Abstract
Inhibition of PKC activity in transformed cells and tumor cells containing activated p21Ras results in apoptosis. To investigate the pro-apoptotic pathway induced by the p21Ras oncoprotein, we first identified the specific PKC isozyme necessary to prevent apoptosis in the presence of activated p21Ras. Dominant-negative mutants of PKC, siRNA vectors, and PKC isozyme-specific chemical inhibitors directed against the PKCδ isozyme demonstrated that PKCδ plays a critical role in p21Ras-mediated apoptosis. An activating p21Ras mutation, or activation of the PI3K Ras effector pathway, increased the levels of PKCδ protein and activity in cells, whereas inhibition of p21Ras activity decreased the expression of PKCδ protein. Activation of the Akt survival pathway by oncogenic Ras required PKCδ activity. Akt activity was dramatically decreased after PKCδ suppression in cells containing activated p21Ras. Conversely, constitutively-activated Akt rescued cells from apoptosis induced by PKCδ inhibition. Collectively, these findings demonstrate that p21Rasthrough its downstream effector PI3K, induces PKCδ expression and that this increase in PKCδ activity, acting through Akt, is required for cell survival. The p21Ras effector molecule responsible for the initiation of the apoptotic signal after suppression of PKCδ activity was also determined to be PI3K. PI3K (p110-CAAX), was sufficient for induction of apoptosis after PKCδ inhibition. Thus, the same p21Ras effector, PI3K, is responsible for delivering both a pro-apoptotic signal, and a survival signal, the latter being mediated by PKCδ and Akt. Selective suppression of PKCδ activity and consequent induction of apoptosis is a potential strategy for targeting of tumor cells containing an activated p21Ras.
The Ras oncogene family is among the most commonly mutated group of genes in human cancer. Its protein products code for three closely-related, p21ras proteins including H-ras, K-ras and N-ras. p21Ras proteins are localized in the inner plasma membrane, bind GDP and GTP and possess an intrinsic GTPase activity. p21Ras proteins function as plasma membrane-bound guanine nucleotide binding proteins and act as molecular switches, thereby regulating signal transduction pathways for hormones, growth factors and cytokine receptors (1). Several downstream effector proteins of p21Ras have been identified which bind preferentially to p21Ras in the GTP-bound state, including Raf, phosphoinositide 3-OH kinase (PI3K), and a family of GDP-GTP exchange factors for the Ral small GTPases (Ral-GDS). Raf proteins, which are proto-oncogene-encoded serine/threonine kinases, activate the MEK-ERK signaling pathway. PI3K activation results in the activation of the anti-apoptotic serine/threonine kinase Akt, among other molecules. Other p21Ras targets include the GTPase-activating proteins, p120GAP and neurofibromin (2). p21Ras proteins were shown to influence proliferation, differentiation, transformation, and apoptosis by relaying mitogenic and growth signals from the membrane into the cytoplasm and the nucleus.
Specific point mutations localized in codons 12, 13, 59, 61, 63, 116, 117, and 146 can lock the p21Ras protein in the active GTP-bound state and permit stimulation of downstream signaling cascades in the absence of extrinsic p21Ras activation. Ras mutations can be found in human malignancies with an overall frequency of 20%. A particularly high incidence of Ras gene mutations has been reported in malignant tumors of the pancreas (80–90%, K-ras), in colorectal carcinomas (30–60%, K-ras), in non-melanoma skin cancer (30–50%, H-ras), and in hematopoietic neoplasia of myeloid origin (18–30%, K-and N-ras) (3).
In addition to its central involvement in cell proliferation, recent studies indicate that the presence of an activated p21Ras protein sensitizes transformed or malignant cells to apoptotic stimuli (4–9). Various signaling pathways have been proposed for this pro-apoptotic activity. Chou et al (10) reported activated p21Ras can cause apoptosis in transformed murine fibroblast cells through activation of the transcription factor NFκB. Another study suggested that the p21Ras/MAP kinase pathway is involved in Ras-specific apoptosis (11). The latter study also found that activating p21Ras mutations increased colon cancer cell sensitivity to 5-FU-induced apoptosis through the negative regulation of gelsolin expression (12). Our previous studies demonstrated suppression of protein kinase C (PKC) activity in cells expressing activated p21Ras rapidly induces apoptosis via FADD/caspase-8 signaling (9). We also found that reactive oxygen species are necessary as downstream effectors of the Ras-mediated apoptotic response to PKC inhibition (7).
There are at least 12 PKC isoforms that are classified into three subfamilies according to the structure of the N-terminal regulatory domain, which determines their sensitivity to the second messengers Ca2+ and diacylglycerol (DAG) (13). Despite the high degree of homology, however, there is a surprising degree of non-redundancy. Thus, individual PKC isoforms mediate different and unique cellular functions in different cell types and different tissues (14). PKCδ belongs to the subfamily of novel isoforms (PKCδ, PKCε, PKCθ and PKCη), which are insensitive to Ca2+. PKCδ is widely regarded as having pro-apoptotic properties (15–17). Caspase activation mediates cleavage of PKCδ which results in release of the active catalytic domain (18, 19). In addition, PKCδ activity is known to initiate a number of pro-apoptotic signals, such as increased expression and stability of p53 (20, 21), mitochondrial cytochrome C release (22, 23) and c-Ab1 activation (24). But recent studies have also shown that PKCδ can protect cells against apoptotic stimuli under certain conditions (25). PKCδ has been reported to regulate B-lymphocyte survival (26). Knock-out experiments have shown that PKCδ-deficient mice have a severely deregulated immune system and develop autoimmue disease (27, 28). Thus, PKCδ activation can serve as pro-apoptotic signal, or as survival signal, to determine cell fate.
The present study examines the mechanism of apoptotic signaling induced by the p21Ras oncoprotein. We found that PKCδ plays a critical role in suppressing p21Ras-mediated apoptosis, and selective inhibition of this isozyme initiates apoptosis in cells containing activated p21Ras. Our data further demonstrate that unregulated Ras activity, through activation of the downstream effector PI3K, up-regulates PKCδ expression and subsequently activates Akt, generating an anti-apoptotic effect and protecting against Ras-mediated apoptosis.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
The activated Ras effectors, Raf-1 (Raf-22W), PI3K (p110-CAAX), Rlf (Rlf-CAAX) and Ral-GDS (RalA-28N, dominant-negative) were cloned into the pBabe puro vector (29) and the HRas effector loop mutants, S35, G37 and C40 (double mutations) and H-ras V12 single mutant (V12) were cloned into the pSG5 vector (30). These vectors were kindly provided by C. Counter (Duke University) and J. Downward (MRC, UK). The pEGFP-PKCδ-KR (dominant-negative PKCδ) vector (31) was kindly provided by Dr. D. Kufe (Dana-Farber Cancer Institute). pEF1α-vAkt and pEF1α-cAkt were kindly provided by Dr. Geoffrey Cooper (Boston University). The GST-NORE CT and FLAG-MST1 CT vectors were generously provided by Dr. J. Avruch (Massachusetts General Hospital). Chemical inhibitors used in this study specific to PKC isozymes, PI3K, p21Rasand MAPK are listed in Table 1. All inhibitors were dissolved in dimethyl sulfoxide for use, and their effects were measured relative to dimethyl sulfoxide (vehicle)-treated controls. The concentration of all inhibitors was optimized to produce greater than 90% inhibition of target molecule activity.
Table 1.
Chemical kinase inhibitors used in this study
| Name | Concentration (µM) |
Target | Vendor |
|---|---|---|---|
| FPT III | 100 | p21Ras | Calbiochem |
| Rottlerin | 20 | PKCδ | Calbiochem |
| Ly294002 | 10 | PI3K | Calbiochem |
| PD98095 | 10 | MAPK | Calbiochem |
| Safingol | 5 | PKCα | Calbiochem |
| Go6976 | 0.3 | PKCα/β | Calbiochem |
| Bisindolylmaleimide I | 10 | pan-PKC | Calbiochem |
| Ly333531 | 0.01 | PKC β1 and β2 | A.G.Scientific |
| HMG | 120 | pan-PKC | Calbiochem |
Cell culture and treatment
NIH/3T3 and Balb cell lines were obtained from ATCC (American Type Culture Collection, Rockville, MD). NIH/3T3-Ras cells were produced by stable transfection of v-Harvey ras, and selected and maintained in 0.5 mg/ml of Geneticin (Gibco BRL, Gaithersburg, MD). Ki-v-ras-Balb (KBalb) cells were produced by stable infection of Balb with retroviral vector stocks containing v-Kirsten ras, and were selected and maintained with Geneticin. The human pancreatic tumor lines Hs 766s, BxPC-3 and MIA PaCa-2, were obtained from ATCC. All cell lines were maintained in Dulbecco Modified Eagle medium (DMEM) (Gibco BRL) supplemented with 2 mM l-glutamine, 100 U/ml of penicillin, and 100 µg/ml of streptomycin (Gibco BRL). Media was additionally supplemented with 10% DCS (NIH/3T3, NIH/3T3-Ha-v-Ras, Balb/3T3, Ki-v-ras-Balb, and Balb-myc), or 10% FBS (BxPC-3, MIA PaCa-2). Cells were cultured at 37°C and 5% CO2.
Cell proliferation assay
Cell proliferation was assessed using an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (Roche, Mannheim, Germany). The number of viable cells growing in a single well on a 96-well microtiter plate was estimated by adding 10 µl of MTT solution (5 mg/ml in phosphatebuffered saline [PBS]). After 4 h of incubation at 37°C, the stain was diluted with 100 µl of dimethyl sulfoxide. The optical densities were quantified at a test wavelength of 550 nm and a reference wavelength of 630 nm on a multiwell spectrophotometer.
siRNA knockdown of PKCδ and PKCα
siRNA duplexes for PKCδ (siRNAs) were obtained from Qiagen (Valencia, CA). The siRNA sequences for targeting PKCδ were PKCδ-siRNA-1 (5'-GAUGAAGGAGGCGCUCAGTT-3') and PKCδ−siRNA-2 (5'-GGCUGAGUUCUGGCUGGA-CTT-3') (32). The corresponding scrambled siRNAs were used as negative control. These siRNA sequences were also cloned into the pRNA6.1-Neo vector with a GFP tag according to the manufacturer's instructions (GenScript, Piscataway, NJ). siRNA for PKCα (PKC-PKCα-V6) was purchased from Upstate (Lake Placid, NY). Transfection of siRNA (oligo) was performed using 50 nM PKCδ siRNA, or the same amount of scrambled siRNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Transfection of plasmid-based siRNA vectors was carried out using the same method. PKCδ protein levels were determined by immunoblot analysis.
