Abstract
Sirtuin 1 (SIRT1) is the most conserved mammalian NAD+-dependent protein deacetylase that has emerged as a key metabolic sensor in various metabolic tissues. In response to different environmental stimuli, SIRT1 directly links the cellular metabolic status to the chromatin structure and the regulation of gene expression, thereby modulating a variety of cellular processes such as energy metabolism and stress response. Recent studies have shown that SIRT1 controls both glucose and lipid metabolism in the liver, promotes fat mobilization and stimulates brown remodeling of the white fat in white adipose tissue, controls insulin secretion in the pancreas, senses nutrient availability in the hypothalamus, influences obesity-induced inflammation in macrophages, and modulates the activity of circadian clock in metabolic tissues. This review focuses on the role of SIRT1 in regulating energy metabolism at different metabolic tissues.
Keywords: SIRT1, energy metabolism, aging, nutrients, metabolic sensor
Introduction
Sirtuins, a family of highly conserved protein modifying enzymes founded by the yeast silent information regulator 2 (Sir2) protein, are NAD+-dependent protein deacetylases and/or ADP-ribosyltransferases [1–4]. Although yeast Sir2 plays an important role in the maintenance of the silent chromatin (reviewed in ref. [5]), sirtuins in other model organisms have been increasingly recognized as crucial regulators of a variety of cellular processes, ranging from energy metabolism, and stress response, to tumorigenesis and aging [6,7].
Among the seven sirtuins that the mammalian genome encodes [8], SIRT1 is the ortholog of the yeast Sir2 protein and a nuclear NAD+-dependant protein deacetylase (Fig. 1). As the most evolutionally conserved mammalian sirtuin, SIRT1 has been a topic of intense research for more than a decade. A wealth of data have shown that this sirtuin is a nuclear metabolic sensor that directly couples the cellular metabolic status (via NAD+) to the chromatin structure and the regulation of gene expression (through deacetylation of histones, transcription factors, and transcription co-factors). Since it plays a vital role in metabolism, development, reproduction, it is not surprising that SIRT1 affects more complex biological phenomena such as aging and disease [7,9,10]. Therefore, understanding the role of SIRT1 in energy metabolism will likely provide insights into therapeutic strategies designed towards metabolic diseases and possibly aging.
Figure 1.
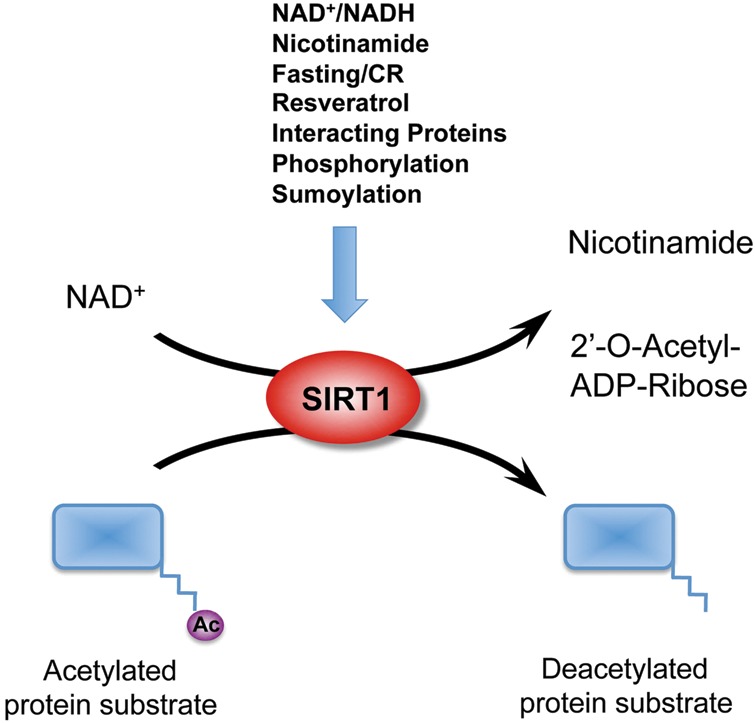
SIRT1 is an NAD+-dependent protein deacetylase SIRT1 splits NAD+ into nicotinamide and ADP-ribose, then transfers the acetyl group from the protein substrate to the 2′-OH group of ribose ring in the ADP-ribose molecule. Nutritional, hormonal, and environmental signals can modulate the deacetylase activity of SIRT1 through changes of the cellular NAD+ levels, alterations in the expression of SIRT1 protein, or posttranslational modifications/interactions on SIRT1 protein.
