Abstract
Long noncoding RNAs (lncRNAs) play a key role in many important areas of epigenetics, stem cell biology, cancer, signaling and brain function. This emerging class of RNAs constitutes a large fraction of the transcriptome, with thousands of new lncRNAs reported each year. The molecular mechanisms of these RNAs are not well understood. Currently, very little structural data exist. We review the available lncRNA sequence and secondary structure data. Since almost no tertiary information is available for lncRNAs, we review crystallographic structures for other RNA systems and discuss the possibilities for lncRNAs in the context of existing constraints.
Keywords: RNA, non-coding, long noncoding RNA, lncRNA, lincRNA, epigenetics, cancer, hormone receptor, secondary structure, structural biology, RNA structure, HOTAIR, MALAT
Introduction
A new class of RNAs has recently emerged as a key player in many rapidly growing areas of research, including epigenetics, hormone signaling, development, stem cell biology, cancer, brain function and plant biology.1–9 The growth of this area has been fueled by recent advances in sequencing technology. These RNAs (long non-coding RNAs, or lncRNAs) are typically 1,000–10,000 residues in length. LncRNAs are often polyadenylated, transcribed by RNA polymerase II and spliced.3,6,7,10–13 While some lncRNAs are found in the cytoplasm, most are localized in the nucleus. Many lncRNAs are associated with histone methylation, chromatin remodeling and subsequent epigenetic effects.14 In the field of epigenetics, the mechanism by which epigenetic factors find their targets remains largely a mystery. The importance of lncRNAs has been underscored in the context of mammalian genomes, where recent evidence suggests that lncRNAs may provide a missing epigenetic link between DNA, histones and methylation factors.15
In humans, over 70% of the genome is actively transcribed.16 In contrast, protein-coding genes constitute only 1–2% of the genome.17 The active transcription of non-protein coding genes gives rise mainly (80–90%) to lncRNAs.18 While some lncRNAs, such as MALAT1, are highly abundant transcripts, many lncRNAs do show low count. However, low transcription levels do not necessarily reflect lack of functionality. Studies on the stability of lncRNAs19 have shown that lncRNAs have stabilities comparable to those of mRNAs (albeit slightly less on average). Here, time scales range from 30 min up to 16 h in the case of MALAT1. Protein half-lives range from 30 min to 2 h.20 We note that transcription rates range from 1–50 kb/min.21 From these data, a picture of the nucleus emerges, where lncRNAs are synthesized in minutes and may persist for hours.
Long noncoding RNAs (lncRNAs) are very broadly defined by two major characteristics: (1) length of the transcript (> 200 nts) and (2) having little or no potential for translation.22 Some lncRNAs ('macroRNAs') achieve incredible lengths, extending beyond 90 kB. Examples include the 108 kB Air and the 91 kB kcnq1ot1.23–25 The term lncRNA is traditionally reserved for regulatory RNAs. LncRNAs are often further divided into categories based on their relative position to neighboring protein-coding genes. Natural antisense transcripts (NATs) are transcribed from the antisense strand of protein-coding genes, overlapping at least one exon. Large intervening noncoding RNAs, as known as long intergenic noncoding RNAs (lincRNAs), are positioned far from protein-coding genes. Intronic noncoding RNAs are uniquely transcribed from intron regions of protein-coding genes either in the sense or antisense direction. Bidirectional lncRNAs are transcribed in the antisense direction in the region of the promoter of a protein-coding gene. An exact estimation of the number of lncRNAs is quite challenging due to their cell-specific, tissue-specific, developmental stage specific and disease-specific expression profiles. The most recent estimates place the number of lncRNAs in humans at ~15,000.26 However, tens of thousands of lncRNAs have been profiled this year alone.27–30
In terms of function, nascent paradigms of lncRNA action include, but are not limited to, critical regulatory roles in embryonic stem cell pluripotency,31 brain function,32–34 subcellular compartmentalization35,36 and chromatin remodeling.3,8,37 Many have been linked to various diseases, such as cancer.38 We note that lncRNAs play key roles in intracellular and extracellular signaling (SRA, Gas5, LINoCR, BC1, BORG and NRON) and stress response (e.g., SAT III, PRINS, npc536, hsr omega transcript, gadd7, Hsr1 and bace1as). More detailed discussion of functional aspects of lncRNAs can be found in several excellent recent review articles.39 Due to the large sizes of intact lncRNAs relative to typical biophysical systems, very few structural studies of these RNAs have been performed.40 By comparison, the high-resolution 3-D structure of the intact ribosome (~5 kB in total) required ~25 y for its solution.41,42
For lncRNAs, the following questions remain unanswered: (1) Are lncRNAs highly structured or disordered? (2) Do they contain globular sub-domains, or are they organized linearly in chains of stem-loops? (3) Do lncRNAs exist in ribonucleoprotein complexes or as isolated RNAs that transiently interact with proteins? (4) Do these molecules contain a compact core, or are they more extended?