Assay of PKCα and PKCδ kinase activities
PKCα and PKCδ activities were measured with an assay kit (Upstate Cell Signaling). After two days of treatment with rottlerin, cells were lysed in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 20 mM MgCl25 mM EGTA, 1 mM othovanadate, 50 µg/ml PSMF and 3 µg/ml aprotinin. PKCα and PKCδ were immunoprecipitated from 200 µg of protein extracts as described above. Immunocomplexes were washed three times with the kinase buffer (20 mM Tris-HCl, 10 mM MgCl2pH 7.5) and then incubated with a PKC-specific peptide substrate, [−32P]ATP, and inhibitors of cAMP-dependent kinase and calmodulin kinase for 10 min at 30°C. 32P incorporated into the substrate was separated from residual 32P using p81 filters and subsequently quantified by scintillation counting. β-actin antibody was used as negative control in immunoprecipitations.
p21Ras activity assays
Cells were cultured in DMEM containing 0.5% serum for 48 h. Then cells were lysed in a buffer containing 25 mM HEPES, pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 0.25% sodium deoxycholate, 10% glycerol, and 25 mM NaF. Protein was normalized to 1 µg/µl, and activated Ras was affinity-precipitated by mixing 1 mg of cell lysate with 10 µg of Raf-RBD-agarose bead conjugate (Upstate Biotechnology, Inc.) for 60 min at 4°C. The conjugates were washed three times in lysate buffer and then separated on a 10% SDS-polyacrylamide gel. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and immunoblotted with a monoclonal p21Ras antibody (BD Transduction Laboratories).
DNA profile analysis
1×105 cells were plated in a 60 mm dish and grown until confluent. Cells were harvested and resuspended with 1 ml of a 35% ethanol/DMEM solution for 5 min at room temperature. Cells were collected and stained with solution containing 50 µg propidium iodide /ml and 25 units RNase/ml in PBS, and incubated in the dark for 30 min at room temperature for flow cytometric analysis.
Immunoblotting analysis
Harvested cells were disrupted in a buffer containing 20 mM Tris (pH 7.4), 0.5% NP-40, and 250 mM NaCl. Total protein (40 µg) was separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes or PVDF membranes. Membranes were blocked overnight and probed with affinity-purified antibodies against p21Ras (BD Transduction Labs), PKCα, β, η, θ, δ (BD Transduction Lab), caspase-3, caspase-9 (Cell Signaling), β-actin (Sigma) Akt, phospho-Akt specific for serine473, Erk1/2, or p-Erk1/2 (Santa Cruz). After washing, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies and visualized using the Amersham enhanced chemiluminescence ECL system.
Cell apoptosis assay
Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end-labeling (TUNEL) assay was used for apoptosis assay. Briefly, cells were fixed with 4% paraformaldehyde in PBS overnight at 4°C. The samples were washed three times in PBS and permeabilized with 0.2% Triton X-100 in PBS for 15 min on ice. After rinsing twice with PBS, the samples were incubated with the TMR red TUNEL reagent (Roche) at 37°C in the dark, according to the manufacturer's instructions. Apoptotic cells were identified by fluorescent microscopy.
Statistical analyses
Results are expressed as mean ± SD. Statistical analysis was performed using Student's t-test, and P-values < 0.05 were considered significant.
RESULTS
Selective down-regulation of PKCδ induces apoptosis in cells with activated p21Ras
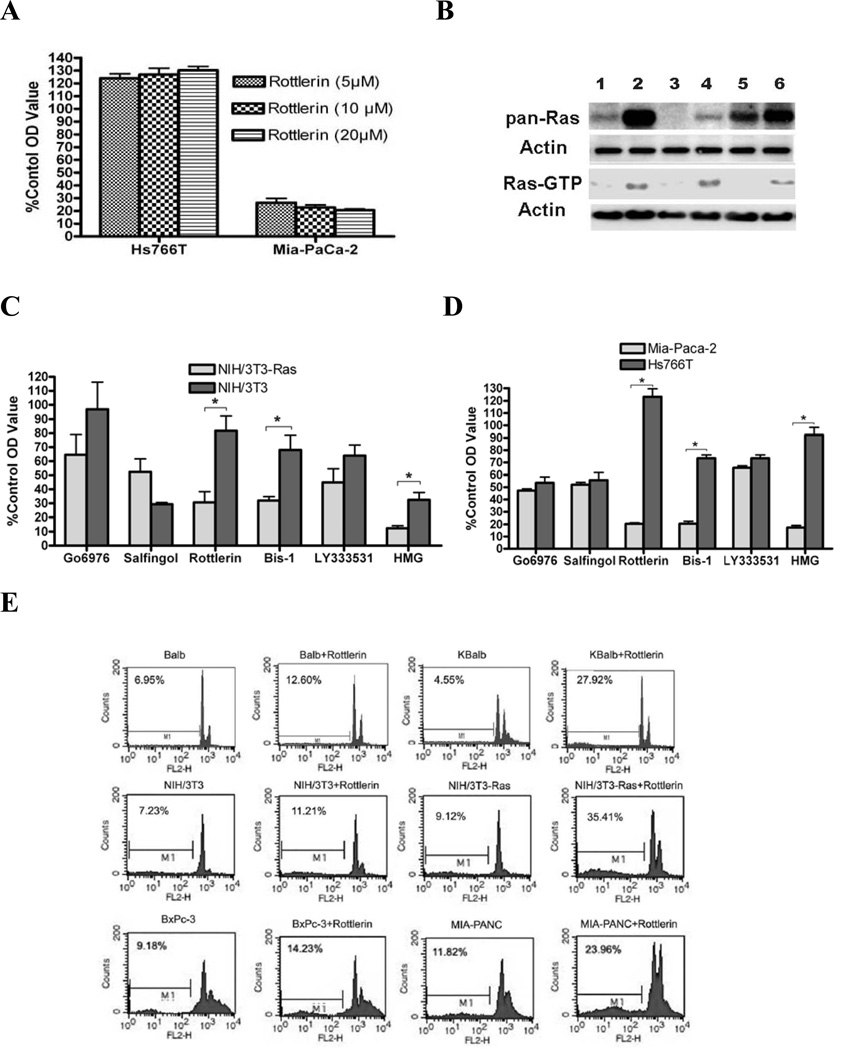
To begin determination of whether a specific PKC isozyme is responsible for suppression of Ras-mediated apoptosis, six isozyme-specific or non-specific PKC inhibitors were used to suppress PKCα, PKCα/β, PKCβ1/2, PKCδ or general PKC activity (Table 1). Optimal maximal effective concentrations of each inhibitor (>90% inhibition of relevant enzyme activity) were determined in pilot experiments (Fig. 1A or data not shown). The elevated activity of p21Ras in the KBalb, the MIA PaCa-2, and NIH/3T3-Ras cell lines, compared their counterpart cell lines (Balb, BxPc-3, and NIH/3T3, respectively), was confirmed by an activated p21Ras pull-down assay (Fig. 1B). Cells were treated for 48 h and cell proliferation was quantitated by an MTT assay. Among the isozymespecific inhibitors, rottlerin dramatically and specifically decreased the proliferation of both MIA PaCa-2 and NIH/3T3-Ras cells (both of which express a mutant, activated p21Ras protein), compared to the corresponding Hs766T cells and NIH/3T3 cells (which contain wild-type p21Ras) (Figs. 1C; 1D). The pan-PKC inhibitor Bis-1 and the DAG antagonist HMG also strongly suppressed the growth of the cells containing activated p21Rasconsistent with our previous studies, while having no significant effect on most of the cell lines which contained a wild-type p21Ras (7, 8). In contrast, rottlerin modestly but consistently stimulated proliferation of those cell lines expressing wild-type p21Ras (Balb and Hs766T cells). Cytofluorometric analysis of PI-stained nuclei showed that rottlerin caused 24–35% apoptosis (manifested as hypodiploid nuclei) in activated-Ras-containing cell lines after 60 h of treatment, whereas the matched cells containing wild-type p21Ras displayed only 12–14% apoptosis (Fig. 1E). PKCδ expression and activity was analyzed in the rottlerin-treated cells to confirm suppression of activity.
Figure 1.
Effects of isozyme-specific and non-specific PKC inhibitors on proliferation or apoptosis of mouse fibroblast cells and human pancreatic carcinoma cells with wild-type or activated p21Ras. Cells were grown to 80% confluence in 96-well plates, then treated with the inhibitors at the concentrations indicated in Table 1. The corresponding solvents at equivalent volumes were used as vehicle controls. After 48 h treatment, cell growth was evaluated by MTT assay or FACS. Results are presented as the means ± SD. Statistical significance was determined using a paired Student’s t-test, and p-values <0.05 were considered significant (*, p<0.01). (A) p21Ras-specific suppression of growth by PKCδ inhibition. Human pancreatic carcinoma cell cultures (Hs766T [wild-type p21Ras] and MiaPaCa-2 [activated p21Ras] were treated with different doses of rottlerin, and cell growth assayed by MTT assay. (B) Assay of cellular p21Ras activity. Nuclear-free lysates containing a total of 400 µg protein from each indicated cell type were used for analysis of Ras activity by Raf-RBD pull-down. Equal loading was demonstrated by re-probing the blot with anti-β-actin Ab. Pan-Ras protein expression levels were also analyzed. Lanes 1 to 6 represent: Balb, KBalb, NIH/3T3, NIH/3T3-Ras, BxPc-3 and MIA PaCa-2 cells respectively. (C) Growth of NIH/3T3 and NIH/3T3-Ras cells treated with isozyme-specific and non-specific PKC inhibitors. (D) Growth of Hs766T and MIA PaCa-2 cells treated with isozyme-specific and non-specific PKC inhibitors. (E) Nuclear DNA content of cells after inhibition of PKCδ. After 60 h treatment with rottlerin, cells were fixed and stained with propidium iodide, and the apoptotic (hypodiploid) fractions (M1 fractions) were evaluated by flow cytometry. Counts refers to cell numbers; FLH2 is a log scale of fluorescence channels. Similar results were observed in three independent experiments.
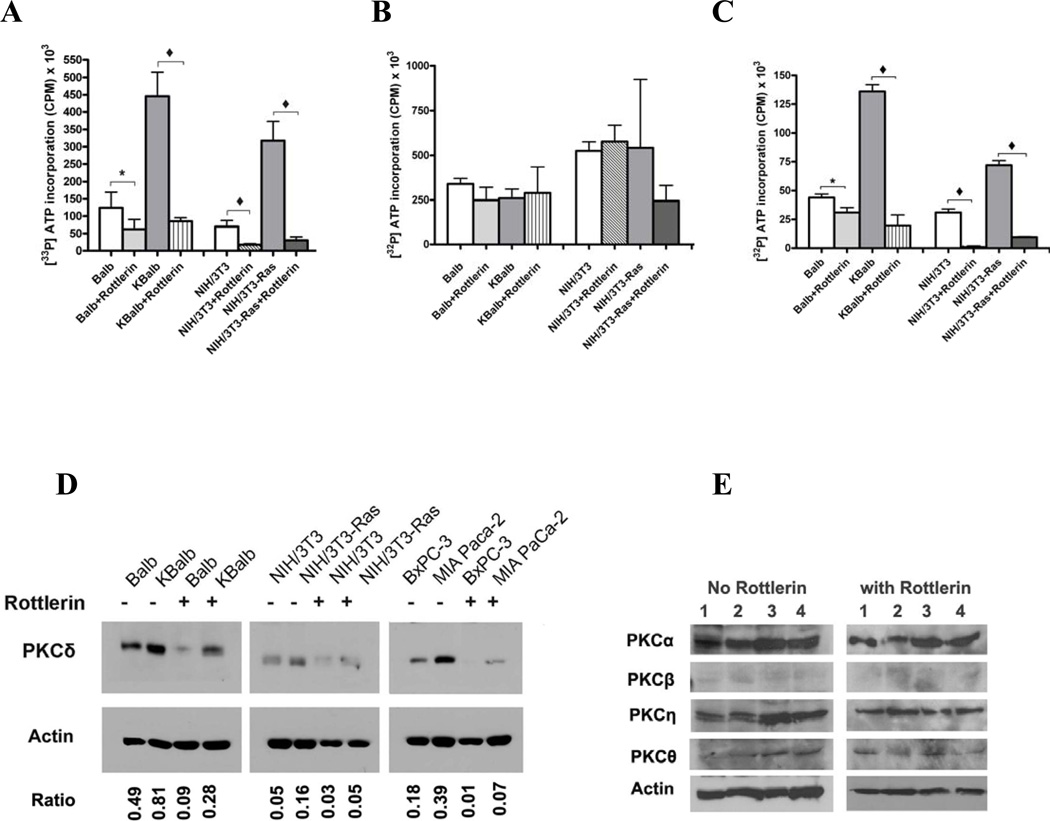
In order to confirm inhibition of PKCδ activity by rottlerin, both in vivo and in vitro kinase assays were performed. For the in vivo assays, cells were treated with rottlerin at 20 µM for 48 h, then lysed and PKCδ was immunoprecipitated by a specific anti-PKCδ antibody. The incorporation of [γ-32P] ATP into a PKC substrate peptide (QKRPSQRSKYL) by the immunoprecipitates was quantitated. Exposure to rottlerin, at the concentrations used in the apoptosis studies described above, blocked greater than 85% of the PKCδ activity in all of the cell lines tested, except Balb (Fig. 2A). In contrast, rottlerin produced only a slight, statistically-insignificant decrease in PKCα activity (Fig. 2B). For the in vitro kinase assay, rottlerin was added to immuno-purified PKCδ protein and incubated for 4 h, and PKCδ kinase activities were assayed using the artificial substrate. Rottlerin directly inhibited 55–90% of PKCδ activity (Fig. 2C),
Figure 2.
