Abstract
Moving the nucleus to an intracellular location is critical to many fundamental cell and developmental processes, including cell migration, differentiation, fertilization, and establishment of cellular polarity. Bridges of SUN and KASH proteins span the nuclear envelope and mediate many nuclear positioning events, but other pathways function independently through poorly characterized mechanisms. To identify and characterize novel mechanisms of nuclear migration, we conducted a nonbiased forward genetic screen for mutations that enhanced the nuclear migration defect of unc-84, which encodes a SUN protein. In Caenorhabditis elegans larvae, failure of hypodermal P-cell nuclear migration results in uncoordinated and egg-laying–defective animals. The process of P-cell nuclear migration in unc-84 null animals is temperature sensitive; at 25° migration fails in unc-84 mutants, but at 15° the migration occurs normally. We hypothesized that an additional pathway functions in parallel to the unc-84 pathway to move P-cell nuclei at 15°. In support of our hypothesis, forward genetic screens isolated eight emu (enhancer of the nuclear migration defect of unc-84) mutations that disrupt nuclear migration only in a null unc-84 background. The yc20 mutant was determined to carry a mutation in the toca-1 gene. TOCA-1 functions to move P-cell nuclei in a cell-autonomous manner. TOCA-1 is conserved in humans, where it functions to nucleate and organize actin during endocytosis. Therefore, we have uncovered a player in a previously unknown, likely actin-dependent, pathway that functions to move nuclei in parallel to SUN-KASH bridges. The other emu mutations potentially represent other components of this novel pathway.
Keywords: nuclear migration, actin regulation, enhancer screen
NUCLEAR migration is a tightly controlled event where the nucleus moves through the cytoplasm to its specific location within a cell. Moving nuclei to specific intracellular locations is critical to many fundamental cell and developmental processes, including mitosis, meiosis, cell migration, differentiation, fertilization, and establishment of cellular polarity (Morris 2000a; Wilhelmsen et al. 2006; Starr and Fridolfsson 2010). While nuclear migration events are essential for proper development in the central nervous system and muscle (Morris et al. 1998; Tsujikawa et al. 2007; Valiente and Marin 2010; Metzger et al. 2012), abnormal nuclear migration can lead to developmental defects and human diseases, such as the smooth brain disease lissencephaly, which results in severe mental retardation and epilepsy (Morris 2000b; Wynshaw-Boris 2007).
The best-characterized mechanism for nuclear migration is how the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex of SUN proteins in the inner nuclear membrane and KASH proteins in the outer nuclear membrane connects the nucleus to the cytoskeleton (Roux et al. 2009; Starr and Fridolfsson 2010). Mutations in either of the Caenorhabditis elegans unc-83 and unc-84 genes disrupt nuclear migration in a number of cells, including embryonic hyp7 precursors, intestinal primordial cells, the distal tip cell of the somatic gonad, the one-cell embryo, and larval hypodermal P cells (Sulston and Horvitz 1981; Malone et al. 1999; Starr et al. 2001; Xiong et al. 2011). In our nuclear-envelope bridging model, the SUN protein UNC-84 is targeted to the inner nuclear membrane with its conserved SUN domain in the perinuclear space and its N terminus in the nucleoplasm, where it can interact with lamin (Malone et al. 1999; Lee et al. 2002; McGee et al. 2006; Tapley et al. 2011). UNC-84 then recruits the KASH protein UNC-83 to the outer nuclear membrane through a direct interaction between SUN and KASH domains in the perinuclear space (Starr et al. 2001; McGee et al. 2006; Mellad et al. 2011). In the cytoplasm, UNC-83 acts as the nuclear-specific cargo adaptor to recruit the microtubule motors kinesin-1 and dynein to the cytoplasmic surface of the nucleus. Kinesin-1 then provides the mechanical forces to move nuclei while dynein mediates bidirectional movements of nuclei to avoid cellular roadblocks (Meyerzon et al. 2009; Fridolfsson and Starr 2010; Fridolfsson et al. 2010). Similar SUN-KASH nuclear-envelope bridges are conserved from yeast to humans (Starr and Fridolfsson 2010; Starr 2011). For example, SUN-KASH bridges recruit microtubule motors to move nuclei in Drosophila (Patterson et al. 2004; Kracklauer et al. 2007), in Schizosaccharomyces pombe (Chikashige et al. 2006), and in mammalian cells (Roux et al. 2009).
Many nuclear migration events rely on mechanisms other than the microtubule motor-based mechanism discussed above. For example, in polarizing fibroblasts, retrograde actin flow is coupled to nuclei by SUN-KASH bridges to move nuclei (Gomes et al. 2005; Luxton et al. 2010). In other cases, nuclear migration is propelled by SUN-KASH–independent mechanisms. Examples include unknown mechanisms used to move nuclei in Arabidopsis root hairs (Chytilova et al. 2000), actomyosin contraction at the rear of nuclei in migrating neurites (Bellion et al. 2005; Schaar and McConnell 2005; Tsai et al. 2007; Schenk et al. 2009; Martini and Valdeolmillos 2010), and kinesin-driven microtubule sliding in developing myotubes (Metzger et al. 2012). Many additional SUN-KASH–independent mechanisms of nuclear migration are likely unknown. Here, we employed a forward genetic screen in C. elegans to identify novel mechanisms of nuclear migration.
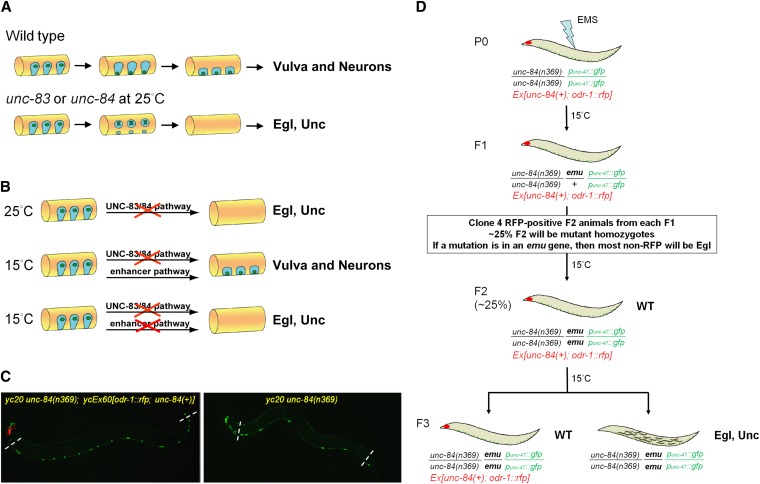
Mutations in unc-83 and unc-84 were originally found in cell-lineage mutant screens where P-cell lineages were missing (Horvitz and Sulston 1980; Sulston and Horvitz 1981). Normally, P-cell nuclei migrate from a lateral position to the ventral cord during the mid-L1 larval stage (Figure 1A) (Sulston 1976; Sulston and Horvitz 1977). After nuclear migration, P cells normally divide and their lineages develop into hypodermal cells, ventral neurons, and vulval cells (Sulston and Horvitz 1977). In unc-83 and unc-84 mutant animals, failure of P-cell nuclear migration leads to the death of P cells, which causes a loss of P-cell lineages and leads to uncoordinated (Unc) and egg-laying–deficient (Egl) phenotypes due to the loss of neurons and vulva, respectively (Sulston and Horvitz 1981; Malone et al. 1999; Starr et al. 2001). As originally described by Sulston and Horvitz, null mutations in unc-83 or unc-84 are temperature sensitive (Sulston and Horvitz 1981). At the restrictive temperature of 25°, <50% of P-cell nuclei migrate to the ventral cord, resulting in Egl and Unc animals (Figure 1B, top line). However, at the permissive temperature of 15°, ∼90% of P-cell nuclei migrate to the ventral cord and the animals have no obvious phenotype (Figure 1B, middle line) (Sulston and Horvitz 1981; Malone et al. 1999; Starr et al. 2001). These observations lead to our central hypothesis—that an uncharacterized pathway is sufficient to facilitate P-cell nuclear migration at 15° in the absence of unc-83 or unc-84 (Figure 1B, bottom line).
Figure 1 .