SIRT1 is a Cellular Metabolic Sensor
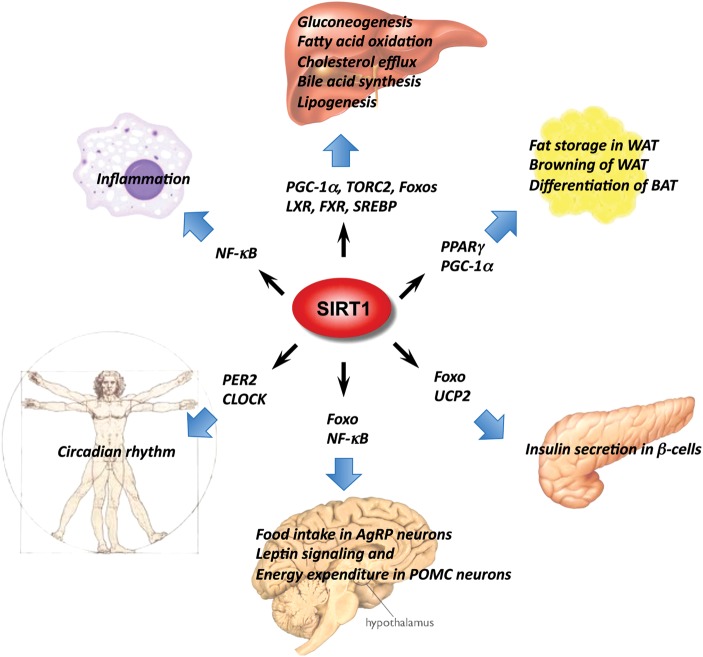
As a protein deacetylase, SIRT1 has a very broad range of protein substrates [7,10] and also shuttles between nucleus and cytosol in response to certain environmental stimuli [11]. However, despite the diversity in protein substrates and subcellular localizations, the activity of SIRT1 is tightly controlled by the cellular levels of one of its substrates, NAD+ (Fig. 1). NAD+ is an essential coenzyme found in all living cells. In metabolism, NAD+ is involved in redox reactions through electron transfer, in which it can readily switch from electron accepting form (oxidizing) NAD+ to electron donating form (reducing) NADH, and vice versa [12]. Therefore, NAD+ and NADH are extensively utilized in an array of metabolic reactions, and the cellular level of NAD+ is an important indicator of the cellular energy status. As a result, SIRT1 provides a molecular link between the cellular energy status and the adaptive transcriptional responses [13]. Due to its ability to modify and control numerous transcription factors and co-factors involved in systemic metabolic homeostasis, SIRT1 is increasingly referred to as a master metabolic regulator (Fig. 2).
Figure 2.
SIRT1 is a master metabolic regulator in different metabolic tissues SIRT1 couples the deacetylation of a number of transcription factors and co-factors to the cleavage of NAD+, an indicator of cellular metabolic status, playing a vital role in metabolism, inflammation, development, and reproduction, which will ultimately affect the processes of aging and disease.
Not surprisingly, the activity of SIRT1 is closely controlled by different environmental cues that can change the cellular NAD+ availability. For instance, a low-energy status that increases cellular levels of NAD+, such as fasting, caloric restriction (CR), and exercise, has been shown to stimulate SIRT1 activity [14–20]. On the other hand, a high-energy status that decreases cellular NAD+ levels, including high-fat diet feeding and acute inflammatory responses, reduces SIRT1 activity [21–24]. In addition, direct manipulation of the biosynthesis and catabolism of NAD+ can also alter cellular NAD+ levels and thereby SIRT1 activity. For example, increasing the activity of Nampt, the rate-limiting enzyme in the salvage pathway of NAD+ biosynthesis, enhances the levels of NAD+ and activity of SIRT1 [25–29], whereas decreasing the activity of Nampt leads to reduced NAD+ levels and SIRT1 activity [30,31]. Consistently, the treatment of cells or animals with nicotinamide mononucleotide or nicotinamide riboside, NAD+ precursors elevates NAD+ levels and SIRT1 activity [21,25,32–34]. Furthermore, deletion of Parp1, an enzyme that creates polymers of ADP-ribose at the expense of NAD+ [35–37], or decrease in the activity of CD38, the major NAD+ glycohydrolase that is present in the inner nuclear membrane [34,38,39], elevates NAD+ levels and promotes the activity of SIRT1. Taken together, these data indicate that SIRT1 is an essential metabolic sensor that can directly adapt the nuclear transcriptional networks to the cellular metabolic state.
In addition to cellular NAD+ levels, the expression and activity of SIRT1 are also under control of an intricate regulatory network at multiple layers in response to nutritional, hormonal, and environmental signals (Fig. 1). This regulatory network functions at different levels and is critical for maintaining a suitable dosage of SIRT1 in response to various environmental stimuli (for detailed reviews, please see [40–42]).
