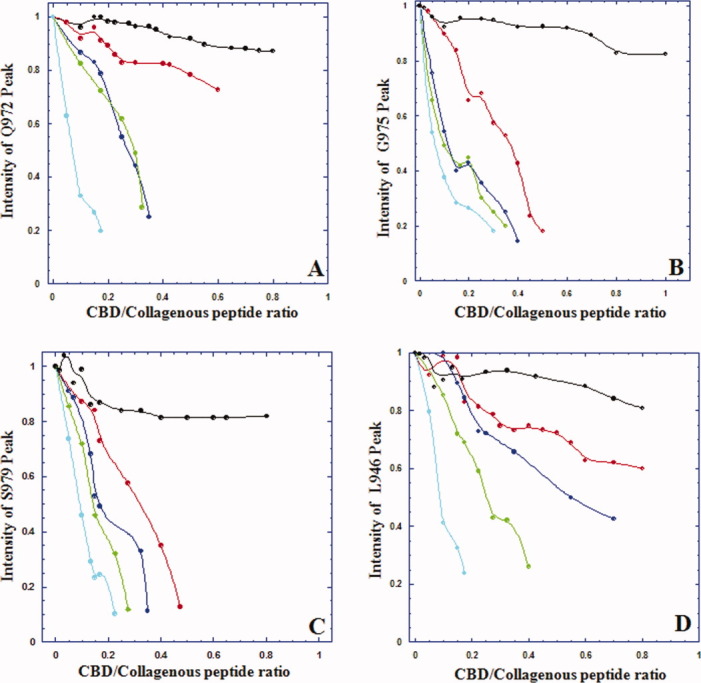
Line broadening effect of a spin label is both distance and concentration-dependent. Amide resonance intensity of (A) Q972, (B) G975, (C) S979, and (D) L946 was plotted as a function of increasing concentrations of minicollagen, that is, [(POG)10]3 (black), [PROXYL-(POG)6POA(POG)3]3 (red), [PROXYL-(POG)5POA(POG)4]3 (blue), [PROXYL-(POG)4POA(POG)5]3 (green), and [PROXYL-(POG)3POA(POG)6]3 (cyan).
Bella16 revealed that the local helical conformation in both the [(POG)2POA]3 and the C-terminal [(POG)3]3 regions of the crystal structure is undertwisted. The degree of rotation about the screw axis symmetry that describes the internal triple helical twist is defined as the helical twist value κ. The κ-value oscillates around an average value of −103° for [(POG)10]3.16 When the κ-value is less than −103°, a triple helix is undertwisted more than the norm. A collagen peptide with Gly → Ala substitution forms triple helices, but with an abrupt undertwisting (κ value ∼−110°) at the substitution site followed by an overtwisting to resume its normal helical conformation.16 Furthermore, the C-terminus of the minicollagen is undertwisted (κ value ∼ −119°), but the N-terminus is overtwisted (κ value ∼ −93°). CBD prefers the undertwisted conformation of the [(POG)2POA]3 region to that of C-terminal [(POG)3]3; however, simultaneous binding of CBD to the C-terminus is still possible.
CBD also targets the C-terminus of the minicollagen
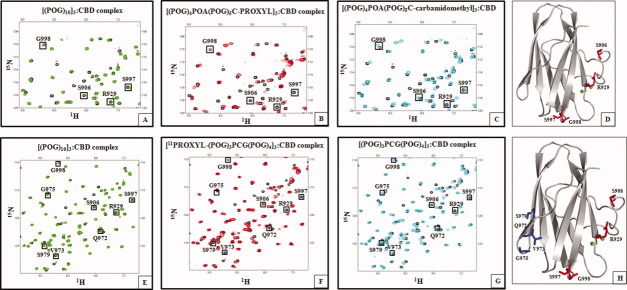
To demonstrate that CBD binds to the C-terminal [(POG)3]3 as well, a collagenous peptide [(POG)4POA(POG)5-PROXYL]3 was synthesized (Supporting Information Fig. 1). [(POG)4POA(POG)5C-PROXYL]3 was titrated with 15N-labeled CBD at molar ratios 0.02:1 to 1.5:1 with increments of 0.02, and the changes in the HSQC spectrum of CBD were monitored. Four residues S906, R929, S997, and G998 disappeared from the HSQC spectrum due to PROXYL [Fig. 4(A,B,D)]. These peaks reappeared upon reducing the complex with ascorbic acid (data not shown). This phenomenon is identical to our previous titration of [GPRG(POG)7C-PROXYL]3 when CBD bound the C-terminus.11 If CBDs were to bind only to the undertwisted Ala site, we would have observed the disappearance of fewer residues. Thus, in addition to targeting the [(POG)2POA]3 region, CBD also binds to the C-terminal [(POG)3]3. As described, the helical confirmation of both the [(POG)2POA]3 region and the C-terminal [(POG)3]3 is undertwisted compared to the norm.16 Our current explanation for why CBD is targeting the undertwisted regions is that the partial undertwisting positions main-chain amide and carbonyl groups to favor hydrogen-bonding interactions with the hydroxyl group of Tyr994. Tyr994 mutation to Phe resulted in 12-fold reduction in binding to minicollagen and the mutation to Ala lost binding capability.8
Figure 4.