P-cell nuclear migration is temperature sensitive in an unc-83 or unc-84 background. (A) A conceptual diagram of the migration of nuclei (green) in P cells (blue) during the mid-L1 larval stage. A lateral view of the mid-body of an L1 animal (orange) is shown; ventral is down. In wild type, P-cell nuclei migrate from a lateral position to the ventral cord; the cytoplasm retracts following the nuclear migration. After migration, P-cell lineages normally develop into ventral neurons and vulva cells. In unc-83 or unc-84 null, mutant animals, failure of P-cell nuclear migration at 25° causes the death of many P cells and leads to egg-laying–deficient (Egl) and uncoordinated (Unc) phenotypes. (B) Our central hypothesis for an enhancer pathway mediating nuclear migration is shown. (Top line) At 25°, P-cell nuclear migration fails in unc-83 or unc-84 null animals. (Middle line) However, at 15°, unc-83 or unc-84 null animals behave normally, suggesting that a second, uncharacterized pathway (the enhancer or emu pathway) is sufficient for nuclear migration at 15°. (Bottom line) When both the unc-83/84 and emu pathways are disrupted, we hypothesized that the resulting animals would be Egl and Unc. (C) punc-47::GFP-labeled GABA neurons were used as a marker of P-cell–derived lineage. On the left, in yc20 unc-84(n369) animals with the unc-84–rescuing extrachromosomal array ycEx60[odr-1::rfp; unc-84(+)], 19 neurons were counted in the ventral cord between the pharynx and the anus (marked by dashed lines). However, failure of P-cell nuclear migration caused loss of GABA neurons as shown on the right where only 8 neurons were seen in a yc20 unc-84(n369) animal. Anterior is left. (D) An F2 clonal screen for emu in unc-84(n369) animals was performed using an extrachromosomal array ycEx60[odr-1::rfp; unc-84(+)] containing an unc-84(+) rescuing construct (see Materials and Methods for details).
To test our hypothesis and to identify and characterize novel nuclear migration pathways, we performed a forward genetic screen for enhancer of the nuclear migration defect of unc-84 (emu) mutations that disrupt P-cell nuclear migration. We isolated eight emu mutations, representing multiple loci, and used whole-genome sequencing (WGS) to determine that one mutation, yc20, is in the toca-1 gene. Transducer of Cdc42-dependent actin assembly (TOCA)-1 protein contains an F-BAR membrane-binding scaffold domain and domains that interact with the Rho family GTPase Cdc42 and the actin-nucleating WASP complex (Ho et al. 2004; Fricke et al. 2009; Giuliani et al. 2009). We conclude that TOCA-1 is the founding member of a novel SUN-KASH–independent pathway for nuclear migration. Furthermore, this study demonstrates that the C. elegans P cell is a powerful model to identify novel and partially redundant mechanisms for nuclear migration.
Materials and Methods
C. elegans strains and genetics
Worms were cultured on OP50-seeded NGM plates at specific temperatures and unless otherwise noted, strains were derived from the N2 strain (Brenner 1974). Integrated punc-47::gfp transgenic animals (oxIs12; strain EG1285) (McIntire et al. 1997) were used as wild-type controls in this study. oxIs12[punc-47::gfp] was crossed into unc-84(n369); ycEx60[odr-1::rfp; unc-84(+)] to create the strain UD87. The odr-1::rfp plasmid was a gift from Noelle L’Etoile [University of California, San Francisco (UCSF)] (L’Etoile and Bargmann 2000). UD87 animals were used to perform the emu forward genetic screen and for backcrosses. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institutes of Health National Center for Research Resources. See Table 1 for genotypes of strains in this study.
Table 1 . Strains used in this study.
| Strain | Genotype |
|---|---|
| CB4856 | A wild-type strain from a divergent population in Hawaii (Koch et al. 2000) |
| EG1285 | lin-15B(n765) oxIs12[punc-47::gfp; lin-15(+)] X (McIntire et al. 1997) |
| MH1654 | unc-83(e1408) V; oxIs12 X (a gift from Min Han, University of Colorado, Boulder) |
| PD4788 | mIs13[pmyo-2::gfp; pes-10::gfp; gut::gfp] I (a gift from Kelly Liu, Cornell University) |
| UD4 | yc3; unc-83(e1408); oxIs12 X |
| UD59 | unc-84(n369); ycIs11[odr-1::gfp; phlh-3::vab-10-ABD::venus; phlh-3::nls::tdTOMATO] |
| UD87 | unc-84(n369) oxIs12 X; ycEx60[odr-1::rfp; unc-84(+)] |
| UD230 | yc22; unc-84(n369) oxIs12 X |
| UD277 | yc3; unc-84(n369) oxIs12 X (10× backcross with UD87) |
| UD278 | yc16; unc-84(n369) oxIs12 X; ycEx60 (10× backcross with UD87) |
| UD279 | yc21; unc-84(n369) oxIs12 X; ycEx60 (10× backcross with UD87) |
| UD283 | yc18; unc-84(n369) oxIs12 X; ycEx60 (10× backcross with UD87) |
| UD284 | yc15; unc-84(n369) oxIs12 X; ycEx60 (10× backcross with UD87) |
| UD346 | toca-1(yc20) unc-84(n369) oxIs12 X; ycEx60 (4× backcross with UD87) |
| UD347 | yc17; unc-84(n369) oxIs12 X; ycEx60 (2× backcross with UD87) |
| UD350 | unc-84(n369) in a mostly CB4856 background (10× backcross with CB4856) |
| UD352 | toca-1(tm2056) unc-84(n369) oxIs12 X; ycEx60 |
| UD354 | toca-2(tm2088) III; unc-84(n369) oxIs12 X; ycEx60 |
| UD355 | toca-1(tm2056) unc-84(n369) oxIs12 X; ycEx60; ycEx189[odr-1::gfp; phlh-3::toca-1::gfp] |
| UD356 | toca-1(tm2056) unc-84(n369) oxIs12 X; ycEx60; ycEx190[odr-1::gfp; pcol-10::toca-1::gfp] |
| UD357 | toca-1(yc20) unc-84(n369) oxIs12 X; ycEx60; ycEx189 |
| UD358 | toca-1(yc20) unc-84(n369) oxIs12 X; ycEx60; ycEx190 |
| UD373 | toca-1(tm2056) unc-84(n369); ycIs11; ycEx60 |
| UD381 | ycIs11[odr-1::gfp; phlh-3::vab-10-ABD::venus; phlh-3::nls::tdTomato] |
For the emu forward genetic screen, standard EMS mutagenesis (Brenner 1974) was performed with UD87 (unc-84(n369) oxIs12[punc-47::gfp] X) animals at 15°. Since ycEx60[odr-1::rfp; unc-84(+)] rescues unc-84, F2 progeny were blindly picked and screened in the F3 generation for Egl, non-RFP animals at 15°. After a candidate mutant was isolated, punc-47::GFP-labeled GABA neurons were counted as a secondary screen.
To determine whether an emu mutation was on the X chromosome, emu; unc-84 (n369) oxIs12[punc-47::gfp] X hermaphrodites were crossed to wild-type males with a GFP-marked pharynx (pmyo-2::gfp or ajm-1::gfp). Male progeny were hemizygous for unc-84(n369) and emu if the emu mutation was on chromosome X. punc-47::GFP-labeled GABA neurons were counted in hemizygous males. If the number of GABA neurons was significantly decreased, we concluded that the emu mutation was on chromosome X.
To assign emu mutations to chromosomes, two-point single-nucleotide polymorphism (SNP) mapping was performed (Fay and Bender 2008). First, UD87 was backcrossed with Hawaiian CB4856 males 10 times to create UD350 that carries the unc-84(n369) null mutation, but otherwise is mostly CB4856, Hawaiian, for nonlinked SNPs. For two-point SNP mapping, unc-84(n369); emu animals were mated with unc-84(n369)-Hawaiian males and Egl F2 progeny were singled onto individual plates. PCR and restriction-enzyme digests for known SNPs were performed on F3 Egl animals and individual emu mutations were assigned to chromosomes.
Assays for quantifying P-cell nuclear migration defects
The oxIs12[punc-47::gfp] marker was used as an assay to label P-cell–derived GABA neurons (McIntire et al. 1997) and to approximate P-cell nuclear migration defects. L4 or young adult worms were mounted on a 2% noble agar pad in 0.5 mM levamisole M9 solution. The number of punc-47::GFP-labeled GABA neurons in the ventral cord was counted using a 10× objective on an epifluorescence compound microscope (DM6000; Leica). The most posterior neuron (DVB) and the seven neurons in the head (out of the plane of the ventral cord) were not counted, as none of them are derived from P-cell lineages (McIntire et al. 1997; Sulston and Horvitz 1977). Thus we counted 19 GABA neurons in wild type, 12 of which were derived from P cells (VD2–VD13) (McIntire et al. 1997; Sulston and Horvitz 1977). A complete loss of P-cell lineages resulted in only 7 GABA neurons in the ventral cord. Images were taken using a CCD camera (DC350 FX; Leica) and analyzed with LAS AF (Leica) software. Statistical analysis was performed by using the two-tailed distribution Student’s t-test function in Excel (Microsoft, Redmond, WA).
To directly follow P-cell nuclei during migration, the plasmid pSL619 (phlh-3::nls::tdTomato) was created using the 3.4-kb hlh-3 promoter (encoding the first 8 aa of hlh-3) (Doonan et al. 2008) and nls::tdTomato sequence. nls::tdTomato was PCR cloned from a mammalian pCAB vector (Spear and Erickson 2012) and the 5′ primer contained an SV40 nuclear localization signal. pSL619 (phlh-3::nls::tdTomato) (2 ng/μl) was coinjected with pSL630 (phlh-3:: vab-10-ABD::venus) (5 ng/μl) and odr-1::gfp (100 ng/μl) into N2 animals to create the extrachromosomal array ycEx202[odr-1::gfp; phlh-3::vab-10-ABD::venus; phlh-3::nls::tdTomato]. The odr-1::gfp plasmid was a gift from Noelle L’Etoile (UCSF) (L’Etoile and Bargmann 2000). ycEx202 was integrated by a UV/TMP method (Gengyo-Ando and Mitani 2000) to create ycIs11, which labeled P-cell nuclei throughout L1. The integrated strain was outcrossed four times to make the strain UD381 (ycIs11 in N2). Subsequently ycIs11 was crossed into unc-84(n369) and toca-1(tm2056) unc-84(n369), to create the strains UD59 and UD373, respectively.
