Abstract
Signal transduction proteins are organized into sensor (input) domains that perceive a signal and, in response, regulate the biological activity of effector (output) domains. We reprogrammed the input signal specificity of a normally oxygen-sensitive, light-inert histidine kinase by replacing its chemosensor domain by a light-oxygen-voltage (LOV) photosensor domain. Illumination of the resultant fusion kinase YF1 reduced net kinase activity by ~1000-fold in vitro. YF1 also controls gene expression in a light-dependent manner in vivo. Signals are transmitted from the LOV sensor domain to the histidine kinase domain via a 40–60° rotational movement within an α-helical coiled coil linker; light is acting as a rotary switch. These signaling principles are broadly applicable to domains linked by α-helices, and to both chemo- and photosensors. Conserved sequence motifs guide the rational design of light-regulated variants of histidine kinases and other proteins.
Keywords: light-oxygen-voltage, Per-Arnt-Sim, photosensor, protein design, reprogramming of signal specificity, signal transduction
Introduction
Perceiving external stimuli and reacting to them are essential for the survival of all living organisms in a changing environment. Proteins involved in signal transduction are constructed in a modular fashion from individual domains1 which fall largely into two groups, output or effector domains that possess biological activity such as catalytic or DNA binding activity, and input or sensor domains that are sensitive to signals such as absorption of light or binding of a chemical ligand. Interactions between sensor and effector domains may allosterically regulate the activity of the effector domain. The recombination of these domains throughout evolution may have been instrumental in the development of ever more sophisticated signaling networks in higher organisms. Moreover, the modularity of these systems allows the design of novel signaling proteins and entire networks that detect and integrate various stimuli. Recent work demonstrates that the activity of proteins such as actin-regulatory proteins2 and mitogen-activated protein (MAP) kinases3 can be reprogrammed by coupling them with novel signal sensing domains.
Signaling proteins containing Per-Arnt-Sim (PAS) sensor domains occur in all kingdoms of life4 and regulate processes as diverse as phototropism in higher plants5, voltage-dependent gating of ion channels in humans6, and nitrogen fixation in rhizobia7. The core of PAS domains adopts a common globular fold in which α-helices are packed on either side of a five-stranded antiparallel β-sheet. The core is often flanked by N- or C-terminal α-helical extensions which are either packed on the core or extend from it8,9. Certain PAS domains bind a cofactor; those that bind flavin nucleotides are referred to as light-oxygen-voltage (LOV) domains10. Different PAS sensor domains detect different signals such as chemical ligands, light and redox potential. Their versatility is also manifest in the wide range of effector domains whose activity they regulate such as kinases, transcription factors and phosphodiesterases. The contrast between the uniformity of sensor domains and the diversity of effector domains to which they are covalently linked argues against structure-based signal transduction mechanisms which depend on specific tertiary contacts between sensor and effector domains. Rather, the mechanism might involve signal-dependent, order-disorder transitions of protein segments, specifically the N-and C-terminal helices8,11, or signal-dependent quaternary structure changes12,13.
Given the natural diversity of effector domains, could PAS domains also be used to construct novel sensor proteins whose effector domains react to desired stimuli? Specifically, are different PAS sensor domains functionally interchangeable? To address these questions, we replaced the heme-binding PAS sensor domain of FixL from Bradyrhizobium japonicum (FixL), which confers oxygen sensitivity on its histidine kinase activity14,15, with the LOV blue light sensor domain of Bacillus subtilis YtvA (YtvA)16. Certain of the resulting fusion proteins retain kinase activity and substrate affinity in vitro fully comparable to FixL, but are regulated by blue light instead of by oxygen. Further, we demonstrate that these fusion proteins are also active in vivo and drive light-dependent gene expression in Escherichia coli. Kinase activity and its regulation by light depend critically on the nature of the linker between the PAS sensor and histidine kinase effector domains. We provide design rules to generate similar light-regulated kinases and other proteins.
Results
Design of Fusion Proteins
B. subtilis YtvA comprises an N-terminal LOV sensor domain and a C-terminal STAS (sulfate transport antisigma factor antagonist) effector domain17, joined by an α-helical linker sequence denoted Jα (Fig. 1A)16. In the dark, its LOV domain binds flavin mononucleotide (FMN) non-covalently. Blue light absorption promotes formation of a covalent bond between the flavin ring and the conserved cysteine 62 within the LOV domain18. This light-activated state thermally decays to the ground, dark state with a time constant of roughly one hour in YtvA16. Although the exact biochemical function of the STAS domain is not known, in response to blue light YtvA enhances transcription from promoters controlled by the general stress regulator σB 19. The isolated LOV photosensor domain of YtvA adopts the common PAS domain fold and forms a tight dimer through a hydrophobic interface13. In our crystal structure of YtvA, the Jα linker forms a C-terminal helix that extends from the core LOV domain. A similar quaternary structure arrangement was found for the heme-binding PAS domain of the oxygen sensor FixL from B. japonicum20,21.
Figure 1.
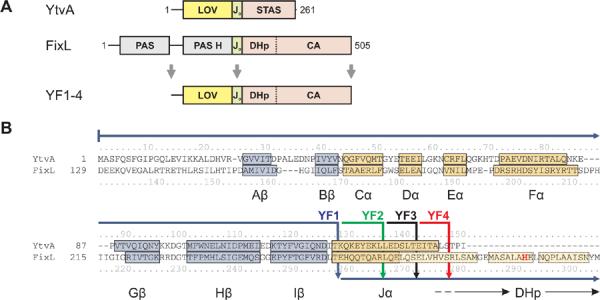
Design of fusion proteins.
A. YtvA comprises a LOV sensor and a STAS effector domain which are connected by the linker Jα. FixL consists of two PAS domains of which the second binds heme (PAS H) and a histidine kinase which comprises phosphoacceptor (DHp) and catalytic (CA) domains. Fusion kinases YF1-4 were obtained by linking the YtvA LOV domain to the FixL histidine kinase.
B. Structure-based sequence alignment between YtvA and FixL. α-helices and β-strands within the LOV/PAS sensor domains are labelled and shown in yellow and blue; lighter shaded colors denote predictions from homology models. The active-site histidine 291 is highlighted in bold red. Solid arrows indicate how the two domains are fused in constructs YF1-4. An alignment spanning the full sequences of YtvA and FixL is given in Suppl. Fig. 1.