Titrations of 15N-CBD with spin-labeled minicollagen molecules demonstrate that CBD binds to C-terminal [(POG)3]3 region as well. A: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no difference in chemical shifts of S906, R929, S997, and G998. B: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)4POA(POG)5C-PROXYL]3:CBD complex (red) at 1:1 molar ratio shows differences in chemical shifts of S906, R929, S997, and G998. C: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)4POA(POG)5C-carbamidomethyl]3:CBD (cyan) at 1:1 molar ratio shows no difference in chemical shifts of S906, R929, S997, and G998. D: Four residues shifted by the spin label of [(POG)4POA(POG)5C-PROXYL]3 are found close to Ca2+-binding site. E: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)10]3:CBD complex (green) at 1:1 molar ratio shows no difference in chemical shifts of S906, R929, Q972, V973, G975, S979, S997, and G998. F: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [11PROXYL-(POG)3PCG(POG)4]3:CBD complex (red) at 1:1 molar ratio shows differences in chemical shifts of S906, R929, Q972, V973, G975, S979, S997, and G998. G: Overlay of the 1H–15N HSQC spectrum of CBD (black) and the 1H–15N HSQC spectrum of the [(POG)3PCG(POG)4]3:CBD complex (cyan) at molar ratio of 1:1 shows no difference in the chemical shifts. H: CBD binds to two regions of [11PROXYL-(POG)3PCG(POG)4]3. At 0.2:1 molar ratio, CBD binds to N-terminal [(POG)3]3 resulting in line broadening of S906, R929, S997, and G998 (red). At higher peptide concentrations (0.3:1–1:1 molar ratio), CBD also binds to C-terminal [(POG)3]3, and additional resonances of Q972, V973, G975, and S979 (blue) disappear.
To demonstrate the CBD's ability to target both the [(POG)2POA]3 region and the C-terminal [(POG)3]3 region, a collagenous peptide [11PROXYL-(POG)3PCG(POG)4]3 modified to accommodate PROXYL group in the middle (11th position) was synthesized (Supporting Information Fig. 1). PROXYL group is covalently joined to the Cys residue. Because of the presence of the bulky PROXYL group, this triple-helical peptide is expected to possess a helical relaxation. The precise degree of undertwisting is not known, but GPX repeats result in a moderate undertwisting as shown in the X-ray data (κ = −105°).16 The bulky PROXYL group will likely induce greater undertwisting than κ = −105°. In addition to the 11 amide resonances either broadened or shifted, 1H–15N HSQC NMR titrations revealed two distinct binding phenomena. At a lower molar ratio (0.2:1), amide resonances corresponding to S906, R929, S997, and G998 disappeared from the HSQC spectrum of CBD (Supporting Information Fig. 7). Then, at a higher molar ratio (0.3:1), additional amide resonances corresponding to Q972, V973, G975, and S979 disappeared from the HSQC spectrum (Supporting Information Fig. 7). Resonances of no other residues are perturbed even at 1:1 molar ratio [Fig. 4(E,F,H)]. To cause the disappearance of four residues (S906, R929, S997, and G998), CBD must initially bind to the N-terminal [(POG)3]3. The subsequent disappearance of resonances Q972, V973, G975, and S979 can be explained if CBD binds to the C-terminal [(POG)3]3. However, the data at low-molar ratio signifies that CBD binds preferentially to the undertwisted N-terminal [(POG)3]3 rather than to the C-terminus.