SIRT1 is a Key Regulator of Hepatic Glucose and Lipid Metabolism
The liver is a central metabolic organ that controls several key aspects of lipid and glucose metabolism in response to nutritional and hormonal signals [43]. It is essential for the maintenance of systemic energy homeostasis in the body. Recent reports have shown that SIRT1 is an important regulator of hepatic glucose metabolism (Fig. 2). For instance, hepatic SIRT1 is a key modulator of gluconeogenesis in response to fasting. It has been shown that during the short-term fasting phase, SIRT1 inhibits TORC2 (also known as CRTC2), a CREB-regulated transcription coactivator that is important for cAMP/CREB-mediated activation of gluconeogenesis genes, leading to reduced gluconeogenesis [44]. Yet the prolonged fasting increases SIRT1-mediated deacetylation and activation of PGC-1α, an essential co-activator for a number of transcription factors, resulting in increased fatty acid oxidation and improved glucose homeostasis [14,45,46]. In addition to TORC2 and PGC-1α, SIRT1 also deacetylates and activates Foxo1, resulting in increased gluconeogenesis [47]. Therefore, the net effect of SIRT1 on the regulation of gluconeogenesis is determined by the complex interaction between multiple factors at different phases of fasting and/or feeding.
Hepatic SIRT1 also plays an important role in hepatic fatty acid metabolism (Fig. 2). For example, it has been shown that adenoviral knockdown of SIRT1 reduces expression of fatty acid β-oxidation genes in the liver of fasted mice [48]. Our lab recently showed that specific deletion of the exon 4 of the mouse hepatic SIRT1 gene, which results in a truncated inactive SIRT1 protein, impairs fatty acid β-oxidation through the PPARα/PGC-1α pathway, thereby increasing the susceptibility of mice to high-fat diet-induced dyslipidemia, hepatic steatosis, inflammation, and endoplasmic reticulum (ER) stress [45]. Consistently, a complete deletion of hepatic SIRT1 by floxing exons 5 and 6 leads to development of liver steatosis even under normal feeding conditions [49]. Conversely, hepatic overexpression of SIRT1 mediated by adenovirus attenuates hepatic steatosis and ER stress, and restores glucose homeostasis in mice [50], confirming the essential role of SIRT1 in maintaining hepatic glucose and lipid homeostasis.
SIRT1 also regulates hepatic cholesterol and bile acid homeostasis through direct modulation of the liver X receptors (LXRs) and farnesoid X receptor (FXR) [51–53]. LXR and FXR are nuclear receptors that function as important cholesterol and bile acid sensors [54]. It has been previously shown that SIRT1 can directly deacetylate LXRs, resulting in increased LXR turnover and enhanced target gene expression [51]. Systemic deletion of SIRT1 in mice results in decreased serum HDL levels [51]. More recently, we and others have shown that the bile sensor FXR is also a target of hepatic SIRT1 in metabolic regulation [52,53]. It appears that SIRT1 is able to regulate the activity of FXR signaling at multiple levels. Firstly, SIRT1 regulates the expression of FXR through interaction with a hepatic nuclear factor HNF1α, which may also involve the co-activator PGC-1α [53]. Secondly, SIRT1 deacetylates FXR, increasing its DNA binding affinity [52]. Thirdly, SIRT1 appears to directly co-activate FXR through its co-activator PGC-1α. Finally, hepatic SIRT1 seems to stimulate the biosynthesis of bile acids, endogenous ligands of FXR, which in turn bind to FXR and induce its transcriptional activity [53]. Interestingly, recent studies have demonstrated that the FXR pathway also positively regulates the translation of SIRT1 protein via p53/miR-34a pathway [55,56]. Therefore, SIRT1 and the FXR signaling pathway mutually interact at multiple levels, coordinately regulating hepatic bile acid and cholesterol homeostasis.
Walker et al. and Ponugoti et al. recently showed that SIRT1 may also regulate hepatic lipid metabolism through deacetylation of SREBPs [57,58], critical regulators of lipogenesis and cholesterolgenesis [59]. These two reports revealed that SIRT1 can directly deacetylate SREBPs, and that SIRT1 activity is important in the fasting-dependent attenuation of SREBPs [57,58]. Consistently, chemical activators of SIRT1 inhibit SREBPs target gene expression in vitro and in vivo, correlating with attenuated liver steatosis in diet-induced and genetically obese mice. In summary, these findings imply that hepatic SIRT1 plays a critical role in the regulation of local and systemic metabolic homeostasis.