Effects of rottlerin on PKCδ and PKCα enzyme activity and protein levels. (A) PKCδ and (B) PKCα kinase activities. The indicated cell lines were treated with rottlerin (20 µM) for 48 h, then harvested for analysis. 400 µg of protein lysates were used for immunoprecipitation by isozyme-specific antibodies, and the immunopurified proteins were assayed using an artificial substrate. (C) PKCδ activity measured by in vitro assay. PKCδ was pulled down from cell lysates by IP and treated with rottlerin (20 µM) for 4 h, and then subjected to an in vitro kinase assay. (D) PKCδ protein levels in cells after rottlerin treatment. 40 µg aliquots of the same protein lysates used in the studies in panels A and B were separated and immunoblotted using an anti-PKCδ monoclonal antibody. Ratio: refers to the value of PKCδ expression/β-actin expression measured by Software BandLead 3.0. (E) PKC isozyme expression levels after treatment with rottlerin. 1×105 cells were plated in each well of a 6-well plate. Cells were treated with rottlerin (20 µM) or with the same volume of vehicle after they reached 80% confluence. After 48 h, cells were harvested, lysed in 50 µl lysis buffer and the lysates separated and subjected to immunoblot analysis using anti-PKC isoform-specific antibodies. Lanes 1 to 4 represent: Balb, KBalb, NIH/3T3 and NIH/3T3-Ras cells respectively. The immunoblots shown in the left and right panels were separated on the same gel and immunoblotted at the same time. Because the samples in the left panel came from untreated cells, whereas the samples in the right panel came from rottlerin-treated cells, and because rottlerin-treatment inhibited cell growth/proliferation, fewer numbers of cells were obtained in these groups, and therefore less protein/lysate was available for assay from the treated group compared to the vehicle control group. The mean activities ±SD were obtained from at least three independent experiments, and assays from immunoprecipitations using an anti-β-actin antibody were used to determine the background activity, which was subtracted. Student’s t-test was used for statistical analysis. (*, p <0.01; ♦, p <0.001).
Cell lines expressing activated p21Ras consistently demonstrated elevated levels of PKCδ activity relative to the corresponding lines containing wild-type p21Ras (Figs. 2A&B). Similarly, total levels of PKCδ protein were elevated in the cell lines containing activated p21Ras relative to the corresponding lines containing wild-type p21Ras (Fig. 2D). The data obtained with the chemical inhibitors of PKC isozymes is consistent with PKCδ being the relevant PKC target for Ras-mediated apoptosis.
Unexpectedly, exposure to rottlerin for 48 h suppressed PKCδ protein levels in all cell lines tested (Fig. 2D), whereas the levels of other PKC isozymes, including PKCα, −β, −η, and −θ, were not changed by rottlerin (Fig. 2E). The magnitude of suppression of PKCδ levels by rottlerin in the cells containing activated p21Ras (3–5 fold) approached the magnitude of the suppression of PKCδ activity in these cells by rottlerin (6–10 fold). Thus, although we can demonstrate direct inhibition of PKCδ activity by rottlerin in in vitro assays, the marked suppression in PKCδ activity observed after 48 h in vivo may be due to suppression of isozyme protein levels as well as direct inhibition of enzyme activity.
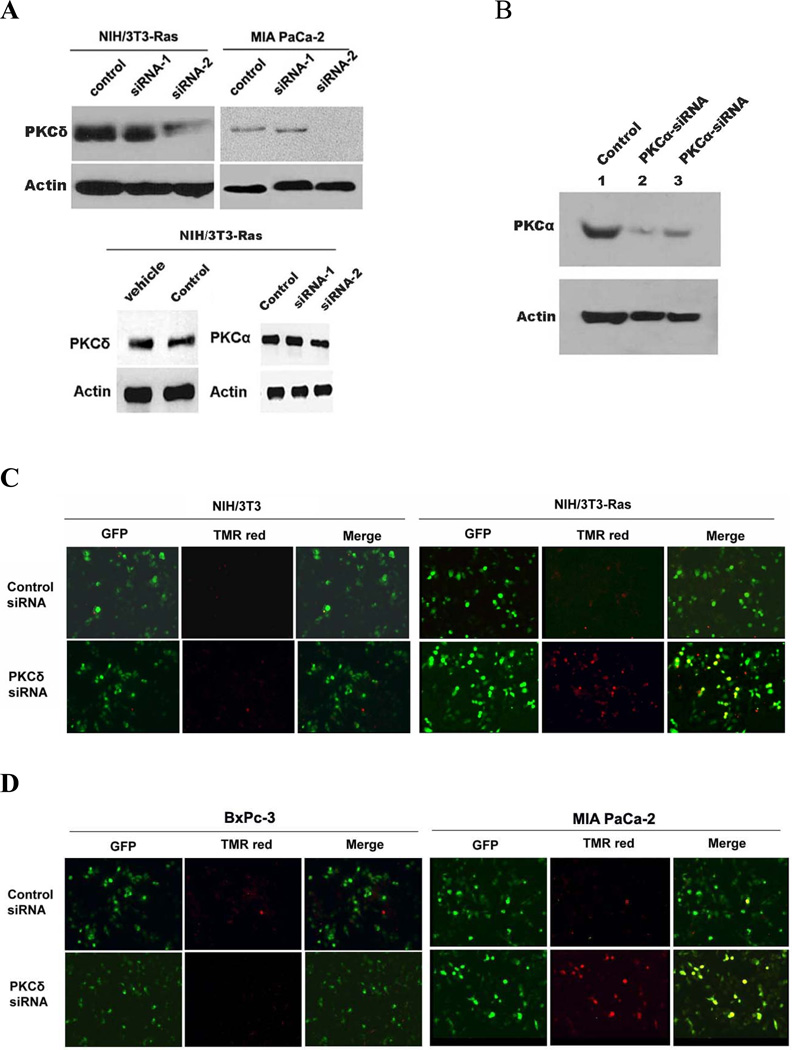
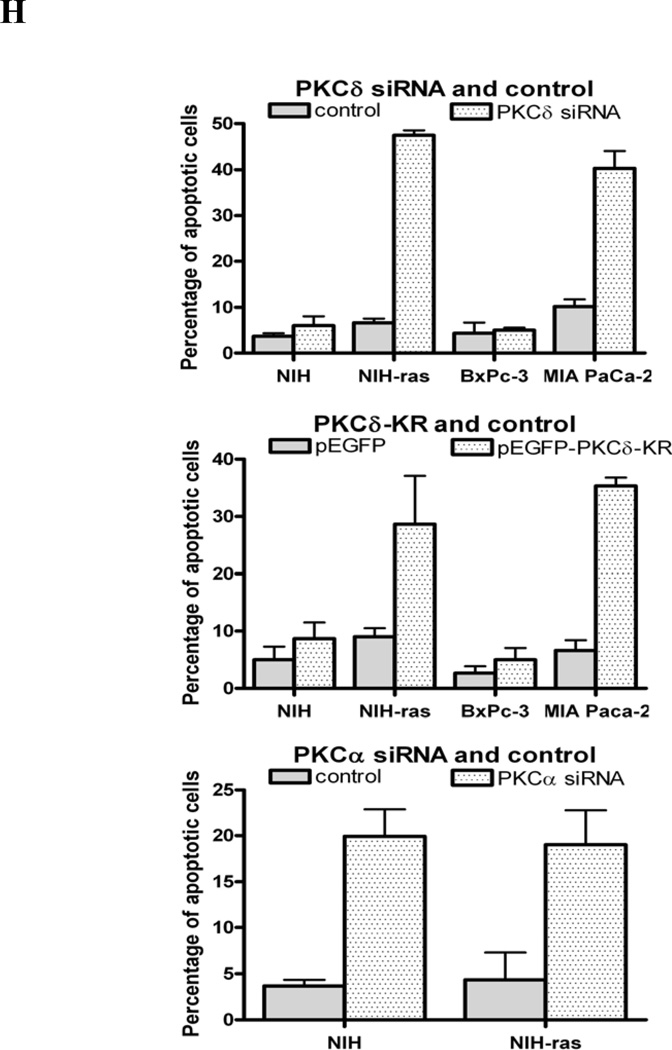
As the specificity of chemical kinase inhibitors is never absolute, we employed two specific genetic techniques to suppress PKCδ activity. At 48 h after transfection of each of two PKCδ-specific hairpin vectors targeted at different PKCδ sequences into matched pairs of cell lines, BxPc-3 / MIA PaCa-2 and NIH/3T3 / NIH/3T3-Ras, immunoblot analysis demonstrated that expression of PKCδ protein was significantly diminished by transfection with the PKCδ-siRNA-2 vector. In contrast, the PKCδ-siRNA-1 vector did not produce significant knockdown of PKCδ protein (Fig. 3A). Because the efficiency of transient transfection in these cells was less than 50%, these analyses likely underestimate the activity of the siRNAs in an individual cell. Control experiments showed no changes of PKCδ by vehicle alone or scrambled siRNA. PKCδ siRNA also had no effect on PKCα expression. Seventy-two hours after transfection with pRNA-U6.1-GFP-control siRNA (scrambled hairpin sequence) or pRNA-U6.1-GFP-PKCδ-siRNA-2, all cell lines were harvested to determine the apoptotic fraction by TUNEL assay. Cells which took up the vector DNA were identified by green fluorescence. TUNEL-positive cells stained red. Superimposition displayed transfected, apoptotic cells as yellow (Figs. 3C&D). For cells transfected with the PKCδ hairpin vector, 40–50% cells were undergoing apoptosis at the 48 h time point, whereas cells transfected with the control scrambled hairpin vector displayed a frequency of apoptosis of less than 10% (Fig. 3H).