FixL is part of a two-component system that regulates the expression of proteins involved in microaerobic respiration, nitrate respiration and nitrogen fixation15. Two PAS domains are linked to a histidine kinase which comprises phosphoacceptor (DHp) and catalytic (CA) subdomains (Fig. 1A). In a two-step reaction, FixL first undergoes autophosphorylation at its conserved histidine 291 in the DHp subdomain and then transfers the phosphate moiety to its cognate, non-covalently-bound, response regulator FixJ14,22. As in other two-component kinases, FixL also exhibits phosphatase activity23. The biological response, i. e. activation of genes involved in nitrogen metabolism, depends on the concentration of phospho-FixJ which in turn reflects the net balance of counteracting kinase and phosphatase activities24. FixL is water-soluble and forms a dimer; autophosphorylation is thought to occur in trans as observed for related histidine kinases25. Under aerobic conditions net kinase activity of FixL is strongly repressed to prevent futile activation of its target genes.
Although the PAS sensor domains of YtvA and FixL bind very different cofactors, FMN and heme, and respond to completely distinct physical and chemical stimuli, their structural similarity could reflect some functional correspondence. Thus they are good candidates for the design of novel proteins controlled by PAS sensor domains. In particular we asked: could the activity of FixL be removed from the control of oxygen and placed under the control of light by coupling its kinase domain to the YtvA LOV sensor domain? We generated such fusion proteins according to a structure-based sequence alignment between the PAS and LOV sensor domains of YtvA and FixL (Fig. 1B). Structural information clearly defines the boundaries of the sensor domains and allows us to precisely align them to each other. To avoid disrupting folded domains, in all fusion proteins YtvA and FixL were linked within their common Jα linker region, which adopts a helical conformation in both isolated sensor domains (Fig. 1B, Table I).
Table I.
In vitro Phosphorylation Assays of Fusion Constructs
| protein | description of fusion | turnover (mol FixJ/(mol kinase·h))a | |
|---|---|---|---|
| dark | light | ||
| YF1 | YtvA (1–127) - FixL (258–505) | 56.4 ± 2.8 | * |
| YF2 | YtvA (1–135) - FixL (266–505) | 67.9 ± 6.5 | * |
| YF3 | YtvA (1–141) - FixL (272–505) | * | * |
| YF4 | YtvA (1–147) - FixL (278–505) | 3.4 ± 0.1 | * |
|
| |||
| YF1 C62A | YtvA (1–127) - FixL (258–505) C62A | 52.9 ± 2.7 | 56.1 ± 2.8 |
| YF1 H291A | YtvA (1–127) - FixL (258–505) H291A | * | * |
|
| |||
| YF1Δ277 | YtvA (1–127) - FixL (258–505), Δ277 | * | * |
| YF1Δ276–277 | YtvA (1–127) - FixL (258–505), Δ276–277 | * | * |
| YF1Δ275–277 | YtvA (1–127) - FixL (258–505), Δ275–277 | 0.36 ± 0.07 | 0.21 ± 0.05 |
| YF1Δ274–277 | YtvA (1–127) - FixL (258–505), Δ274–277 | * | * |
|
| |||
| YF1 I1 | YtvA (1–127) - FixL (258–505), 268↓H | * | 0.27 ± 0.05 |
| YF1 I2 | YtvA (1–127) - FixL (258–505), 268↓HS | * | * |
| YF1 I3 | YtvA (1–127) - FixL (258–505), 268↓HSD | * | * |
| YF1 I4 | YtvA (1–127) - FixL (258–505), 268↓HSDG | 37.2 ± 1.7 | 41.5 ± 1.8 |
| YF1 I5 | YtvA (1–127) - FixL (258–505), 268↓HSDGL | * | * |
| YF1 I6 | YtvA (1–127) - FixL (258–505), 268↓HSDGLQ | * | * |
| YF1 I7 | YtvA (1–127) - FixL (258–505), 268↓HSDGLQQ | 4.2 ± 0.2 | * |
| YF1 I8 | YtvA (1–127) - FixL (258–505), 268↓HSDGLQQM | * | 0.11 ± 0.03 |
Activities of fusion constructs in FixJ phosphorylation assays.
Asterisks indicate activities below the detectable limit of 0.04 mol FixJ/(mol kinase·h).
Fusion Proteins Display Light-dependent Kinase Activity
The fusion protein YF1 covalently connects residues 1–127 of YtvA with 258–505 of FixL (Fig. 1B). Incorporation of the FMN cofactor and the ability of YF1 to undergo the characteristic reversible LOV photoreaction were confirmed by absorption spectroscopy (Suppl. Fig. 2). Sedimentation equilibrium centrifugation at protein concentrations between 0.5 and 7 μM showed that in solution YF1 forms a single species with an apparent molecular weight of (78.4 ± 10.4) kD corresponding to a dimer (data not shown; exp. monomer weight 42.1 kD). We estimate that its dissociation equilibrium constant is less than 0.5 μM.
We measured the autophosphorylation activity of YF1 by radiography using γ-32P-ATP as a substrate. The initial reaction velocity in the dark of (0.080 ± 0.003) h−1 at 23 °C was decreased under saturating illumination with white light to (0.020 ± 0.001) h−1. The autophosphorylation activity of the parent enzyme FixL is greatly stimulated by the presence of the response regulator FixJ even though FixJ does not directly participate in this reaction26. In all subsequent experiments we therefore monitored enzyme activity and regulation in turnover assays in which multiple rounds of FixJ phosphorylation are catalyzed, whose kinetics reflect the net balance of counteracting kinase and phosphatase activities (Fig. 2A). At the beginning of the reaction no phospho-FixJ is present and therefore the initial time-course of phosphorylation is dominated by the forward kinase reaction. We evaluated the initial velocity of phosphate incorporation into FixJ and report these values as kinase activities. In the dark, YF1 phosphorylated FixJ with an initial velocity of (56.4 ± 2.8) mol FixJ/(mol kinase·h) (Fig. 2B). In the following, we drop the two mol terms and report turnover activities in units of h−1. Under constant illumination net kinase activity was strongly suppressed to the point where it was too low to be quantified. Based on the minimum phosphorylation activity of 0.04 h−1 we can reliably measure in these turnover assays, we estimate that light absorption causes a more than 1000-fold reduction of net kinase activity in YF1.