To demonstrate that PROXYL caused the line broadening and Ala or Cys residues did not, three more control peptides, [(POG)4POA(POG)5]3, [(POG)4POA(POG)5C-carbamidomethyl]3, and [(POG)3PCG(POG)4]3, that lack the PROXYL groups were synthesized, and NMR titrations were repeated [Figs. 2(F) and 4(C,G), respectively]. The titration results were nearly identical with those of [(POG)10]3. None of amide resonances except the 11 amide resonances in the collagen-binding cleft either line broaden or shifted even at a 1:1 (minicollagen:CBD) molar ratio. The minicollagen binds to the collagen-binding cleft, and PROXYL causes the additional residues in CBD to line broaden.
To examine if CBD binds a single site (the undertwisted site in the middle or the C-terminus of minicollagen) to form 1:1 complex, or CBD binds both sites simultaneously to form 1:2 complex, dynamic light scattering (DLS) experiments were performed. DLS experiments provided the stoichiometries of collagen:CBD complexes to be 1:1. The hydrodynamic radius of [(POG)4POA(POG)5-PROXYL]3:CBD and [11PROXYL-(POG)3PCG(POG)4]3:CBD was 3 nm, and the apparent molecular weight of the complex was 43 ± 1 kDa, which is similar to those observed for [(POG)10]3:CBD complex (Table 2). Other complexes also exhibited similar values. CBD binds to either one of the available undertwisted sites in minicollagen but does not occupy both sites to form a 1:2 complex.
Table II.
Molecular Weight and Molecular Dimension of Minicollagen:CBD Complexs: Hydrodynamic Radius (RH), Apparent Molecular Weight (Mw), Radius of Gyration (Rg) and Maximum Diameter (Dmax)
| DLS | SAXS | |||
|---|---|---|---|---|
| Minicollagen:CBD complexes | RH (nm) | Mw (Da) | Rg (Å) | Dmax (Å) |
| [(POG)10]3:CBD | 3 ± 0.1 | 43 ± 1 | 22.62 ± 0.04 | 93 |
| [PROXYL-(POG)6POA(POG)3]3:CBD | 3 ± 0.2 | 43 ± 1 | 24.67 ± 0.09 | 87 |
| [PROXYL-(POG)5POA(POG)4]3:CBD | 3 ± 0.2 | 43 ± 1 | 21.08 ± 0.02 | 90 |
| [PROXYL-(POG)4POA(POG)5]3:CBD | 3 ± 0.3 | 43 ± 1 | 25.48 ± 0.08 | 92 |
| [(POG)4POA(POG)5C-carbamidomethyl]3:CBD | 3 ± 0.1 | 43 ± 1 | 24.45 ± 0.14 | 85 |
| [PROXYL-(POG)3POA(POG)6]3:CBD | 3 ± 0.1 | 43 ± 1 | 24.97 ± 0.14 | 94 |
| [(POG)4POA(POG)5C-PROXYL]3:CBD | 3 ± 0.3 | 43 ± 1 | 24.09 ± 0.16 | 85 |
| [(POG)4POA(POG)5]3:CBD | 3 ± 0.1 | 43 ± 1 | 24.67 ± 0.10 | 84 |
| [11PROXYL-(POG)3PCG(POG)4]3:CBD | 3 ± 0.2 | 43 ± 1 | 21.97 ± 0.11 | 96 |
| [(POG)3PCG(POG)4]3:CBD | 3 ± 0.3 | 43 ± 1 | 23.59 ± 0.05 | 90 |
Small angle X-ray scattering experiments
The three-dimensional molecular shapes of the minicollagen:CBD complexes were constructed using SAXS measurements. The main advantage of SAXS measurements is that the experiments are performed in solution under near physiological conditions.29 In our previous work, three-dimensional molecular shapes were used to demonstrate asymmetric binding of CBD to the C-terminal [(POG)3]3 of minicollagen.11 The molecular shapes were constructed for complexes of CBD and six different minicollagen molecules. In all cases, CBD binds to the [(POG)2POA]3 region preferentially to the C-terminal [(POG)3]3 [Fig. 5(A–F)]. For example, the docking model for [(POG)4POA(POG)5]3:CBD constructed using the crystal structure of CBD interacting with the [(POG)2POA]3 region of the minicollagen fits the envelope well [Fig. 5(B)]. Although the NMR results demonstrated that CBD also binds to the C-terminal [(POG)3]3 of [(POG)4POA(POG)5-PROXYL]3, CBD predominantly binds to the [(POG)2POA]3 region of the peptide [Fig. 5(E,F)].
Figure 5.