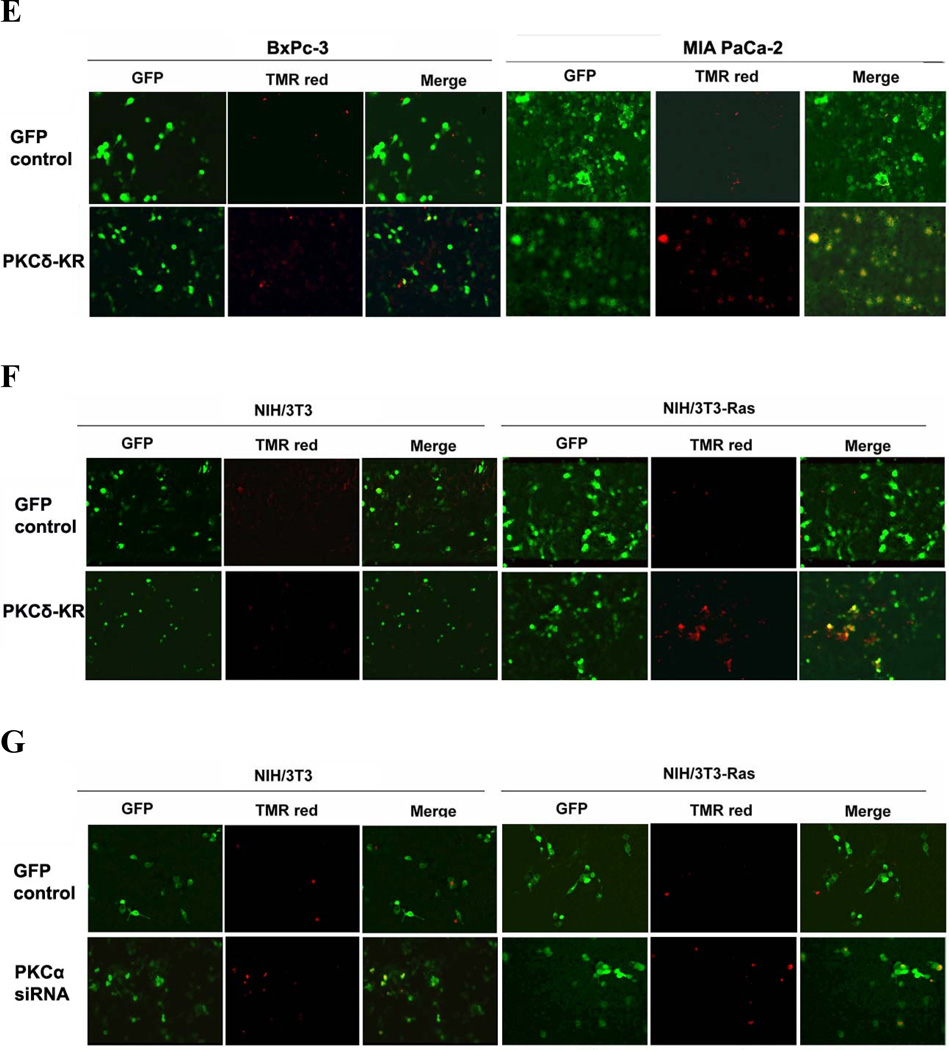
Figure 3.
Effect of PKCδ inhibition on the viability of cells expressing activated p21Ras. (A) Immunoblot analysis of PKCδ, PKCα or β-actin expression in NIH/3T3-Ras and MIA PaCa-2 cells transfected with PKCδ siRNA-1 or -2. A total of 50 nM of PKCδ siRNAs were used. Transfections with a scrambled siRNA or with vehicle alone were used as negative controls. In all experiments, transfections with scrambled siRNA or with vehicle alone yielded identical results, and in subsequent experiments only the scrambled siRNA transfection control is shown. (B) Knock-down activity of PKCα siRNA or scrambled siRNA (control) on NIH/3T3 (lanes 1 and 2) and NIH/3T3-Ras cells (lane 3) evaluated by immunoblotting cell lysates for PKCα or β-actin. NIH/3T3-Ras (C) and MIA PaCa-2 (D) were transfected with pRNA-U6.1-GFP scrambled siRNA hairpin vector (control) or pRNA-U6.1-GFP-PKCδ siRNA-2 hairpin vector. After 72 h, apoptosis was detected by TUNEL assay. NIH/3T3 and BxPc-3 were used as control cell lines, respectively. Shown are 200x magnifications. MIA PaCa-2 cells (E) and NIH/3T3-Ras cells (F) were transfected with a dominant-negative, kinase-dead PKCδ vector. After 72 h, apoptosis was detected by TUNEL assay. BxPc-3 and NIH/3T3 were used as control cell lines (respectively). Shown are 200x magnifications. (G) NIH/3T3 and NIH/3T3-Ras cells were co-transfected with PKD-PKCα siRNA and pEGFP or scrambled siRNA and pGEFP. After 72 h, apoptotic cells were assayed by TUNEL reagent. (H) Quantitation of apoptotic cells in the transfected populations, with error bars indicating SD. Top panel: PKCδ siRNA; middle panel: PKCδ-KR; bottom panel: PKCα siRNA. Data shown are representative of at least three independent experiments.
Similar results were obtained when a dominant-negative, kinase-dead PKCδ mutant protein was used as an alternative method of blocking specifically PKCδ activity in cells expressing an activated p21Ras or wild-type p21Ras (Figs. 3E&F). Transfection of NIH/3T3-Ras cells with the dn-PKCδ vector produced a 30–40% fraction of cells with a hypodiploid (apoptotic) DNA content, compared to a less than 5–10% apoptotic fraction in cells expressing a wild-type p21Ras (Fig. 3H). Transfection of the empty vector as a control generated no significant apoptosis above background levels. The induction of apoptosis by the competitive expression of dominant-negative PKCδ with a single-base mutation, rendering it catalytically inactive, also demonstrates that it is the kinase activity of PKCδ that is required for the survival of cells expressing activated p21Rasrather than a non-catalytic function of the molecule. We studied the effects of knockdown of PKCa (via expression of a PKCα-siRNA) as a specificity control. Forty-eight hours after transfection of PKD-PKCαV6, the levels of PKCα protein were significantly decreased in both NIH/3T3 and NIH/3T3-Ras cells (Fig. 3B). Analysis of apoptosis demonstrated that PKCα inhibition by PKCα-siRNA induced apoptosis in approximately 20% of both NIH/3T3 and NIH/3T3-Ras cells, with no selective toxicity for cells containing an activated p21Ras (Figs. 3G&H). This finding was consistent with results of the MTT assay (Fig. 1C & D). Collectively, the data demonstrate that the PKCδ isozyme plays a critical survival role in p21Ras-induced apoptosis.
PKCδ inhibition induces mitochondrial apoptotic pathways in cells expressing an activated p21Ras
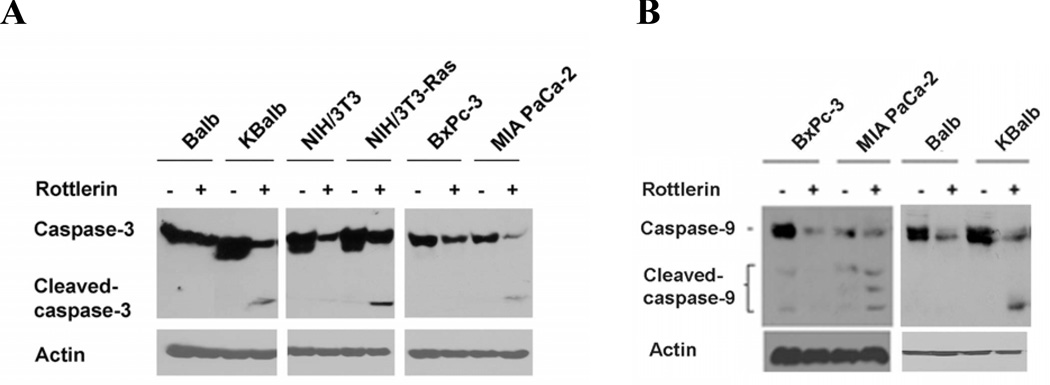
To further characterize the molecular mechanisms of p21Ras–mediated apoptotic pathways, we investigated the influence of mitochondrial apoptotic signaling when PKCδ activity is suppressed by assay of caspase-3 and caspase-9 activation. Immunoblot analysis demonstrated that exposure to rottlerin activated both procaspase-3 and procaspase-9 exclusively in the cells expressing activated p21Ras (Figs. 4A&B). For caspase-3, the full-length protein (35 kDa) and the large cleavage fragment (17 kDa) were detected (Fig. 4A); three activation fragments from caspase-9 zymogen (35, 17 & 10 kDa) were detected (Fig. 4B).
Figure 4.
Inhibition of PKCδ induces the cleavage of caspase-3 and caspase-9 in cells expressed activated p21Ras. All the cells were treated with 20 µM rottlerin for 60 h. Thereafter, cells were harvested, and cell lysates were prepared and subjected to immunoblot analysis to detect the levels of caspase-3/cleaved caspase-3 (A) and caspase-9/cleaved caspase-9 (B) and β-actin. A representative blot from duplicate experiments, producing similar results, is shown.
The PI3K Ras effector pathway is sufficient to sensitize cells to apoptosis by PKCδ inhibition
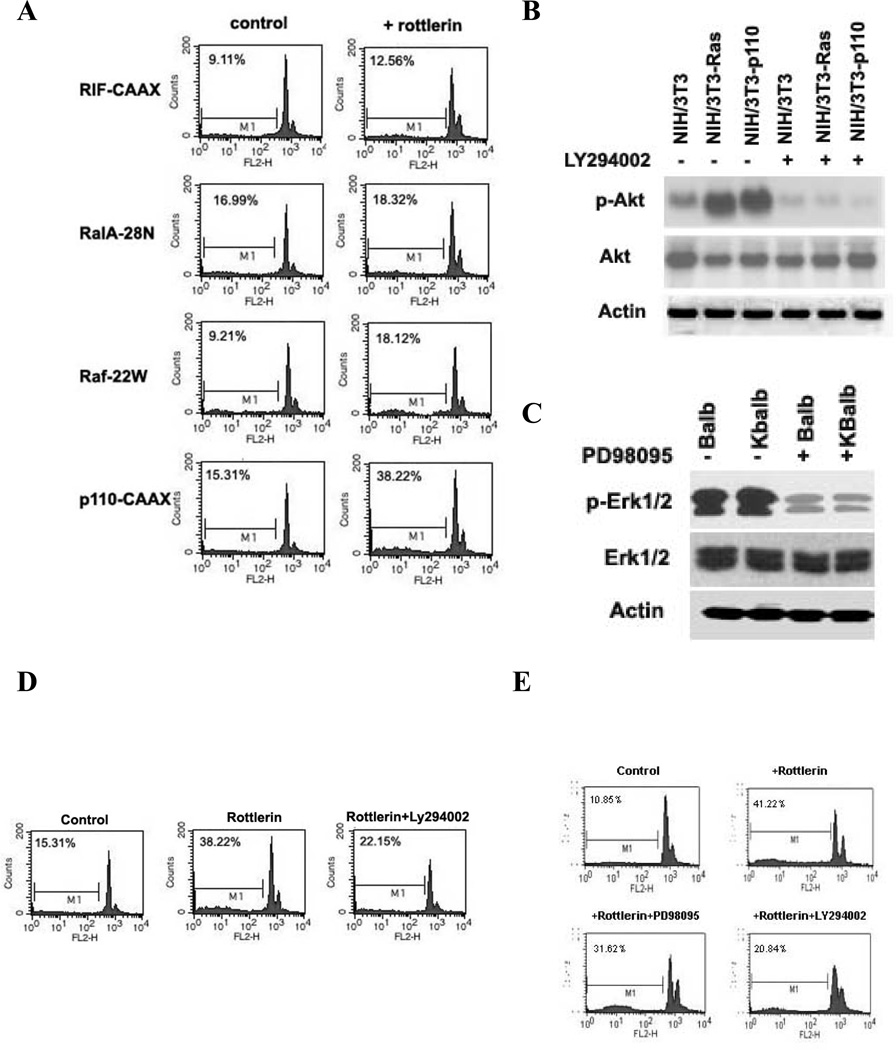
Although previous studies have demonstrated that either constitutive expression of activated p21Ras or acute increases in endogenous p21Ras activity stimulate apoptosis following inhibition of PKC activity in multiple types and lineages of cells, the roles of specific p21Ras downstream effectors in the process have never been determined. In general, three major effector pathways activated by Ras have been defined: Raf1/MAPK, Ral-GDS, and PI3K. To begin to address this question, p21Ras effector loop mutants, consisting of the activating Ras mutation (V12) and a second mutation (S35, G37, or C40) were employed. The three RasV12 mutants (S35, G37, and C40) differ in their ability to bind to p21Ras effectors (Raf, Ral-GEFs, and the p110 subunit of PI3K, respectively) (29). Cells were treated with 20 µM rottlerin for 60 h, and subjected to flow cytometric analysis. All three p21Ras downstream effector-loop mutants stimulated apoptosis to some extent after inhibition of PKCδ, although expression of the C40 mutant consistently generated the greatest amount of apoptosis (data not shown).