SIRT1 is an Important Modulator of Maturation and Remodeling of Adipose Tissues
Adipose tissues, including white adipose tissue (WAT) and brown adipose tissue (BAT), are important metabolic tissues involved in fat storage, body insulation, and body temperature regulation. They are also major endocrine organs functioning to modulate systemic energy metabolism. For example, WAT-derived hormones, such as leptin, adiponectin, and resistin, control energy balance, glucose regulation, and fatty acid catabolism. Adipose tissues originate from the differentiation of lipoblasts, and one of the primary factors involved in adipose tissue differentiation is the nuclear receptor PPARγ [60]. SIRT1 has been shown to repress PPARγ in WAT, thereby suppressing the expression of adipose tissue markers, such as the mouse aP2 gene ([61], Fig. 2). Consistently, genetic ablation of SIRT1 in adipose tissues leads to increased adiposity and insulin resistance [62]. The treatment of mice on a high-fat diet with resveratrol, a polyphenol that activates SIRT1 in cells directly or indirectly [63–67], has also been shown to protect against high-fat-induced obesity and metabolic derangements [68–70]. These findings demonstrate that SIRT1 acts in concert with lipid sensing transcription factors, such as PPARγ, to adapt gene transcription in WAT to changes in systemic nutrient levels.
In addition to WAT, previous studies have also implicated a role of SIRT1 in the differentiation and function of BAT, an important non-shivering thermogenesis organ that is essential for survival of non-shivering animals and neonates. It appears that SIRT1 can modulate BAT through both cell autonomous and non-cell autonomous mechanisms. On the one hand, SIRT1 in BAT may directly promote BAT differentiation through repression of the MyoD-mediated myogenic gene expression signature and stimulation of PGC-1α-mediated mitochondrial gene expression [71]. On the other hand, SIRT1 in propiomelanocortin (POMC) expressing neurons is able to selectively control perigonadal WAT-to-BAT-like remodeling to increase energy expenditure in female mice [72]. A recent study indicates that SIRT1 can also regulate the brown remodeling of WAT in response to cold exposure by deacetylation of PPARγ [73]. Qiang et al. showed that SIRT1-dependent deacetylation of PPARγ is required to recruit the BAT program coactivator Prdm16 to PPARγ, leading to selective induction of BAT genes and repression of WAT genes [73]. Therefore, it seems that SIRT1 differentially modulates the activity of PPARγ signaling in response to different environmental stimuli in WAT. SIRT1 inhibits the PPARγ signaling through modulation of local acetylation status of histones and recruitment of co-repressor NCoR during the fasting response [61], yet directly enhances the PPARγ signaling through deacetylation of the nuclear receptor itself in response to cold exposure [73].
SIRT1 Regulates Pancreatic Insulin Secretion in Response to Nutritional and Hormonal Signals
Pancreatic β cells are specialized endocrine cells located in the islets of Langerhans in pancreas. They are systemic metabolic sensors that synthesize and store insulin, and release insulin in response to the increase in blood glucose levels. Destruction of these cells is the leading cause of type 1 diabetes mellitus, and their dysfunction partially contributes to the pathogenesis of type 2 diabetes [74–76]. SIRT1 has been shown to be a positive regulator for pancreatic insulin secretion, which in turn triggers glucose uptake and utilization. For example, increased dosage of SIRT1 specifically in pancreatic β cells improves glucose tolerance and enhances insulin secretion in response to glucose in mice [77], whereas systemic deletion of SIRT1 impairs glucose-stimulated insulin secretion [78]. In both studies, SIRT1 has been shown to promote insulin secretion through transcriptional repression of uncoupling protein 2 (UCP2) (Fig. 2). Furthermore, mice treated with resveratrol display an increased capability to secrete insulin in β cells in response to glucose [79]. In line with these observations, activation of SIRT1 by its activators in animals protects against high-fat-induced obesity and insulin resistance [68–70], and modest overexpression of SIRT1 has a protective effect against high-fat induced glucose intolerance [80,81]. Collectively, these observations indicate that SIRT1 is a vital regulator of pancreatic insulin secretion in response to the nutrient availability.
SIRT1 in Central Control of Metabolic Homeostasis
The brain plays a critical role in the regulation of systemic energy homeostasis. Through assessment and integration of peripheral metabolic, endocrine, and neuronal signals, central nervous circuits are able to orchestrate a modulating program on both behavioral patterns and peripheral metabolism to adapt acute and chronic energy requirements [82,83].
Among several key brain areas involved in the regulation of energy balance, the hypothalamus/pituitary axis is the primary structure that interprets adiposity and nutrient-related inputs. Recent studies have implied that SIRT1 may play a role in this central energy regulatory area. For instance, it has been shown that both CR and fasting enhance SIRT1 expression and activity in the hypothalamus [84,85]. Mice lacking SIRT1 in the brain show specific anterior pituitary cell defects and fail to mediate changes in pituitary signaling and physical activity in response to CR [86], while brain-specific SIRT1 transgenic mice display enhanced neural activity in the hypothalamus [85]. These findings suggest that SIRT1 in the brain may function as a link between the hypothalamus/pituitary hormones and animal metabolic status.