Mechanistic studies of lncRNAs have the potential to be more challenging than ribosomes, because lncRNAs are not as highly conserved nor as highly expressed. Nonetheless, RNA molecules are well known to utilize a wide spectrum of functional elements either at their sequence, secondary or tertiary level. RNA interference and RNA silencing leverage sequence specificity to control gene expression. Riboswitch RNAs regulate gene expression via secondary structure. The ribosome uses its complex tertiary structure to synthesize proteins, in a manner analogous to a protein-based molecular machine. LncRNAs may or may not use aspects of each of these three mechanisms to regulate gene expression. In light of the tremendous variety of lncRNAs, it is possible that all three of these mechanisms are employed by lncRNA systems. A great deal of useful information can be produced using modern structural biology techniques In this review, we provide a summary of current knowledge of sequence and structural features of eukaryotic lncRNAs. Although studies of lncRNA tertiary structure have yet to be performed, we examine known crystallographic structures of other RNAs and explore the possibilities that might occur in lncRNA systems.
Sequence Elements in lncRNAs
Some lncRNAs rely on Watson-Crick base pairing for functional activity. This may be in the form of 'perfect' pairing, where a stretch of the lncRNA forms a continuous sequence of Watson-Crick base pairs with another RNA molecule, such as an mRNA. There are also lncRNAs that implement 'imperfect' pairing, where a stretch of Watson-Crick pairs is interspersed with non-Watson-Crick base pairs. Finally, evidence exists for regions of lncRNAs directly interacting with DNA. In some cases, it has been suggested that base pairing between the RNA and DNA occurs, while in other cases, triple helix mechanisms have been proposed.43
miRNA-sequestering lncRNAs
These lncRNAs provide alternative miRNA binding sites to regulate expression levels of protein-coding genes post-transcriptionally. Linc-MD1, involved in muscle differentiation, acts as a competing endogenous RNA (ceRNA), sequestering miR-133 and miR-135 from their target genes.44
Half-STAU1-binding site RNAs (1/2-sbsRNAs)
This lncRNA binds to 3′-UTRs via Alu elements in a process known as STAU1-mediated mRNA decay.7 This event involves imperfect base pairing between the Alu element of the lncRNA and the Alu element of the mRNA. The interaction is recognized by the dsRNA-binding protein STAU1 and results in degradation of the mRNA.
Antisense lncRNAs
These lncRNAs may bind to mRNAs, regulating their splicing. The long noncoding Zeb2NAT transcript originates antisense to the 5′ splice site of mRNA Zeb2. The transcript is known to prevent splicing of this mRNA region, preserving the internal ribosome entry site for efficient translation.45
Upstream lncRNAs
These lncRNAs may form triplex complexes with DNA promoter regions. One such lncRNA originates upstream of the dihydrofolate reductase gene (DHFR) in humans. Here, upstream transcription is initiated at the alternative minor promoter site, resulting in the decreased occupancy of transcription factors at the major promoter and subsequent repression of gene expression.46 Moreover, the noncoding RNA product that originated upstream was found to interact directly with a major DNA promoter site, forming a purine-purine-pyrimidine triplex.
Secondary Elements of lncRNA Structure
In addition to sequence, RNA secondary and tertiary structural motifs often play a central role in the mode of action of RNA, be it specific binding, allosteric, catalytic or structural. While few structural studies of lncRNAs exist, we describe below the hints of structure that have been uncovered to date.