The Ras-effector loop mutants are not completely specific in their activation of a single Ras effector pathway, and each activates all three pathways to some degree. In order to more clearly identify the down-stream effector of p21Ras relevant to Ras-mediated apoptosis, we activated single effector pathways using expression vectors for activated PI3K (p110-CAAX), Raf (Raf-22W), and Ral-GEF (RIF-CAAX) into NIH/3T3 cells. The dominant-negative RalA-22N vector was used as control. Constitutive activation of the PI3K pathway (transfection of p110-CAAX) was capable of inducing apoptosis after PKCδ inhibition (apoptotic frequency: 38.22%). In contrast, the other Ras effectors Raf and Ral-GEF induced little apoptosis in response to PKCδ suppression (Fig. 5A).
Figure 5.
Activation of apoptosis by individual p21Ras-effector pathways. (A) NIH/3T3 cell lines stably expressing the p21Ras downstream effectors Raf-22W (Raf-1), P110-CAAXC (PI3K), RIF-CAAX (RIF), or RalA-28N (as a negative control) were treated with rottlerin (20 µM) for 60 h and analyzed for the apoptotic fraction. (B) immunoblot analysis of Akt and p-Akt expression in NIH/3T3, NIH/3T3-Ras and NIH/3T3-p110CAAX cells after 48 h treatment with 10 µM LY294002. (C) Erk1/2 and p-Erk1/2 expression in Balb and KBalb cells after 48 h treatment with 10 µM PD98095. The left and right sides of immunoblot 5B and of immunoblot 5C were separated on the same gels, transferred to the same filters and immunoblotted together. (D) Suppression of apoptosis by inhibition of PI3K activity. NIH/3T3 cells stably expressed p110-CAAX were pre-treated for 30 minutes with Ly294002 (10 µM) before rottlerin (20 µM) was added. The apoptotic (hypodiploid) fractions were evaluated 60 h later. (E) KBalb cells were pre-treated with vehicle solvent, LY294002 (10 µM), or PD98095 (10 µM) and then treated with rottlerin (20 µM). The apoptotic (hypodiploid) fractions were evaluated 60 h later. Cells were fixed and stained with propidium iodide. DNA fragmentation was analyzed by flow cytometry using the FL2-H channel. Data shown are representative of at least three independent experiments.
Yes, (In this revision, gels 5C has been rerun, as requested by Reviewer.)
To independently confirm that the downstream p21Ras effector PI3K plays a critical role in Ras-mediated apoptosis, a PI3K-specific inhibitor (LY294002) and a MAPK inhibitor (PD98095) were employed. In dose-ranging studies, we evaluated concentrations of LY and PD from 1.0 to 60 µM, assaying for efficiency of target enzyme inhibition and for toxicity. Even at 60 µM concentrations of LY294002 or PD98095, only modest and non-statistically-significant levels of apoptosis were observed. At 10 µM concentrations, there were no signs of toxicity or apoptosis (data not shown). Their effectiveness in suppressing the relevant target pathways at 10 µM concentrations was confirmed by immunoblot (Figs. 5B&C). A NIH/3T3 cell line stably-expressing p110-CAAX was pre-treated with Ly294002 (10 µM) for 30 min, and then rottlerin (20 µM) was added for 60 h. In the presence of the PI3K inhibitor, the apoptosis induced by rottlerin decreased nearly 50% (Fig. 5D). Similarly, in KBalb cells, LY294002 blocked 50% of the apoptosis induced by PKCδ inhibition (Fig. 5E). In cells which contained an activated p21Rasbut not those with activation of the single PI3K-effector pathway, the MAPK inhibitor PD98095 also suppressed p21Ras-dependent apoptosis to some extent, but to a substantially lesser degree than did PI3K-inhibition. Collectively, these data demonstrate that the pro-apoptotic signal manifested during Ras-mediated apoptosis following PKCδ inhibition is mediated mainly through the downstream effector PI3K, although the MAPK effector pathway may contribute to some extent in this process.
The anti-apoptotic Akt Ras-effector pathway is regulated by PKCδ
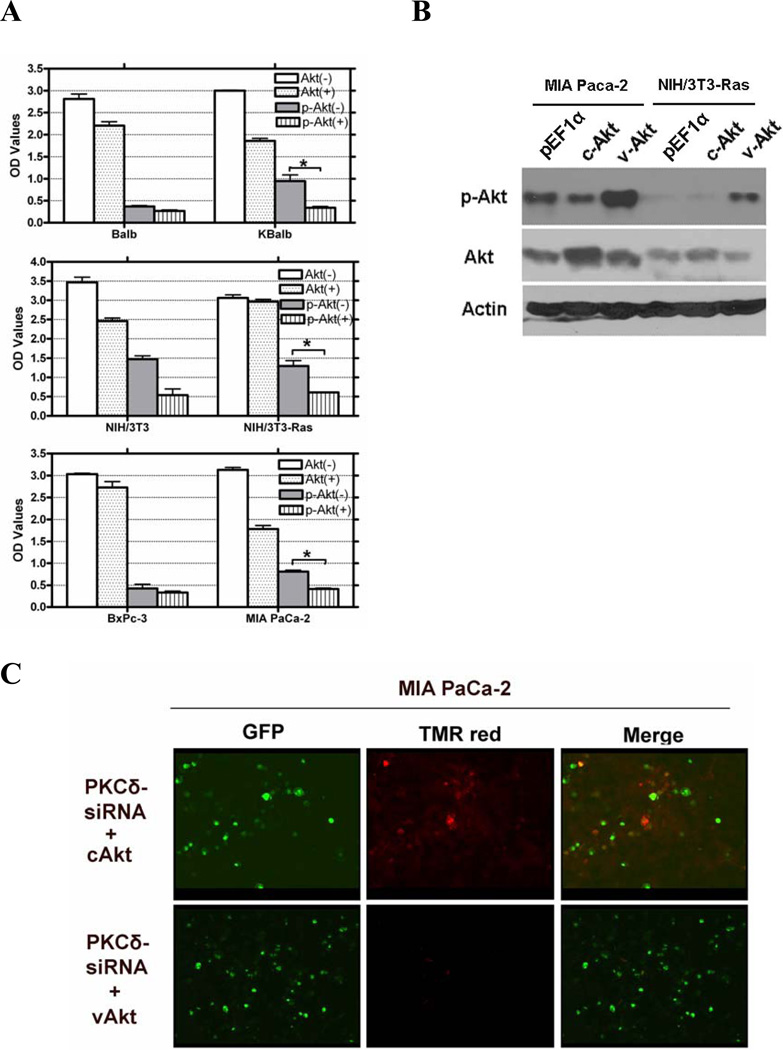
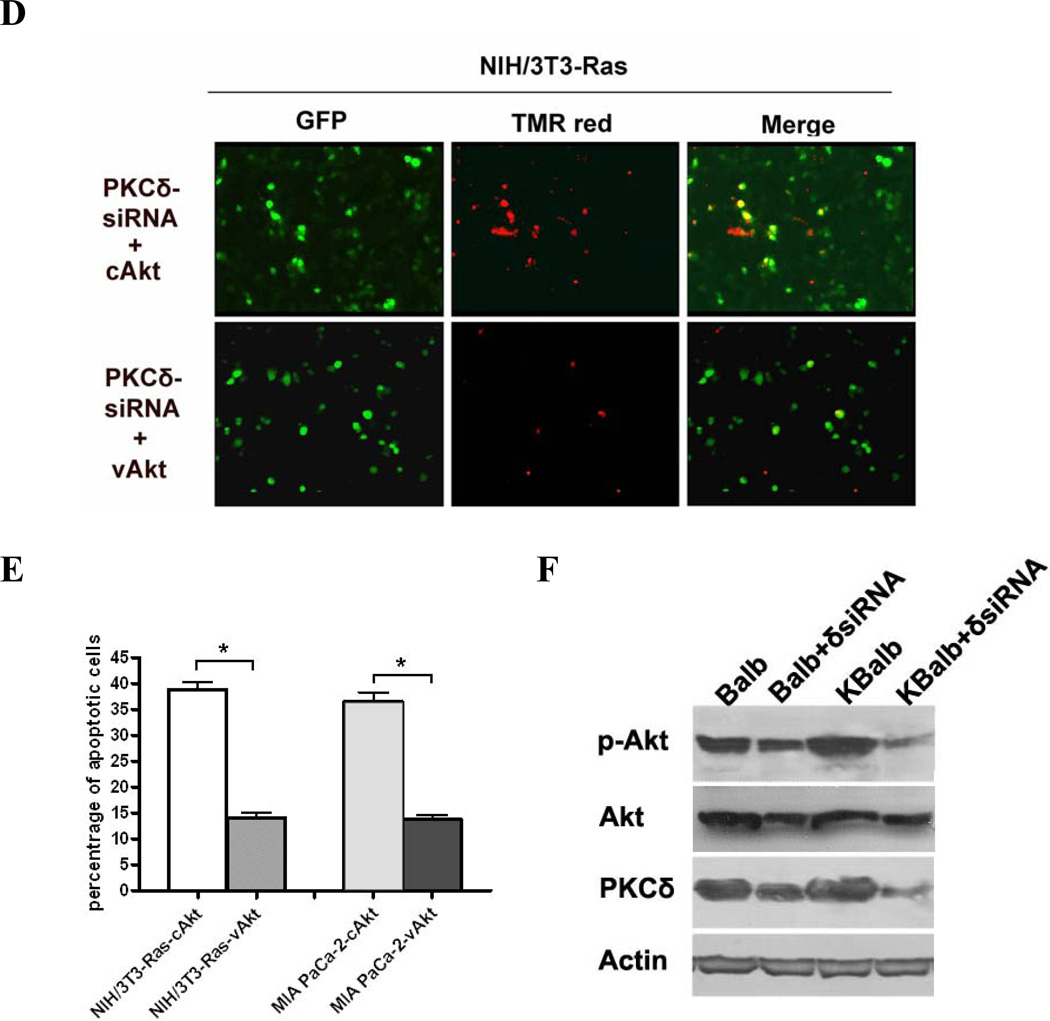
One direct downstream target of the PI3K pathway are the members of the Akt kinase family, which have well-characterized pro-survival and anti-apoptotic activities, mediated through regulation of the mitochondrial apoptotic machinery and the expression of genes involved in this process. To determine the role of Akt in p21Ras-mediated apoptosis, we first examined Akt levels and activity in three paired cell lines with wild-type or mutant Ras alleles. These cells were treated rottlerin for 48 h or left untreated. Akt activity, as assessed by quantitation of serine473-phosphorylated Akt, was down-regulated approximately 50% by rottlerin in all three cell lines expressing activated Ras (Fig. 6A). In contrast, no significant changes in levels of p-Akt were induced in the cell lines expressing wild-type Ras. Exposure to rottlerin also suppressed total Akt levels to a modest extent in some cell lines, but this effect was independent of the presence of activated p21Ras. If the suppression of Akt activation by PKCδ suppression in activated Ras-containing cells were responsible for the resulting apoptosis, enforced expression of activated Akt should be able to effect rescue from apoptosis. MIA Paca-2 and NIH/3T3-Ras were co-transfected with a PKCδ-hairpin vector (which efficiently suppressed PKCδ levels in these experiments by at least 85%, see Fig. 3A for an example) and a constitutively-activated Akt (vAkt) expression vector, or a cAkt expression vector as a control. Akt activity in these cells was confirmed by immunoblotting for phosphorylated (activated) Akt (Fig. 6B). In both cell lines containing activated Ras, expression of vAkt reversed approximately 70–80% of the apoptosis induced by