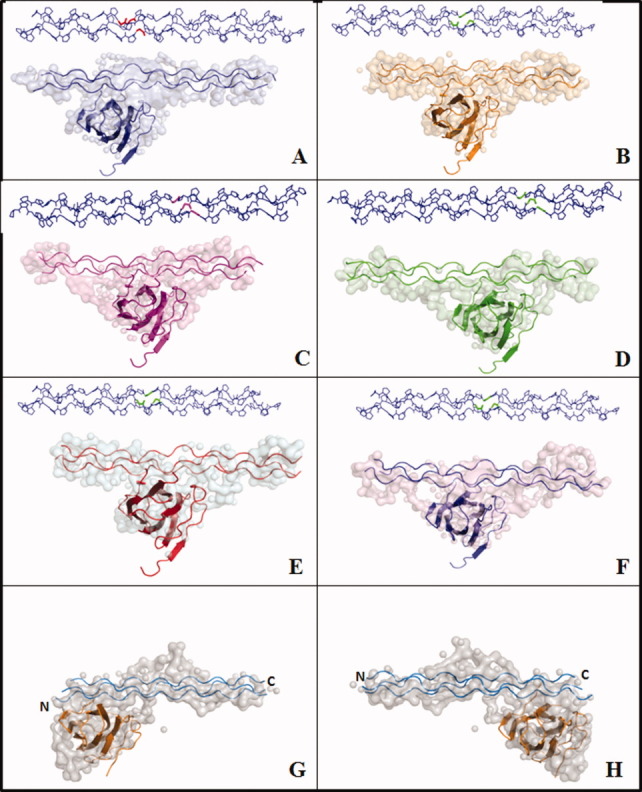
Structures of minicollagen:CBD complexes derived from SAXS profiles demonstrate that CBD binds to [(POG)2POA]3 region. A: [PROXYL-(POG)3POA(POG)6]3:CBD complex, (B) [PROXYL-(POG)4POA(POG)5]3:CBD complex, (C) [PROXYL-(POG)5POA(POG)4]3:CBD complex, (D) [PROXYL-(POG)6POA(POG)3]3:CBD complex, (E) [(POG)4POA(POG)5C-PROXYL]3:CBD complex, and (F) [(POG)4POA(POG)5C-carbamidomethyl]3:CBD. The Gly → Ala mutation sites are highlighted. G, H: Two probable binding modes of [11PROXYL-(POG)3PCG(POG)4]3:CBD complex.
Structures derived from SAXS profiles for [11PROXYL-(POG)3PCG(POG)4]3:CBD [Fig. 5(G,H)] indicate an additional density in the middle that could be attributed to the PROXYL group. The SAXS-derived three-dimensional shape of [11PROXYL-(POG)3PCG(POG)4]3:CBD complex superimposes well with either NMR-derived complexes, that is, CBD binding to the N-terminal [(POG)3]3 or to the C-terminal [(POG)3]3 (Fig. 5(G,H)].
Little conformational change of 15N-minicollagen upon CBD binding
The studies thus far suggest that CBD scans collagen for undertwisted regions. Upon binding to the less-structured regions, does it actively unwind collagen? Active unwinding by CBD would facilitate collagenolysis. Two collagenous peptides selectively labeled with 15N-Gly near the N- or near C-terminus of [(POG)10]3 were synthesized (Supporting Information Table 2, peptides A and B), and the structural changes due to the binding of unlabeled CBD were monitored using 1H–15N HSQC titration.
The 15N-Gly-labeled peptides exhibited two distinct cross peaks in the 1H–15N HSQC spectra [Fig. 6(A,B)]. These cross peaks correspond to monomer and trimer conformations assigned in earlier NMR studies.13,14,27,28 The Gly residue closer to the terminal triplets exhibited both monomer and trimer peaks in the HSQC spectrum, whereas Gly residue in the middle of the triple helix exhibited a strong trimer cross peak.13,14 If CBD is to bend or to cause any unwinding of the triple helix upon binding, we expected the cross peak corresponding to the triple helix to line broaden and disappear on the course of titration and the cross peak corresponding to the single chain to intensify. However, during the course of the titration, CBD does not cause any changes on the 1H–15N HSQC spectra of the collagenous peptides. Thus, binding to C-terminal [(POG)3]3, CBD imposes little structural changes to the triple helix.
The triple-helical peptide with a helical relaxation selectively labeled with 15N-Gly either near the N- or C-termini (Supporting Information Table 2 peptides C and D) was titrated with unlabeled CBD. Cross peaks corresponding to monomer and triple-helix were identified on the HSQC spectra [Fig. 6(C,D)]. The titration of unlabeled CBD induces little change in the intensity of either monomer or trimer cross peak. Even upon binding to the minicollagen with undertwisted regions, CBD does not initiate any further unwinding.