Double stem loops in chromatin remodeling
Many lncRNAs have been shown to play an important role in chromatin remodeling. Large-scale identification of functional lncRNAs has resulted from their association with chromatin proteins.13,47 For example, a CLIP-seq investigation of RNA associated with the SFRS1 splicing factor uncovered > 6,000 spliced noncoding RNAs with unknown function. While the full functional repertoire of lncRNAs remains to be delineated,48 it is clear that lncRNAs play a critical role in chromatin remodeling, often acting in trans via association with chromatin modifying enzymes.47 Lee and coworkers identified > 9,000 lncRNAs in mouse embryonic stem cells, which interact with the polycomb repressive complex, PRC2.49 EMSA analysis of a number of PRC2-interacting lncRNAs suggested that binding occurs through EZH2, one of four polycomb proteins domains of PRC2. The remaining three domain proteins of PRC2 are thought to further tighten this interaction. The large number of identified lncRNAs, including RepA/Xist, HOTAIR and Air, suggest the presence of certain common features across the PRC2-interacting family of lncRNAs. In addition, lncRNAs can also associate with the LSD1/CoREST/REST complex, critical in H3K4 demethylation. The lincRNA HOTAIR is an excellent example of this bifunctionality.3
HOTAIR is ~2.2kB in length and regulates the gene expression of HoxD genes and a number of other genes by recruiting the LSD1 and PRC2 histone modification complexes to targeted loci. Deletion experiments on HOTAIR narrowed the interaction sites down to two modular regions which are responsible for these interactions: (1) a 5′ 300 nt region that binds PRC2, and (2) a 646 nt region located downstream at the 3′-end, responsible for binding to LSD1. This raises the question: while two key motifs are located at the 5′ and 3′-ends of HOTAIR, does the remainder of the sequence have functional importance? The intervening sequence may provide the required distance in terms of spatial organization between two interaction sites. Alternatively, it may contain motifs necessary for targeting, or comprise additional protein binding motifs required for activity, yet to be found. It has been suggested that a double stem-loop RNA motif is present in a PRC2-binding region. Binding motifs for LSD1 have not been identified.49,50
Similar to HOTAIR, an A-repeat subregion of Xist, known to bind PRC2, has long been thought to form multiple double stem-loop structures, encoded by the repeat sequences located in this region.51 Recent detailed chemical probing investigations were not consistent with the proposed arrangements of secondary structure elements and suggested a more complex secondary fold, comprising elongated helical subdomains.52 Until an additional number of detailed structural/mechanistic investigations for PRC2-interacting lncRNAs are performed, the critical RNA motifs for association with PRC2 will not be clear.
Cloverleaf elements in 3′end processing and in brain evolution
A cloverleaf secondary architecture similar to that found in tRNA has been found in many different regions of long noncoding RNAs. One of its roles has been assigned to 3′-end maturation of the lncRNAs transcripts involved in subcellular organization.53 These include the MALAT1 and NEAT1 lncRNAs, involved in the formation of nuclear speckles and paraspeckles, respectively. Non-canonical end maturation of the MALAT1 ncRNA involves a cloverleaf secondary element at its 3′-end.35 This subregion is the most conserved element in both the MALAT1 and NEAT1 sequences, adopting tRNA-like architecture. This structural element is responsible for recruiting RNase P (involved in maturation of tRNA molecules) for cleavage and generation of mature MALAT1 transcripts. The remaining cleaved fragment is further processed by RNase Z/tRNA nucleotidyl transferase to yield a tRNA-like transcript (mascRNA), which is further shuttled to the cytoplasm. Interestingly, the mature 3′-end of MALAT1 sequence comprises only a relatively short and genomically-encoded stretch of poly(A) region, suggested to be stabilized by two conserved U-rich motifs, located upstream. The details of this interaction are currently unknown. The same mechanism of 3′- end processing has been determined for the NEAT1_v2 transcript, generating small and independent tRNA-like molecule as well, named menRNA.54
Interestingly, cloverleaf architecture has been found in the subregion of another lncRNA, HAR1 (human accelerated region), associated with neocortex development.55 This region covers 118 nucleotides, which are highly conserved across vertebrates (2 nt change between chicken and chimpanzee), but more divergent in humans (18 mutations relative to chimpanzee).56,57 Rapid changes in sequence homology between chimpanzee and human have been associated with human brain evolution. In vitro structure probing experiments on human (hHAR) and chimpanzee HAR (cHAR) regions showed distinct secondary folds.55 In chimpanzee, cHAR adopts a rather unstable and extended hairpin architecture; in humans, hHAR folds into a cloverleaf-type element, consisting of a 4-way junction. The authors have mentioned that the chimpanzee sequence could possibly adopt a cloverleaf architecture, but may require a stabilizing factor, the necessity of which is likely to be diminished in human HAR.