knock-down of PKCδ. Expression of a non-activated, wild-type Akt, however, did not similarly rescue the cells (Figs. 6C, D&E). To further support a role for Akt in Ras-mediated apoptosis induced by PKCδ inhibition, Akt activity was analyzed after PKCδ knock-down. Balb and KBalb cells were treated with a PKCδ siRNA or a scrambled siRNA with a fluorescein tag (serving as control and to quantitate transfection efficiency). After 48 h, cells were harvested and p-Akt expression was analyzed. p-Akt expression was dramatically decreased when PKCδ expression was knocked-down by PKCδ siRNA in KBalb cells (Fig. 6F). For reasons not yet elucidated, we found PKCδ-siRNA was consistently not as effective in downregulating PKCδ in Balb cells as it was in Balb cells containing a mutated, activated p21Ras (KBalb cells) (e.g.62% suppression in Balb compared to 83% suppression in KBalb). Accordingly, the effects of PKCδ downregulation on p-AKT levels cannot be directly compared between Balb and K-Balb cells in these studies. Collectively, these studies indicate that activation of Akt is required for survival of cells expressing activated Ras, that PKCδ is required for the activation of Akt by Ras, and that induction of Ras-mediated apoptosis by suppression of PKCδ is effected through interference with this anti-apoptotic Ras effector pathway.
Figure 6.
Regulation of AKT activity by PKCδ. (A) Treatment with rottlerin reduces Akt activity in cells expressing activated Ras. All cells were treated with 20 µM rottlerin for 60 h in 96-well plates. Total Akt and p-Akt levels were assayed using a SuperArray Case ELISA kit (*, p<0.005). (+): treated with 20 µM rottlerin; (−): treated with vehicle (control); (B) pAkt protein levels in MIA PaCa-2 and NIH/3T3-Ras cells, assayed by immunoblotting with an antibody specific for phosphorylated serine473 Akt, or for total Akt, or for β-actin. Both cell lines were transfected with pEGFP and with pEF1α empty vector, or pEF1α-cAKT, or pEF1α-vAkt. GFP expression was used to normalize for transfection efficiency. (C & D) Activated Akt rescues cells from apoptosis induced by PKCδ knock-down. MIA PaCa-2 and NIH/3T3-Ras cells were cultured on 4-well chamber slides and were co-transfected with pRNA-U6.1-GFP-PKCδ hairpin vector and with pEF1α-vAkt or pEF1α-cAkt vectors. The PKCδ-hairpin vector efficiently suppressed PKCδ levels in these experiments by at least 85% (e.g., see Fig. 3A). After 72 h, cells were fixed with 4% paraformaldehyde for 1 h at room temperature, and then subjected to TUNEL assay. Shown are 200x magnifications. (E) Quantitation of apoptotic cells in the transfected populations (*, p < 0.001). (F) Immunoblot of lysates from Balb or KBalb cells treated with PKCδ-specific siRNA or scrambled, control siRNA. Antibodies against PKCδ, total Akt, phospho-serine473 Akt, and β-actin were used. The results are representative of at least two independent experiments.
A pro-apoptotic p21Ras effector, NORE1 (33), which binds to Ras-GTP and is a member of the RASSF family of tumor suppressors, has been described by Eckfeld (34). To determine whether activation of this Ras-effector pathway plays any role in Ras-mediated apoptosis induced by suppression of PKCδ, we employed vectors expressing the fragment of NORE1 which binds to its apoptotic effector MST1 (aa 358–413), or the carboxy-terminal non-catalytic segment of MST1 (aa 307–487) as GST fusion peptides, each of which has been demonstrated to block formation of the NOR-MST1 apoptotic complex (33). Neither peptide attenuated the apoptosis induced by suppression of PKCδ (data not shown).
p21Ras up-regulates PKCδ protein levels post-transcriptionally
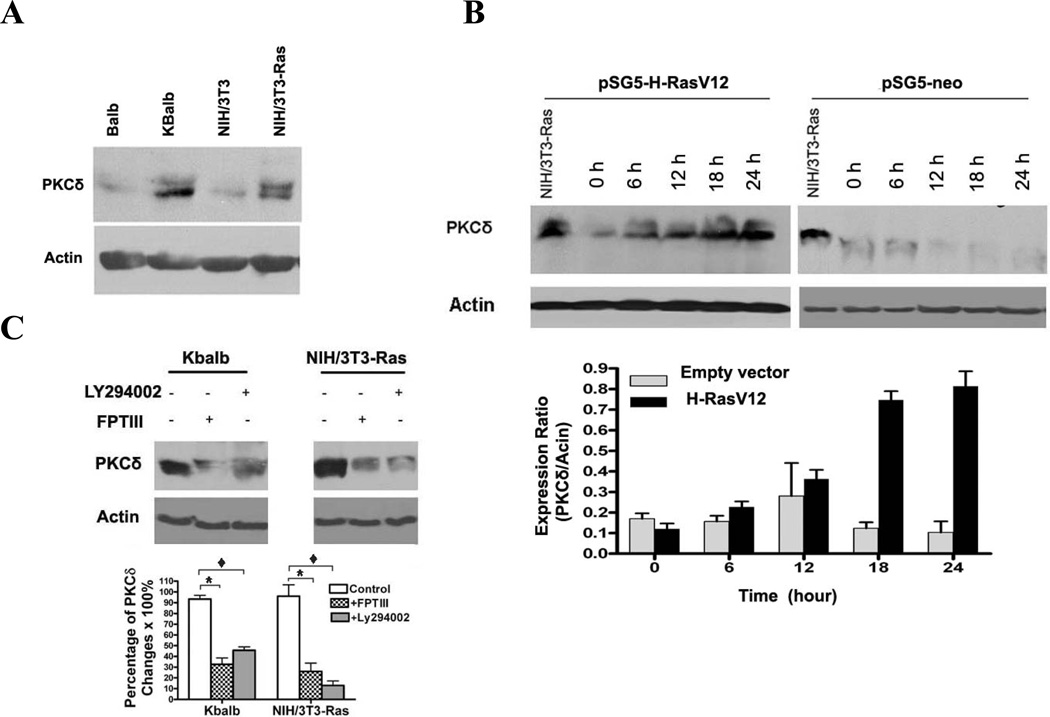
We observed that cell lines expressing an activated p21Ras invariably expressed substantially more PKCδ protein than did their wild-type p21Ras-expressing counterparts (Fig. 7A), suggesting that p21Ras activity may upregulate PKCδ protein expression as well as activity (Fig. 2D&2E). To test this hypothesis, NIH/3T3 cells were transfected with pSG5-H-(V12)Ras and PKCδ protein levels were assayed over time. PKCδ protein levels increased rapidly after transfection, reaching peak levels at 18 h (Fig. 7B). In contrast, transfection with the empty pSG5 vector had no effect on PKCδ protein levels. Conversely, when p21Ras activity, or PI3K activity, in KBalb and NIH/3T3-Ras cells was inhibited by the Ras inhibitor FPT III or the PI3K inhibitor LY294002, respectively, PKCδ protein levels fell (Fig. 7C). This regulation of PKCδ by p21Ras is not at the level of transcription, as PKCδ transcript levels, assessed by quantitative RT-PCR, did not vary as a function of p21Ras activity (data not shown).
Figure 7.
Regulation of PKCδ levels by p21Ras. (A) Immunoblot analysis of PKCδ expression in matched cell line pairs NIH/3T3 and NIH/3T3-Ras; and Balb and Kbalb. Blots were stripped and reprobed for β-actin. (B) (Upper panel) NIH/3T3 cells were transfected with the pSG5-H-ras vector and harvested at the time points indicated. The NIH/3T3-Ras cell line was used as positive control. (Lower panel) The relative levels of PKCδ expression were quantitated by Software BandLead 3.0. (C) Effects of a p21Ras inhibitor and a PI3K inhibitor on PKCδ expression. (Upper panel) NIH/3T3-Ras cells were treated with FPT inhibitor III (100 µM) or with Ly294002 (10 µM) for 48 h, then lysed and subjected to immunoblot assay for PKCδ. Blots were then stripped and reprobed for p21Ras and β-actin. (Lower panel) Quantitative measurement of changes in PKCδ Levels. Data shown are representative of at least three independent experiments (*, p <0.01, and ♦, p < 0.05).