To perform P-cell nuclei counting assays, transgenic animals carrying ycIs11 were synchronized at the beginning of the L1 stage by starvation in S-basal media for 36, 30, or 24 hr at 15°, 20°, or 25°, respectively (Stiernagle 2006). The early L1 larval arrest was released by the addition of food (Escherichia coli OP50) for 16, 13, or 10 hr at 15°, 20°, or 25°, respectively. Late L1 larvae, after the completion of nuclear migration, were collected by centrifugation, anesthetized with 0.5 mM levamisole, and mounted on a 2% agar pad. The positions of tdTOMATO-labeled nuclei were examined using a 63× Plan Apo 1.40 NA objective on an epifluorescence compound microscope (DM6000; Leica). Nuclei that failed to migrate could be seen at a lateral position on either side of the animals.
Whole-genome sequencing and analysis
emu; unc-84(n369) oxIs12[punc-47::gfp] X; ycEx60[unc-84(+); odr-1::rfp] homozygous animals were grown at 15° and washed with M9 from NGM plates. A worm pellet of ∼200 μl was mixed with lysis buffer [100 mM NaCl, 100 mM Tris (pH 8.5), 50 mM EDTA, 1% SDS, 1% BME, 100 μg/ml Proteinase K] and incubated at 60° for 60 min. After phenol/chloroform extraction and RNase A (20 μg/ml) treatment at 30° for 30 min, genomic DNA was purified by ethanol precipitation. For WGS, DNA libraries were made using the next-generation DNA library kit (New England Biolabs, Beverly, MA). The DNA library from each mutant strain was subjected to WGS on an Illumina GAII sequencing platform at the University of California (UC) Davis Genome Center Core Facility, using paired-end 85-nt reads. Raw sequencing files were analyzed using MAQGene software (Bigelow et al. 2009). The MAQGene output data were imported into Excel (Microsoft) to filter out background mutations and identify candidates for the emu mutation.
RNA interference tests
dsRNA was made from PCR templates amplified from Ahringer library clones (Fraser et al. 2000; Kamath et al. 2003) or genomic DNA, using T7 RNApol (Promega, Madison, WI) as described (Fire et al. 1998). For toca-1(RNAi), Ahringer library clone X-1B09 was used. dsRNA was microinjected into the gonads of L4 or young adult UD87 hermaphrodites as previously described (Fire et al. 1998). F1 progeny grown at 15° and produced between 24 and 48 hr postinjection were collected and their GABA neurons were counted as above.
TOCA-1::GFP rescue and localization
toca-1 was expressed specifically in P cells with the promoter of hlh-3 to assay the ability for wild-type toca-1 to rescue toca-1(yc20). toca-1 genomic DNA containing all exons and introns except for the first intron was amplified by PCR from fosmid WRM0623aA03 from the Moerman fosmid library (Source BioScience LifeSciences), using primers ods1546 (GGTACCCCCATGAACGACAGTTGCAGTTGGGACCAGCTATGGGACCAACAAGGTACTCTCA) and ods1547 (GGTACCGACTTTTCCGAACCAGGTGA). The toca-1 PCR product was cloned into the KpnI site of pPD95.79 (Addgene) to make pSL614, a promoterless construct to express GFP fused to the C terminus of TOCA-1. Next, 3.4 kb of the promoter for hlh-3, which is expressed specifically in embryonic and larval P cells, was obtained from pRD1 (Doonan et al. 2008) and cloned into the SmaI site of pSL614 to make phlh-3::toca-1::gfp (pSL626). Alternatively, 1.2 kb of the promoter for col-10, which is expressed in embryonic and larval hypodermal cells, was obtained from pOS12 (Spencer et al. 2001) and cloned into the SphI and SmaI sites of pSL614 to make pcol-10::toca-1::gfp (pSL627). phlh-3::toca-1::gfp (2 ng/μl) or pcol-10::toca-1::gfp (5 ng/μl) was microinjected with odr-1::gfp (100 ng/μl) as a marker into the gonads of young adult hermaphrodites (Mello and Fire 1995). Multiple, independent stable transgenic lines were isolated and GABA neurons were counted as above. TOCA-1::GFP images were acquired using a 63× objective on a Leica DM6000 microscope as above.
RT-PCR and Western analysis
Total RNA from N2, UD87, toca-1(tm2056) unc-84(n369), and toca-1(yc20) unc-84(n369) animals was extracted using the RNeasy kit (Qiagen, Valencia, CA) and cDNA was made using an oligo(dT)12–18 primer (Invitrogen, Carlsbad, CA). RT-PCR was performed as previously described (Fridolfsson et al. 2010). For Western analysis, total worm lysate was made, separated by SDS–PAGE, and transferred to a membrane as described (Tapley et al. 2011). Anti–TOCA-1 rabbit serum (1:1000) (Giuliani et al. 2009) and monoclonal anti–α-tubulin mouse antibody (1:1000) (Clone DM 1A; Sigma, St. Louis) were used as primary antibodies. IRDye 680LT donkey anti-rabbit IgG (1:5000) and IRDye 800CW donkey anti-mouse IgG (1:5000) (LI-COR Biosciences) were used as secondary antibodies. Images of Western blotting were scanned by the Odyssey Infrared Imaging System (LI-COR Biosciences).
Results
Forward genetic screens for enhancers of the nuclear migration defect of unc-83 or unc-84
Mutations in unc-83 or unc-84 disrupt P-cell nuclear migration in a temperature-sensitive manner and are not additive (Sulston and Horvitz 1981). In unc-83 or unc-84 mutant animals raised at 25°, P-cell nuclear migration fails, leading to the death of P cells and a loss of neurons and the vulva, which leads to Unc, Egl animals (Figure 1, A and B) (Sulston and Horvitz 1981). However, if unc-83 or unc-84 null mutant animals are raised at 15°, ∼90% of P-cell nuclei migrate normally and adults have no obvious phenotypes (Figure 1B) (Sulston and Horvitz 1981; Malone et al. 1999; Starr et al. 2001). Since most alleles of unc-83 and unc-84 are molecular nulls (Malone et al. 1999; Starr et al. 2001), the process of P-cell nuclear migration itself is temperature sensitive in an unc-83 or unc-84 mutant background. We therefore hypothesized that there is an unknown pathway(s) that functions in parallel to the unc-83/84 pathway to move P-cell nuclei (Figure 1B, middle line).
To test our hypothesis, we carried out genetic screens for enhancer of the nuclear migration defect of unc-84 (emu) mutations. We reasoned that simultaneous mutations in both the unc-84 and the parallel pathway would synthetically lead to a strong P-cell nuclear migration defect at 15°, resulting in Egl, Unc animals missing P-cell lineages (Figure 1B, bottom line). We screened for emu mutations, using a powerful synthetic approach (Fay et al. 2002). In this approach, we mutagenized an unc-84(n369) starting strain containing an extrachromosomal array carrying an unc-84 rescuing construct (McGee et al. 2006) and marked with odr-1::rfp (L’Etoile and Bargmann 2000). After mutagenesis, F2 animals potentially homozygous for unc-84 and an emu mutation, but also carrying the unc-84 rescuing transgene and therefore phenotypically unc-84(+), were individually plated. In the F3 generation, non-RFP animals (mutant for unc-84) were compared to RFP-positive siblings carrying the extrachromosomal unc-84 rescuing array. A candidate emu mutation that acted synthetically with unc-84 was isolated when the non-RFP animals were Egl, but the RFP-positive siblings were not Egl (Figure 1D). About 4500 mutagenized genomes were screened in this manner and seven emu mutations (yc15–18 and -20–22) were isolated. An eighth mutation, yc3, was isolated in an independent screen in unc-83(e1408) and crossed into unc-84(n369). This rate of about one emu mutant animal for every ∼650 genomes screened was similar to rates observed in other enhancer and synthetic screens in C. elegans (Ferguson and Horvitz 1989; Rocheleau et al. 2002).