However, it turns out that the role of SIRT1 in the central control of whole body energy homeostasis is more complicated than previously expected (Fig. 2). In the hypothalamus, the anorexigenic POMC expressing neurons and the orexigenic agouti-related protein (AgRP) expressing neurons are the major regulators of feeding and energy expenditure [87]. The POMC neurons produce satiety peptides thereby inhibiting food intake after feeding, while the AgRP neurons promote feeding in response to fasting and CR. It appears that SIRT1 displays distinct functions in these two neuron populations. On the one hand, inhibition of hypothalamic SIRT1 activity increases acetylation of FOXO1, resulting in increased POMC and decreased AgRP expressions, thereby decreasing food intake and body weight gain [84]. In line with these observations, AgRP neuron-specific deletion of SIRT1 decreases AgRP neuronal activity, thereby alleviating the inhibitory tone on the POMC neurons, resulting in decreased food intake and body weight [88]. On the other hand, specific deletion of SIRT1 in POMC neurons in mice causes a blunted response to leptin signaling and reduced energy expenditure, leading to hypersensitivity to diet-induced obesity [72]. Although the physiological significance of these distinct functions of SIRT1 is still not clear, these studies confirm that SIRT1 is an essential element in the periphery-central feedback circuits that mediate normal responses to nutrient availability. Consistent with these notions, central administration of small molecule SIRT1 activator has shown promise in controlling of diet-induced obesity. For example, long-term intracerebroventricular infusion of resveratrol normalizes hyperglycemia and greatly improves hyperinsulinemia in mice with diet-induced obesity and diabetes [89]. In summary, SIRT1 activity appears to be an important player in the central regulation of nutrient sensing.
SIRT1 is a Key Transcriptional Regulator of Inflammation
Macrophage activation and infiltration into resident tissues is known to mediate local inflammation and is a hallmark of metabolic syndrome [90–92]. This infiltration-induced local inflammation has been increasingly recognized as a causal factor leading to the development of the cluster of diseases surrounding metabolic syndrome [93].
Over the past few years, SIRT1 has been identified as an important repressor of inflammation in multiple tissues/cells including the macrophage [94–97]. For example, in mice, modest overexpression of SIRT1 leads to suppression of the inflammatory response, whereas whole-body insufficiency of SIRT1 induces systemic inflammation upon high-fat diet challenge [80,98,99]. Furthermore, deletion of SIRT1 in hepatocytes results in increased local inflammation under high-fat diet [45]. Several recent studies indicate that the beneficial effect of SIRT1 on metabolic disorders is due in part to its ability to suppress the activity of NF-κB, the master regulator of cellular inflammatory response, in macrophages ([100], Fig. 2). It has been shown that SIRT1 deacetylates the RelA/p65 subunit of NF-κB at lysine 310, leading to decreases in the NF-κB transcriptional activity, thereby reducing production of proinflammatory cytokines and anti-apoptotic genes [100]. In line with this notion, moderate overexpression of SIRT1 in mice leads to reduced NF-κB activity [80], while knockdown of SIRT1 in the mouse macrophage cell line RAW264.7 and in intraperitoneal macrophages increases LPS-stimulated TNFα secretion [96]. Moreover, it has been shown that cigarette smoke decreases cellular SIRT1 protein levels, causing increased acetylation and activation of NF-κB proinflammatory response in human macrophages [94]. Using a macrophage-specific knockout mouse (Mac-SIRT1KO), our group has recently provided in vivo evidence that SIRT1 deacetylates the nuclear RelA/p65 subunit of NF-κB and attenuates NF-κB-mediated gene transcription, predisposing mice to the development of insulin resistance and metabolic disorders [97]. Together, these findings demonstrate that SIRT1 activity in macrophages directly regulates immune responses and suggests that activators of SIRT1 may play an important therapeutic role in the treatment of chronic inflammatory diseases.
Interestingly, the activity and expression of SIRT1 are also under control of systemic inflammation. For instance, it has been shown that interferon gamma (IFNγ), a pro-inflammatory cytokine, represses the transcription of SIRT1 through class II transactivator, thereby disrupting metabolism and energy expenditure [101]. Moreover, TNFα has been shown to induce cathepsin B-mediated cleavage and inactivation of SIRT1 in chondrocytes [102]. More importantly, high-fat diet feeding in mice induces the cleavage of SIRT1 protein in adipose tissue through the inflammation-activated caspase-1 [62]. Therefore, SIRT1 and inflammatory signals mutually interacts at various levels, and SIRT1 is an essential molecular link between nutrient, inflammation, and metabolic dysfunction.
