Abstract
A variety of ribonucleoprotein (RNP) granules form in eukaryotic cells to regulate the translation, decay, and localization of the encapsulated messenger RNA (mRNAs). The work here examined the assembly and function of two highly conserved RNP structures, the processing body (P body) and the stress granule, in the yeast Saccharomyces cerevisiae. These granules are induced by similar stress conditions and contain translationally repressed mRNAs and a partially overlapping set of protein constituents. However, despite these similarities, the data indicate that these RNP complexes are independently assembled and that this assembly is controlled by different signaling pathways. In particular, the cAMP-dependent protein kinase (PKA) was found to control P body formation under all conditions examined. In contrast, the assembly of stress granules was not affected by changes in either PKA or TORC1 signalling activity. Both of these RNP granules were also detected in stationary-phase cells, but each appears at a distinct time. P bodies were formed prior to stationary-phase arrest, and the data suggest that these foci are important for the long-term survival of these quiescent cells. Stress granules, on the other hand, were not assembled until after the cells had entered into the stationary phase of growth and their appearance could therefore serve as a specific marker for the entry into this quiescent state. In all, the results here provide a framework for understanding the assembly of these RNP complexes and suggest that these structures have distinct but important activities in quiescent cells.
EUKARYOTIC cells contain a number of membrane-bound compartments that partition the cytoplasm into distinct functional units. Proteins that act in similar pathways are often localized to the same compartment whereas those with competing activities are sequestered within different environments. Interestingly, recent data suggest that particular proteins and RNAs are also concentrated in what can be thought of as nontraditional compartments that lack a boundary membrane. These ribonucleoprotein (RNP) complexes, or granules, are more dynamic in nature and are found in both the nucleus and the cytoplasm of the cell (Anderson and Kedersha 2006; Mao et al. 2011; Weber and Brangwynne 2012). The formation of these granules can be induced by a variety of cues, including an exposure to stress or specific developmental transitions. In some cases, the underlying reasons for this reorganization of protein and RNA are known. For example, the polar granules present in germ cells store maternal mRNAs that are translated following fertilization (Schisa et al. 2001; Leatherman and Jongens 2003). However, for most RNP granules, the physiological role of the larger aggregate-like structures remains unclear. Nonetheless, the prevalence and evolutionary conservation of these complexes suggests that they serve important functions in the cell.
Two of the better-characterized cytoplasmic RNPs are the processing bodies (P bodies) and stress granules that form in response to a variety of stress conditions. These particles contain translationally repressed messenger RNA (mRNAs) and a partially overlapping set of protein constituents (Kedersha and Anderson 2002; Anderson and Kedersha 2009; Balagopal and Parker 2009). Since a number of factors important for protein translation are also found in stress granules, these structures have been suggested to be sites of mRNA storage (Yamasaki and Anderson 2008). In contrast, P bodies were originally identified as cytoplasmic foci containing proteins important for mRNA decay (Sheth and Parker 2003; Eulalio et al. 2007a). Although this initially led to speculation that these foci were sites of mRNA turnover, more recent studies have found that this decay proceeds normally in cells lacking the larger P body complexes (Stoecklin et al. 2006; Decker et al. 2007; Eulalio et al. 2007b). As a result, the biological functions associated with P body foci remain unclear. However, an intriguing possibility has been suggested by studies demonstrating that P bodies contain proteins that do not appear to have a direct role in mRNA decay. These proteins include the phosphatase, calcineurin, and the catalytic subunits of the cAMP-dependent protein kinase (PKA) (Tudisca et al. 2010; Kozubowski et al. 2011). P bodies may therefore carry out specific functions that are dictated by the particular proteins present within these cytoplasmic structures. These functions may vary depending upon the particular cell type and stress condition used to induce the foci.
A significant body of work has linked the induction of both P bodies and stress granules to the inhibition of protein synthesis, but less is known about the mechanisms regulating the subsequent formation of the larger aggregate-like assemblies (Franks and Lykke-Andersen 2008). These latter structures appear to form by a self-assembly process that involves the prion-like domains present in a number of granule proteins (Gilks et al. 2004; Decker et al. 2007; Reijns et al. 2008). Some insight into the regulation of this latter process was provided by a recent study of the P bodies that form in response to glucose deprivation in Saccharomyces cerevisiae (Ramachandran et al. 2011). This work showed that the inactivation of the PKA signaling pathway was both a necessary and a sufficient condition for P body foci formation. PKA directly phosphorylates Pat1, a conserved core constituent of these RNP structures, and thereby disrupts Pat1 interactions with a number of P body components, including the RNA helicase Dhh1 (Ramachandran et al. 2011). In contrast, defects in other nutrient-sensing pathways, including those involving the TORC1 or Snf1 protein kinases, did not have a significant effect upon P body formation. This work also suggested that P body foci were important for the long-term survival of cells that had entered into the stationary phase of growth. In particular, mutants that were defective for foci formation lost viability more rapidly during this period of quiescence (Ramachandran et al. 2011). This latter result is of interest in light of other work indicating that as much as 20% of the yeast proteome might relocalize to cytoplasmic foci when cells enter into this G0-like resting state (Narayanaswamy et al. 2009; Noree et al. 2010). Since this phenomenon might not be restricted to yeast (An et al. 2008; Noree et al. 2010), the concentration of material at discrete sites in the cytoplasm may be generally important for the biology of the quiescent cell. Determining the underlying reasons for this relocalization of protein will therefore be critical for a complete understanding of the physiology of the eukaryotic cell.
In S. cerevisiae, PKA is an essential component of one of the key signaling pathways responsible for coordinating cell growth with nutrient availability (Bahn et al. 2007; Zaman et al. 2008). This pathway also involves the GTP-binding Ras proteins Ras1 and Ras2 and is thought to respond, either directly or indirectly, to the levels of glucose present within cells (Santangelo 2006; Slattery et al. 2008; Zaman et al. 2009). The active, GTP-bound forms of the Ras proteins interact with the adenylyl cyclase Cyr1 and stimulate the production of cAMP (Field et al. 1990; Suzuki et al. 1990). This cyclic nucleotide is then bound by Bcy1, the regulatory subunit of the PKA enzyme, leading to the subsequent release of the active catalytic subunits; the basal state of PKA is an inactive heterotetramer made up of two catalytic and two regulatory subunits (Uno et al. 1982; Toda et al. 1987a; Taylor et al. 2008). These catalytic subunits are then free to phosphorylate their respective targets and thereby influence cell growth (Budovskaya et al. 2005). The existing genetic data suggest that this Ras/PKA pathway might play an important role in regulating the entry into stationary phase. For example, mutants that inactivate this pathway result in a growth arrest that resembles stationary phase (Iida and Yahara 1984; Schneper et al. 2004). Conversely, cells with constitutively elevated levels of PKA activity fail to arrest normally in stationary phase when nutrients are limiting (Broek et al. 1985; Broach 1991). The above results with Pat1 suggest that the PKA-mediated control of P body formation is one important component of this regulation of stationary-phase biology.
In this study, we examined the regulation of P body and stress granule assembly in response to a variety of environmental cues, including several that can induce both of these RNP foci. This work demonstrated that the PKA pathway has a general role in the regulation of P body foci formation as mutants with constitutive PKA signaling were defective for P body assembly in all conditions tested. In contrast, stress granule formation was not influenced by changes in either PKA or TORC1 signalling activity. The results here also demonstrate that both types of RNP foci are present in stationary-phase cells and provide further support for a role for P bodies in the long-term survival of these quiescent cells. Finally, we show that P bodies and stress granules form at different times during batch culture growth and that stress granules in particular appear only after cells enter into stationary phase. Therefore, stress granule formation could serve as a useful marker for cell entry into this quiescent state. In all, this work indicates that P bodies and stress granules form independently of one another and that each assembly pathway is regulated by distinct signaling mechanisms.
Materials and Methods
Yeast strain construction and growth conditions
The yeast strains used are listed in Table S1. Standard Escherichia coli and yeast growth conditions and media were used throughout this study. The yeast rich growth medium, Yeast extract- Peptone- Adenine- Dextrose (YPAD), and the minimal Yeast Minimal (YM) and SC media have been described (Kaiser et al. 1994; Chang et al. 2001; Ramachandran and Herman 2010). The carbon starvation experiments were performed in SC- or YPA-based media lacking glucose for strains with and without plasmids, respectively. For ethanol stress, cells were grown in synthetic complete dextrose (SCD) medium and then transferred to SCD containing 6 or 15% ethanol. Similar transfers were made for the other stress conditions used in this study, including 0.5% NaN3, 3 mM H2O2, and 1 M KCl. The incubation time for each stress was typically 10–15 min for the induction of P bodies and 30 min for stress granules. As noted previously, stress granules were not induced by an exposure to 1 M KCl or 3 mM H2O2, and P bodies did not form following a 46° heat stress (our unpublished observations; Teixeira et al. 2005; Buchan et al. 2008, 2010, 2011; Grousl et al. 2009). To assess P body or stress granule formation in stationary phase, cells were inoculated into either YPAD or SCD media and then grown with agitation at 30° for the indicated number of days.