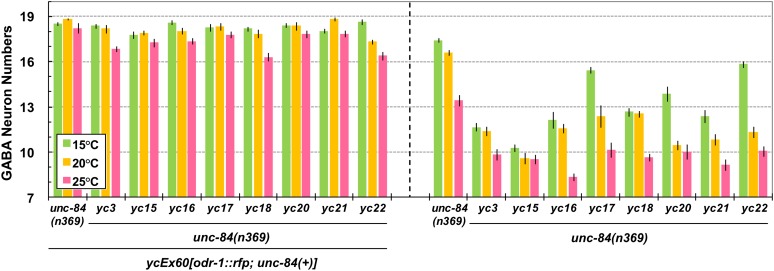
Since Egl can result from defects other than a failure of P-cell nuclear migration, a secondary screen was performed to directly count cells derived from P-cell lineages. punc-47::GFP is expressed in 19 GABA neurons in the ventral cord, 12 of which are derived from P cells (Figure 1C) (Sulston and Horvitz 1977; McIntire et al. 1997). Mutants with significantly fewer GABA neurons at 15° (Figure 1C, right) than the unc-84(n369) starting strain were saved as emu mutant strains. The strongest mutants had only 10–14 GABA neurons per animal at 15°, similar to unc-84 mutants at 25°, and significantly fewer than the 18.5 neurons in the starting background at 15° (Figure 2 and Table 2).
Figure 2 .
emu mutations enhance the nuclear migration defect of unc-84. GABA neuron numbers were used to quantify P-cell nuclear migration defects. All strains are in the unc-84(n369) background. The animals counted for the data on the left half contained an unc-84(+) rescuing array, so they were effectively wild type for unc-84 and mutant for the emu indicated (yc3, -15–18, and -20–22). The data on the right are from emu; unc-84(n369) double mutants. All emu; unc-84 double mutants had significantly fewer GABA neurons compared to unc-84 alone at 15°, 20°, and 25° (t-test; P < 0.0001 for all comparisons to unc-84 mutants with no emu mutation). Sample sizes are all >20 L4 or young adult animals, and error bars show the standard error of the mean (SEM).
Table 2 . Number of GABA neurons (±SEM) of emu mutations.
| 15° | 20° | 25° | |
|---|---|---|---|
| unc-84 screen | |||
| Wild type | 18.5 ± 0.1 | 18.8 ± 0.1 | 18.3 ± 0.4 |
| yc3 | 18.4 ± 0.2 | 18.2 ± 0.3 | 16.9 ± 0.2 |
| yc15 | 17.8 ± 0.2 | 18.0 ± 0.1 | 17.3 ± 0.3 |
| yc16 | 18.7 ± 0.1 | 18.1 ± 0.2 | 17.4 ± 0.2 |
| yc17 | 18.3 ± 0.2 | 18.3 ± 0.3 | 17.8 ± 0.2 |
| yc18 | 18.2 ± 0.2 | 17.9 ± 0.3 | 16.3 ± 0.3 |
| yc20 | 18.4 ± 0.1 | 18.4 ± 0.3 | 17.9 ± 0.2 |
| yc21 | 18.1 ± 0.1 | 18.9 ± 0.1 | 17.9 ± 0.2 |
| yc22 | 18.6 ± 0.2 | 17.3 ± 0.2 | 16.4 ± 0.3 |
| unc-84(n369) | 17.4 ± 0.2 | 16.6 ± 0.2 | 13.4 ± 0.4 |
| yc3, unc-84 | 11.7 ± 0.3 | 11.4 ± 0.3 | 9.8 ± 0.4 |
| yc15, unc-84 | 10.3 ± 0.2 | 9.6 ± 0.4 | 9.5 ± 0.3 |
| yc16, unc-84 | 12.1 ± 0.6 | 11.6 ± 0.3 | 8.3 ± 0.3 |
| yc17, unc-84 | 15.4 ± 0.2 | 12.4 ± 0.8 | 10.2 ± 0.5 |
| yc18, unc-84 | 12.7 ± 0.3 | 12.5 ± 0.2 | 9.6 ± 0.2 |
| yc20, unc-84 | 13.9 ± 0.5 | 10.5 ± 0.3 | 10.0 ± 0.5 |
| yc21, unc-84 | 12.4 ± 0.4 | 10.8 ± 0.5 | 9.1 ± 0.4 |
| yc22, unc-84 | 15.8 ± 0.2 | 11.3 ± 0.4 | 10.1 ± 0.4 |
| unc-83 screen | |||
| unc-83(e1408) | 18.2 ± 0.2 | 17.1 ± 0.3 | 13.9 ± 0.3 |
| yc3, unc-83 | 11.8 ± 0.4 | 9.7 ± 0.3 | 8.6 ± 0.2 |
Genetic characterization of emu mutations
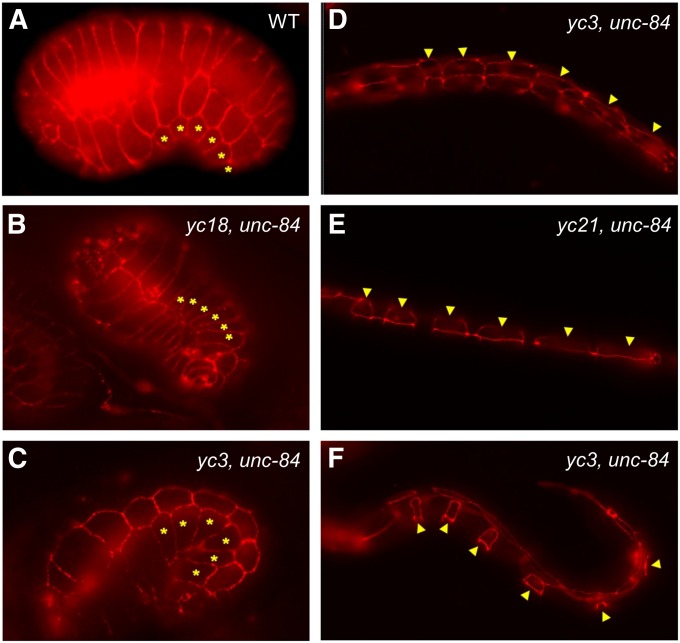
None of the eight emu mutant strains had a nuclear migration defect in unc-84 rescued animals at 15° (Figure 2, left). These data suggest that the emu mutants have mutations in genes that are synthetic to the unc-84 pathway at 15° and likely represent genes that previous screens for nuclear migration mutants would have missed. The yc3 mutation also enhanced the nuclear migration defect of unc-83(e1408). yc3; unc-83(e1408) double-mutant animals had only 11.8 ± 0.4, 9.7 ± 0.3, and 8.6 ± 0.2 GABA neurons (±SEM) in the ventral cord at 15°, 20°, or 25°, respectively, compared to 18.2 ± 0.2, 17.1 ± 0.3, and 13.9 ± 0.3 GABA neurons in unc-83(e1408) single-mutant animals (Table 2). Furthermore, toca-1(RNAi) (yc20 is a mutation in toca-1, see below) also enhanced the nuclear migration defect of unc-83 at 15° (17.7 ± 0.1 GABA neurons; n = 94; t-test; P = 0.012), suggesting that the emu mutations function in parallel to the SUN-KASH bridge pathway, rather than in an UNC-83–independent function of UNC-84. To test whether any emu mutations had caused defects in P-cell lineages prior to nuclear migration, we counted P cells in comma-stage embryos or young L1 larvae. No P-cell lineage defects were observed prior to the middle of the L1 larval stage when nuclear migration would normally occur (Figure 3). In all emu single or emu; unc-84 double mutants, 6 P cells (if only one side could be observed) or 12 P cells (both sides) were present (Figure 3). We therefore concluded that the emu phenotypes occur after the proper numbers of P cells were born and that they function normally through embryogenesis.
Figure 3 .
emu mutations do not disrupt P-cell specification. The morphology of P cells is shown in (A) a wild-type embryo, (B) a yc18; unc-84(n369) embryo, (C) a yc3; unc-84(n369) embryo, (D) a yc3; unc-84(n369) early-L1 larva, (E) a yc21; unc-84(n369) mid-L1 larva, and (F) a yc3; unc-84(n369) mid-L1 larva prior to the onset of nuclear migration. P-cell boundaries were marked using MH27 immunostaining to mark adherens junctions. Asterisks or arrowheads mark six P cells on one ventral–lateral side of the embryo or L1 larvae. Sample size of each mutant strain is between 18 and 50. A variety of stages were observed for each of the eight emu mutants; all worms had normal P-cell shapes.
Next, we aimed to determine whether the emu mutations were dominant or recessive and to clean up their mutagenized backgrounds. All eight emu mutant lines acted as recessive mutations since heterozygous emu/+ animals in an unc-84 homozygous background were neither Egl nor Unc at 15°. Also, no decrease in the number of GABA neurons was observed in heterozygous animals compared to unc-84(n369) animals alone raised at 15° (data not shown). To remove unlinked mutations that were a result of mutagenesis, the eight emu mutant lines in the unc-84 rescuing array background were outcrossed to unc-84(n369) animals.