CBD unidirectionally binds to undertwisted sites in the triple-helical collagen. CBD may help break up the collagen fibril, but it does not unwind the triple helix. Targeting undertwisted regions of tropocollagen may circumvent the energy barrier required for unwinding the triple helices. When CBD was subcutaneously or intraperitoneally injected, the molecule was delivered prominently to the end plates of vertebral discs, near the growth plates of tibia and fibula, and also to skin.5–7 The molecule could be used as a drug-delivery vehicle to unload its payload to the most blood accessible collagen that is undergoing remodeling, thus rich in undertwisted regions.
Materials and Methods
15N-labeled protein production
The s3b (CBD) (Gly893-Lys1008) derived from Clostridium histolyticum class I collagenase (ColG) was expressed as a glutathione S-transferase (GST)-fusion protein. The GST-tag was cleaved off by thrombin, and CBD was purified as described previously.3 Uniform 15N isotope labeling was achieved using Tanaka minimal medium-containing 40 mM15NH4Cl. The labeling efficiency was estimated to be 99.6% by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS).
Peptides
(POG)10 was purchased from Peptide Institute (Osaka, Japan). Other peptides were synthesized by a standard N-(9-fluorenyl) methoxycarbonyl (Fmoc)-based strategy on Rink-amide resins (Novabiochem, Darmstadt, Germany). N-terminal spin labeling was performed on the resin by the treatment with five equivalents of 3-carboxy-PROXYL (Aldrich), 1-hydroxybenzotriazole, and diisopropylcarbodiimide in N,N-dimethylformamide at room temperature for 2 h. Peptide cleavage and deprotection steps were performed by treatment with a standard trifluoroacetic acid (TFA) scavenger cocktail (TFA: m-cresol: thioanisole: water: triisopropylsilane = 82.5: 5: 5: 5: 2.5, v/v). Spin labeling at Cys residues was performed using 3-(2-iodoacetamido)-PROXYL (IPSL, Sigma-Aldrich). Briefly, a 10M excess of IPSL dissolved in ethanol was added to the same volume of 10 mg/mL peptide in 0.1M Tris–HCl (pH 8.8), 5 mM ethylenediaminetetraacetic acid. After reacting at room temperature for 1 h, the reaction was quenched by adding excess dithiothreitol. All peptides were purified by reversed-phase HPLC using a Cosmosil 5C18 AR-II column (Nacalai Tesque, Kyoto, Japan) and characterized by MALDI-TOF-MS. All the measured masses agreed with the expected values. The chemical structures of the peptides are shown in Supporting Information Figure 1.
Circular dichroism spectroscopy
The triple helical conformation and the stability of the collagenous peptides were verified using CD spectroscopy (Supporting Information Figs. 2 and 3). CD spectra were recorded with a J-820 CD spectropolarimeter (JASCO Co., Hachioji, Japan) equipped with a Peltier thermocontroller, using a 0.5-mm quartz cuvette and connected to a data station for signal averaging. All the peptide samples were dissolved in water (1 mg/mL) and stored at 4°C for 24 h. The spectra are reported in terms of ellipticity units per mole of peptide residues [θ]mrw. CD spectra were recorded at three different temperatures (4, 10, and 16°C). All the peptides exhibited a signature CD spectral profile that consists of a maximum at 225 and a minimum near 200 nm (Supporting Information Fig. 2). Thermostability of the triple helix was monitored by the [θ]225 values of each peptide with increasing temperature at the rate of 0.3°C/min (Supporting Information Fig. 3).
NMR spectroscopy
NMR experiments were performed on a Bruker 700 MHz spectrometer equipped with cryoprobe™. All the NMR titration experiments were carried out at 16°C ± 0.5°C. The temperature range recommended for the cryoprobe is between 15 and 60°C. Hence, the temperature that is at least 11°C lower than the melting temperatures (Tm; Supporting Information Fig. 3 and Table 1) of the collagenous peptides was chosen. The concentration of the protein was 0.1 mM in 50 mM Tris–HCl (pH 7.5) containing 100 mM NaCl and 20 mM CaCl2. The titration of a highly concentrated (4 mM) peptide stock minimized the dilution effect. Aliquots of collagenous peptide were added to the protein and equilibrated for 5 min before acquiring 1H–15N HSQC spectra. The pH of the NMR samples, monitored during the titration, exhibited no significant shift in the pH (within ±0.2 U).