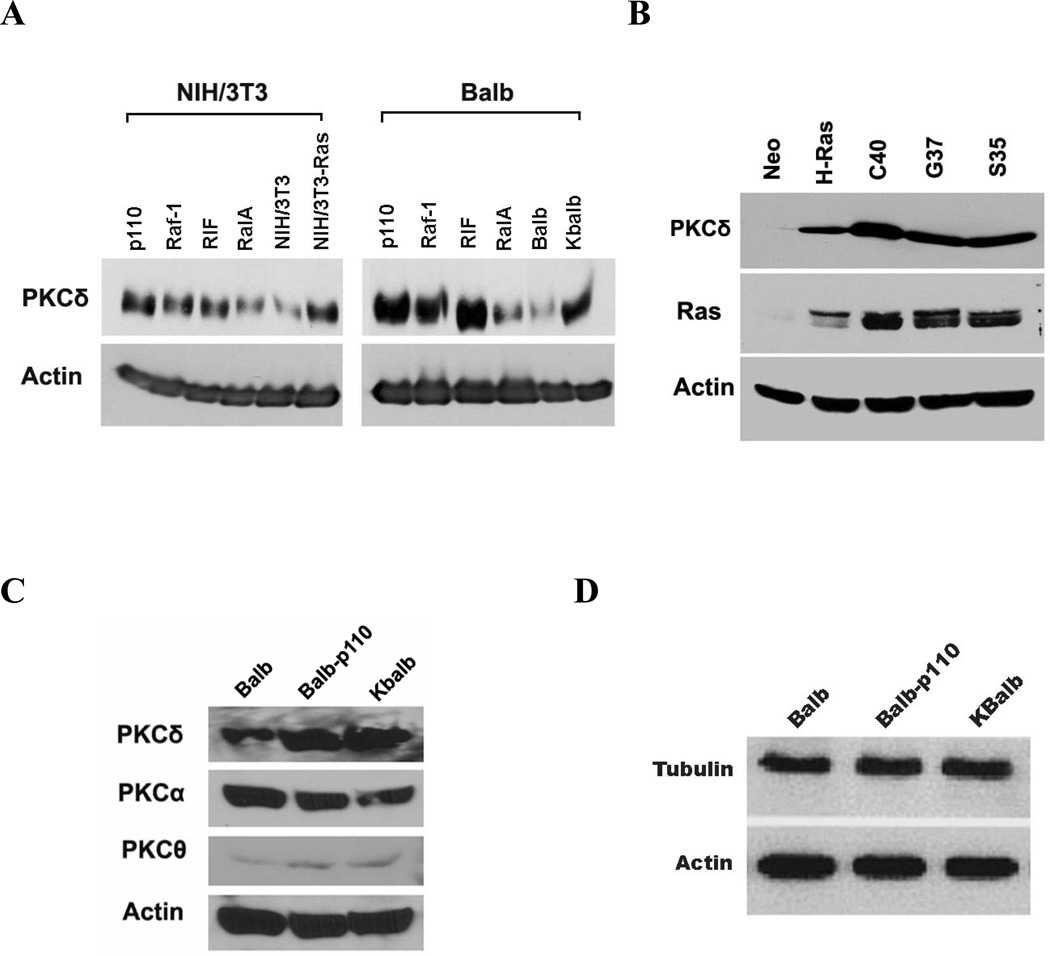
To determine which p21Ras effector pathway mediated the upregulation of PKCδ protein, p110-CAAX, Raf-22w, RIF-CAAX, and RalA-28N expression vectors were each stably transfected into two different cell lines, and PKCδ protein levels were quantitated (Fig. 8A). The p110-CAAX expression vector was consistently the most potent inducer of PKCδ expression in both Balb and NIH/3T3 cells, although activation each of the other two effector pathways could also induce PKCδ protein expression to a variable extent. In agreement, the C40 p21Ras-effector loop mutant, which activates predominantly the PI3K pathway, was the most potent of the effector mutants for induction of PKCδ protein levels (Fig. 8B). The expression of other isozymes of PKC (−α and −θ), β-actin and tubulin were not changed by expression of p21Ras or PI3K (Fig. 8C&D). Similarly, levels of p53, cyclin D1 and p16 were not changed by expression of the p21Ras or PI3K (data not shown).
Figure 8.
Effect of isolated p21Ras effector pathways on PKC expression. Immunoblots of PKCδ, β-actin, Ras, PKCα, PKCθ, or tubulin in: (A) NIH/3T3-Ras cells or NIH-3T3 cells stably-expressing the indicated Ras downstream effectors or empty vector as control (left panel) and KBalb cells or Balb cells stably-expressing the indicated Ras downstream effectors or empty vector as control (right panel); (B) Balb cells stably-expressing Ras-effector loop mutants; (C & D) Balb cells stably-expressing PI3K catalytic domain p110CAAX or KBalb Cells. A total of 50 µg of protein was separated on a 10% SDS-PAGE for each sample. The results are representative of at least three independent experiments.
DISCUSSION
The p21Ras family members comprise critical molecular switches transducing signals to diverse downstream pathways, ultimately controlling such processes as proliferation, cytoskeletal integrity, apoptosis, cell adhesion, and cell migration (35–38). p21Ras and Ras-related proteins are frequently deregulated in cancers, leading to invasion and metastasis and enhancement of survival by activation of anti-apoptotic pathways. Paradoxically, several studies have demonstrated that enforced, high-level expression of oncogenic p21Ras can induce a permanent growth arrest in normal cells, mimicking natural senescence (39, 40). Activation of the Raf-1/MEK/p38MAPK pathway is thought to be essential for oncogenic p21Ras-induced senescence (40, 41). Additionally, inactivation of the ARF/p53 tumor suppressor pathway in mouse fibroblasts and skin keratinocytes, or inactivation of the p16/Rb tumor suppressor pathway in human fibroblasts, can bypass p21Ras-induced senescence, suggesting that cellular fate resulting from oncogenic p21Ras signaling is dependent upon the cellular context and the integration of tumor suppressor signals (39, 42). Our laboratory and others have established p21Ras as a modulator of apoptosis in transformed cells and malignant cells, and also in normal cells. Through FADD, caspase 8, and downstream effector reactive oxygen species (ROS), p21Ras sensitizes cells to apoptosis induced by PKC inhibition (7)(43, 44). However, the specific molecular mechanism by which oncogenic p21Ras mediates apoptosis in the setting of inhibition of PKC activity remains incompletely understood. As different PKC isozymes may have opposing functions with respect to cellular proliferation, differentiation, and apoptosis, a more complete understanding of this process required identification of the specific PKC isozyme required for survival of cells expressing activated p21Rasalong with elucidation of the specific p21Ras downstream effectors evoking the apoptotic outcome.
In this study, we demonstrate that PKCδ activity is required to prevent the induction of apoptosis in cells expressing activated p21Ras. It is noteworthy that p21Ras activity, and in particular activation of the PI3K pathway, up-regulates PKCδ protein levels, thus positively reinforcing an anti-apoptotic, protective response to p21Ras dysregulation in the cell. Conversely, when this induction is prevented by siRNA knockdown of PKCδ, programmed cell death is initiated. Pro-apoptotic activity engendered by activated Ras has been described in a number of other systems (45), and isoform differences in the proapoptotic effects of Ras have been reported, with K-ras sensitizing cells to gamma irradiation-induced apoptosis, but H-Ras exerting a protective effect (46). Activated K-Ras has been reported to sensitize cells to the pro-apoptotic effects of PKC agonists, though phosphorylation of serine residue 181 of p21Rasand its translocation to intracellular membranes (47). K-ras, but not H-ras was reported to bind to, and potentially inactivate or sequester, the anti-apoptotic proteins Bcl-2 and Bcl-XL (48). In contrast to these observations of isotype-specific differences in p21Ras functions, we have previously reported, and confirm herein, that both activated K-ras and H-ras sensitize cells to apoptosis induced by inhibition of PKCδ activity, which prevents their obligate activation of the Akt survival pathway.
PKCδ has been reported to both positively and negatively regulate apoptotic programs (49–52). These findings have generated conflicting hypotheses as to the role of PKCδ in the control of cell proliferation and survival. The normal phenotype of PKCδ-null mice demonstrates that PKCδ is not required for appropriate control of cell proliferation during normal development (28, 53). In contrast, PKCδ may be suborned during cellular transformation and become necessary for one or more components of the malignant phenotype. Inhibition of PKCδ was reported to suppress the metastatic potential of breast cancer cells (54) and to reduced their survival (55). Similar results were reported using non-small cell lung cancer cells (56). Our findings support this hypothesis. We find that PKCδ functions as a survival signal in a variety of cells with dysregulated activation of p21Ras. PKCδ expression is upregulated in response to p21Ras activity, primarily through PI3K activation, and is required for the survival of these cell lines. However, PKCδ is not required for the survival or proliferation of the counterparts of these cell lines containing wild-type p21Raswhether or not they are transformed, and indeed suppression of PKCδ actually leads to a small but reproducible increase in the proliferation or normal cells.
Activated Akt is a well-established survival factor, exerting anti-apoptotic activity by suppressing the release of cytochrome c from the mitochondria (57). Activated Akt also inactivates the pro-apoptotic factors BAD and procaspase-9 by direct phosphorylation (58). A number of reports have suggested relationships between PKC activity and Akt activation. One recent study showed that PKCδ contributes to the phosphorylation of Ebp1 in PC12 cells (59). Phosphorylation of Ebp1 is required for its association/interaction with active nuclear Akt, and the Akt/Ebp1 complex mediates an anti-apoptotic effect in intact cells. A very recent study suggested that both PKCδ and PKCε can negatively regulate Akt activity in mouse keratinocytes (60), although this study utilized only chemical inhibitors lacking absolute specificity. Gonzalez-While PKCα has been shown to promote the dephosphorylation and inactivation of Akt in prostate cancer cells (61), another study demonstrated that phosphorylation of PKCδ protects glioma cells from TRAIL-induced apoptosis by activation of Akt (62). In our study, we prove that Akt activity can be regulated by PKCδ (Fig. 6F), and that cells in which PKCδ has been selectively suppressed by treatment with PKCδ-siRNA can be rescued from apoptosis by enforced expression of a constitutively-activated Akt (Fig. 6C). These data suggest that oncogenic p21Ras protein, through PI3K and PKCδ, induces Akt activity, initiating an anti-apoptotic signaling cascade which is required for their survival. However, whether PKCδ alone is the major regulator of Akt activity under all conditions remains to be elucidated, as the levels of p-Akt were only slightly changes after knocking-down PKCδ in cells containing wild-type (normal) Ras (Fig. 6F). There are two possible explanations for the differences observed on PKCδ actions of Akt between normal cells and those containing activated p21Ras-: the first is the relative differences in efficiency of transient transfection, while the second is that the robust regulation of Akt activity by PKCδ requires the involvement of an activated p21Ras protein.
It is noteworthy that the PI3K effector pathway, in addition to generating the anti-apoptotic signal mediated through PKCδ and activation of Akt (and upregulation of PKCδ levels and activity), is also responsible for the pro-apoptotic signal delivered in Ras-transformed cells, which is uncovered and made manifest by inhibition of PKCδ. We show that isolated activation of the PI3K pathway is sufficient to render cells susceptible Ras-mediated apoptosis, and conversely, that inhibition of the PI3K pathway is sufficient to protect cells from apoptosis initiated by suppression of PKCδ.
In conclusion, this study significantly extends our understanding of the mechanism underlying p21Ras-mediated apoptosis by identifying the molecules required to redirect p21Ras signaling from a proliferative/transforming outcome towards instead an apoptotic fate. Furthermore, elucidation of the particular PKC isozyme necessary for survival of cells transformed by p21Ras suggests that selective suppression of PKCδ activity, and the consequent induction of apoptosis, is a potential strategy for targeting of tumor cells containing an activated p21Ras GTPase.