Figure 2.
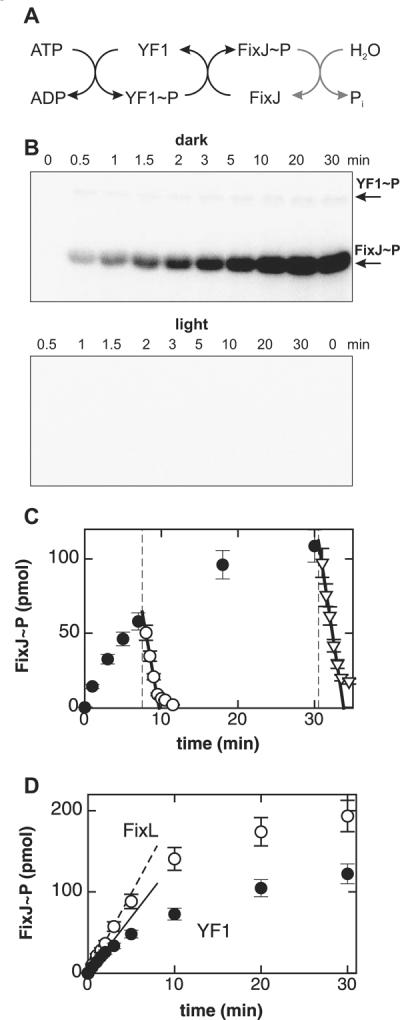
Kinase activity of fusion kinase YF1.
A. In consecutive steps, YF1 autophosphorylates and then transfers the phosphoryl group to the response regulator FixJ.
B. YF1 phosphorylates FixJ in the dark but is strongly inhibited in the light. Reactions were started by adding γ-32P-ATP and aliquots were taken at indicated times. Samples were separated on a SDS gel and incorporated phosphate monitored by 32P-radiography. Each lane contained 15 pmol YF1 and 380 pmol FixJ.
C. Light absorption switches YF1 from kinase to phosphatase activity. Phosphorylation assays were started in the dark as in panel B (●). After 7.5 and 30.5 min aliquots of the reaction mix were exposed to light (dashed lines) which caused phospho-FixJ levels to decrease with initial velocities of (120 ± 20) (◯) and (140 ± 7) mol FixJ/(mol kinase·h) (▽).
D. Comparison of reactions catalyzed by YF1 (●) and FixL (◯). Lines denote initial reaction velocities and correspond to turnovers of (56.4 ± 2.8) h−1 and (80.2 ± 3.6) h−1 for YF1 and FixL, respectively.
In principle, any or all of the elementary steps in the overall reaction scheme could be regulated by light (Fig. 2A). To address whether YF1 also possesses phosphatase activity, we performed kinase assays in the dark as before, but took aliquots after 7.5 and 30.5 min at which times appreciable amounts of phospho-FixJ had accumulated. Upon exposure to light, the amount of phospho-FixJ in these aliquots decreased sharply with initial velocities of (120 ± 20) and (140 ± 7) mol FixJ/(mol kinase·h), respectively (Fig. 2C). The observation that these velocities depend only weakly on the initial amount of phospho-FixJ argues for a process that is enzyme-catalyzed by the YF1/FixJ complex; for uncatalyzed hydrolysis these velocities would be proportional to the initial concentration of phospho-FixJ. Furthermore, under similar reaction conditions phospho-FixJ is chemically quite stable; the lifetime of the closely related phospho-FixJ from Sinorhizobium meliloti is on the order of two hours27.
As a control, we determined the activity of the parent enzyme FixL under the same reaction conditions. In vivo, FixL occurs in the ferrous state (FeII); binding of oxygen to the ferrous deoxy state inhibits kinase activity. In vitro, FixL is routinely assayed in its ferric (FeIII) state which has activity similar to the ferrous deoxy form22. The net kinase activity of ferric FixL can be greatly reduced by adding cyanide28. Under our reaction conditions, ferric FixL phosphorylated FixJ with an initial velocity of (80.2 ± 3.6) h−1 which is faster by a factor of 1.4 ± 0.1 than the reaction catalyzed by YF1 in the dark (Fig. 2D). Addition of cyanide led to an approximately 1000-fold decrease of net kinase activity of FixL to (0.086 ± 0.014) h−1. These values are comparable with previous studies under similar reaction conditions that reported a maximum initial velocity of 81 h−1 and a more than 2700-fold reduction in the presence of cyanide28.
We measured the net kinase activity of YF1 in the dark at varying substrate concentrations. Catalysis followed Michaelis-Menten kinetics with apparent KM values of (33 ± 2) μM and (1.4 ± 0.4) μM for ATP and FixJ, respectively (Suppl. Table I). In the case of FixL, the KM values were (206 ± 13) μM for ATP and (1.6 ± 0.4) μM for FixJ which agree closely with values for the orthologous FixL from S. meliloti26.
Kinase activity of YF1 has thus been placed under the control of light without incurring significant losses in kinase activity or alteration in substrate affinities. In the dark state, YF1 possesses net kinase activity closely similar to its parent FixL. In the light state, its phosphatase activity is greatly enhanced and YF1 is converted to a net phosphatase.
Light Inactivation is Linked to LOV Photocycle
To address whether the light dependence of YF1 activity is indeed mediated by its LOV domain, we conducted several control experiments.
First, we verified that the activity of the parent enzyme FixL is independent of light (not shown).
Second, we generated a point mutant of YF1 in which the active-site cysteine 62 in the LOV domain is replaced by alanine which abolishes the normal LOV photochemistry (Suppl. Fig. 2). Net kinase activity of YF1 C62A was identical under dark ((52.9 ± 2.7) h−1) and light conditions ((56.1 ± 2.8) h−1) and corresponded to that noted above for YF1 in the dark ((56.4 ± 2.8) h−1). Replacement of the phosphoacceptor histidine 291 within the DHp domain by alanine abolished phosphorylation of FixJ both in the dark and light.
Third, we explored whether light absorption might simply cause global unfolding and thus inactivation of YF1. Light absorption induces a 5–10% loss of the circular dichroism (CD) signal of YF1 at 208 nm, similar to observations for a plant phototropin LOV domain (Suppl. Fig. 3)8. This indicates a partial loss or rearrangement of α-helical structure elements upon light absorption but rules out global unfolding of YF1. Global unfolding is also incompatible with the observations noted above, that the autophosphorylation activity of isolated YF1 is only moderately suppressed in the light and that the YF1/FixJ complex displays greatly enhanced phosphatase activity in the light.