SIRT1 in the Regulation of Circadian Rhythm
Circadian rhythm refers to an endogenous entrainable 24-hour oscillation of any biological process in all living entities on Earth. This circadian rhythm depends on internal clocks that work in part through chromatin modification and epigenetic control of gene expression [103]. In mammals, the circadian clock is largely controlled by negative-feedback loops mediated by the heterodimeric transcription factors CLOCK-BMAL1 and their transcriptional targets, including the PER and CRY proteins that in turn directly repress CLOCK-BMAL1 activity, as well as REV-ERB and ROR nuclear receptors that control BMAL1 expression [104]. Intriguingly, the circadian clock has been recently associated with cellular metabolism. For example, circadian disruption in mice has been linked to metabolic dysfunctions [105,106], while high-fat diet feeding alters both behavioral and molecular circadian rhythms [107,108].
Chromatin modification has been shown to play an important role in the regulation of circadian gene expression. For example, the CLOCK protein itself is a transcriptional activator that functions as a histone acetyltransferase [109]. Moreover, SIRT1 has also been linked to the regulation of the circadian rhythm (Fig. 2). Early studies have shown that the expression and/or activity of SIRT1 are cyclically regulated by the circadian clock [110,111]. SIRT1 then interacts with CLOCK-BMAL1 to directly regulate the amplitude and duration of circadian clock-controlled gene expression through deacetylation of PER2 and/or BMAL1 [110,111]. However, how the circadian clock modulates SIRT1 expression/activity remained unclear in these studies. Two later studies discovered that the circadian regulation of SIRT1 activity is due to circadian oscillations of the cellular NAD+ levels [112,113]. It has been found that Nampt is a direct transcriptional target of CLOCK-BMAL1. Together, these studies add a new feedback loop in the circadian clock that involves CLOCK-BMAL1, Nampt, NAD+, and SIRT1, providing an important connection between physiological rhythm and cellular metabolism.
Although oscillations in the cellular NAD+ levels have been limited to the cyclical regulation of SIRT1 activity [112,113], an earlier study pointed towards an alternative mechanism [111]. Nakahata et al. [111] showed that the activity of immuno-purified endogenous SIRT1 proteins display a circadian oscillating pattern when measured in vitro with fixed amount of exogenous NAD+, suggesting that post-translational modifications or protein–protein interactions also play a role in the circadian regulation of SIRT1 activity. SIRT1 has been shown to be phosphorylated by several kinases in cells [114–116]. In particular, phosphorylation of SIRT1 by DYRK1A, an essential clock component that governs the rhythmic phosphorylation and degradation of CRY2 protein [117], activates its NAD+-dependent deacetylase activity in response to various environmental stresses [118]. SIRT1 also has several interacting partners that can directly inhibit or activate its activity [119–122]. Therefore, exploring the possible role of these factors in the circadian regulation of SIRT1 activity may provide novel insights into its function in circadian rhythm.
SIRT1 is a Key Regulator of Systemic Insulin Sensitivity
The important roles that SIRT1 plays in metabolism, inflammation, and circadian rhythm in various metabolic organs strongly suggest that SIRT1 is vital in regulation of whole-body insulin sensitivity, a physiological condition that is tightly associated with development of metabolic syndrome, a cluster of metabolic abnormalities including obesity, type 2 diabetes, dyslipidemia, fatty liver, and a pro-inflammatory and prothrombotic state [123–125]. Consistent with this notion, SIRT1 has been shown to directly regulate pancreatic insulin secretion, which in turn helps to improve systemic insulin sensitivity [77]. It has also been shown that SIRT1 stimulates mitochondrial fatty acid oxidation genes through PGC-1α in skeletal muscle, thereby promoting insulin sensitization [126]. Moreover, SIRT1 can improve systemic insulin sensitivity through interaction with AMP-activated protein kinase (AMPK), another essential metabolic sensor. On the one hand, SIRT1 directly deacetylates the AMPK protein kinase liver kinase B1, increasing the phosphorylation and activity of AMPK [127,128]. On the other hand, activation of AMPK by its synthetic activator, AICAR, increases cellular NAD+/NADH ratio, activating SIRT1 and increasing fatty acid oxidation and mitochondrial biogenesis [19,20,67]. In addition, it has been shown that SIRT1 directly regulates the secretion of adiponectin from adipocytes through deacetylation of FoxO1 [129], which can then help to improve insulin sensitivity in the liver and muscle [130,131].
SIRT1 also appears to be involved in the insulin downstream signaling pathway. It has been shown that SIRT1 represses the expression of PTP1B, a negative regulator of insulin signaling, thereby improving insulin sensitivity, particularly under insulin-resistant conditions [132]. A few cell culture studies also indicate that SIRT1 can interact with insulin receptor substrates, such as IRS-1 and IRS-2, thereby modulating their phosphorylation or acetylation status and improving insulin signaling [133].