To compare the severity of the P-cell nuclear migration defects of emu mutant lines, the number of GABA neurons in the ventral cord was quantified at 15°, 20°, and 25° (Figure 2). At 15°, the permissive temperature for unc-84(n369), emu; unc-84(n369) double mutants had significantly fewer GABA neurons than did unc-84 mutants alone, which had 17.4 ± 0.2 (±SEM) on average. yc15 was the strongest enhancer of unc-84 at 15°; occasionally (5 of 62) only 7 GABA neurons were found in yc15; unc-84 double mutants (10.3 ± 0.2 on average). Since 7 of the 19 GABA neurons originate from lineages other than P cells, 7 GABA neurons in the ventral cord represent a complete loss of P-cell lineages and likely a complete failure of P-cell nuclear migration. emu; unc-84 double mutants were also cultured at 20° and 25° to see how emu mutations enhance unc-84 P-cell nuclear migration defects at restrictive temperatures for unc-84(n369). At both 20° and 25°, emu; unc-84 double mutants had significantly fewer GABA neurons compared to unc-84(n369) single mutants (Figure 2, right). At 20°, there was a range of averages for the double mutants from 9.6 to 12.5 [compared to unc-84(n369) single mutants, which had 16.6 ± 0.2 on average], but emu; unc-84(+) mutants had near wild-type numbers of GABA neurons. In many cases (yc3, yc17, yc20, and yc22) the phenotypes of the emu; unc-84 doubles were significantly more severe at 20° than at 15° (t-test; P < 0.01) (Figure 2, right). At 25°, all eight emu; unc-84 double mutants had significantly fewer GABA neurons than unc-84 single mutants, which had 13.41 ± 0.37 on average. However, at 25°, some emu single mutants [in the presence of the unc-84(+) rescuing array] had a mild phenotype of their own when compared to wild type (Figure 2, left). In summary, unc-84 P-cell nuclear migration defects were significantly enhanced by all eight emu mutations at 15°, 20°, and 25°.
Identification of the molecular lesion in the emu mutation yc20
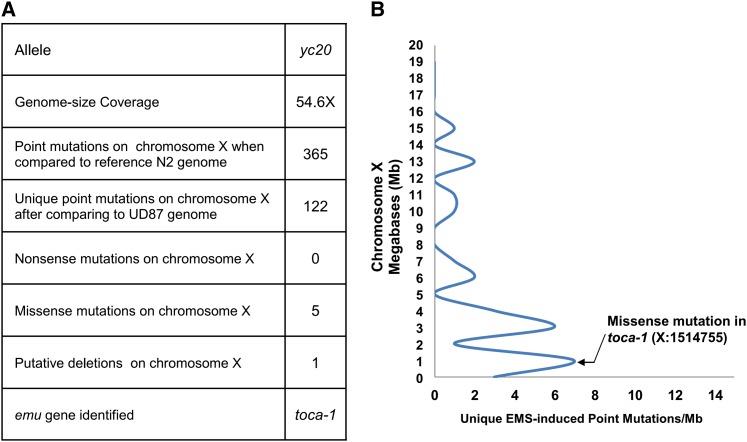
To identify molecular lesions underlying emu mutations, we undertook a WGS approach. The C. elegans genome is ∼100 Mb, making next-generation WGS well suited to rapidly and cost-effectively identify molecular lesions (Sarin et al. 2008; Doitsidou et al. 2010; Zuryn et al. 2010). To narrow our focus for the WGS data analysis, we mapped yc15, -16, -20, and -21 to chromosome X and yc3 and -18 to chromosome I (see Materials and Methods). We sequenced the yc20 genome to 54.6× coverage (Figure 4A). In addition, we have sequenced other genomes derived from mutagenesis of the UD87 starting strain, which allowed us to filter out changes in the yc20 genome that were also found in other emu strains that originated from UD87 when compared to the reference N2 genome. Using the MAQGene software (Bigelow et al. 2009) to analyze the sequence data and filter out background mutations, we identified 122 unique mutations on chromosome X in the yc20 genome. This list included only 5 predicted missense mutations and no predicted nonsense mutations (Figure 4A). We then analyzed the WGS data to further map yc20 on chromosome X. The backcrossed yc20 strain was expected to have a cluster of G/C to A/T transitions from the EMS mutagenesis linked to the emu locus (Zuryn et al. 2010). There was a cluster of such transitions toward the left end of chromosome X in the yc20 genome (Figure 4B). This clustering approach is based on a small number of mutations and is therefore subject to false positives. Nonetheless, it is supportive of the hypothesis that yc20 maps to the left end of chromosome X. One predicted missense mutation in the gene toca-1 (transducer of Cdc42-dependent actin assembly) was found in the region of chromosome X implicated by the mapping data for yc20. Thus, toca-1 was an excellent candidate for the yc20 mutation.
Figure 4 .
Whole-genome sequencing (WGS) and mapping of yc20. (A) Summary of yc20 WGS. (B) The yc20 mutation was mapped to the toca-1 region. Each peak represents a cluster of G-to-A or C-to-T mutations identified by WGS characteristic of EMS mutagenesis. Since yc20 was backcrossed after mutagenesis, it is expected that the remaining EMS-induced mutations would be linked to yc20 on chromosome X.
TOCA-1 is conserved across animals and contains an F-BAR domain that binds to membranes, an HR domain that recruits the Rho-family GTPase Cdc42, and an SH3 domain that binds the WASP/WAVE complex (Ho et al. 2004; Giuliani et al. 2009). Both Cdc42 and WASP/WAVE regulate actin filament polymerization. The point mutation in toca-1(yc20) is a C-to-T mutation that is predicted to cause an Ala-to-Val amino acid change in the putative F-BAR domain at position 100 of the TOCA-1 protein. Thus, the yc20 mutation may disrupt TOCA-1 function and actin dynamics at membranes.
Disruption of toca-1 enhances the unc-84 nuclear migration defect
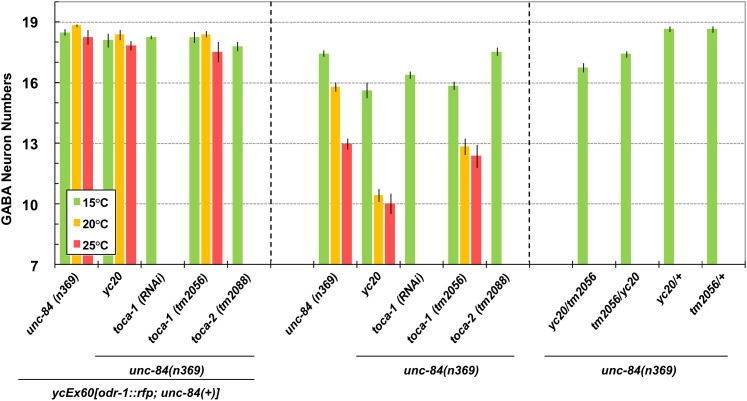
To further confirm that TOCA-1 functions to move P-cell nuclei, additional alleles of toca-1 were examined in an unc-84 mutant background. Like yc20, both toca-1(RNAi) and the deletion allele toca-1(tm2056) had significant P-cell nuclear migration defects in an unc-84(n369) background at 15° (Figure 5, center), but no phenotype in the presence of the unc-84(+) transgene (Figure 5, left). These yc20 data are slightly different from those in Figure 2 because they were obtained at different times. These data show that toca-1 is an emu gene. toca-1 is partially redundant to toca-2 (Giuliani et al. 2009) and no phenotype has previously been reported for a single mutation in either gene, although the double mutant blocks endocytosis of yolk granules in oocytes, resulting in sterility (Giuliani et al. 2009). toca-2(tm2088); unc-84(n369) animals did not have a significant nuclear migration defect (Figure 5, center), suggesting that the role of toca-1 in P-cell nuclear migration is not redundant with that of toca-2.
Figure 5 .
toca-1 RNAi or a deletion allele enhance unc-84. (Left) yc20, toca-1(RNAi), toca-1(tm2056), and toca-2(tm2088) have similar average GABA neuron numbers in comparison with unc-84(n369) ycEx60[odr-1::rfp; unc-84(+)] animals at all temperatures. (Center) At 15°, yc20, toca-1(RNAi), and toca-1(tm2056) showed significantly lower GABA neuron numbers (15.6 ± 0.4, 16.4 ± 0.2, and 15.9 ± 0.2, respectively) than unc-84 (17.5 ± 0.2) (t-test; P < 0.0001). However, at 15° toca-2(tm2088) and unc-84 showed similar GABA neuron numbers (17.5 ± 0.2 vs. 17.5 ± 0.2). At 20°, yc20 and toca-1(tm2056) showed significantly fewer GABA neuron numbers (10.5 ± 0.3 and 12.9 ± 0.4, respectively) than unc-84 (15.8 ± 0.2) (t-test; P < 0.0001). At 25°, yc20 showed significantly lower average GABA neuron numbers (10.0 ± 0.5) than unc-84 (13.0 ± 0.3) (t-test; P < 0.0001) but toca-1(tm2056) showed similar numbers to unc-84 (12.4 ± 0.6 vs. 13.0 ± 0.3). (Right) Both yc20/tm2056 and tm2056/yc20 (male X chromosome/hermaphrodite X chromosome) trans-heterozygous animals showed significantly lower average GABA neuron numbers (16.8 ± 0.2 and 17.4 ± 0.2, respectively) than yc20/+ and tm2056/+ (18.7 ± 0.1 and 18.7 ± 0.2, respectively) (t-test; P < 0.0001). Sample sizes are all >20 L4 or young adult animals, and error bars show the standard error of the mean (SEM).