Fourth, YF1 was illuminated through a (430 ± 10) nm interference filter which reduced light output to 240 μW. Using this illumination protocol, the fraction of YF1 in the dark state diminished in a first-order reaction with a time constant of (58.4 ± 1.0) s (Fig. 3A, open circles). Net kinase activity of YF1 decayed approximately twice as fast, with a time constant of (27.1 ± 1.0) s (Fig. 3A, closed symbols). Recovery of YF1 after photobleaching occurred with a complex time course over several hours, as indicated by absorption spectroscopy (Fig. 3B). Approximately 70% of the absorption signal recovered with a time constant of (5900 ± 25) s, followed by a slower phase. Complex recovery kinetics have also been observed for natural LOV histidine kinases29. The long recovery time enabled us to determine the kinase activity of YF1 as recovery proceeded. Net kinase activity was regenerated in a sigmoid time course (Fig. 3B), and at 12 h after light absorption, activity had recovered to 95% of that of the dark state. The presence of a pronounced lag phase suggests that photorecovery proceeds through an intermediate state with little or no kinase activity.
Figure 3.
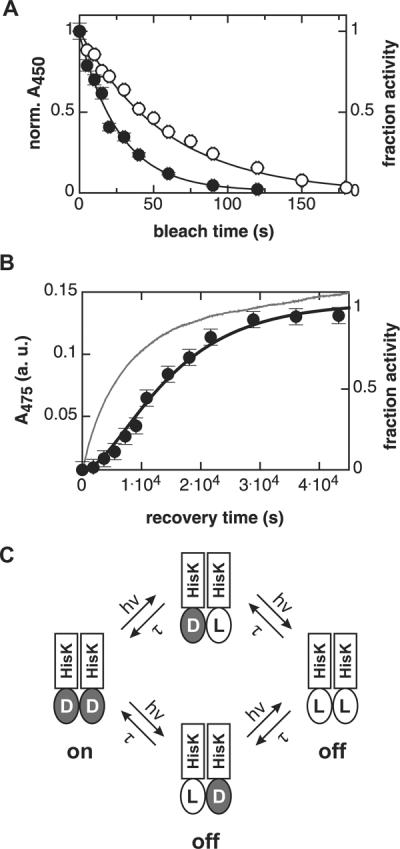
Kinase activity of YF1 depends on LOV photocycle.
A. Upon illumination with 430 nm light (240 μW), the fraction of YF1 in the dark state as determined by absorption spectroscopy (◯) decays with a first-order time constant of (58.4 ± 1.0) s. YF1 kinase activity (●) decays with a time constant of (27.1 ± 1.0) s.
B. Recovery kinetics of YF1 after saturating photobleaching. 70% of the absorption signal (thin line) recovers with a time constant of (5900 ± 25) s, followed by a slower phase. In contrast, kinase activity (●) displays sigmoid recovery kinetics which can be described by the model shown in panel C (thick line).
C. Model for light inactivation of net kinase activity in dimeric YF1. D and L denote LOV domains in their dark and light states, respectively. Maximal kinase activity requires both YF1 LOV domains in the dark state. Photobleaching of one or both LOV domains strongly diminishes kinase activity. Analysis of YF1 recovery data according to this model yields a microscopic time constant τ of (10800 ± 1600) s.
A minimal model to account for both recovery and photobleaching data is shown in Fig. 3C. In this model, we assume that full kinase activity of YF1 requires both LOV domains to be in their dark state, and that after photobleaching, the two LOV monomers of YF1 independently relax to their dark state with the microscopic time constant τ. The value of τ of (10800 ± 1600) s is consistent with the recovery kinetics of the absorption signal after photobleaching of YF1 (Fig. 3B). According to the model, the absorption signal should recover with an observable time constant τ/2 = (5400 ± 800) s which agrees well with the measured value of (5900 ± 25) s. The model also accounts for the observation that upon partial photobleaching, YF1 kinase activity diminishes twice as fast as the absorption signal (Fig. 3A). Despite the simplicity of the model, it quantitatively explains the experimental data remarkably well. However, we cannot rule out other, more complex reaction schemes.
In summary, these experiments demonstrate that the effect of light on YF1 activity is mediated by its LOV domain, is fully reversible, and requires that YF1 be a dimer.
Kinase Activity and Regulation Depend on Properties of the Domain Linker
To address how the properties of the linker between the LOV sensor and kinase effector domains affect enzyme activity and regulation by light, we generated a number of protein variants which differ in that region. In a first series, we varied the site within the Jα helix at which the two domains are fused but retained the same linker length as in YF1. Variants YF2–4 successively derive more residues from FixL as indicated in Fig. 1B and Table I. Variant YF3 was inactive, but YF2 and YF4 showed light inactivation of net kinase activity as seen for YF1 albeit with quantitatively different activities in the dark of (67.9 ± 6.5) h−1 and (3.4 ± 0.1) h−1, respectively (Suppl. Fig. 4).
A second series of variants contains successive deletions of amino acids 274–277 at the C-terminus of the Jα linker sequence of YF1 (Fig. 1B, Table I). Kinase activity in variants with 1, 2 or 4 residue deletions in the linker was below the detectable limit both in the dark and light (Fig. 4A). Construct YF1Δ275–277 showed low activity of about 0.3 h−1 but regulation by light was abolished.
Figure 4.
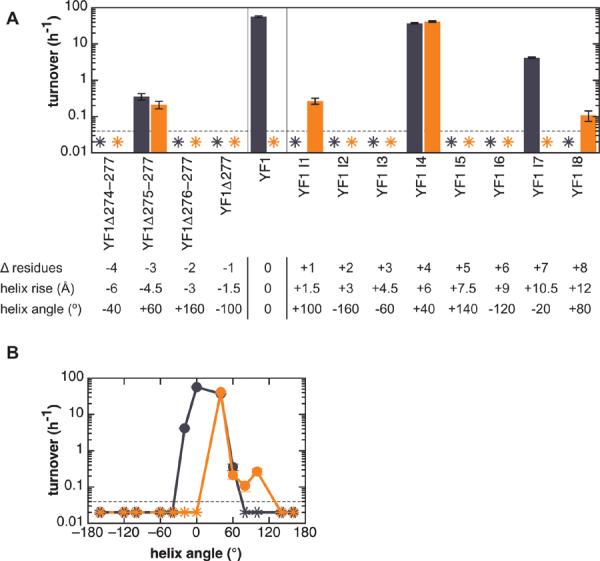
A. Activity of fusion kinase variants in turnover assays. Blue bars denote activity in the dark and orange bars under constant illumination. Asterisks indicate activities below the detectable limit of 0.04 h−1 (dashed line). The table lists changes in helix angle and rise in the variants relative to YF1 in a canonical α-helix structure. Closely similar values are obtained when calculating these parameters based on a coiled coil model.