Dynamic light-scattering experiments
DLS data were collected using a DynaPro-E equipped with a temperature-controlled microsampler on the samples of CBD, collagenous peptides, and CBD:minicollagen complexes in 10 mM Tris–HCl (pH 7.5) containing 100 mM NaCl and 20 mM CaCl2. Protein samples were spun at 10,000 rpm for 10 min and filtered through 0.02-μm Whatman syringe directly into a 50-μL quartz cuvette. For each experiment, 20 measurements were recorded. The mean hydrodynamic radius (RH), standard deviation, polydispersity, and percent of peak area were analyzed using a Dynamics V6 (Protein Solutions). The hydrodynamic radius and molecular weight estimations were calculated from time-dependent fluctuations induced by Brownian motion as described.30
Small-angle X-ray scattering experiments
Small-angle X-ray solution scattering (SAXS) data were collected on solutions of CBD, collagenous peptides, and minicollagen:CBD complexes in 10 mM Tris–HCl (pH 7.5), 100 mM NaCl, and 20 mM CaCl2 at the SAXS/WAXS setup located at the 5-ID-D beamline of the DND-CAT Synchrotron Research Center at the Advanced Photon Source (APS), in the Argonne National Laboratory (Argonne, IL). The monochromatic radiation source (10 keV, λ = 1.2398 Å) was the APS Undulator A insertion device using an Si-111 monochromator, with 1:1 horizontal focusing and higher harmonic rejection from a Rh-coated mirror, and beam-defining slits set at 0.3 mm vertical and 0.25 mm horizontal. A 1.6-mm diameter capillary flow cell with a flow rate of 4 μL/s was used to capture four frames with exposure time of 10 s. The SAXS detector used was a Mar165 scintillator fiber-optic-coupled CCD detector and covered the momentum transfer range 0.005 < q < 0.198 Å−1, where q = 4π sin θ/λ (2θ is the scattering angle). The WAXS detector was a custom-made Roper scintillator fiber-optic-coupled CCD detector and covered 0.191 < q < 1.8 Å−1 S.31
All scattering data were acquired at 10°C. Radiation damage is the major obstacle for obtaining structural information from biological samples. Collecting data at this temperature not only minimized radiation damage but also prevented condensation around the flow cell. The four scattering patterns from each detector were averaged and merged with the rejection of outlying scans. For further analysis, the program IGOR Pro 5.5A (WaveMetrics) was used. The scattering profiles of the protein, peptide, and their complexes were obtained after subtracting the buffer profiles. The reduced scattering data were plotted as scattering intensity I(Q) versus Q [Supporting Information Fig. 4(A)]. The radius of gyration, Rg, was obtained from the Guinier approximation by linear least squares fitting in the QRg < 1 region, where the forward scattering intensity I(0) is proportional to the molecular weight of the protein complex. An indirect Fourier transformation of the I(Q) data, using GNOM,32 provided the pair-distance distribution function P(r) in the real space [Supporting Information Fig. 4(B)]. Where P(r) intersects with x-axis represents the maximum diameter, Dmax. The molecular envelopes were constructed from the SAXS data after ab initio calculation with the program GASBOR.33 √(χ2) calculated at the end of each GASBOR run indicates the agreement between the calculated scattering curve and the experimental curve. √(χ2) values calculated for the minicollagen:CBD complexes described in this manuscript range between 0.82 and 1.27. The P(r) ranges varied to generate various (>10) molecular envelopes using GASBOR. No symmetry restraints were applied to any of the shape reconstructions. The 10 highest scoring ab initio models that satisfied NMR results were chosen and averaged using DAMAVER.34 Atomic models were docked into ab initio envelopes with the program SUBCOMB.35
Docking model
The minicollagen:CBD complex was generated from the Protein Data Bank entries of 1NQD for CBD and 1CAG for the collagenous peptide (Ala mutation in 15th position). Other minicollagen molecules were generated by modifying 1CAG using fragments derived from the [(POG)10]3 structure (1K6F) to obtain the complex, the soft-docking algorithm BiGGER was used.36 Solutions were filtered using NMR titration data, and the highest scoring model that satisfied both NMR and SAXS data was chosen. The manual adjustments were aided by the use of MIFit.37
Acknowledgments
The authors are grateful to K. Hamada and H. Kamio for help in peptide synthesis and characterization and Dr. Chisato M. Yamazaki for acquiring the CD data. The authors also thank Dr. Steven Weigand for his help with SAXS experiments. SAXS experiments were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) located at Sector 5 of the Advanced Photon Source (APS). We thank Dr. James Hinton for carefully reading the manuscript.