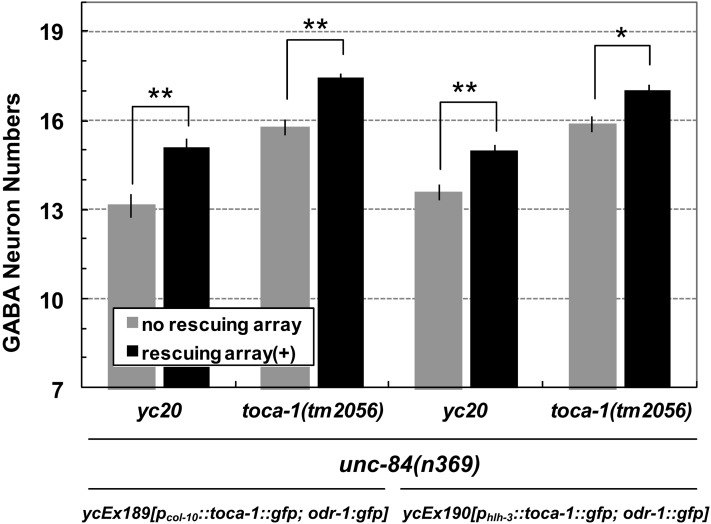
The above data are consistent with the hypothesis that yc20 is an allele of toca-1. Two experiments were performed to further test this hypothesis. First, we performed a complementation test. yc20/tm2056 heterozygous animals in an unc-84(n369) background raised at 15° had only 16.8 ± 0.2 GABA neurons on average (Figure 5, right). This was significantly fewer than that in either yc20/+ unc-84(n369) or tm2056/+ unc-84(n369) animals, which were similar to those in the unc-84(n369) background alone (Figure 5, right). Second, we tested whether wild-type toca-1 could rescue the nuclear migration defect of the yc20 mutation. Wild-type toca-1 was expressed in P cells prior to nuclear migration from two independent promoters: the pan-hypodermal promoter of col-10 (Spencer et al. 2001) and the P-cell–specific promoter of hlh-3 (Doonan et al. 2008). The yc20unc-84(n369) nuclear migration defect was rescued in transgenic animals expressing either pcol-10::toca-1::gfp or phlh-3::toca-1::gfp. The average number of GABA neurons significantly increased from 13.2 ± 0.4 to 15.1 ± 0.3 in the pcol-10::toca-1::gfp rescued lines and from 13.59 ± 0.28 to 15.0 ± 0.2 in the phlh-3::toca-1::gfp rescued animals (Figure 6). Moreover, the P-cell–specific rescue of the phlh-3::toca-1::gfp transgene suggested that TOCA-1 functions to move P-cell nuclei in a cell-autonomous manner. Based on these data, we concluded that yc20 mutation is in toca-1.
Figure 6 .
yc20 is a mutation in toca-1. The number of GABA neurons counted in each indicated genotype is shown. Solid bars indicate the presence of ycEx189[pcol-10::toca-1::gfp; odr-1::gfp] that is broadly expressed in all hypodermal cells (left) or ycEx190[phlh-3::toca-1::gfp; odr-1::gfp] that is specifically expressed in P cells (right). **P < 0.001, t-test; *P < 0.003, t-test. Sample sizes are all >30 L4 or young adult animals, and error bars show the standard error of the mean (SEM).
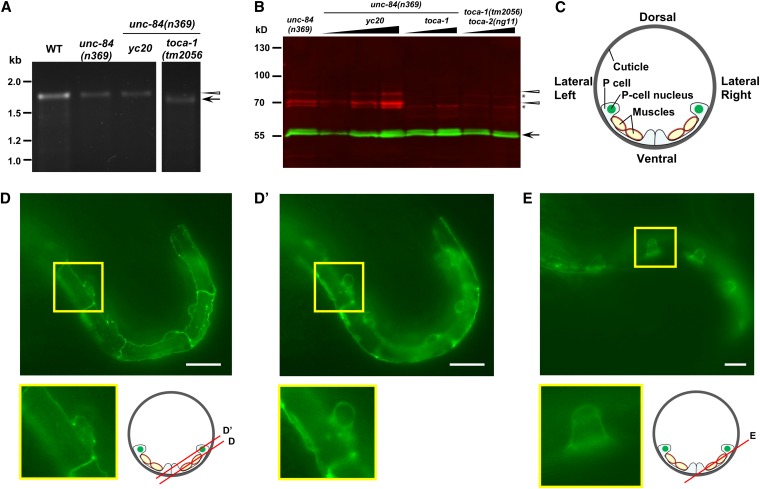
To further explore the nature of the toca-1(yc20) allele, we examined toca-1 mRNA and TOCA-1 protein levels in toca-1(yc20) animals. Using RT-PCR, full-length mRNA with a missense mutation from Ala to Val at residue 100 was detected in toca-1(yc20) animals (Figure 7A). Moreover, a slightly smaller mRNA was detected by RT-PCR in the deletion allele toca-1(tm2056) (Figure 7A). TOCA-1 protein was visualized using a polyclonal antibody against TOCA-1 residues 362–592 (Giuliani et al. 2009). In both wild-type and toca-1(yc20) animals grown at either 15° or 25°, TOCA-1 was detected as a doublet of ∼70 kDa on Western blots. However, no TOCA-1 bands were detected in toca-1(tm2056) animals (Figure 7B). Based on these data, we concluded that toca-1(yc20) unc-84(n369) animals made full-length toca-1 mRNA and protein with an Ala-to-Val mutation at residue 100 that is responsible for the defect in nuclear migration.
Figure 7 .
toca-1 expression and localization. (A) RT-PCR from mRNA amplified a 1.7-kb band of the expected size (arrowhead) in N2, unc-84(n369) and toca-1(yc20) unc-84(n369) animals. A 1.6-kb band of the expected size (arrow) was amplified from toca-1(tm2056) unc-84(n369) RNA. (B) On Western blots, anti–TOCA-1 antibodies recognized a doublet (red, marked with arrowheads) of the expected size of ∼70 kDa in both unc-84(n369) and toca-1(yc20) unc-84(n369) animals. As expected, both bands of the TOCA-1 doublet were absent from toca-1(tm2056) unc-84(n369) and toca-1(tm2056) toca-2(ng11) animals. Nonspecific bands were marked with asterisks. α-Tubulin (55-kDa band in green, marked with an arrow) was used as loading control. (C) Schematized section of an early L1 animal. Underneath a sheath of cuticle, P cells are the epidermis that covers the lateral-to-ventral side of the animal. P cells are very thin where they are over muscles. P-cell nuclei localize to the lateral side before the nuclear migration occurs. (D and E) TOCA-1::GFP expression in P cells of early (D and D′) and mid-L1 (E) animals. Two planes of focus as diagramed in the cartoon, one at the lateral surface (D) and one farther inside (D′), are shown for the same animal. Insets are enlargements of the indicated boxes. Bars: 10 μm.
To examine the subcellular localization of TOCA-1 in larval P cells, TOCA-1::GFP was expressed specifically in P cells of L1 wild-type animals, using the promoter phlh-3. In early L1 larval stage, we found that TOCA-1::GFP localization was clearly decorating the cell–cell contacts with few noticeable punctate spots (Figure 7D) and was also concentrated at the front end (close to the ventral cord) and the rear end (lateral side) of cytoplasm (Figure 7D′). We also found TOCA-1::GFP was surrounding the P-cell nuclei (seen as dark holes) at the rear end of P cells (Figure 7D′). Later in the mid-L1 larval stage just prior to P-cell nuclear migration, TOCA-1::GFP similarly localized to the cell–cell contact boundaries and to the region of both the front and the rear ends (Figure 7E). The localization of TOCA-1::GFP suggests its function could occur at the sites of cell–cell contacts, at the ventral-cord front end, or at the lateral rear end of P cells in the vicinity of the nucleus.