Secondary structures in steroid receptor chemistry
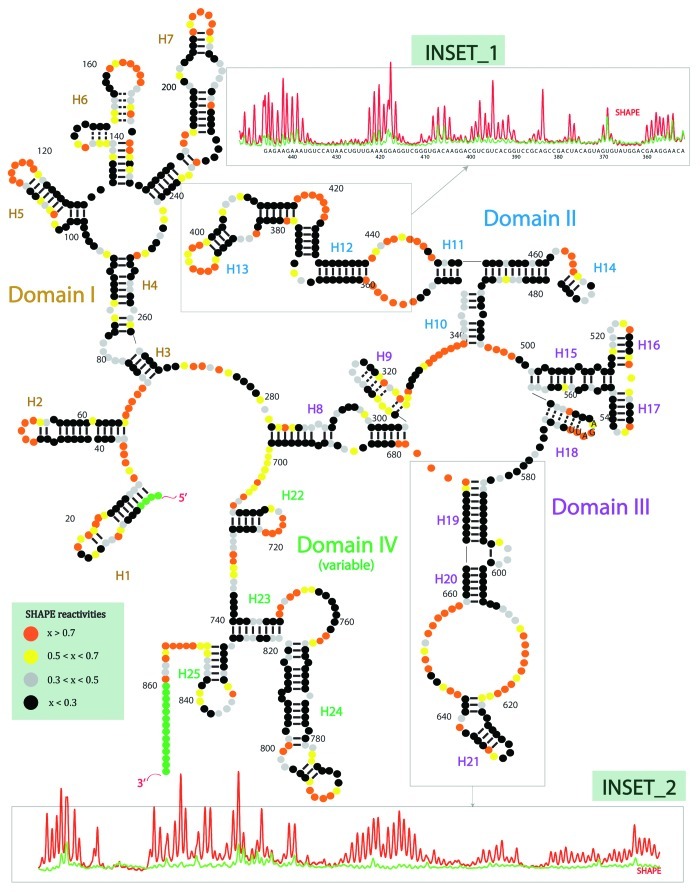
We have recently performed extensive chemical and enzymatic investigations of another long noncoding RNA, the steroid receptor RNA activator or, SRA.58 This study produced the first experimentally determined secondary structure of an intact human lncRNA to our knowledge (Fig. 1). SRA co-activates several steroid receptors (e.g., ER, AR, TR, GR, RAR), and it is known to interact directly with many proteins (e.g., SHARP, SLIRP, DAX-1, TR), suggesting that it may play a scaffolding role in the transcription complex. SRA has also been shown to interact with CTCF.59 This RNA was one of the first lncRNAs discovered in humans. Our biochemical probing revealed a complex 2D architecture, comprising four major subdomain regions. The identified secondary elements range from small modular helical regions to complex multiway junctions. In total, SRA contains 25 helical segments, 16 terminal loops, 15 internal loops and 5 junction regions. We have also noticed that purine-rich sequences are highly conserved and often located in single-stranded regions such as terminal, internal and junction loops. The same trend in structural preference is generally observed for rRNA. The vast majority of helices in our structure were validated by covariance analysis using multiple sequence alignment across vertebrates.