Consistently, genetic deletion of SIRT1 in adipose tissues [62], myeloid cells [97], or liver [45,134], or systemic loss of a single-copy of the SIRT1 gene [96,97], leads to the development of insulin resistance and signs of metabolic syndrome upon high-fat diet feeding, whereas modest overexpression of SIRT1 protects animals against high-fat-induced glucose resistance [80,81]. In line with these observations, activation of SIRT1 by its activators such as the resveratrol and SRT1720 in animals protects against high-fat-induced obesity and insulin resistance [68–70]. Therefore, SIRT1 could be an important therapeutic target for insulin-resistance-associated metabolic syndrome.
Conclusions
The alterations of hepatic lipid metabolism, pancreatic insulin secretion, fat maturation and remodeling, central nutrient sensing, inflammatory response, circadian gene expression, as well as insulin sensitivity in SIRT1 knockout and transgenic mouse models suggest that SIRT1 is an essential regulator of systemic energy homeostasis, and that pharmacological modulation of SIRT1 could be of interest to control diseases associated with obesity. Consistent with this notion, small-molecule activators of sirtuins, such as the resveratrol and SRT1720, have shown promise as therapeutic agents for the treatment of metabolic diseases [63,70,135]. However, whether these small-molecule drugs are direct activators of SIRT1 that function to prevent obesity and diabetes is still a topic of intense debate [64–67]. Therefore, further research on the mechanism-based drug design, such as chemical compounds designed based on the phosphorylation mimetics of SIRT1 [118,136], might help to develop the next generation of SIRT1-activating molecules.
Funding
The work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to X.L. (Z01 ES102205).
Acknowledgements
I thank members of the Li laboratory for critical reading of the manuscript.
References
- 1.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 2.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 5.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 6.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 7.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 9.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–587. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 12.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 14.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashida S, Arimoto A, Kuramoto Y, Kozako T, Honda S, Shimeno H, Soeda S. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol Cell Bio. 2010;339:285–292. doi: 10.1007/s11010-010-0391-z. [DOI] [PubMed] [Google Scholar]
- 17.Graham TE, Saltin B. Estimation of the mitochondrial redox state in human skeletal muscle during exercise. J Appl Physiol. 1989;66:561–566. doi: 10.1152/jappl.1989.66.2.561. [DOI] [PubMed] [Google Scholar]
- 18.Chabi B, Adhihetty PJ, O'Leary MF, Menzies KJ, Hood DA. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. J Appl Physiol. 2009;107:1730–1735. doi: 10.1152/japplphysiol.91451.2008. [DOI] [PubMed] [Google Scholar]
- 19.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 23.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, et al. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thakur BK, Lippka Y, Dittrich T, Chandra P, Skokowa J, Welte K. NAMPT pathway is involved in the FOXO3a-mediated regulation of GADD45A expression. Biochem Biophys Res Commun. 2012;420:714–720. doi: 10.1016/j.bbrc.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Skokowa J, Lan D, Thakur BK, Wang F, Gupta K, Cario G, Brechlin AM, et al. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat Med. 2009;15:151–158. doi: 10.1038/nm.1913. [DOI] [PubMed] [Google Scholar]
- 30.van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- 31.van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Xu TY, Guan YF, Tian WW, Viollet B, Rui YC, Zhai QW, et al. Nicotinamide phosphoribosyltransferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Ann Neurol. 2011;69:360–374. doi: 10.1002/ana.22236. [DOI] [PubMed] [Google Scholar]
- 34.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 36.Sassone-Corsi P. SIRT1/PARP-1 functional interplay. Cell Cycle. 2009;8:1649. doi: 10.4161/cc.8.11.8638. [DOI] [PubMed] [Google Scholar]
- 37.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun. 2006;349:353–359. doi: 10.1016/j.bbrc.2006.08.066. [DOI] [PubMed] [Google Scholar]
- 39.Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, Chini EN. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 40.Zschoernig B, Mahlknecht U. SIRTUIN 1: regulating the regulator. Biochem Biophys Res Commun. 2008;376:251–255. doi: 10.1016/j.bbrc.2008.08.137. [DOI] [PubMed] [Google Scholar]
- 41.Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Revollo JR, Li X. The ways and means that fine tune Sirt1 activity. Trends Biochem Sci. 2012 doi: 10.1016/j.tibs.2012.12.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J Inherit Metab Dis. 1991;14:407–420. doi: 10.1007/BF01797914. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dominy JE, Jr, Lee Y, Gerhart-Hines Z, Puigserver P. Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta. 2010;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 48.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci. 2010;6:682–690. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Xu S, Giles A, Nakamura K, Lee JW, Hou X, Donmez G, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25:1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 52.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purushotham A, Xu Q, Lu J, Foley JF, Yan X, Kim DH, Kemper JK, et al. Hepatic deletion of SIRT1 decreases hepatocyte nuclear factor 1alpha/farnesoid X receptor signaling and induces formation of cholesterol gallstones in mice. Mol Cell Biol. 2012;32:1226–1236. doi: 10.1128/MCB.