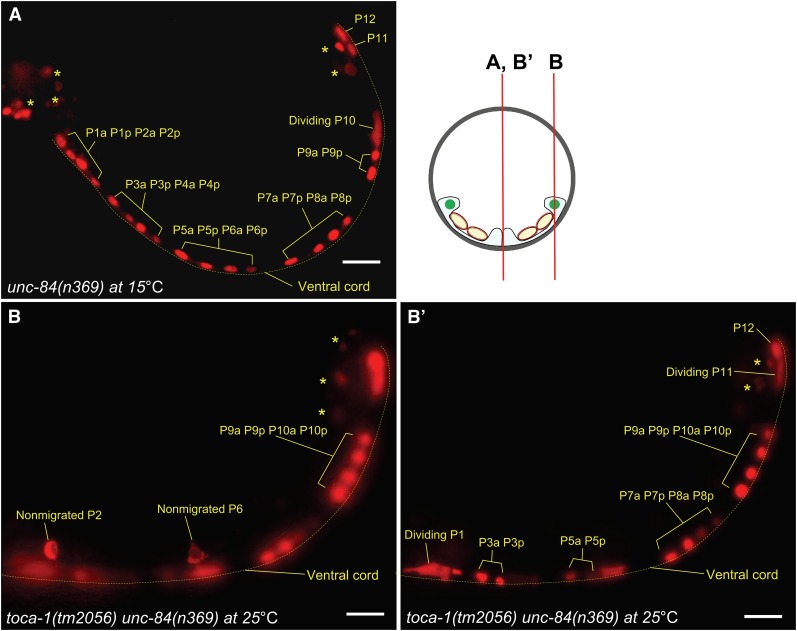
Finally, to more directly analyze the defects of P-cell nuclear migration in toca-1unc-84 double mutants, we developed a P-cell nuclear counting assay, using an integrated array, ycIs11, expressing tdTOMATO in nuclei of P cells and their descendants throughout the L1 stage (see Materials and Methods) (Figure 8). Nuclear migration of 12 P cells starts at the mid-L1 stage and ends at the late L1 stage (Sulston 1976; Sulston and Horvitz 1977). We therefore reasoned that in late L1 animals, nuclei that failed to migrate would be seen at the lateral sides of late L1 larvae and <12 P-cell nuclei or their divided nuclei would be seen in the ventral cord. In wild-type N2 animals, almost all P-cell nuclei migrated normally at 15°, 20°, and 25° (only 2 of 2232 nuclei failed to migrate; Table 3). Furthermore, toca-1(tm2056) single-mutant animals had no P-cell nuclear migration defects (Table 3). In contrast, unc-84(n369) animals had only 11.7 ± 0.1 (average ± SEM), 10.5 ± 0.2, or 9.5 ± 0.3 nuclei that migrated normally at 15°, 20° or 25°, respectively (Figure 8A and Table 3). As expected, toca-1(tm2056) unc-84(n369) animals had only 11.7 ± 0.1, 9.7 ± 0.2, or 6.9 ± 0.3 nuclei that migrated at 15°, 20°, or 25°, respectively (Table 3). Therefore, the toca-1 (tm2056) mutation significantly enhanced the defects of P-cell nuclear migration in unc-84(n369) animals, at least at 20° (t-test; P < 0.01) and 25° (t-test; P < 0.0001).
Figure 8 .
P-cell nuclear migration defects in toca-1(tm2056) unc-84(n369) are shown. (A) tdTOMATO labels nuclei in unc-84(n369) animals at 15° at the late L1 stage. Non–P-cell nuclei are marked by asterisks. The ventral cord is marked by the dotted line. (B) Two nuclei that failed to migrate to the ventral side in a toca-1(tm2056) unc-84(n369) L1 larva at 25° are shown. Migrated nuclei and their descendants can be seen in the ventral cord in B′.
Table 3 . toca-1 enhances the P-cell nuclear migration phenotype of unc-84.
| 15° | 20° | 25° | |
|---|---|---|---|
| Wild type | 12.00 ± 0.0 (n = 45)a | 12.0 ± 0.0 (n = 50) | 12.00 ± 0.0 (n = 91) |
| unc-84(n369) | 11.7 ± 0.1 (n = 39) | 10.5 ± 0.2 (n = 73) | 9.5 ± 0.3 (n = 66) |
| toca-1(tm2056) | 12.0 ± 0.0 (n = 21) | 12.00 ± 0.0 (n = 17) | 12.0 ± 0.0 (n = 30) |
| toca-1(tm2056) unc-84(n369) | 11.7 ± 0.1 (n = 100) | 9.7 ± 0.2 (n = 56) | 6.9 ± 0.3 (n = 35) |
P-cell nuclei that migrated normally were counted using an nls::tdTomato marker expressed in P-cell nuclei. The means ± SEM are shown.
Discussion
Overcoming genetic redundancy is a major hurdle to the study of many cell and developmental processes, including nuclear migration. For example, <20% of predicted genes in C. elegans are thought to play essential roles (Kamath et al. 2003; Sonnichsen et al. 2005). However, one recent study found that 38% of the predicted genes in the C. elegans genome have clear human orthologs (Shaye and Greenwald 2011). Thus, there are many genes in the C. elegans genome that are clearly conserved to humans, but have no known function on their own. It is likely that many or most of these conserved, nonessential genes function redundantly to other pathways. Identification of such genes requires relatively difficult synthetic or enhancer screens, such as the one employed here.
Our goal here was to identify novel mechanisms employed to move nuclei. Previous studies in multiple systems demonstrated that SUN and KASH proteins function at the nuclear envelope to connect and move nuclei along the cytoskeleton (Roux et al. 2009; Starr and Fridolfsson 2010). The screen presented here was designed to identify novel mechanisms that function in parallel to SUN-KASH bridges. Our model system, C. elegans larval P-cell nuclear migration, is better suited for identifying such pathways than embryonic dorsal hyp7 precursors where the mechanisms of UNC-84 and UNC-83 were elucidated (McGee et al. 2006; Meyerzon et al. 2009; Fridolfsson et al. 2010; Fridolfsson and Starr 2010; Tapley et al. 2011). SUN-KASH bridges are absolutely required for nuclear migration in the embryonic hypodermis at all temperatures. However, the process of P-cell nuclear migration is temperature sensitive in the background of SUN (unc-84) or KASH (unc-83) mutants (Sulston and Horvitz 1981; Malone et al. 1999; Starr et al. 2001), allowing us to identify emu mutations that enhance P-cell nuclear migration defects.
The temperature-sensitive nature of nuclear migration defects in unc-84 or unc-83 mutants led to our hypothesis that an unknown pathway or pathways work, in part, redundantly to the SUN-KASH pathway. In support of our hypothesis and fulfilling our goal, we identified eight, independently isolated, mutant lines that carry mutations in genes that appear to function parallel to the SUN-KASH pathway. We determined that one of these mutations is in the toca-1 gene. Thus, our screen was successful in identifying conserved, nonessential genes that function to move nuclei. The remaining emu mutations are likely to be a continuing source of interesting genes that also function in parallel to unc-84 to move nuclei.
We quantified nuclear migration in the emu mutants by two different assays. For most of our experiments, we counted L4/young adult GABA neurons that were derived from P-cell lineages and concluded that fewer GABA neurons represented failed nuclear migrations. To more directly observe P cells in L1 larvae, we employed a P-cell–specific nuclear marker. The nuclear migration defect of toca-1(tm2056) unc-84(n369) appeared less severe using the L1 larval P-cell nuclear marker (Figure 8 and Table 3) than when the GABA neuron assay was used (Figure 2 and Table 2). There are two possible reasons to explain this discrepancy. First, toca-1 could play an important role in the GABA lineage after nuclear migration, such as in a polarized cell division in the ventral cord. We favor an alternative, where our assay missed defects in nuclear migration. Because we waited until all P-cell nuclear migration was completed before scoring, we would have missed a significant delay in P-cell nuclear migration, which could lead to a later loss of the P-cell lineage. Therefore, further live imaging will be required to determine the specific effects of the loss of toca-1 on P-cell lineages.
The P-cell defect quantified by both of our assays is likely to represent a nuclear migration defect, not a more general cell migration defect. rho-1(dn) mutants, which disrupt cell migration, were previously shown to lead to ∼60% of L2 larvae with lateral P-cell derivatives (Spencer et al. 2001). In our observations of GABA neurons in L2 larvae, we very rarely (eight neurons in 149 L2 toca-1unc-84 double-mutant animals) observed misplaced neurons at the lateral sides, suggesting we are not studying a general defect in cell migration and can genetically separate nuclear migration from cell migration.
toca-1 is conserved from nematodes to humans, but does not have a phenotype on its own (Giuliani et al. 2009). It was previously shown that toca-1 functions redundantly to toca-2 to mediate the essential process of endocytosis of yolk proteins in oocytes (Giuliani et al. 2009). Here, we showed that toca-1, but not toca-2, functions redundantly to unc-84 at 15° to move P-cell nuclei. Four observations are consistent with the conclusion that the nuclear migration defect in the yc20 mutant isolated in the emu screen is caused by a missense mutation in TOCA-1: (1) The yc20 mutation mapped to a region of chromosome X using EMS-linked mutations; (2) WGS identified a missense mutation in toca-1; (3) toca-1(RNAi) and the toca-1(tm2056) deletion allele both enhanced the nuclear migration defect of unc-84(n369) to a similar extent to that of the yc20 allele; and (4) most importantly, the yc20 nuclear migration defect was rescued with a wild-type copy of toca-1 from a transgene expressed specifically in P cells. We therefore conclude that yc20 is an allele of toca-1.