Strains carrying the MET3-RAS2val19 allele were grown in medium containing 500 μM methionine to repress expression from the MET3 promoter. Expression from this promoter was induced by transferring cells to a medium that lacked methionine. For constructs under the control of the CUP1 promoter, expression was induced by adding CuSO4 to a final concentration of 100 μM. Expression from the GAL1 promoter was induced by growing cells overnight in SC medium with 2% raffinose and then shifting cells to SC medium containing 1% raffinose and 2% galactose for 3–4 hr. To generate strains with integrated GFP-tagged versions of Pat1-SS, Pat1-AA, and Pat1-EE, the respective integrative plasmids were linearized within the HIS3 locus and transformed into a pat1Δ strain. The PAT1 locus was deleted where indicated with a PCR-based strategy (Baudin et al. 1993). The presence of the null allele in each of the commercial deletion strains was confirmed with a PCR analysis.
Plasmid construction
The plasmids used in this study are listed in Table S2. The MET3-RAS2val19 plasmids, pPHY795 and pPHY796, have been described (Howard et al. 2001; Ramachandran and Herman 2010). Plasmids expressing Dcp2-GFP (pRP1175), Edc3-mCherry (pRP1574), Edc3-mCherry/Pbp1-GFP (pRP1944), Pab1-GFP/Edc3-mCherry (pRP1657), and Pab1-GFP/Dcp2-mCherry (pRP1660) constructs were kindly provided by Roy Parker (Coller and Parker 2005; Decker et al. 2007). The GFP-PAT1 fusion constructs were made in the vector pRS413 (Ramachandran et al. 2011). The PAT1 variants described in the study were constructed by site-directed mutageneses performed with the Transformer mutagenesis kit (Clontech). The HIS3-marked, single-copy plasmids with the PAT1 locus were constructed in pRS413; the encoded protein was tagged with a single myc epitope at its N terminus. The plasmid vectors used for the copper-inducible expression of the HA epitope-tagged versions of the Tpk proteins have been described (Deminoff et al. 2006).
Fluorescence microscopy
Cells expressing the appropriate fusion construct(s) were grown to early log phase (0.3–0.6 OD600 units/ml) before exposure to the indicated stress regimen or drug treatment. The cells were then collected by centrifugation, resuspended in the appropriate medium, and spotted onto microscope slides (Budovskaya et al. 2005). The samples were then imaged with a ×100/1.4 numerical aperture Plan-Apo objective lens on a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY) equipped with a Nikon cooled digital camera DS-QI1 and appropriate filter sets (DIC, GFP, mCherry). For the quantitation shown, the data in all cases represent the average of two or more experiments; in each of these experiments at least 100 cells were examined. For the colocalization experiments, the final merged images were created with the Image J software package. For the analysis of RNP foci in stationary phase, cells were incubated in either YPAD or SCD media for the indicated number of days.
Stationary-phase viability assays
Cells were grown in YM glucose minimal medium for the appropriate number of days, collected by centrifugation, and resuspended in water at a final concentration of 5 OD600 equivalents/ml. The growth assays were performed by spotting 5 μl of this cell suspension, and fivefold serial dilutions thereafter, onto the appropriate growth medium and incubating the plates at 30° for 2–3 days.
Protein analysis
Protein samples for Western blotting were prepared with a glass-bead lysis method, separated on 7.5–10% SDS-polyacrylamide gels, and transferred to nitrocellulose membranes as described (Budovskaya et al. 2002, 2004). The membranes were then probed with the appropriate primary and secondary antibodies. The Supersignal chemiluminescent substrate (Pierce) was subsequently used to detect the reactive bands. Cell extracts for immunoprecipitation were prepared by resuspending log-phase cells in lysis buffer [25 mM Tris⋅HCl (pH 7.4), 140 mM NaCl, 0.1% Tween-20, 1 mM PMSF] and lysing by agitation with glass beads. Myc- and GST-tagged proteins were immunoprecipitated with the appropriate monoclonal antibodies (Cell Signaling), and the immunoprecipitates were collected on protein A-Sepharose beads (GE Healthcare). To assess the PKA phosphorylation status of Pat1 in vivo, this protein was immunoprecipitated from cell extracts prepared under denaturing conditions as described (Budovskaya et al. 2005; Deminoff et al. 2009). Protease and phosphatase inhibitors were present at all steps. The level of PKA phosphorylation was then assessed by Western blotting with the anti-PKA substrate antibody (Cell Signaling) as described (Chang et al. 2004; Deminoff et al. 2006).
In vitro kinase assays
The substrate proteins were immunoprecipitated, treated with λ phosphatase (NEB), and then washed three times with wash buffer as described (Deminoff et al. 2009; Ramachandran et al. 2011). The precipitated substrate was then incubated with the indicated Tpk protein and 10 μCi [γ-32P]ATP in a 40-ml reaction (50 mM potassium phosphate, 5 mM NaF, 10 mM MgCl2, 4.5 mM DTT, and both protease and phosphatase inhibitors). The proteins were separated by SDS-polyacrylamide gel electrophoresis, and the level of phosphorylation was assessed by autoradiography.
Results
Presence of PKA signaling activity is sufficient to trigger P body disassembly
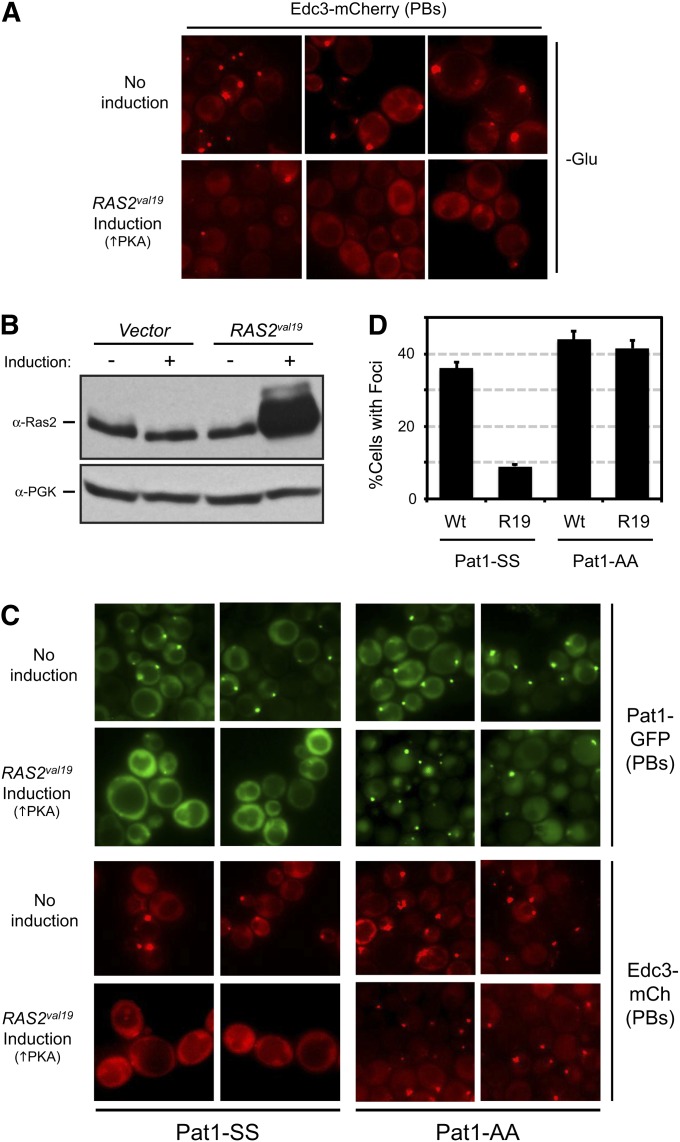
Our goal here was to test whether the ectopic presence of Ras/PKA signaling activity was sufficient to trigger P body disassembly in the absence of glucose addition. For this analysis, we engineered cells to express the dominant RAS2val19 allele in glucose-free conditions. The protein encoded by this allele exhibits diminished GTPase activity and results in constitutively elevated levels of PKA activity (Toda et al. 1985). For these experiments, RAS2val19 was placed under the control of the inducible promoter from the MET3 gene; expression from this promoter is induced when methionine is absent from the growth medium (Cherest et al. 1987; Howard et al. 2002). Log-phase cells expressing Edc3-mCherry and/or Pat1-GFP reporters were transferred to a glucose-free medium for 10 min to allow P body foci to form. The cells were then moved to a medium that also lacked methionine to elicit expression from the RAS2val19 locus. Under these conditions, we found that the majority of pre-existing P bodies were disassembled after RAS2val19 induction (Figure 1, A and B). In contrast, the transfer of the wild-type strain to this methionine-minus medium did not have a significant effect upon P body number. Since our previous work identified Pat1 as a critical target for PKA, we tested whether Pat1 phosphorylation was required for the observed P body disassembly. For this analysis, the inducible MET3-RAS2val19 allele was introduced into strains expressing GFP-tagged versions of either the wild-type Pat1 (Pat1-SS) or the nonphosphorylatable Pat1-AA variant. The Pat1-AA protein has alanine residues replacing the two serines that are phosphorylated by PKA, Ser-456 and Ser-457 (Ramachandran et al. 2011). These cells also contained the Edc3-mCherry reporter. As above, we found that the P bodies in cells with the wild-type Pat1 were efficiently dispersed following the induction of RAS2val19 expression (Figure 1C). However, this disassembly was effectively blocked in cells containing the Pat1-AA variant (Figure 1, C and D). Therefore, the presence of PKA activity was sufficient to trigger the disassembly of already formed P bodies in a Pat1-dependent manner.