Glossary
Abbreviations
- CBD
collagen-binding domain
- CD
circular dichroism
- DLS
dynamic light scattering
- HSQC NMR
heteronuclear single quantum coherence nuclear magnetic resonance
- MALDI-TOF-MS
matrix-assisted laser desorption/ionization-time of flight mass spectrometry
- MMP
matrix metalloproteinase
- O
4-hydroxyproline
- PROXYL
2,2,5,5-tetramethyl-l-pyrrolidinyloxy
- SAXS
small-angle X-ray scattering
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Holzer LA, Holzer G. Injectable collagenase clostridium histolyticum for Dupuytren's contracture. N Engl J Med. 2009;361:2579–2580. [PubMed] [Google Scholar]
- 2.Matsushita O, Jung CM, Minami J, Katayama S, Nishi N, Okabe A. A study of the collagen-binding domain of a 116-kDa Clostridium histolyticum collagenase. J Biol Chem. 1998;273:3643–3648. doi: 10.1074/jbc.273.6.3643. [DOI] [PubMed] [Google Scholar]
- 3.Matsushita O, Koide T, Kobayashi R, Nagata K, Okabe A. Substrate recognition by the collagen-binding domain of Clostridium histolyticum class I collagenase. J Biol Chem. 2001;276:8761–8770. doi: 10.1074/jbc.M003450200. [DOI] [PubMed] [Google Scholar]
- 4.Nishi N, Matsushita O, Yuube K, Miyanaka H, Okabe A, Wada F. Collagen-binding growth factors: production and characterization of functional fusion proteins having a collagen-binding domain. Proc Natl Acad Sci USA. 1998;95:7018–7023. doi: 10.1073/pnas.95.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katikaneni R, Ponnapakkam T, Suda H, Miyata S, Sakon J, Matsushita O, Gensure RC. Treatment for chemotherapy-induced alopecia in mice using parathyroid hormone agonists and antagonists linked to a collagen binding domain. Int J Cancer. 2012;131:E813–E821. doi: 10.1002/ijc.27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponnapakkam T, Katikaneni R, Miller E, Ponnapakkam A, Hirofumi S, Miyata S, Suva LJ, Sakon J, Matsushita O, Gensure RC. Monthly administration of a novel PTH-collagen binding domain fusion protein is anabolic in mice. Calcif Tissue Int. 2011;88:511–520. doi: 10.1007/s00223-011-9485-1. [DOI] [PubMed] [Google Scholar]
- 7.Ponnapakkam T, Katikaneni R, Nichols T, Tobin G, Sakon J, Matsushita O, Gensure RC. Prevention of chemotherapy-induced osteoporosis by cyclophosphamide with a long-acting form of parathyroid hormone. J Endocrinol Invest. 2011;34:e392–e397. doi: 10.3275/7864. [DOI] [PubMed] [Google Scholar]
- 8.Wilson JJ, Matsushita O, Okabe A, Sakon J. A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 2003;22:1743–1752. doi: 10.1093/emboj/cdg172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philominathan ST, Matsushita O, Gensure R, Sakon J. Ca2+-induced linker transformation leads to a compact and rigid collagen-binding domain of Clostridium histolyticum collagenase. FEBS J. 2009;276:3589–3601. doi: 10.1111/j.1742-4658.2009.07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sides CR, Liyanage R, Lay JO, Jr, Philominathan ST, Matsushita O, Sakon J. Probing the 3-D structure, dynamics, and stability of bacterial collagenase collagen binding domain (apo- versus holo-) by limited proteolysis MALDI-TOF MS. J Am Soc Mass Spectrom. 2012;23:505–519. doi: 10.1007/s13361-011-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philominathan ST, Koide T, Hamada K, Yasui H, Seifert S, Matsushita O, Sakon J. Unidirectional binding of clostridial collagenase to triple helical substrates. J Biol Chem. 2009;284:10868–10876. doi: 10.1074/jbc.M807684200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berisio R, Vitagliano L, Mazzarella L, Zagari A. Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)(10)](3) Protein Sci. 2002;11:262–270. doi: 10.1110/ps.32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Siegel DL, Fan P, Brodsky B, Baum J. Direct NMR measurement of folding kinetics of a trimeric peptide. Biochemistry. 1996;35:4306–4313. doi: 10.1021/bi952270d. [DOI] [PubMed] [Google Scholar]
- 14.Li MH, Fan P, Brodsky B, Baum J. Two-dimensional NMR assignments and conformation of (Pro-Hyp-Gly)10 and a designed collagen triple-helical peptide. Biochemistry. 1993;32:7377–7387. doi: 10.1021/bi00080a007. [DOI] [PubMed] [Google Scholar]