The abbreviations used are
- MAPK
mitogen-activated protein kinase
- BIM-1
bisindolylmaleimide I
- HMG
1-O-hexadecyl-2-O-methyl-rac-glycerol
- FPT III
(E,E)-[2-Oxo-2-[[(3,7,11-trimethyl-2,6,10-dodecatrienyl)oxy]amino]ethyl]phosphonic Acid, (2,2-Dimethyl-1-oxopropoxy) methyl Ester, Na
- PKC
Protein kinase C
Footnotes
We thank Drs. C. Counter (Duke University), J. Downward (MRC, UK). D. Kufe (Dana-Farber Cancer Institute), J. Avruch (Massachusetts General Hospital), and G. Cooper (Boston University) for generously providing reagents. This work was supported by grant support from the Philip Morris External Research Program, the National Cancer Institute (CA 108100 and CA112102), and the Karin Grunebaum Cancer Research Foundation (DVF).
REFERENCES
- 1.Wiesmuller L, Wittinghofer F. Cell Signal. 1994;6(3):247–267. doi: 10.1016/0898-6568(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 2.Adjei AA. J Natl Cancer Inst. 2001;93(14):1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 3.Downward J. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 4.Hiraragi H, Michael B, Nair A, Silic-Benussi M, Ciminale V, Lairmore M. J Virol. 2005;79(15):9449–9457. doi: 10.1128/JVI.79.15.9449-9457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimmaiah KN, Easton J, Huang S, Veverka KA, Germain GS, Harwood FC, Houghton PJ. Cancer Res. 2003;63(2):364–374. [PubMed] [Google Scholar]
- 6.Shao J, Sheng H, DuBois RN, Beauchamp RD. J Biol Chem. 2000;275(30):22916–22924. doi: 10.1074/jbc.M002235200. [DOI] [PubMed] [Google Scholar]
- 7.Liou JS, Chen CY, Chen JS, Faller DV. J Biol Chem. 2000;275(50):39001–39011. doi: 10.1074/jbc.M007154200. [DOI] [PubMed] [Google Scholar]
- 8.Liou JS, Chen JS, Faller DV. J Cell Physiol. 2004;198(2):277–294. doi: 10.1002/jcp.10409. [DOI] [PubMed] [Google Scholar]
- 9.Chen CY, Faller DV. J Biol Chem. 1996;271(5):2376–2379. doi: 10.1074/jbc.271.5.2376. [DOI] [PubMed] [Google Scholar]
- 10.Chou CK, Liang KH, Tzeng CC HG, Chuang JI, Chang TY, HS L. Life Sci. 2006;78(16):1823–1829. doi: 10.1016/j.lfs.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 11.Jung JW, Cho SD, Ahn NS, Yang SR, Park JS, Jo EH, Hwang JW, Jung JY, Kim SH, Kang KS, Lee YS. Cancer Lett. 2005;225(2):199–206. doi: 10.1016/j.canlet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 12.Klampfer L, Swaby LA, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Oncogene. 2005;24(24):3932–3941. doi: 10.1038/sj.onc.1208552. [DOI] [PubMed] [Google Scholar]
- 13.Parker PJ, Murray-Rust J. J Cell Sci. 2004;117(Pt 2):131–132. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- 14.Jaken S, Parker PJ. Bioessays. 2000;22(3):245–254. doi: 10.1002/(SICI)1521-1878(200003)22:3<245::AID-BIES6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Santiago-Walker AE, Fikaris AJ, Kao GD, Brown EJ, Kazanietz MG, Meinkoth JL. J Biol Chem. 2005;280(37):32107–32114. doi: 10.1074/jbc.M504432200. [DOI] [PubMed] [Google Scholar]
- 16.Basu A. J Cell Mol Med. 2003;7(4):341–350. doi: 10.1111/j.1582-4934.2003.tb00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodie C, Blumberg PM. Apoptosis. 2003;8(1):19–27. doi: 10.1023/a:1021640817208. [DOI] [PubMed] [Google Scholar]
- 18.Brodie C, Kuperstein I, Acs P, Blumberg PM. Brain Res Mol Brain Res. 1998;56(1–2):108–117. doi: 10.1016/s0169-328x(98)00035-7. [DOI] [PubMed] [Google Scholar]
- 19.Ghayur T, Hugunin M, Talanian RV, Ratnofsky S, Quinlan C, Emoto Y, Pandey P, Datta R, Huang Y, Kharbanda S, Allen H, Kamen R, Wong W, Kufe D. J Exp Med. 1996;184(6):2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P. J Neurosci. 2006;26(4):1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas T, White D, Hui L, Yoshida K, Foster DA, Bargonetti J. J Biol Chem. 2004;279(11):9970–9977. doi: 10.1074/jbc.M306979200. [DOI] [PubMed] [Google Scholar]
- 22.Majumder S, Chakraborty AK, Bhattacharyya A, Mandal TK, Basak DK. Indian J Exp Biol. 1997;35(2):162–167. [PubMed] [Google Scholar]
- 23.Basu A, Mohanty S, Sun B. Biochem Biophys Res Commun. 2001;280(3):883–891. doi: 10.1006/bbrc.2000.4209. [DOI] [PubMed] [Google Scholar]
- 24.Sun C, Zong Z, Wang Y, Wang Z, Yu B. Hua Xi Kou Qiang Yi Xue Za Zhi. 2000;18(4):237–239. [PubMed] [Google Scholar]
- 25.Zhang J, Liu N, Zhang J, Liu S, Liu Y, Zheng D. J Cell Biochem. 2005;96(3):522–532. doi: 10.1002/jcb.20535. [DOI] [PubMed] [Google Scholar]
- 26.Mecklenbrauker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A. Nature. 2004;431(7007):456–461. doi: 10.1038/nature02955. [DOI] [PubMed] [Google Scholar]
- 27.Mecklenbrauker I, Saijo K, Zheng NY, Leitges M, Tarakhovsky A. Nature. 2002;416(6883):860–865. doi: 10.1038/416860a. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, Nakayama KI. Nature. 2002;416(6883):865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Cell. 1997;89(3):457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 30.Peyssonnaux C, Provot S, Felder-Schmittbuhl MP, Calothy G, Eychene A. Mol Cell Biol. 2000;20(19):7068–7079. doi: 10.1128/mcb.20.19.7068-7079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bharti A, Kraeft SK, Gounder M, Pandey P, Jin S, Yuan ZM, Lees-Miller SP, Weichselbaum R, Weaver D, Chen LB, Kufe D, Kharbanda S. Mol Cell Biol. 1998;18(11):6719–6728. doi: 10.1128/mcb.18.11.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida K, Wang HG, Miki Y, Kufe D. Embo J. 2003;22(6):1431–1441. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Curr Biol. 2002;12(4):253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 34.Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. Cancer Res. 2004;64(23):8688–8693. doi: 10.1158/0008-5472.CAN-04-2065. [DOI] [PubMed] [Google Scholar]
- 35.Cowley S, Paterson H, Kemp P, Marshall CJ. Cell. 1994;77(6):841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 36.Stice LL, Forman LW, Hahn CS, Faller DV. Exp Cell Res. 2002;275(1):17–30. doi: 10.1006/excr.2002.5482. [DOI] [PubMed] [Google Scholar]
- 37.Nakada M, Niska JA, Tran NL, McDonough WS, Berens ME. Am J Pathol. 2005;167(2):565–576. doi: 10.1016/S0002-9440(10)62998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Nat Cell Biol. 2005;7(8):837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 39.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 40.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Genes Dev. 1998;12(19):3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, Sun P. Mol Cell Biol. 2002;22(10):3389–3403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brookes S, Rowe J, Ruas M, Llanos S, Clark PA, Lomax M, James MC, Vatcheva R, Bates S, Vousden KH, Parry D, Gruis N, Smit N, Bergman W, Peters G. Embo J. 2002;21(12):2936–2945. doi: 10.1093/emboj/cdf289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CY, Liou J, Forman LW, Faller DV. J Biol Chem. 1998;273(27):16700–16709. doi: 10.1074/jbc.273.27.16700. [DOI] [PubMed] [Google Scholar]
- 44.Chen CY, Juo P, Liou JS, Li CQ, Yu Q, Blenis J, Faller DV. Cell Growth Differ. 2001;12(6):297–306. [PubMed] [Google Scholar]
- 45.Cox AD, Der CJ. Oncogene. 2003;22(56):8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 46.Choi BY, Choi HS, Ko K, Cho YY, Zhu F, Kang BS, Ermakova SP, Ma WY, Bode AM, Dong Z. Nat Struct Mol Biol. 2005;12(8):699–707. doi: 10.1038/nsmb960. [DOI] [PubMed] [Google Scholar]
- 47.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. Mol Cell. 2006;21(4):481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Romero F, Martinez AC, Camonis J, Rebollo A. Embo J. 1999;18(12):3419–3430. doi: 10.1093/emboj/18.12.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe T, Ono Y, Taniyama Y, Hazama K, Igarashi K, Ogita K, Kikkawa U, Nishizuka Y. Proc Natl Acad Sci U S A. 1992;89(21):10159–10163. doi: 10.1073/pnas.89.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao L, Ramsay K, Jaken S. Cell Growth Differ. 1994;5(11):1185–1194. [PubMed] [Google Scholar]
- 51.Li W, Jiang YX, Zhang J, Soon L, Flechner L, Kapoor V, Pierce JH, Wang LH. Mol Cell Biol. 1998;18(10):5888–5898. doi: 10.1128/mcb.18.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pal S, Claffey KP, Dvorak HF, Mukhopadhyay D. J Biol Chem. 1997;272(44):27509–27512. doi: 10.1074/jbc.272.44.27509. [DOI] [PubMed] [Google Scholar]
- 53.Leitges M, Mayr M, Braun U, Mayr U, Li C, Pfister G, Ghaffari-Tabrizi N, Baier G, Hu Y, Xu Q. J Clin Invest. 2001;108(10):1505–1512. doi: 10.1172/JCI12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiley SC, Clark KJ, Goodnough M, Welch DR, Jaken S. Cancer Res. 1999;59(13):3230–3238. [PubMed] [Google Scholar]
- 55.McCracken MA, Miraglia LJ, McKay RA, Strobl JS. Mol Cancer Ther. 2003;2(3):273–281. [PubMed] [Google Scholar]
- 56.Clark AS, West KA, Blumberg PM, Dennis PA. Cancer Res. 2003;63(4):780–786. [PubMed] [Google Scholar]
- 57.Whang YE, Yuan XJ, Liu Y, Majumder S, Lewis TD. Vitam Horm. 2004;67:409–426. doi: 10.1016/S0083-6729(04)67021-X. [DOI] [PubMed] [Google Scholar]
- 58.Downward J. Semin Cell Dev Biol. 2004;15(2):177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 59.Ahn JY, Liu X, Liu Z, Pereira L, Cheng D, Peng J, Wade PA, Hamburger AW, Ye K. Embo J. 2006;25(10):2083–2095. doi: 10.1038/sj.emboj.7601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Sampat K, Hu N, Zakari J, Yuspa SH. J Biol Chem. 2006;281(6):3237–3243. doi: 10.1074/jbc.M512167200. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Guerrico AM, Meshki J, Xiao L, Benavides F, Conti CJ, Kazanietz MG. J Biochem Mol Biol. 2005;38(6):639–645. doi: 10.5483/bmbrep.2005.38.6.639. [DOI] [PubMed] [Google Scholar]
- 62.Okhrimenko H, Lu W, Xiang C, Ju D, Blumberg PM, Gomel R, Kazimirsky G, Brodie C. J Biol Chem. 2005;280(25):23643–23652. doi: 10.1074/jbc.M501374200. [DOI] [PubMed] [Google Scholar]