Figure 1 .
The presence of Ras/PKA signaling activity was sufficient to trigger P body disassembly. (A) The ectopic expression of the constitutively active Ras2val19 protein resulted in the dispersal of Edc3-containing foci. Cells expressing the Edc3-mCherry protein were transferred to a glucose-minus medium containing 250 mM methionine for 10 min to induce P body formation. The cells were then moved to a medium that also lacked methionine for 2 hr to induce expression from the MET3-RAS2val19 locus and then were examined by fluorescence microscopy. Three representative images are shown for both the control and the induced conditions. (B) Increased Ras2 protein expression was detected in cells containing the MET3-RAS2val19 locus. Protein extracts were prepared from cells containing either a control vector or a MET3-RAS2val19 plasmid, and the level of Ras2 protein was assessed by Western blotting with an anti-Ras2 antibody. (C and D) The RAS2val19-triggered disassembly of P bodies was blocked in cells containing the nonphosphorylatable variant Pat1-AA. The disassembly of preformed P bodies was assessed as described above in cells expressing GFP-tagged versions of either the wild-type Pat1-SS or the nonphosphorylatable Pat1-AA. The microscopy data are shown in C, and the quantitation of these data in D. The data shown in the graph represent the average of at least two experiments (n = 100) with the Edc3-mCh reporter.
All three PKA isoforms influence P body formation
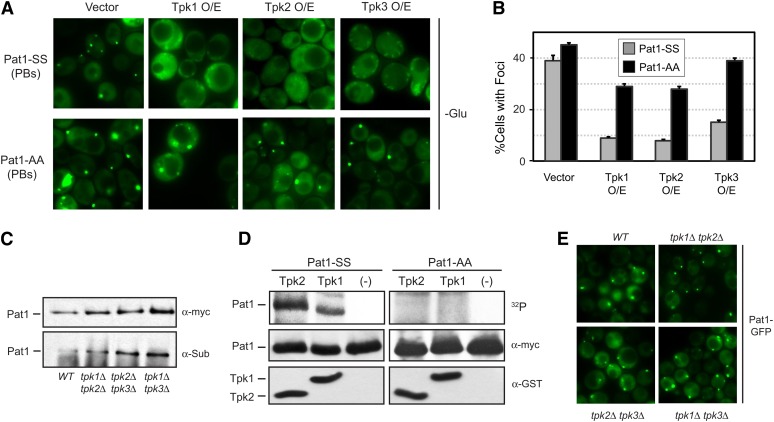
S. cerevisiae has three genes, TPK1–3, that encode a catalytic subunit of the PKA enzyme (Toda et al. 1987b). Although each of these genes is sufficient for viability, there are data suggesting that the different isoforms have distinct functions (Robertson and Fink 1998; Pan and Heitman 2002; Ptacek et al. 2005). Moreover, a recent study has indicated that the Tpk2 and Tpk3 proteins are themselves recruited to P body foci in response to glucose deprivation (Tudisca et al. 2010). Therefore, we tested here whether each of the isoforms could influence P body assembly. Consistent with this possibility, we made the following observations. First, we found that the overexpression of any single Tpk protein was able to efficiently inhibit P body formation (Figure 2, A and B). This inhibition was suppressed in each case by the introduction of the nonphosphorylatable Pat1-AA. Second, we found that Pat1 was phosphorylated in mutants lacking any two Tpk isoforms, suggesting that all three Tpk proteins were capable of phosphorylating Pat1 (Figure 2C). This in vivo phosphorylation was assessed with an antibody that specifically recognizes the PKA phosphorylated form of Pat1 (Chang et al. 2004; Ramachandran et al. 2011). Third, we found that both Tpk1 and Tpk2 were able to phosphorylate the Pat1 protein in vitro (Figure 2D). The epitope-tagged Tpk3 protein had very low enzymatic activity, and we were unable to assess its ability to phosphorylate Pat1 in these assays. Finally, we found that P bodies were able to form in double-mutant cells lacking any two of the Tpk proteins (Figure 2E). In all, these results indicated that each of the three Tpk enzymes was able to phosphorylate Pat1 and thereby regulate P body assembly in S. cerevisiae.
Figure 2 .
All three isoforms of the S. cerevisiae PKA were able to influence P body formation. (A and B) The overexpression of any one of the three Tpk proteins was sufficient to inhibit P body formation. Cells containing either the control vector or a plasmid with the indicated TPK gene were transferred to a glucose-minus medium for 10 min to induce P body formation. The cells also expressed either the Pat1-SS or Pat1-AA protein, as indicated. The microscopy data are presented in A and the quantitation of these data in B. (C) The PKA site in Pat1 was phosphorylated in cells lacking any two of the three Tpk isoforms. Pat1 was immunoprecipitated from log-phase cells with the indicated genotypes, and the level of in vivo PKA phosphorylation was assessed by Western blotting with an antibody that specifically recognizes the phosphorylated form of Pat1 (α-Sub). (Top) Western blot showing the total amount of the myc-tagged Pat1 present. (D) Pat1 was phosphorylated in vitro by the Tpk1 and Tpk2 isoforms of PKA. The myc-tagged Pat1 proteins were immunoprecipitated from log-phase cells, treated with phosphatase, and then incubated with the indicated GST-tagged Tpk protein and [γ-32P]ATP. The bottom two panels are Western blots indicating the total amount of the respective proteins present in the reaction mixtures. “(-)” indicates the “no kinase” control. (E) P body formation in mutants containing only a single Tpk isoform. Cells with the indicated genotypes were transferred to a medium lacking glucose for 10 min, and P body formation was then assessed by fluorescence microscopy.
PKA activity inhibits the P body formation induced by a variety of stress conditions
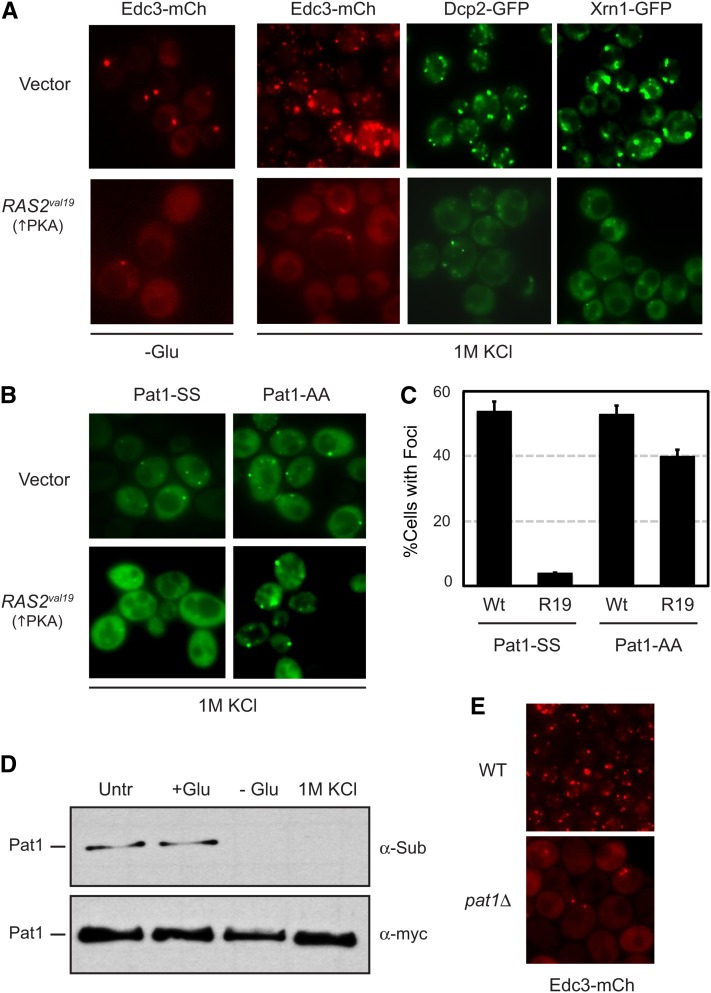
P body foci form in response to a number of stress conditions in addition to glucose deprivation (Teixeira et al. 2005; Izawa et al. 2007; Yoon et al. 2010; Buchan et al. 2011). Although previous work indicated that PKA activity is influenced by glucose availability, it is not clear how the other conditions affect the Ras/PKA signaling pathway (Santangelo 2006; Zaman et al. 2008; Ramachandran and Herman 2010). Therefore, we tested whether the presence of the Ras2val19 protein would inhibit the P bodies induced by these other stresses. We first examined the foci that form in response to a hyperosmotic stress. For these experiments, we examined cells after a 10-min exposure to media containing 1 M KCl. The foci were visualized with three different P body reporters, Edc3, Dcp2, and Xrn1. Dcp2 is the catalytic subunit of the main mRNA decapping enzyme in eukaryotes, and Xrn1 is the primary exonuclease in the 5′-to-3′ mRNA decay pathway (Bashkirov et al. 1997; Ingelfinger et al. 2002; Van Dijk et al. 2002; Eystathioy et al. 2003; Sheth and Parker 2003; Cougot et al. 2004). For each reporter, we found that the localization to cytoplasmic foci was greatly diminished in the presence of the Ras2val19 protein (Figure 3A). To test whether this inhibition was dependent upon the phosphorylation of Pat1, we introduced the RAS2val19 allele into cells expressing GFP-tagged versions of either the wild-type Pat1 (Pat1-SS) or Pat1-AA. Although the presence of RAS2val19 inhibited foci formation with the wild-type protein, this inhibition was effectively suppressed in cells containing Pat1-AA (Figure 3, B and C). In addition, we found that the level of PKA phosphorylation on Pat1 was dramatically reduced following a 10-min exposure to 1 M KCl (Figure 3D). This reduction was similar in magnitude to that observed upon glucose starvation and did not occur in cells containing the RAS2val19 allele (Figure 3D; Supporting Information, Figure S1). Consistent with a role for Pat1, we also found that the number of P body foci induced by this hyperosmotic stress was reduced in pat1Δ cells (Figure 3E).