- 15.Perumal S, Antipova O, Orgel JP. Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis. Proc Natl Acad Sci USA. 2008;105:2824–2829. doi: 10.1073/pnas.0710588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bella J. A new method for describing the helical conformation of collagen: dependence of the triple helical twist on amino acid sequence. J Struct Biol. 2010;170:377–391. doi: 10.1016/j.jsb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Toyoshima T, Matsushita O, Minami J, Nishi N, Okabe A, Itano T. Collagen-binding domain of a Clostridium histolyticum collagenase exhibits a broad substrate spectrum both in vitro and in vivo. Connect Tissue Res. 2001;42:281–290. doi: 10.3109/03008200109016842. [DOI] [PubMed] [Google Scholar]
- 18.Nerenberg PS, Salsas-Escat R, Stultz CM. Do collagenases unwind triple-helical collagen before peptide bond hydrolysis? Reinterpreting experimental observations with mathematical models. Proteins. 2008;70:1154–1161. doi: 10.1002/prot.21687. [DOI] [PubMed] [Google Scholar]
- 19.Salsas-Escat R, Nerenberg PS, Stultz CM. Cleavage site specificity and conformational selection in type I collagen degradation. Biochemistry. 2010;49:4147–4158. doi: 10.1021/bi9021473. [DOI] [PubMed] [Google Scholar]
- 20.Stultz CM. Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J Mol Biol. 2002;319:997–1003. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 21.Fields GB. A model for interstitial collagen catabolism by mammalian collagenases. J Theor Biol. 1991;153:585–602. doi: 10.1016/s0022-5193(05)80157-2. [DOI] [PubMed] [Google Scholar]
- 22.Nagase H, Fushimi K. Elucidating the function of non catalytic domains of collagenases and aggrecanases. Connect Tissue Res. 2008;49:169–174. doi: 10.1080/03008200802151698. [DOI] [PubMed] [Google Scholar]
- 23.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 24.Kuivaniemi H, Tromp G, Prockop DJ. Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels. Hum Mutat. 1997;9:300–315. doi: 10.1002/(SICI)1098-1004(1997)9:4<300::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Philominathan ST, Matsushita O, Jordan JB, Sakon J. 1H, 13C and 15N resonance assignments of Ca2+ bound collagen-binding domain derived from a clostridial collagenase. Biomol NMR Assign. 2008;2:127–129. doi: 10.1007/s12104-008-9102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan P, Li MH, Brodsky B, Baum J. Backbone dynamics of (Pro-Hyp-Gly)10 and a designed collagen-like triple-helical peptide by 15N NMR relaxation and hydrogen-exchange measurements. Biochemistry. 1993;32:13299–13309. doi: 10.1021/bi00211a043. [DOI] [PubMed] [Google Scholar]
- 27.Xiao J, Madhan B, Li Y, Brodsky B, Baum J. Osteogenesis imperfecta model peptides: incorporation of residues replacing Gly within a triple helix achieved by renucleation and local flexibility. Biophys J. 2011;101:449–458. doi: 10.1016/j.bpj.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Brodsky B, Baum J. NMR conformational and dynamic consequences of a gly to ser substitution in an osteogenesis imperfecta collagen model peptide. J Biol Chem. 2009;284:20660–20667. doi: 10.1074/jbc.M109.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struct Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Proteau A, Shi R, Cygler M. Curr Protoc Protein Sci. 2010 doi: 10.1002/0471140864.ps1710s61. Chapter 17, Unit 17.10. [DOI] [PubMed] [Google Scholar]
- 31.Weigand S, Stillwell B, Guise W, Quintana J, Keane D. Advances in X-ray Analysis. International centre for diffraction data. 2009;52:58–68. [Google Scholar]
- 32.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 33.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volkhov VV, Svergunb DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozin MB, Svergun DI. A software system for rigid-body modelling of solution scattering data. J Appl Cryst. 2000;33:775–777. [Google Scholar]
- 36.Palma PN, Krippahl L, Wampler JE, Moura JJ. BiGGER: a new (soft) docking algorithm for predicting protein interactions. Proteins. 2000;39:372–384. [PubMed] [Google Scholar]
- 37.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.