Figure 1. The first experimentally determined secondary structure of an intact lncRNA, to our knowledge. The steroid receptor RNA activator (SRA) lncRNA contains 4 subdomains, and 25 helices. The structure was determined using four methods of chemical (SHAPE, in-line, DMS) or enzymatic (RNase V1) probing. Covariance analysis based on multiple sequence alignment across vertebrates was used to help validate the structure. In SHAPE probing (selective 2’-hydroxyl acylation analyzed by primer extension), high reactivity corresponds to high mobility and low likelihood for base pairing; low reactivity corresponds to low mobility and high likelihood for base pairing. Orange, high SHAPE reactivity. Yellow, medium reactivity. Grey, low reactivity. Black, no reactivity. Insets: Red, SHAPE reactivity capillary electrophoresis trace for lncRNA. Green, raw blank trace.
Previously Studied Tertiary Structures of Other RNA Systems
Three-dimensional structures of lncRNAs have not been attempted to date. Here, we review previously solved structures of other RNAs and discuss these in the context of lncRNAs. Prior to 2000, the set of three-dimensional RNA structures roughly consisted of tRNA, various ribozyme and aptamer RNAs, the group I and group II introns, RNA helices and quadruplexes, portions of the bacterial ribosome and components of the spliceosome. The initial high-resolution structures of the ribosome published in 2000 spurred a large number of additional RNA crystallographic studies. These include many different riboswitch RNAs, TLS RNA, RNase P, the signal recognition particle, the HIV-1 frame-shifting element, regions of telomerase RNA, as well as many other ribosome constructs.60 The ribosome and introns remain the only large RNA (> 200 nts) high-resolution crystallographic structures solved to date.
The group I and group II introns are isolated compact RNAs characterized by numerous RNA helices capped with RNA stem-loops (Fig. 2A).61,62The helices are connected through various junctions. Tertiary contacts between helices, loops and junctions also exist.
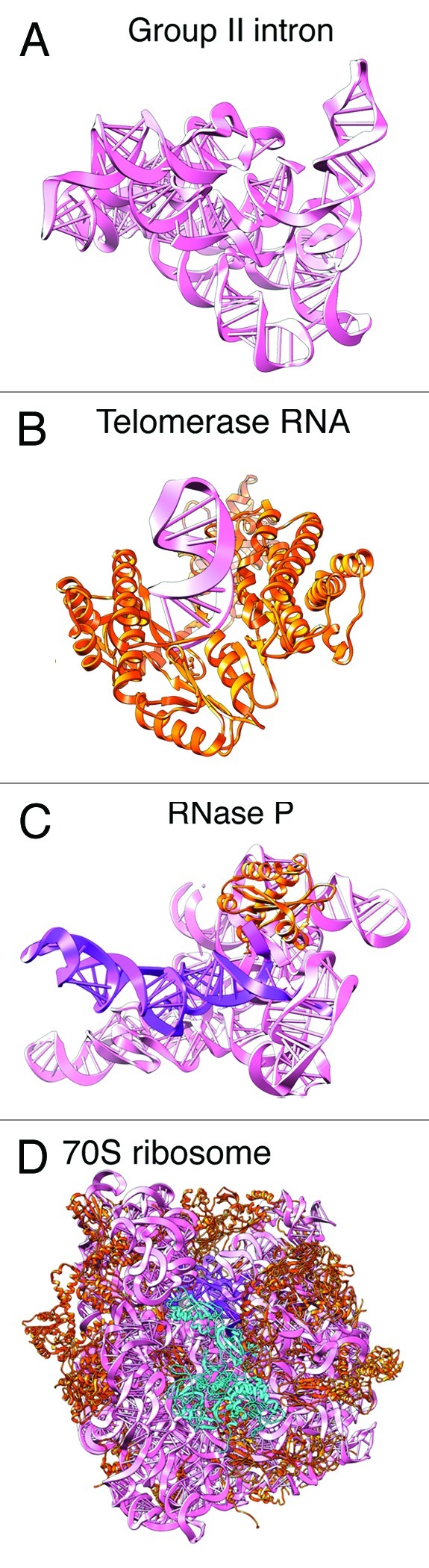
Figure 2. Examples of RNA tertiary structures solved by crystallography. (A) Group II intron, solved by Pyle, Toor and coworkers.62 The intron is a highly-structured isolated RNA with compact core. (B) Telomerase RNA solved by Skordalakes and coworkers.63 (C) RNase P solved by Mondragon and coworkers.65 RNase P is a highly structured RNA with a single protein binding domain. (D) Ribosome, Ramakrishnan and coworkers.66 The ribosome is a highly structured and highly compact RNA complex containing ~50 proteins that help stabilize the RNA structure. The ribosome contains a limited number of factor binding sites. Different factors bind to the same binding sites, regulating protein synthesis.