Figure 3 .
Ras/PKA signaling activity inhibited P body formation in response to hyperosmotic stress. (A) Osmotic stress-induced P body formation was inhibited in cells expressing the constitutively active Ras2val19 protein. Cells transformed with either a control vector or a plasmid containing the RAS2val19 locus were grown to mid-log phase and transferred to a medium either lacking glucose (−Glu) or containing 1 M KCl for 10 min. P body formation for each of the indicated reporters was then assessed by fluorescence microscopy. (B) The PKA-mediated inhibition of P body formation was suppressed by the presence of the nonphosphorylatable Pat1-AA. P body formation in response to 1 M KCl was assessed as above for pat1Δ cells expressing GFP-tagged versions of either Pat1-SS or Pat1-AA. (C) Quantification of the data shown in B. The graph indicates the fraction of cells with Pat1-containing foci after a 10-min exposure to 1 M KCl. (D) The PKA-dependent phosphorylation on Pat1 was rapidly diminished upon exposure to 1 M KCl. Cells expressing a myc-tagged Pat1 were grown to mid-log phase and then transferred for 10 min to the same medium (SC glucose; +Glu) or media either lacking glucose (−Glu) or containing 1 M KCl. The level of PKA phosphorylation was assessed by Western blotting with the anti-PKA substrate antibody (α-Sub). Untr, the extract prepared from the untreated control culture. (E) The frequency of P body formation following an osmotic stress was diminished in cells lacking Pat1. Wild-type and pat1Δ cells expressing the Edc3-mCh reporter were exposed to a medium containing 1 M KCl for 10 min and then examined by fluorescence microscopy.
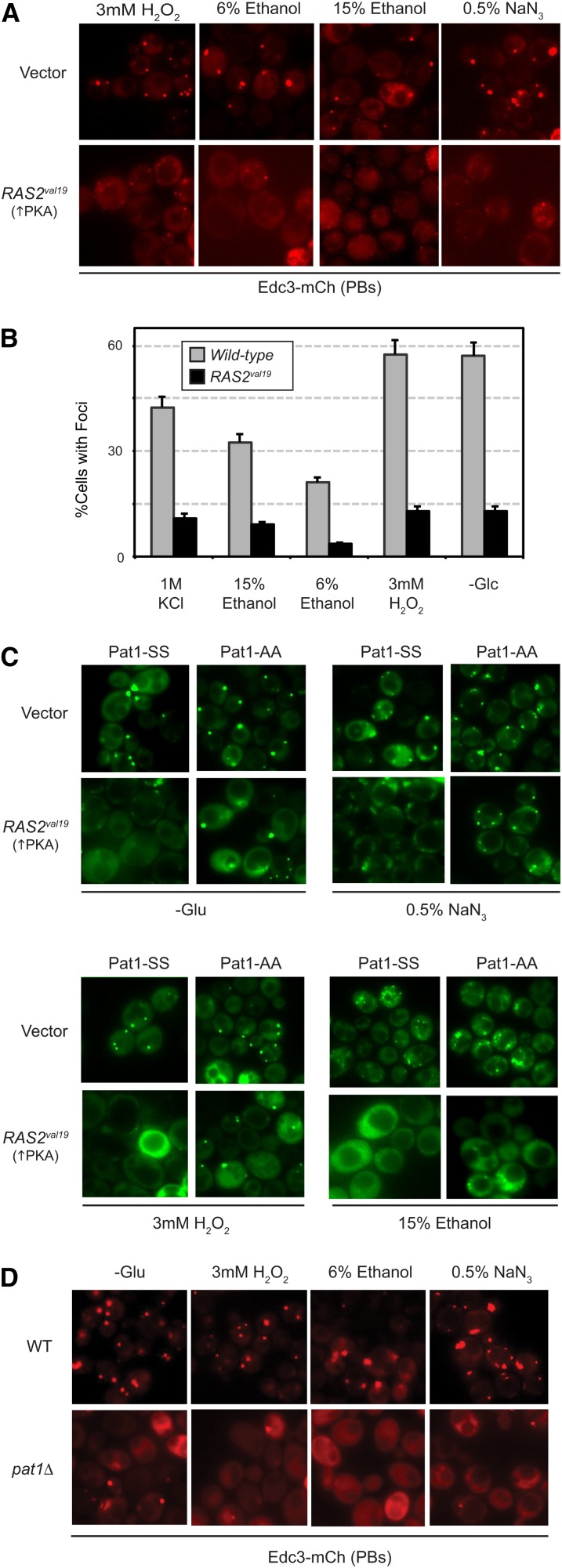
To determine if PKA generally inhibited P body formation, we examined the effects of expressing the Ras2val19 protein in cells exposed to 3 mM H2O2, 6 and 15% ethanol, and 0.5% NaN3. All of these stress conditions have been shown to induce P body formation. In each case, we found that the number of P bodies was significantly diminished in the presence of elevated PKA activity (Figure 4, A–C). Moreover, these PKA inhibitory effects were effectively suppressed by the presence of the Pat1-AA variant (Figure 4C; Figure S2A). Finally, the number of Edc3-mCherry foci induced by these different conditions was greatly reduced in cells lacking the Pat1 protein (Figure 4D; Figure S2B). Altogether, these data indicate that PKA signaling activity has a general role in the regulation of P body foci formation in S. cerevisiae.
Figure 4 .
The presence of elevated PKA signaling activity inhibited P body induction in response to a variety of stress conditions. (A and B) Expression from the RAS2val19 locus inhibited the P body formation brought on by a number of stress conditions. Wild-type cells expressing the Edc3-mCh reporter were transformed with either a RAS2val19 plasmid or a control vector and exposed to the indicated stress for 10 min. P body formation was then assessed by fluorescence microscopy. The microscopy data are presented in A and the quantitation of these data in B. The 1 M KCl microscopy data are shown in Figure 3A. (C) The presence of the nonphosphorylatable Pat1-AA diminished the inhibitory effects of the Ras2val19 protein. P body foci formation was assessed after a 10-min exposure to the indicated stress in pat1Δ cells expressing GFP-tagged Pat1-SS or Pat1-AA. The cells contained either a control vector or a RAS2val19 plasmid, as indicated. A graph showing the quantitation of these data are presented in Figure S2A. (D) P body formation in response to a variety of stresses required the presence of the Pat1 protein. P body formation was assessed in wild-type and pat1Δ cells after a 10-min exposure to the indicated conditions. A graph showing the quantitation of these data is presented in Figure S2B.