The genetics show that TOCA-1 functions in parallel to the SUN-KASH nuclear-envelope bridge. To our knowledge, this is the first report of a role for TOCA-1 in nuclear migration. TOCA-1 is conserved across animals and contains an F-BAR domain that is thought to bind curved membranes and two domains that recruit cdc42 and N-WASP (Ho et al. 2004; Giuliani et al. 2009). How TOCA-1 functions in nuclear migration remains unclear. One possibility is that because the yc20 lesion is predicted to change an alanine in the F-BAR domain to a valine, the mutation could affect TOCA-1 localization to membranes. However, TOCA-1::GFP did not obviously associate with the nuclear envelope. A second possibility is that the toca-1(yc20) mutation disrupts actin dynamics by disrupting the function of the cdc42 and/or N-WASP/WAVE interaction domains. In support of this hypothesis, depletion of the actin nucleator Arp2/3 or WAVE leads to the loss of one to two GABA neurons at 25°, suggesting a weak P-cell nuclear migration defect (Xiong et al. 2011). A third possibility is that TOCA-1 is functioning through its known roles in membrane tubulation and clathrin-mediated endocytosis (Ho et al. 2004; Kakimoto et al. 2006; Bu et al. 2009, 2010; Fricke et al. 2009; Giuliani et al. 2009). In Drosophila, Rab-mediated endocytosis and vesicular transport contribute to nuclear positioning in the developing eye (Houalla et al. 2010). Future experiments will help clarify the mechanism of TOCA-1 in P-cell nuclear migration. Molecular characterization of the additional emu mutations from this screen should significantly increase our general understanding of the mechanisms of nuclear migration.
Acknowledgments
We thank David Fay at University of Wyoming; Min Han at University of Colorado, Boulder; Laura Herndon at Albert Einstein College of Medicine; and JoAnne Engebrecht, Lesilee Rose, Noelle L’Etoile, Li-Min Hao, Shih-Yu Chen, Hsuan-Chung Ho, and members of the Starr laboratory at University of California (UC), Davis, for helpful discussions. We thank Dawei Lin, Ian Korf at UC Davis, Kelly Lu at Cornell University, and Henry Bigelow and Oliver Hobert at Columbia University for help analyzing whole-genome sequencing data sets. We thank Shohei Matani of the Tokyo Women’s Medical University for the deletion strains, Giorgio Scita at the University of Milan, Italy for TOCA-1 antibody, and Ryan Doonan for the hlh-3 promoter. Some strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health (NIH)-National Center for Research Resources. This study was supported by NIH grant R01 GM073874 (to D.A.S.).
Footnotes
Communicating editor: K. Kemphues
Literature Cited
- Bellion A., Baudoin J. P., Alvarez C., Bornens M., Metin C., 2005. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25: 5691–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow H., Doitsidou M., Sarin S., Hobert O., 2009. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat. Methods 6: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W., Chou A. M., Lim K. B., Sudhaharan T., Ahmed S., 2009. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem. 284: 11622–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W., Lim K. B., Yu Y. H., Chou A. M., Sudhaharan T., et al. , 2010. Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation and vesicle motility: implications for endocytosis. PLoS ONE 5: e12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., et al. , 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69 [DOI] [PubMed] [Google Scholar]
- Chytilova E., Macas J., Sliwinska E., Rafelski S. M., Lambert G. M., et al. , 2000. Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11: 2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan R., Hatzold J., Raut S., Conradt B., Alfonso A., 2008. HLH-3 is a C. elegans Achaete/Scute protein required for differentiation of the hermaphrodite-specific motor neurons. Mech. Dev. 125: 883–893 [DOI] [PubMed] [Google Scholar]
- Fay D., Bender A., 2008. SNPs: introduction and two-point mapping. WormBook 25: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Keenan S., Han M., 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1989. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., et al. , 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Fricke R., Gohl C., Dharmalingam E., Grevelhorster A., Zahedi B., et al. , 2009. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr. Biol. 19: 1429–1437 [DOI] [PubMed] [Google Scholar]
- Fridolfsson H. N., Starr D. A., 2010. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H. N., Ly N., Meyerzon M., Starr D. A., 2010. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 338: 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Mitani S., 2000. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269: 64–69 [DOI] [PubMed] [Google Scholar]
- Giuliani C., Troglio F., Bai Z., Patel F. B., Zucconi A., et al. , 2009. Requirements for F-BAR proteins TOCA-1 and TOCA-2 in actin dynamics and membrane trafficking during Caenorhabditis elegans oocyte growth and embryonic epidermal morphogenesis. PLoS Genet. 5: e1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E. R., Jani S., Gundersen G. G., 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121: 451–463 [DOI] [PubMed] [Google Scholar]
- Ho H. Y., Rohatgi R., Lebensohn A. M., Le M., Li J., et al. , 2004. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 118: 203–216 [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houalla T., Shi L., van Meyel D. J., Rao Y., 2010. Rab-mediated vesicular transport is required for neuronal positioning in the developing Drosophila visual system. Mol. Brain 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T., Katoh H., Negishi M., 2006. Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 281: 29042–29053 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Koch R., van Luenen H. G., van der Horst M., Thijssen K. L., Plasterk R. H., 2000. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 10: 1690–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer M. P., Banks S. M., Xie X., Wu Y., Fischer J. A., 2007. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1: 75–85 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Starr D., Cohen M., Liu J., Han M., et al. , 2002. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell 13: 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Etoile N. D., Bargmann C. I., 2000. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25: 575–586 [DOI] [PubMed] [Google Scholar]
- Luxton G. W., Gomes E. R., Folker E. S., Vintinner E., Gundersen G. G., 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329: 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. J., Fixsen W. D., Horvitz H. R., Han M., 1999. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126: 3171–3181 [DOI] [PubMed] [Google Scholar]
- Martini F. J., Valdeolmillos M., 2010. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J. Neurosci. 30: 8660–8670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee M. D., Rillo R., Anderson A. S., Starr D. A., 2006. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell 17: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., Jorgensen E. M., 1997. Identification and characterization of the vesicular GABA transporter. Nature 389: 870–876 [DOI] [PubMed] [Google Scholar]
- Mellad J. A., Warren D. T., Shanahan C. M., 2011. Nesprins LINC the nucleus and cytoskeleton. Curr. Opin. Cell Biol. 23: 47–54 [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation. Methods Cell Biol. 48: 451–482 [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E. S., et al. , 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M., Fridolfsson H. N., Ly N., McNally F. J., Starr D. A., 2009. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 136: 2725–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R., 2000a. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148: 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R., Efimov V. P., Xiang X., 1998. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 8: 467–470 [DOI] [PubMed] [Google Scholar]
- Morris R., 2000b. A rough guide to a smooth brain. Nat. Cell Biol. 2: E201–E202 [DOI] [PubMed] [Google Scholar]
- Patterson K., Molofsky A. B., Robinson C., Acosta S., Cater C., et al. , 2004. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau C. E., Howard R. M., Goldman A. P., Volk M. L., Girard L. J., et al. , 2002. A lin-45 raf enhancer screen identifies eor-1, eor-2 and unusual alleles of Ras pathway genes in Caenorhabditis elegans. Genetics 161: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Crisp M. L., Liu Q., Kim D., Kozlov S., et al. , 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA 106: 2194–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., O’Meara M. M., Pe’er I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B. T., McConnell S. K., 2005. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA 102: 13652–13657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk J., Wilsch-Brauninger M., Calegari F., Huttner W. B., 2009. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc. Natl. Acad. Sci. USA 106: 16487–16492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- Spear P. C., Erickson C. A., 2012. Apical movement during interkinetic nuclear migration is a two-step process. Dev. Biol. 370: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A. G., Orita S., Malone C. J., Han M., 2001. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98: 13132–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., 2011. KASH and SUN proteins. Curr. Biol. 21: R414–R415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Fridolfsson H. N., 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26: 421–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Hermann G. J., Malone C. J., Fixsen W., Priess J. R., et al. , 2001. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128: 5039–5050 [DOI] [PubMed] [Google Scholar]
- Stiernagle T., 2006 Maintenance of C. elegans, pp. 1–11 in WormBook, ed. The C. elegans Research Community WormBook, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Sulston J. E., 1976. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275: 287–297 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82: 41–55 [DOI] [PubMed] [Google Scholar]
- Tapley E. C., Ly N., Starr D. A., 2011. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol. Biol. Cell 22: 1739–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. W., Bremner K. H., Vallee R. B., 2007. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10: 970–979 [DOI] [PubMed] [Google Scholar]
- Tsujikawa M., Omori Y., Biyanwila J., Malicki J., 2007. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc. Natl. Acad. Sci. USA 104: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M., Marin O., 2010. Neuronal migration mechanisms in development and disease. Curr. Opin. Neurobiol. 20: 68–78 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K., Ketema M., Truong H., Sonnenberg A., 2006. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119: 5021–5029 [DOI] [PubMed] [Google Scholar]
- Wynshaw-Boris A., 2007. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin. Genet. 72: 296–304 [DOI] [PubMed] [Google Scholar]
- Xiong H., Mohler W. A., Soto M. C., 2011. The branched actin nucleator Arp2/3 promotes nuclear migrations and cell polarity in the C. elegans zygote. Dev. Biol. 357: 356–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Le Gras S., Jamet K., Jarriault S., 2010. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]