An important component of the telomerase complex is telomerase RNA, which is directly bound to telomerase reverse transcriptase and acts as template for nucleotide additions of telomeric regions (Fig. 2B).63 Human telomerase RNA is ~450 nts. In yeast, it was shown that the est1p binding domain on the RNA can be moved around to other locations within the RNA while maintaining function, suggesting regions of telomerase RNA are highly flexible.64
RNase P is a ribonuclease responsible for cleaving a precursor sequence from tRNA. The structure is dominated by the RNA (Fig. 2C).65 A small protein component increases the affinity of tRNA to RNase P. This RNA is highly structured and compact, similar to the group I and group II introns.
The ribosome is the universally conserved molecular machine responsible for protein synthesis (Fig. 2D).66 In bacteria, the ribosome consists of two subunits. The small subunit (30S) contains a ~1.5 kB RNA (16S rRNA) and ~20 different proteins. The large subunit (50S) contains a ~3 kB RNA (23S rRNA), a ~120 nt RNA (5S rRNA) and ~35 different proteins. The two subunits fit together, producing a large cavity between the two, through which the tRNAs enter and exit. Most of the protein factors bind to the ribosome at the GTP-associated center or at one of the three tRNA binding sites. For example, both elongation factors EF-Tu and EF-G bind to the same location on the ribosome at different stages of the elongation cycle. The scaffold of ribosome structure is RNA, while many proteins interspersed throughout the ribosome, providing structural stability to the overall architecture. Functional factors, which help the ribosome proceed through protein synthesis in a GTP-dependent manner, come on and off the ribosome at various stages of protein synthesis.
Macromolecular Complexes and lncRNA Quaternary Interactions
So far, the main evidence for lncRNA participation in quaternary complex formation has been obtained for NEAT1 transcripts. Here, two NEAT1 isoforms (NEAT_V1: 3.7kB and NEAT_V2: 22.7 kB for human) are expressed from the same promoter with similar expression levels.67 Both isoforms are involved in the formation of specific nuclear compartments called paraspeckles. These are ribonucleoprotein complexes characterized by three distinct proteins (e.g., p54, PSF and PSP1), which all contain RNA-binding motifs.68,69 The proposed model of paraspeckle association relies on the initial NEAT_V2 association with PSF and p54, followed by subsequent recruitment of PSP1 and NEAT1_V1.67 Depletion of either NEAT_V2, p54 or PSF1 results in paraspeckle disintegration; however, depletion of PSP1 did not affect the architecture. Based on immuno-hybridization and in situ hybridization electron microscopy studies, paraspeckle association requires association of multiple NEAT1 transcripts, creating a fiber-like network.70 Interestingly, using DNA probes specific for subregions of NEAT1, the 5′ and 3′ ends of NEAT1 were localized to the periphery of paraspeckles, while the central regions of longer NEAT1 (NEAT1_v2) transcripts were localized to the inner part of these bodies. The NEAT1 arrangements in the inner region appear to be distributed uniformly. The expression of a shorter isoform of NEAT1 (NEAT1_v1), lacking 3′end, cannot rescue the paraspeckle formation. The following observations suggest the presence of certain functional elements in RNA molecule. Paraspeckles are also formed with a relatively fixed diameter, but vary in their length. Mouse NEAT1 transcripts (shorter by 2 kB compared with human) have a 9% diameter reduction relative to human, leading the authors to conclude that the length of NEAT1 transcript might be also the limiting factor in this arrangement. However, it remains unclear how the final NEAT1 arrangement in paraspeckles is accomplished. There are number of possibilities here. This can be the direct result of RNA-RNA interactions between NEAT1 transcripts to create a complex macromolecular platform. Alternatively, proteins can serve as bridges to connect multiple NEAT1_V2 transcripts, especially, knowing the fact that p54 is able to form heterodimers with PSF and PSP1 proteins.68,71
Perspectives on lncRNA Structure and Mechanism
lncRNAs are not likely to exist in ribosome-like ribonucleoprotein complexes
As the ribosome is the only RNA structure > 1 kB solved to date, we ask the question, are lncRNA systems similar to ribosomes in their structural composition? We note that our recent structural study of the SRA lncRNA revealed RNA secondary structure similar to the ribosome in its overall architecture. This study, which used multiple forms of extensive chemical probing combined with multiple sequence analysis, demonstrated that SRA has 4 sub-domains with numbers of helices and loops comparable (in proportional terms) to a ribosome subunit. We currently have no information on the tertiary structure of this lncRNA or other lncRNAs. In addition, we do not know if lncRNAs exist in ribonucleoprotein complexes (RNPs) or as isolated RNAs.