Assembly of stress granules is not influenced by PKA-signaling activity
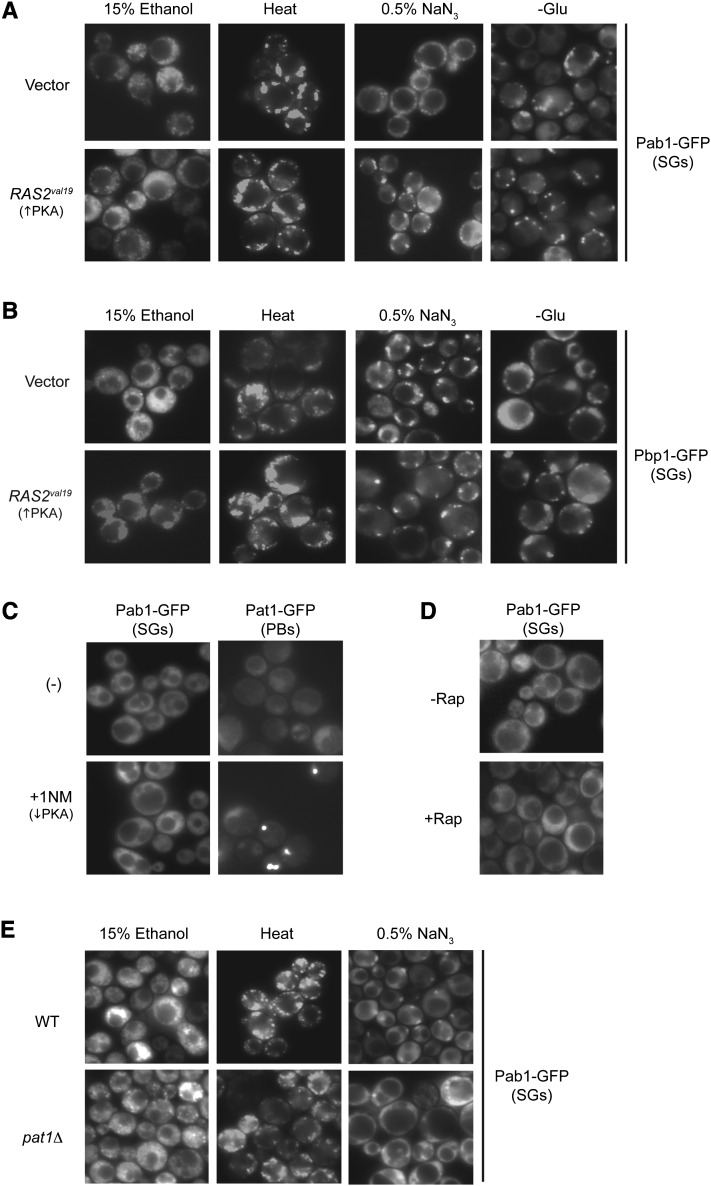
A second evolutionarily conserved RNP complex, known as the stress granule, is also induced by several of the above conditions (Hoyle et al. 2007; Buchan et al. 2008, 2011; Grousl et al. 2009; Kato et al. 2011). Here, we tested whether stress granule assembly was also controlled by PKA activity. For these studies, stress granules were induced by a 30-min exposure to either 15% ethanol, 0.5% NaN3, a 46° heat stress, or glucose deprivation. GFP-tagged versions of Pab1 and Pbp1 were used to mark stress granule foci. Pab1 is the yeast poly(A)-binding protein, and Pbp1 is the homolog of the human ataxin-2 protein that is the product of the spinocerebellar ataxia type 2 gene, SCA2 (Sachs et al. 1986; Imbert et al. 1996; Ralser et al. 2005). An induction of stress granules was observed for each of these conditions, but the final gross morphology of the fluorescent RNP structures varied for each stress (Figure 5, A and B). However, in contrast to our results with P bodies, these stress granules were present in similar numbers in cells either expressing RAS2val19 or lacking Bcy1, the regulatory subunit of the PKA enzyme (Figure 5, A and B; Figure S3). In addition, stress granules were not induced by the inactivation of either the Ras/PKA or the TORC1 signalling pathways (Figure 5, C and D). To inactivate the PKA pathway, we used a strain that expressed an analog-sensitive version of Tpk1 as its sole source of PKA activity (Bishop et al. 2001; Stephan et al. 2009). The encoded Tpk1as protein is rapidly inactivated by the membrane-permeable inhibitor 1NM-PP1 (Stephan et al. 2009). As noted previously, the inactivation of the PKA pathway resulted in the formation of P body foci (Figure 5C) (Ramachandran et al. 2011). To inactivate the TORC1 pathway, the cultures were treated with the macrolide rapamycin. Finally, we found that stress granules were able to form at essentially wild-type levels in cells lacking the Pat1 protein for all inducing conditions except NaN3 (Figure 5E). In this latter case, we found that pat1Δ cells had ∼20–30% fewer foci than the wild-type strain (see also Buchan et al. 2011). Altogether, these results indicated that stress granule assembly is not regulated by PKA signaling activity in S. cerevisiae.
Figure 5 .
Stress granule formation was not influenced by Ras/PKA signaling activity. (A and B) Elevated Ras/PKA activity did not inhibit stress granule formation in response to a number of inducing conditions. Wild-type cells expressing either Pab1-GFP (A) or Pbp1-GFP (B) were transformed with either a control vector or a RAS2val19 plasmid. Stress granule formation was induced by a 30-min exposure to a 46° heat stress (Heat) or media either lacking glucose or containing 15% ethanol or 0.5% NaN3, as indicated. (C) The down-regulation of PKA signaling did not result in stress granule formation. Plasmids with GFP-tagged versions of either Pab1 or Pat1 were introduced into a yeast strain (PHY4697) with an analog-sensitive version of TPK1, tpk1as. The cells were then treated for 30 min with 5 μM 1NM-PP1 to inhibit the encoded Tpk1as protein, and foci formation was assessed by fluorescence microscopy. (D) Stress granule formation was not induced upon inactivation of the TORC1 signalling pathway. Wild-type cells expressing the Pab1-GFP fusion protein were treated for 20 min with 200 ng/ml rapamycin. (E) Stress granule formation in pat1Δ cells. Wild-type and pat1Δ cells were exposed to the indicated stresses for 30 min, and Pab1-GFP foci were visualized by fluorescence microscopy.
Presence of P body foci is correlated with the long-term survival of stationary-phase cells
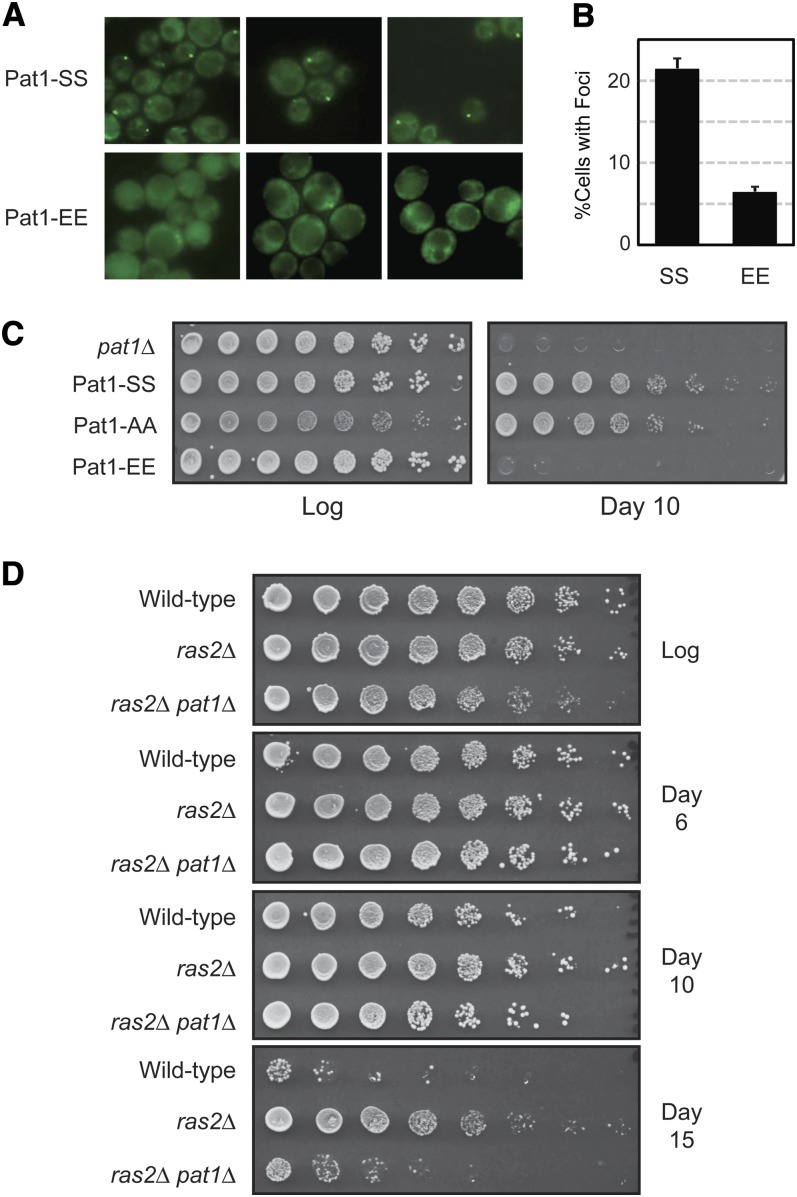
Our previous work indicated that P body foci formation might be required for the long-term survival of stationary-phase cells (Ramachandran et al. 2011). This survival has been used as a model for the study of eukaryotic aging, and the length of time that the quiescent yeast cells remain viable has been referred to as the chronological life span, or CLS (Kennedy 2008). To examine this link with P bodies further, we asked whether the presence of a phosphomimetic form of Pat1, Pat1-EE, would affect foci formation and/or the survival of stationary-phase cells. Pat1-EE has glutamic acid residues replacing the two serines that are normally phosphorylated by PKA (Ramachandran et al. 2011). The presence of a glutamic acid can functionally substitute for a phosphorylated serine residue (Thorsness and Koshland 1987; Pearlman et al. 2011), and Pat1-EE could therefore act as a constitutively phosphorylated version of Pat1. Consistent with this latter possibility, we found that cells containing the Pat1-EE variant had significantly fewer P body foci than wild-type cells after 5 days of growth (Figure 6, A and B; Figure S4A). The presence of Pat1-EE also resulted in fewer P body foci in response to all of the other stress conditions tested (Figure S4, B and C). Moreover, the foci that were present in the Pat1-EE cells were generally much smaller than those in the wild type. Strikingly, the decrease in foci number correlated with a diminished rate of stationary-phase survival as the cells containing Pat-EE rapidly lost viability during this period of quiescence (Figure 6C). Two observations indicated that these results were not due to Pat1-EE being simply a misfolded and nonfunctional polypeptide. First, a Western blot analysis showed that the Pat1-EE variant was present at wild-type levels in cell extracts (Figure S4D). Second, we had shown previously that the overexpression of Pat1-EE results in a growth arrest that is similar to that seen with the wild-type protein (Ramachandran et al. 2011). Pat1 has been shown to act as a translational repressor, and this overexpression result therefore indicates that Pat1-EE retains at least some functions of the wild-type protein (Coller and Parker 2005). Therefore, the presence of the Pat1-EE variant resulted in both a diminished number of P body foci and a decreased rate of survival during the stationary phase of growth.