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 61.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chalkiadaki A, Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16:180–188. doi: 10.1016/j.cmet.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 64.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 65.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 70.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci USA. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12:78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talchai C, Lin HV, Kitamura T, Accili D. Genetic and biochemical pathways of beta-cell failure in type 2 diabetes. Diabetes Obes Metab. 2009;11(Suppl. 4):38–45. doi: 10.1111/j.1463-1326.2009.01115.x. [DOI] [PubMed] [Google Scholar]
- 75.Accili D, Ahren B, Boitard C, Cerasi E, Henquin JC, Seino S. What ails the beta-cell. Diabetes Obes Metab. 2010;12(Suppl. 2):1–3. doi: 10.1111/j.1463-1326.2010.01296.x. [DOI] [PubMed] [Google Scholar]
- 76.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 78.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vetterli L, Brun T, Giovannoni L, Bosco D, Maechler P. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E beta-cells and human islets through Sirt1 dependent mechanism. J Biol Chem. 2010;286:6049–6060. doi: 10.1074/jbc.M110.176842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, et al. SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 83.Horvath TL, Diano S, Tschop M. Brain circuits regulating energy homeostasis. Neuroscientist. 2004;10:235–246. doi: 10.1177/1073858403262151. [DOI] [PubMed] [Google Scholar]
- 84.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One. 2009;4:e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 88.Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, et al. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010;30:11815–11825. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramadori G, Gautron L, Fujikawa T, Vianna CR, Elmquist JK, Coppari R. Central administration of resveratrol improves diet-induced diabetes. Endocrinology. 2009;150:5326–5333. doi: 10.1210/en.2009-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 92.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suganami T, Ogawa Y. Adipose tissue macrophages: their role in adipose tissue remodeling. J Leukoc Biol. 2010;88:33–39. doi: 10.1189/jlb.0210072. [DOI] [PubMed] [Google Scholar]
- 94.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, et al. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29:1363–1374. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh da Y, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, Purushotham A, et al. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010;30:4712–4721. doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Purushotham A, Xu Q, Li X. Systemic SIRT1 insufficiency results in disruption of energy homeostasis and steroid hormone metabolism upon high-fat-diet feeding. FASEB J. 2012;26:656–667. doi: 10.1096/fj.11-195172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li P, Zhao Y, Wu X, Xia M, Fang M, Iwasaki Y, Sha J, et al. Interferon gamma (IFN-gamma) disrupts energy expenditure and metabolic homeostasis by suppressing SIRT1 transcription. Nucleic Acids Res. 2012;40:1609–1620. doi: 10.1093/nar/gkr984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dvir-Ginzberg M, Gagarina V, Lee EJ, Booth R, Gabay O, Hall DJ. Tumor necrosis factor alpha-mediated cleavage and inactivation of SirT1 in human osteoarthritic chondrocytes. Arthritis Rheum. 2011;63:2363–2373. doi: 10.1002/art.30279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 104.Wijnen H. Circadian rhythms. A circadian loop asSIRTs itself. Science. 2009;324:598–599. doi: 10.1126/science.1174132. [DOI] [PubMed] [Google Scholar]
- 105.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 108.Barnea M, Madar Z, Froy O. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology. 2009;150:161–168. doi: 10.1210/en.2008-0944. [DOI] [PubMed] [Google Scholar]
- 109.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 110.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 111.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, et al. Phosphorylation regulates SIRT1 function. PloS one. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PloS one. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS one. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol. 2010;30:1757–1768. doi: 10.1128/MCB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 121.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 122.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Arterioscler Thromb Vasc Biol. 2004;24:e13–e18. doi: 10.1161/01.ATV.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 124.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 125.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 126.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 130.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 131.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 133.Zhang J. The direct involvement of SirT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J Biol Chem. 2007;282:34356–34364. doi: 10.1074/jbc.M706644200. [DOI] [PubMed] [Google Scholar]
- 134.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Minor RK, Baur JA, Gomes AP, Ward TM, Csiszar A, Mercken EM, Abdelmohsen K, et al. SRT1720 improves survival and healthspan of obese mice. Sci Rep. 2011;1:70. doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo X, Kesimer M, Tolun G, Zheng X, Xu Q, Lu J, Sheehan JK, et al. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]