B. Activity of linker variants as a function of helix angle relative to YF1. Variants with helix angles between −20° to +60° showed kinase activity in the dark; variants with angles between +40° to +100° were active in the light.
In a third series of variants, YF1 I1–8, up to eight additional amino acids were inserted into the Jα linker region (Table I). To reduce the chance of incorporating deleterious residues, the insertion site and the nature of the amino acids introduced were determined based on sequence comparison to Caulobacter crescentus LovK kinase29 (Suppl. Fig. 5). Insertion of 2, 3, 5 or 6 amino acids led to variants YF1 I2, YF1 I3, YF1 I5 and YF1 I6 that were inactive in the light and the dark (Fig. 4A). Variant YF1 I7 had higher net kinase activity in the dark ((4.2 ± 0.2) h−1) than in the light (< 0.04 h−1), as in the parent YF1. Variant YF1 I4 had activity in the dark of (37.2 ± 1.7) h−1 comparable to YF1, but this activity was independent of light. Lastly, YF1 I1 and YF1 I8 with 1 or 8 extra amino acids showed inverse dependence on light. Although with much lower activity than YF1, both variants were more active in the light (around 0.1–0.3 h−1) than in the dark (< 0.04 h−1). That is, they were light-activated kinases.
Taken together, the deletion and insertion variants display a striking heptad periodicity of kinase activity and light regulation (Fig. 4A). All pairs of fusion proteins that differ in their linker length by seven residues, as for example YF1 and YF1 I7, or YF1 I1 and YF1 I8, show remarkably similar activity patterns. Moreover, the length of the domain linker is an important design parameter that can be used to invert the regulatory behavior; insertion of one amino acid into the linker region of YF1 replaced net light-inactivated kinase activity by net light-activated kinase activity.
In Vivo Activity of Fusion Kinases
As discussed above, FixL activates expression of genes involved in nitrogen metabolism in an oxygen-dependent manner. Could we reengineer this regulatory network and use our fusion kinases to regulate gene expression in response to light in vivo?
The activity of fusion kinases in E. coli was determined in β-galactosidase assays using a reporter construct which expresses LacZ under the control of the B. japonicum FixK2 promoter to which phospho-FixJ binds15. In the absence of FixJ only low levels of β-galactosidase activity around 15 Miller Units/(mL·min) were observable (Table II). Upon introduction of B. japonicum FixJ alone into E. coli, expression of the LacZ reporter was constitutively activated with β-galactosidase activities of around 3800 Miller Units/(mL·min). Constitutive activation of a related FixJ pathway has been ascribed to phosphorylation of FixJ either enzymatically by an endogenous E. coli histidine kinase or non-enzymatically by intracellular acyl phosphates23.
Table II.
In vivo Assay of Fusion Kinase YF1
| expression plasmid | β-galactosidase activity (Miller Units/(mL·min)) | |
|---|---|---|
| dark | light | |
| YF1 only | 16 ± 3 | 15 ± 4 |
| FixJ only | 3730 ± 250 | 3910 ± 50 |
| YF1 / FixJ | 2860 ± 360 | 42 ± 16 |
| YF1 C62A / FixJ | 800 ± 250 | 650 ± 250 |
Constitutive activation of the FixJ pathway precludes measurements of kinase activity of our fusion kinases in E. coli. However, our in vitro experiments showed that light switches YF1 from net kinase to phosphatase activity (Fig. 2C). Might the phosphatase activity of YF1 in its light state be sufficient to counteract constitutive phosphorylation of FixJ in E. coli? Upon introduction of both YF1 and FixJ, we observed β-galactosidase activity of (2860 ± 360) Miller Units/(mL·min) in the dark (Table II). In the light, β-galactosidase activity was suppressed by 70-fold to (42 ± 16) Miller Units/(mL·min). When YF1 was replaced by the photoinactive YF1 mutant C62A, β-galactosidase activity was independent of light (Table II), consistent with the in vitro kinase activity for this mutant (Suppl. Fig. 4).
We conclude that the fusion kinase YF1 is active in vivo and regulates gene expression in a light-dependent manner, in accord with our in vitro results. Interestingly, in E. coli regulation of gene expression by light appears to be mediated mainly by the phosphatase activity of YF1.
Discussion
The Linker between the Sensor and Effector Domains Forms an α-Helical Coiled Coil
In the structure of the isolated YtvA LOV domain13, the C-terminal Jα linker forms an α-helix. Although the structure of the histidine kinase domain of FixL is not known, that of the homologous histidine kinase HK853 from Thermotoga maritima shows that the N-terminus of the DHp subdomain also forms an α-helix30. We propose that the linker between the sensor and effector domains within our fusion proteins is formed by uniting these two shorter helices into a long, continuous, signaling α-helix31.