Figure 6 .
The presence of the phosphomimetic Pat1-EE resulted in fewer P body foci and a reduced survival in stationary phase. (A and B) Cells with the Pat1-EE variant had fewer P body foci. The number of P body foci after 5 days of growth in YPAD medium was assessed for cells expressing GFP-tagged versions of either Pat1-SS or Pat1-EE. Three representative images are shown for cells containing either Pat1-SS or Pat1-EE. The microscopy data are shown in A and the quantitation of these data in B. (C) Cells expressing the Pat1-EE variant had a diminished CLS. The number of viable cells present during log-phase growth and after 10 days in a minimal YM glucose medium was determined by plating out increasing dilutions of these cultures. (D) Deletion of the PAT1 locus suppressed the extended CLS of a ras2Δ mutant. Serial dilutions of strains with the indicated genotypes were plated out at the indicated times of culture growth in a YM glucose minimal medium.
We also tested whether Pat1 was required for the extended CLS observed for mutants with diminished levels of Ras/PKA signaling activity. For these experiments, we used a ras2Δ yeast strain that remains viable because of the presence of the wild-type RAS1 locus. As reported previously, we found that this strain survived significantly longer than the isogenic wild type when in stationary phase (Figure 6D) (Longo 2003). However, this increased survival was largely lost upon deletion of the PAT1 locus (Figure 6D). Cumulatively, these data are consistent with a role for Pat1, and perhaps for P body foci, in the long-term survival of stationary-phase cells.
P body foci persist throughout the stationary phase of growth
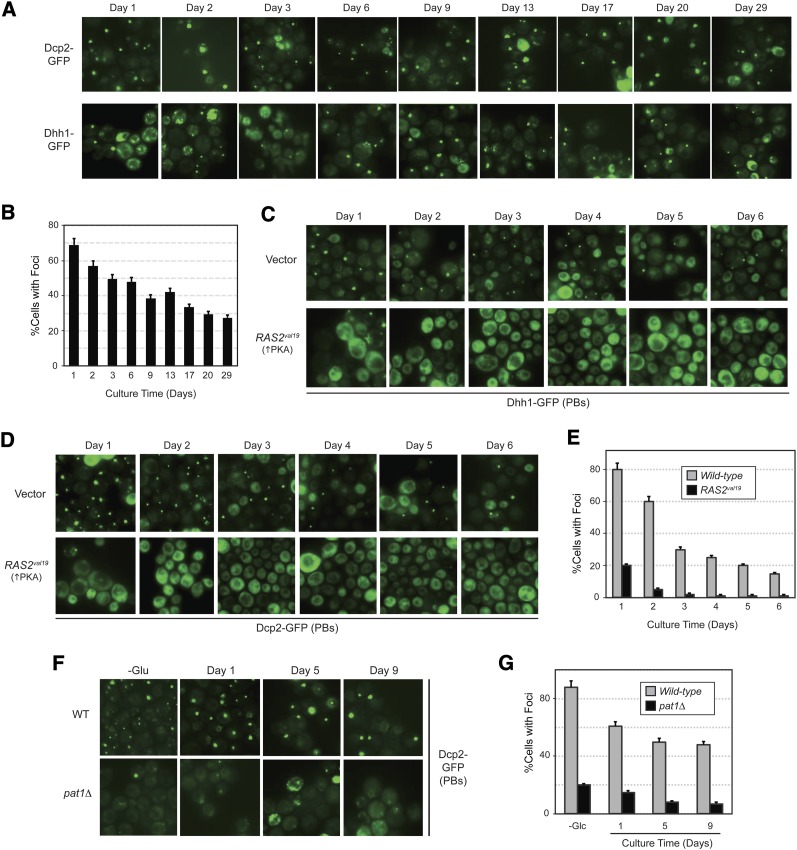
Despite their potential role in stationary-phase survival, it was not known whether P bodies were present in these quiescent cells. Previous studies have most likely examined cells that had not yet entered into stationary phase (Teixeira et al. 2005; Brengues and Parker 2007). To address this question, we examined cells expressing various P body markers at different times during a 4-week period of growth. The fraction of cells containing foci at any time was assessed by fluorescence microscopy. With each reporter, P bodies were initially detected after ∼24 hr of growth and were found to persist throughout the entire incubation period (Figure 7, A and B; Figure S5, A and B). The time of P body induction observed is consistent with previous reports indicating that these RNP complexes begin to form when glucose is almost depleted from the medium (see below) (Teixeira et al. 2005). As with the other stress conditions, we found that the number of stationary-phase foci was severely decreased in both RAS2val19 and pat1Δ mutant cells (Figure 7, C–G). These data therefore indicated that P body foci are present throughout the stationary phase of growth and that the induction of these RNP granules is regulated by the PKA signaling pathway.
Figure 7 .
P body foci are present and persist throughout the stationary phase of growth. (A) P body foci are present in stationary-phase cells. The frequency of P body formation was determined for cells with the integrated Dcp2-GFP and Dhh1-GFP reporters at the indicated times of growth in YPAD cultures. (B) A graph showing the number of Dcp2 foci at the indicated culture times; the relevant microscopy data are presented in A. (C and D) The presence of the constitutively active Ras2val19 protein resulted in fewer P body foci. P body foci were visualized by fluorescence microscopy after the indicated number of days of growth for wild-type cells with either a control vector or a RAS2val19 plasmid. (E) A graph showing the quantitation of the Dcp2-GFP microscopy data presented in D. (F and G) The presence of the Pat1 protein was required for the formation of stationary-phase P body foci. The fraction of cells with P body foci was determined for wild-type and pat1Δ cells after the indicated number of days of growth in YPAD. The results for a 10-min starvation for glucose are shown for comparison purposes (−Glu). The microscopy data are shown in F and the quantitation of these data in G.
Stress granule foci form specifically upon entry into stationary phase
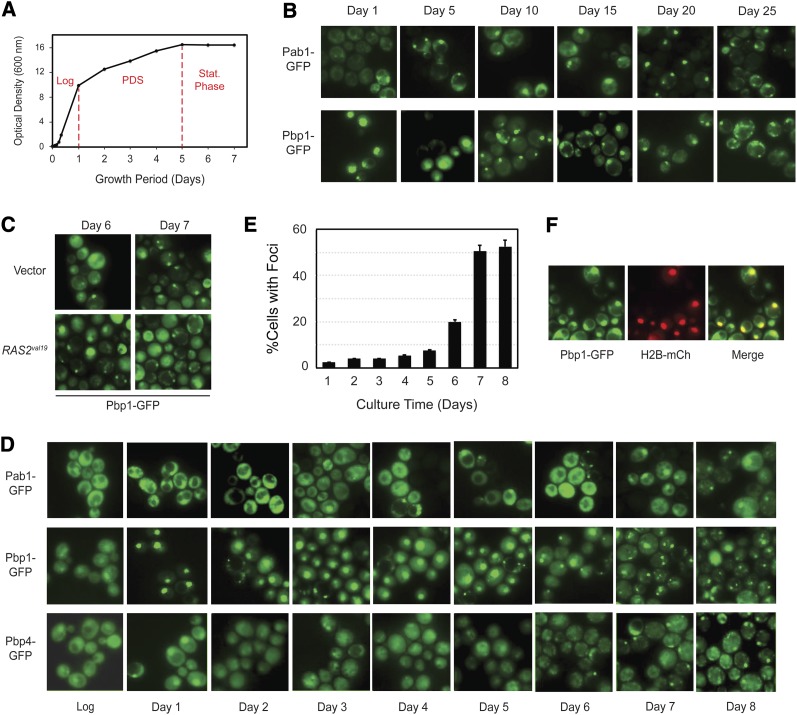
We also examined the temporal pattern of stress granule appearance in cells that were grown into stationary phase on glucose-based media. Under these batch culture conditions, yeast cells proceed through a number of distinct growth transitions (Figure 8A). During the initial or logarithmic phase, the cells grow rapidly via fermentation of the available glucose. This fermentative growth occurs even though oxygen is available to the cells and continues for ∼24 hr (Diaz-Ruiz et al. 2009; Castelli et al. 2011). When the glucose is depleted, the cells alter their metabolism and subsequently grow by respiration on the ethanol that was produced during the prior period of fermentation. This switch in carbon source utilization is known as the diauxic shift and the time of respiratory growth as the post-diauxic shift growth period (PDS) (Figure 8A) (Gray et al. 2004). This latter period of slower growth continues for another 4–5 days until the ethanol is also used up. At this time, the cells enter the true stationary phase of growth (Werner-Washburne et al. 1993; Herman 2002).
Figure 8 .