Both the YtvA LOV domain and the histidine kinase HK853 form parallel dimers, which in HK853 is stabilized by an N-terminal four-helix bundle comprising its DHp subdomain30. The striking heptad periodicity of kinase activity and regulation in the domain linker variants (Fig. 4A) argues that a coiled coil structural element might be crucial for regulating kinase activity in response to light. This coiled coil would be formed by parallel dimerization of the signaling helices. We therefore examined the sequences of PAS kinases for the characteristic features of coiled coils. From the SMART database we retrieved 3194 protein sequences in which a PAS domain immediately precedes a DHp subdomain32. The conserved H-box motif containing the phosphoacceptor histidine in the DHp subdomain could be readily identified in all sequences, as could a novel, highly conserved DIT motif at the junction between the core of the PAS domain and the Jα helix (Fig. 5). With two key residues now fixed (the D of the DIT motif at the PAS – Jα junction and the phosphoacceptor H in the DHp subdomain), precise alignment of the domain linkers of the different kinases was possible. Strikingly, the lengths of these linkers were not uniformly distributed but fell into two distinct classes: those with 7n residues between the D and H, and those with 7n + 2 residues, where n = 3, 4, 5 or 6 (Suppl. Fig. 6). Sequence alignment of the two classes separately revealed that although the sequences of the linker are only weakly conserved, both clearly show the heptad periodicity of hydrophobic residues characteristic of α-helical coiled coils (Fig. 5;33). Within each class, insertion of 7, 14 or 21 residues within the linker would introduce 2, 4 or 6 helical turns respectively and more importantly, preserve the heptad periodicity. The two classes show a remarkably similar pattern of hydrophobicity within their linkers (Fig. 5) but differ at the N-terminus of their DHp domains. In the 7n+2 class, this region is two residues longer and rich in glycine and charged amino acids. In the absence of detailed structural information on members of the two classes, we can only speculate on how they differ structurally. However, two additional residues could readily be accommodated by deviations from ideal α-helical and coiled coil structure34. It is not clear whether the 7n and 7n+2 sequence classes also differ in a fundamental functional property. We note that a similar sequence pattern was observed within HAMP domains, which are often found N-terminal to membrane-associated histidine kinases35.
Figure 5.
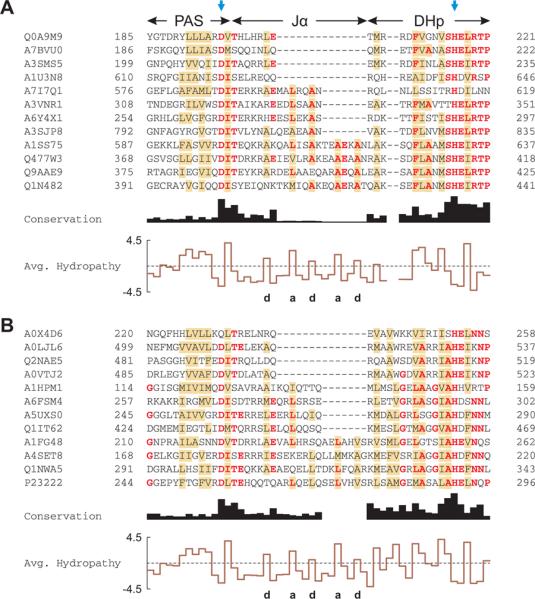
Multiple sequence alignment of PAS histidine kinases.
A. Alignment of PAS histidine kinases that have 7n residues between the C-terminus of the PAS domain and the active-site histidine, indicated by the blue arrows. Out of 1811 sequences analyzed, 12 are shown and labeled with their UniProt identifiers. Residues conserved in more than half of all 1811 sequences are shown in bold red; brown shading denotes columns with more than 50% hydrophobic residues. The C-terminus of the PAS domains displays a highly conserved DIT consensus motif. The linker between the PAS and DHp domains shows the pattern of hydrophobic residues (labeled a and d) characteristic of α-helical coiled coils. Two gap positions have been arbitrarily inserted into the alignment to facilitate comparison with panel B.
B. Alignment of PAS histidine kinases that have 7n+2 residues between the PAS domain and the active-site histidine. Out of 960 sequences analyzed, 12 are shown; B. japonicum FixL is in the bottom row (P23222). In comparison to panel A, the DHp domain differs in sequence and length around the phosphoacceptor histidine.
We conclude that the linker between sensor and histidine kinase domains is largely α-helical and forms a coiled coil in the dimer, as judged by the heptad periodicity of kinase activity and regulation in our fusion proteins, linker length and hydrophobicity.
Transmission of Signal from the Sensor to the Histidine Kinase Domain
This structural model readily accounts for the strong dependence of net kinase activity on insertions or deletions of one or more residues in the linker. Each insertion or deletion within a monomeric α-helix causes a change of 100° in helix angle and of 1.5 Å in helix rise (Fig. 4A). Within a dimeric coiled coil, each insertion or deletion would in addition alter the heptad periodicity and thus the structure and/or stability of the coiled coil, and reposition the YtvA-LOV sensor domain with respect to the DHp phosphoacceptor and CA catalytic subdomains with potentially profound effects on enzyme activity and regulation. However, variants of fusion proteins which differ in length by seven residues (such as YF1 and YF1 I7) would retain the heptad periodicity of the coiled coil and a closely similar relative angular orientation of the LOV and histidine kinase domains, although the distance between these domains would change by about 10 Å (Fig. 4A). Such insertions of seven residues are evident in natural PAS kinases which exhibit a wide range of linker lengths (Suppl. Fig. 6A).
We conclude that the primary effect on the regulation of kinase activity by light arises from the relative angular orientation of the sensor and effector domains, as established by the structure of the coiled coil that links them. However, the length of the linker and its exact sequence exert secondary effects, as indicated by the range of activity in kinase variants YF1-4 (Suppl. Fig. 4).
How could signals originating in absorption of light by the FMN chromophore be transmitted to and along α-helices? Absorption of a photon leads to small tertiary and quaternary structural changes in the isolated LOV domain of YtvA, in particular in the region of the conserved DIT motif (see Fig. 6b of13). The aspartate and threonine residues of this motif directly couple the core of the LOV domain to the Jα helix, by a salt bridge between the side chains of D125 and K96 and a hydrogen bond between the side chain of T127 and the backbone amide of W103. Due to their large persistence length, at the molecular level α-helices behave as rigid rods and are well suited to transmit conformational changes over long distances to remote sites31, even in monomeric proteins. Within a dimer, signal-dependent perturbation of a coiled coil would generate a quaternary structural change that could readily propagate over long distances.
Several models for signal transduction along α-helices have been proposed, primarily in the context of integral membrane proteins that act as chemosensors. Transmission of signals may depend on translational, piston-type, pivot-type or rotational movements of helices, or a combination thereof36. Do these models apply here? Linker variants with helix angles relative to YF1 between −20° to +60° showed some kinase activity in the dark; variants with helix angles between +40° and +100° showed activity in the light (Fig. 4B). Thus, both in the dark and in the light, activity spanned a range of helix angles of 60° to 80°, but the dark and light activity curves are displaced with respect to each other by 40° to 60°. Displacement of the activity curves enables the generation of either light-inactivated systems (here, with helix angles between −20° and 0°), systems whose activity is insensitive to light (angles between +40° and +60°) or light-activated systems (angles between +80° and +100°). These data suggest that the major effect of light is to induce relative rotational motion between the LOV sensor and histidine kinase effector domains, probably through perturbation of the coiled coil. Light is acting as a rotary switch. Translational, piston-type or pivot-type movements, if present, are secondary.