Stress granules appeared in cells after entry into stationary phase. (A) A representative growth curve for a wild-type strain with the integrated Pbp1-GFP reporter. The red lines delimit the log, PDS, and stationary phases of growth. (B) Stress granule foci were present in stationary-phase cells. The subcellular localization of Pab1-GFP and Pbp1-GFP was assessed by fluorescence microscopy after the indicated days of growth in the rich growth medium YPAD. (C) Stress granule formation was not affected by the presence of the RAS2val19 locus. The subcellular localization of a Pbp1-GFP reporter was assessed after the indicated number of days of growth for cells containing either a control vector or a RAS2val19 plasmid. (D and E) Stress granule foci did not appear in significant numbers until after cells had entered into the stationary phase of growth. The subcellular localization of the indicated reporters was assessed in cells at daily intervals of growth in a rich medium. The graph in E shows the fraction of cells with a Pbp1-GFP focus after the indicated days of growth. (F) Pbp1 was found in the nucleus during the PDS period of growth. Cells expressing Pbp1-GFP were transformed with a plasmid encoding the nuclear marker, histone H2B-mCh, and the colocalization of the two reporters was assessed after 3 days of culture growth.
Using a variety of GFP-tagged reporters, we found that stress granules were present in stationary-phase cells and that these foci appeared to persist for as long as the cells remained in this quiescent state (Figure 8B). As with the other inducing conditions, the number of these stress granules was not significantly diminished by either the presence of the Ras2val19 protein or the absence of Pat1 (Figure 8C; Figure S6A). In addition, the stress granule reporters tested, Pbp1, Pbp4, and Lsm12, did not colocalize with the Edc3-mCh marker for P bodies, suggesting that these two foci were distinct structures in these resting cells (Figure S6B). In all, we found that the stress granule and P body reporters failed to colocalize in 92.0% (±7.1%) of those cells that had both types of these foci. However, the important point to emphasize here is that the stress granule foci did not appear in significant numbers until after 6–7 days of culture incubation, a time when the cells appear to have already entered into the stationary phase of growth (Figure 8, D and E; Figure S6C). This temporal pattern of localization is illustrated here with the stress granule constituent Pbp1. Whereas Pbp1 is distributed throughout the cell during the fermentative, or log phase, of growth, this protein is found predominantly in the nucleus during the PDS growth period (Figure 8, D and F). Upon the cessation of growth, this protein relocalizes to discrete foci in the cytoplasm of the cells (Figure 8, D and E). Other stress granule reporters, including Pab1, Pbp4, Pub1, and Lsm12, were also found in foci after day 6 or 7 of culture incubation (Figure 8D; Figure S6C). These data therefore indicate that stress granules form specifically after cells enter into the stationary phase of growth.
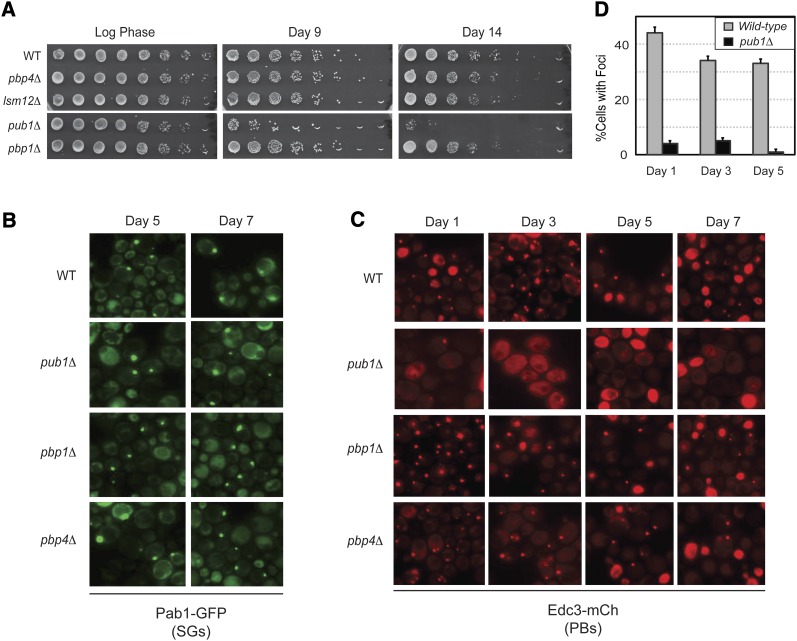
We also examined the CLS of strains lacking different stress granule constituents, including the pbp1Δ, pbp4Δ, lsm12Δ, and pub1Δ mutants (Buchan and Parker 2009). Of these strains, only the pub1Δ mutant exhibited a significantly shorter CLS than the wild-type control (Figure 9A). This survival phenotype was apparently not due to a defect in stress granule formation as all of these mutant strains had wild-type levels of Pab1-containing foci (Figure 9B). However, we found that pub1Δ cells contained a dramatically reduced number of P body foci, a defect that was specific to this particular mutant (Figure 9, C and D). This latter result was somewhat surprising as pub1Δ mutants were not defective for P body formation under other induction conditions, including glucose deprivation (Buchan et al. 2008, 2011) (Figure S7). The defect observed here suggests that Pub1 may have additional functions in stationary-phase cells, and it will be interesting to determine the nature of this activity. In any case, the findings with the pub1Δ mutant were consistent with the above assertion that P body foci play an important role in the long-term survival of stationary-phase cells.
Figure 9 .
Mutants lacking the stress granule protein Pub1 have a diminished CLS and fewer stationary-phase P body foci. (A) pub1Δ mutants exhibited a decreased rate of survival during stationary phase. The number of viable cells present at the indicated times of growth was determined by plating out increasing dilutions of the cultures as described in Materials and Methods. (B and C) The subcellular localization of a stress granule (B: Pab1-GFP) or P body (C: Edc3-mCh) marker was assessed by fluorescence microscopy after the indicated days of growth. (D) Quantitation of the P body foci data for the wild-type and pub1Δ strains shown in C.
Discussion
In this study, we examined the assembly and function of two representative RNP complexes, the P body and stress granule, in S. cerevisiae. These complexes are structurally related and are induced by similar types of stress conditions (Kedersha et al. 2005; Buchan and Parker 2009). However, despite these similarities, the data here indicate that these two RNP granules are independently assembled and that this assembly is controlled by different signaling pathways. In particular, we found that the PKA pathway has a general role in the regulation of P body foci formation. Constitutive PKA signaling inhibited P body formation under all conditions tested, and, in each case, this inhibition was reversed by the presence of the nonphosphorylatable Pat1-AA variant. All three isoforms of the PKA enzyme, Tpk1–3, were found to inhibit P body formation. Consistent with a key role for Pat1, we also found that P body assembly was less efficient in cells carrying the phosphomimetic variant Pat1-EE. Finally, the ectopic presence of PKA activity was found to be sufficient to trigger the disassembly of preformed P bodies in glucose-starved cells. In all, these data point to a key regulatory role for the PKA-mediated phosphorylation of Pat1 in the P body assembly process. For each stress condition, this control could be due to changes in the activity of either PKA itself or the particular phosphatase(s) responsible for the dephosphorylation of Pat1. Ongoing efforts in the laboratory aim to discern between these two possibilities for each of the induction conditions.
In contrast to these results with P bodies, we found no evidence of a role for PKA in the control of stress granule assembly. Neither the presence of constitutive PKA activity nor the inactivation of this pathway had any significant effect upon stress granule formation. This was true for all conditions tested, including those that can induce both types of RNP foci. Thus, stress granule assembly is apparently regulated by other, as-yet-unidentified, signaling pathways. The results also point out that stress granules, for the most part, form normally in cells, such as the RAS2val19 and pat1Δ mutants, that are defective for P body assembly. Stress granule formation therefore was not generally dependent upon a pre-existing P body template, an observation that is consistent with studies of these granules in mammalian cells (Ohn et al. 2008; Anderson and Kedersha 2009). However, it should be noted that our results conflict with a prior study that found that the presence of stress granules in yeast cells was dependent upon P body formation in glucose-limiting conditions (Buchan et al. 2008). Although we do not yet know the underlying reason for these different observations, it is relevant to point out that glucose deprivation was the least robust condition for stress granule formation. Subtle changes in media composition and strain background had a significant effect on the efficiency of stress granule induction, and either could be responsible for the differences reported in these two studies (Nissan and Parker 2008; Takahara and Maeda 2012). Nonetheless, the key points here are that stress granule formation is not regulated by PKA activity and that stress granule and P body assembly appear to occur independently in S. cerevisiae.
The data here also show that P body and stress granule foci are present in stationary-phase cells and that these complexes persist throughout this period of quiescence. However, each of these RNP structures is induced at a very different time. P bodies form as glucose is depleted from the growth medium, and these foci are then present throughout both the PDS and stationary phases of growth. The presence of these foci in the PDS period indicates that P bodies can form in yeast cells that are actively dividing. In contrast, stress granules did not appear in significant numbers until the cells had stopped dividing and entered into stationary phase. The induction of these stress granules was again not dependent upon the prior presence of P body foci. In addition, we found that reporters for P body and stress granule foci did not colocalize in stationary-phase cells, a result consistent with a recent study of stationary-phase RNP foci containing the Hos2 deacetylase enzyme (Liu et al. 2012). In all, these results emphasize again the independent nature of the two RNP assembly pathways and further point out that the appearance of stress granules might serve as a marker for cell entry into stationary phase. There are very few useful reporters for this transition, and it will be important to determine the underlying reasons for the induction of stress granule foci that occurs at this time.
The final point that we would like to make concerns one of the key outstanding questions in the RNP field, that of the physiological function of the larger P body assemblies. Interestingly, the data here provide further support for a model positing that the presence of P bodies is important for the long-term survival of stationary-phase yeast cells (Ramachandran et al. 2011). In particular, this work has identified a direct correlation between the number of P body foci and the length of time that cells survive in this quiescent state. For example, we found here that cells either carrying the phosphomimetic variant Pat1-EE or lacking the stress granule protein Pub1 had fewer P body foci and a correspondingly shorter CLS. In addition, previous work showed that the introduction of the nonphosphorylatable Pat1-AA was able to restore both P body foci and a more-normal CLS to mutants harboring the constitutively active RAS2val19 allele (Ramachandran et al. 2011). The underlying reasons for this correlation are not yet clear as the precise physiological activity of the larger P body foci in quiescent cells has not been determined. The foci could be responsible for the removal of particular deleterious activities from the cytoplasm or for the storage of proteins (or RNAs) necessary for the subsequent resumption of growth. Finally, it is interesting to speculate that stress granules are also important for stationary-phase survival as these RNP granules appear specifically upon entry into this quiescent state. Unfortunately, we were unable to test this possibility as all of the mutants examined here were proficient for stress granule formation. Obtaining answers to these and related questions are major goals for the field at large and should provide important insights into the physiological roles of the various RNP foci that are found in quiescent cells.
Supplementary Material
Acknowledgments
We thank James Broach, Tien-Hsien Chang, James Hopper, and Roy Parker for strains and plasmids used in this study; Jian-Qiu Wu for the use of his fluorescence microscope; and members of the Herman laboratory for their comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM65227) to P.K.H.
Footnotes
Communicating editor: J. Heitman
Literature Cited
- An S., Kumar R., Sheets E. D., Benkovic S. J., 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320: 103–106 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., 2006. RNA granules. J. Cell Biol. 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Bahn Y. S., Xue C., Idnurm A., Rutherford J. C., Heitman J., et al. , 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5: 57–69 [DOI] [PubMed] [Google Scholar]
- Balagopal V., Parker R., 2009. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V. I., Scherthan H., Solinger J. A., Buerstedde J. M., Heyer W. D., 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Buzko O., Shokat K. M., 2001. Magic bullets for protein kinases. Trends Cell Biol. 11: 167–172 [DOI] [PubMed] [Google Scholar]
- Brengues M., Parker R., 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., 1991. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7: 28–33 [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., et al. , 1985. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell 41: 763–769 [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Parker R., 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R., 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Nissan T., Parker R., 2010. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol. 470: 619–640 [DOI] [PubMed] [Google Scholar]
- Buchan J. R., Yoon J. H., Parker R., 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Hama H., Dewald D. B., Herman P. K., 2002. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 277: 287–294 [DOI] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., Herman P. K., 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279: 20663–20671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K., 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli L. M., Lui J., Campbell S. G., Rowe W., Zeef L. A., et al. , 2011. Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated. Mol. Biol. Cell 22: 3379–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. W., Howard S. C., Budovskaya Y. V., Rine J., Herman P. K., 2001. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics 157: 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. W., Howard S. C., Herman P. K., 2004. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15: 107–116 [DOI] [PubMed] [Google Scholar]
- Cherest H., Kerjan P., Surdin-Kerjan Y., 1987. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5′ non-coding region to that of MET25. Mol. Gen. Genet. 210: 307–313 [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R., 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B., 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R., 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff S. J., Howard S. C., Hester A., Warner S., Herman P. K., 2006. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics 173: 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff S. J., Ramachandran V., Herman P. K., 2009. Distal recognition sites in substrates are required for efficient phosphorylation by the cAMP-dependent protein kinase. Genetics 182: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz R., Uribe-Carvajal S., Devin A., Rigoulet M., 2009. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim. Biophys. Acta 1796: 252–265 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E., 2007a. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E., 2007b. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E. K., Seraphin B., Cougot N., et al. , 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Xu H. P., Michaeli T., Ballester R., Sass P., et al. , 1990. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science 247: 464–467 [DOI] [PubMed] [Google Scholar]
- Franks T. M., Lykke-Andersen J., 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., et al. , 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., et al. , 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68: 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frydlova I., Vasicova P., Janda F., et al. , 2009. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122: 2078–2088 [DOI] [PubMed] [Google Scholar]
- Herman P. K., 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5: 602–607 [DOI] [PubMed] [Google Scholar]
- Howard S. C., Chang Y. W., Budovskaya Y. V., Herman P. K., 2001. The Ras/PKA signaling pathway of Saccharomyces cerevisiae exhibits a functional interaction with the Sin4p complex of the RNA polymerase II holoenzyme. Genetics 159: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S. C., Budovskaya Y. V., Chang Y. W., Herman P. K., 2002. The C-terminal domain of the largest subunit of RNA polymerase II is required for stationary phase entry and functionally interacts with the Ras/PKA signaling pathway. J. Biol. Chem. 277: 19488–19497 [DOI] [PubMed] [Google Scholar]
- Hoyle N. P., Castelli L. M., Campbell S. G., Holmes L. E., Ashe M. P., 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H., Yahara I., 1984. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J. Cell Biol. 98: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert G., Saudou F., Yvert G., Devys D., Trottier Y., et al. , 1996. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 14: 285–291 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D. J., Luhrmann R., Achsel T., 2002. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Izawa S., Kita T., Ikeda K., Miki T., Inoue Y., 2007. Formation of cytoplasmic P-bodies in sake yeast during Japanese sake brewing and wine making. Biosci. Biotechnol. Biochem. 71: 2800–2807 [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Kato K., Yamamoto Y., Izawa S., 2011. Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 28: 339–347 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P., 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30: 963–969 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., et al. , 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B. K., 2008. The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J. Intern. Med. 263: 142–152 [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Aboobakar E. F., Cardenas M. E., Heitman J., 2011. Calcineurin colocalizes with P-bodies and stress granules during thermal stress in Cryptococcus neoformans. Eukaryot. Cell 10: 1396–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman J. L., Jongens T. A., 2003. Transcriptional silencing and translational control: key features of early germline development. Bioessays 25: 326–335 [DOI] [PubMed] [Google Scholar]
- Liu I. C., Chiu S. W., Lee H. Y., Leu J. Y., 2012. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell 23: 1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V. D., 2003. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 38: 807–811 [DOI] [PubMed] [Google Scholar]
- Mao Y. S., Zhang B., Spector D. L., 2011. Biogenesis and function of nuclear bodies. Trends Genet. 27: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O’Connell J. D., et al. , 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106: 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T., Parker R., 2008. Analyzing P-bodies in Saccharomyces cerevisiae. Methods Enzymol. 448: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C., Sato B. K., Broyer R. M., Wilhelm J. E., 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P., 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J., 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22: 3981–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman S. M., Serber Z., Ferrell J. E., Jr, 2011. A mechanism for the evolution of phosphorylation sites. Cell 147: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., et al. , 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Ralser M., Albrecht M., Nonhoff U., Lengauer T., Lehrach H., et al. , 2005. An integrative approach to gain insights into the cellular function of human ataxin-2. J. Mol. Biol. 346: 203–214 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Herman P. K., 2010. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 187: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K., 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M. A., Alexander R. D., Spiller M. P., Beggs J. D., 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L. S., Fink G. R., 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95: 13783–13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D., 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45: 827–835 [DOI] [PubMed] [Google Scholar]
- Santangelo G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298 [DOI] [PubMed] [Google Scholar]
- Schneper L., Duvel K., Broach J. R., 2004. Sense and sensibility: nutritional response and signal integration in yeast. Curr. Opin. Microbiol. 7: 624–630 [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R., 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M. G., Liko D., Heideman W., 2008. Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae. Eukaryot. Cell 7: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K., 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 106: 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P., 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Choe H. R., Nishida Y., Yamawaki-Kataoka Y., Ohnishi S., et al. , 1990. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc. Natl. Acad. Sci. USA 87: 8711–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T., Maeda T., 2012. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell. 47: 242–252 [DOI] [PubMed] [Google Scholar]
- Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., et al. , 2008. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim. Biophys. Acta 1784: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R., 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., Koshland D. E., Jr, 1987. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 262: 10422–10425 [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., et al. , 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40: 27–36 [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., et al. , 1987a. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M., 1987b. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50: 277–287 [DOI] [PubMed] [Google Scholar]
- Tudisca V., Recouvreux V., Moreno S., Boy-Marcotte E., Jacquet M., et al. , 2010. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur. J. Cell Biol. 89: 339–348 [DOI] [PubMed] [Google Scholar]
- Uno I., Matsumoto K., Ishikawa T., 1982. Characterization of cyclic AMP-requiring yeast mutants altered in the regulatory subunit of protein kinase. J. Biol. Chem. 257: 14110–14115 [PubMed] [Google Scholar]
- Van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., et al. , 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. C., Brangwynne C. P., 2012. Getting RNA and protein in phase. Cell 149: 1188–1191 [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M., Braun E., Johnston G. C., Singer R. A., 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57: 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S., Anderson P., 2008. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Choi E. J., Parker R., 2010. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Schneper L., Slonim N., Broach J. R., 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.