In this model, the tight dimer interface of the LOV sensor domain provides an anchor point around which the histidine kinase domains can rotate. The displacement of the dark and light activity curves by 40 to 60° suggests that light absorption induces a 40 to 60° rotation of the histidine kinase domain relative to the LOV sensor domain within the dimeric fusion kinase, accompanied by partial helix unfolding or untwisting of coiled coil regions, as indicated by our CD measurements. Light-induced rotational movements of the histidine kinase domains might alter the geometry of their active site, or affect the binding of the response regulator FixJ to the DHp subdomain. In principle, any or all of the rate coefficients associated with each elementary step in the overall kinase, phosphotransfer and phosphatase reactions could thereby be modulated by light (Fig. 2A). It is interesting to note that both YF1 and FixL represent kinases which are inactivated by their signals, light and oxygen, respectively. Intriguingly, all natural LOV histidine kinases characterized to date show the opposite regulatory behavior, i.e. increased kinase activity upon light absorption29,37. A more detailed understanding has to await structural information on YF1 or related sensor kinases.
Models for ligand-dependent signal transduction involving helix rotation have been invoked for related membrane-bound chemosensors38,39. In particular, a recent study suggests signal-dependent rotation of two helices within the parallel four-helix bundle of a HAMP domain by about 26° each, generating a movement of 52° relative to each other35,40. For the dimeric, quorum-sensing LuxPQ receptor, a signaling mechanism involving a 40° rotation of one subunit was advanced41.
Design of Novel, Light-regulated Proteins
Chimeric histidine kinases with altered signaling properties have been constructed before based on seminal work by Inouye and coworkers42,43,44. In these studies, change of signal specificity of integral membrane receptors was achieved by replacement of their cytoplasmic domains. Our data show that this general approach is not restricted to membrane proteins but also applicable to soluble proteins. Indeed, a chemosensor domain (here, for oxygen) can be functionally replaced by a photosensor (here, for blue light). The signaling principles are therefore generally applicable to proteins in which the sensor and effector domains are linked by α-helices, and to both chemo- and photosensors.
In an elegant study, Laub and coworkers very recently demonstrated that the substrate specificity of related histidine kinases primarily resides within their DHp domains45. The output of these histidine kinases could thus be reprogrammed to phosphorylate different response regulators either by replacing their DHp domains or by mutating as few as three residues. Our data show that the input specificity can similarly be reprogrammed, thus enabling complete control of the activity of sensor histidine kinases.
By directly replacing the oxygen-sensing PAS domain in FixL by the YtvA LOV photosensor domain, we generate the light-inactivated histidine kinase YF1 that retains the high catalytic efficiency and substrate affinity of its parent enzyme but responds to a different input signal. It did not prove necessary to screen a large library of fusion proteins. Although our successful initial design was facilitated by sequence and structural homology between the FixL and YtvA sensor domains, our design strategy is readily applicable to a large group of histidine kinases and other proteins as the sequence analysis shows (Fig. 5). Even in the absence of detailed structural information, the C-terminus of many PAS domains can now be reliably defined based on the conserved DIT sequence motif. We note that this DIT motif was the site at which YtvA and FixL were fused to generate the light-inactivated histidine kinase YF1 (Fig. 1B). We therefore suggest that functional fusion proteins in other systems could be readily generated, that light sensitivity could thereby be conferred on kinases and other, desired effector domain activities, and that successful design does not necessarily require screening big libraries of different constructs.
Although PAS domains can be identified by sequence analysis, in most cases the small molecule or metabolite to which they respond is unknown, often despite much biochemical effort. One prominent example is human PAS kinase46. Replacement of such putative chemosensor PAS domains with known LOV photosensor domains puts the activity of their associated effector domain under the control of a known and readily manipulable signal, light. Using light as a stimulus, the roles of these proteins and the signaling pathways in which they are involved could be identified.
LOV domains can also be used to regulate the activity of proteins not naturally associated with PAS domains. By fusing a LOV photosensor domain to the N-terminus of a Trp repressor, DNA binding was rendered light-sensitive although the effect was modest, with a 5.6 fold enhancement47.
We expect that similar design strategies will lead to the generation of further light-controlled proteins. The use of light as a signal affords excellent spatiotemporal and noninvasive control over the signaling process. Moreover, as LOV domains employ the ubiquitous cofactor FMN they can be readily genetically encoded and used for in vivo studies, as we have shown. Synthetic light-regulated proteins are also of considerable interest in the study of fast protein dynamics, for example by time-resolved X-ray crystallography48.
Materials and methods
Molecular Biology and Protein Purification
Genes encoding YtvA, FixL and FixJ were amplified via PCR and cloned into pET28c vectors. Fusion constructs were generated by fusion PCR, overlap-extension PCR and site-directed mutagenesis. Proteins were purified similar to the protocol described in13. Concentrations were determined using extinction coefficients of 12500 M−1 cm−1 at 450 nm for the fusion proteins, 1.26·105 M−1 cm−1 at 395 nm for FixL and 4860 M−1 cm−1 at 280 nm for FixJ.
Phosphorylation Assays
Kinase activity assays were conducted by adapting established procedures28. Experiments were done in reaction buffer (50 mM Tris/HCl pH 8.0, 50 mM KCl, 100 μM MnCl2, 5% (v/v) ethylene glycol) at (23 ± 1) °C. For the assay of the fusion proteins, 5 mM β-mercaptoethanol was added. Phosphorylation reactions were started by adding 1 mM γ-32P ATP/MgCl2. Aliquots were mixed with half the volume of stop buffer (0.5 M Tris/HCl pH 6.8, 0.2 M NaCl, 40 mM EDTA, 30% (v/v) glycerol, 4% (v/v) SDS, 2% (v/v) β-mercaptoethanol) and visualized on SDS gels. Radioactivity in dried gels was quantified using a Phosphorimager. The standard phosphorylation reaction mixture contained 2 μM kinase and 50 μM FixJ. Kinase experiments were conducted in the dark (i.e. dim red light) or under constant illumination from a fiber optics lamp (white light, 136 mW power).
In vivo Experiments
All in vivo experiments were conducted in E. coli Tuner cells (ΔlacZY). Fusion proteins were expressed in tandem with FixJ from pET28-based plasmids. Net kinase activity was assayed using a pRK290-derived plasmid which carried the reporter gene LacZ under the control of the B. japonicum FixK2 promoter. β-galactosidase activities were quantified as described and represent the averages of at least five independent cultures49. Cell cultures were either kept in the dark or constantly illuminated (white light, ca. 1 mW) and otherwise treated identically.
Multiple Sequence Alignment
Sequences of proteins comprising both PAS and DHp domains were retrieved from the SMART database32 and aligned with CLUSTAL W50. Average hydropathies were calculated according to51. Evaluation of the sequence alignment was done with custom Perl scripts.
Supplementary Material
Acknowledgements
We thank Drs. Hauke Hennecke and Hans-Martin Fischer (ETH Zürich) and Dr. Michael Sadowsky (University of Minnesota) for supplying bacterial strains and plasmids. Drs. Tobin Sosnick, Sean Crosson and Michael Elowitz (CalTech) are acknowledged for providing materials, advice and comments on the manuscript. We gratefully appreciate the use of facilities of Drs. Francisco Bezanilla, Sean Crosson, Phoebe Rice and Tobin Sosnick. Laura Satkamp provided advice on fusion PCR, and Clark Hyde helped with lamp power measurements. We thank all members of the Moffat lab for discussion. Supported by NIH grant GM036452 to K.M.
Abbreviations
- LOV
light-oxygen-voltage
- PAS
Per-Arnt-Sim
Footnotes
Authors' contributions: AM and KM designed research. AM and RAA performed research and analyzed data. AM and KM wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–8. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 3.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–43. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 4.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 6.Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–55. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 7.David M, Daveran ML, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–83. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 8.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–4. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 9.Key J, Hefti M, Purcell EB, Moffat K. Structure of the Redox Sensor Domain of Azotobacter vinelandii NifL at Atomic Resolution: Signaling, Dimerization, and Mechanism. Biochemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 10.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 11.Lee BC, Pandit A, Croonquist PA, Hoff WD. Folding and signaling share the same pathway in a photoreceptor. Proc Natl Acad Sci U S A. 2001;98:9062–7. doi: 10.1073/pnas.111153598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa H, Lee DS, Watanabe M, Sagami I, Mikami B, Raman CS, Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–93. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- 13.Möglich A, Moffat K. Structural Basis for Light-dependent Signaling in the Dimeric LOV Domain of the Photosensor YtvA. J Mol Biol. 2007;373:112–26. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilles-Gonzalez MA, Ditta GS, Helinski DR. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–2. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 15.Fischer HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–86. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Losi A, Polverini E, Quest B, Gärtner W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–34. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aravind L, Koonin EV. The STAS domain - a link between anion transporters and antisigma-factor antagonists. Curr Biol. 2000;10:R53–5. doi: 10.1016/s0960-9822(00)00335-3. [DOI] [PubMed] [Google Scholar]
- 18.Salomon M, Eisenreich W, Durr H, Schleicher E, Knieb E, Massey V, Rudiger W, Muller F, Bacher A, Richter G. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc Natl Acad Sci U S A. 2001;98:12357–61. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbar S, Gaidenko TA, Kang CM, O'Reilly M, Devine KM, Price CW. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J Bacteriol. 2001;183:1329–38. doi: 10.1128/JB.183.4.1329-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A. 1998;95:15177–82. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Key J, Moffat K. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry. 2005;44:4627–35. doi: 10.1021/bi047942r. [DOI] [PubMed] [Google Scholar]
- 22.Gilles-Gonzalez MA, Gonzalez G, Perutz MF, Kiger L, Marden MC, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–73. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 23.Weinstein M, Lois AF, Ditta GS, Helinski DR. Mutants of the two-component regulatory protein FixJ of Rhizobium meliloti that have increased activity at the nifA promoter. Gene. 1993;134:145–52. doi: 10.1016/0378-1119(93)90088-k. [DOI] [PubMed] [Google Scholar]
- 24.Russo FD, Silhavy TJ. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993;1:306–10. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:11057–61. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuckerman JR, Gonzalez G, Dioum EM, Gilles-Gonzalez MA. Ligand and oxidation-state specific regulation of the heme-based oxygen sensor FixL from Sinorhizobium meliloti. Biochemistry. 2002;41:6170–7. doi: 10.1021/bi025628w. [DOI] [PubMed] [Google Scholar]
- 27.Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–15. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- 28.Gilles-Gonzalez MA, Caceres AI, Sousa EH, Tomchick DR, Brautigam C, Gonzalez C, Machius M. A proximal arginine R206 participates in switching of the Bradyrhizobium japonicum FixL oxygen sensor. J Mol Biol. 2006;360:80–9. doi: 10.1016/j.jmb.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 29.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–6. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. Embo J. 2005;24:4247–59. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anantharaman V, Balaji S, Aravind L. The signaling helix: a common functional theme in diverse signaling proteins. Biol Direct. 2006;1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 34.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 35.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–40. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 36.Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127:447–50. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–3. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 38.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr., Kim SH. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–7. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 39.Cochran AG, Kim PS. Imitation of Escherichia coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science. 1996;271:1113–6. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 40.Inouye M. Signaling by transmembrane proteins shifts gears. Cell. 2006;126:829–31. doi: 10.1016/j.cell.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–9. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 43.Jin T, Inouye M. Transmembrane signaling. Mutational analysis of the cytoplasmic linker region of Taz1-1, a Tar-EnvZ chimeric receptor in Escherichia coli. J Mol Biol. 1994;244:477–81. doi: 10.1006/jmbi.1994.1746. [DOI] [PubMed] [Google Scholar]
- 44.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–2. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 45.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–54. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci U S A. 2001;98:8991–6. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci U S A. 2008;105:10709–14. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat K. Time-resolved biochemical crystallography: a mechanistic perspective. Chem Rev. 2001;101:1569–81. doi: 10.1021/cr990039q. [DOI] [PubMed] [Google Scholar]
- 49.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 50.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