To compare with the ribosome, we note that it consists of a few long RNAs complexed with many unique (i.e., non-identical) proteins. The total number of protein-coding genes in the human genome is estimated to be ~21,000.72 As most proteins reside in the cytoplasm, we can reasonably estimate that the number of proteins in the nucleus, Nprotein, nucl < 21,000. Many thousands of lncRNAs have been identified, with most residing in the nucleus. As a very conservative estimate, we use NlncRNA, nucl > 3,000, giving NlncRNA,nucl/Nprotein,nucl > 1/7. We note that many lncRNAs are as large as or larger than the ribosome. In the case of the ribosome itself, the ratio of RNA molecules to protein molecules is NrRNA/Nrp ~1/25 for each subunit. Thus, even if every single unique protein encoded in the human genome formed a complex with a lncRNA, we would still not expect lncRNAs to be similar in structural composition to ribosomes. There are not enough unique proteins to form ribosome-like complexes (with ~25 unique proteins) for each lncRNA. Using an optimistic estimate that 1 in 10 of all proteins binds to a lncRNA, this would still provide less than 1 unique protein per lncRNA. Therefore, we conclude that lncRNAs in the nucleus are not likely to exist in ribosome-like RNP complexes. A few lncRNAs could theoretically exist in ribosome-like complexes. These complexes would be more likely to exist in the cytoplasm. The following possibilities remain: (1) lncRNAs exist in RNP complexes with many repeats of a few proteins, (2) lncRNAs exist in complexes with only a few proteins or (3) lncRNAs exist as isolated RNAs that transiently bind proteins as needed for function. We note that in the case of (1), to produce a similar protein density (number of proteins per RNA) to ribosomes, we would require ~10 proteins bound per 1 kB of lncRNA (e.g., 90 identical proteins bound to MALAT-1). While (1) is certainly possible, we favor (2) or (3).
The large diversity of lncRNAs may produce complexes similar in overall form to telomerase RNA, RNase P or the group I and II introns
While we suspect lncRNA complexes are not similar to ribosomes, we cannot rule out similarity to RNase P, telomerase RNA or the group I and group II introns. In the case of an ‘RNase P-like’ complex, the lncRNA would be highly structured and compact, containing a main protein binding site, where various proteins bind (Fig. 3A). Alternatively, the lncRNA could be decentralized without a compact core. It may contain several distinct protein-binding sites and act as flexible structural tether, as suggested for the telomerase RNA (Fig. 3B).64 The lncRNA could also be a stand alone, highly structured RNA, similar to the group I and group II introns. In this case, the lncRNA may transiently bind proteins as needed. Finally, another possibility is a highly disordered RNA, containing loosely organized protein binding domains (Fig. 3C). Our experimentally determined secondary structure of SRA is highly organized and more suggestive of a structure with characteristics of Figure 3A. We enumerate various combinations of secondary structure and tertiary structure in Table 1, with column 1 corresponding to Figure 3A, column 6 corresponding to Figure 3B, and columns 6–7 possibly corresponding to Figure 3C.
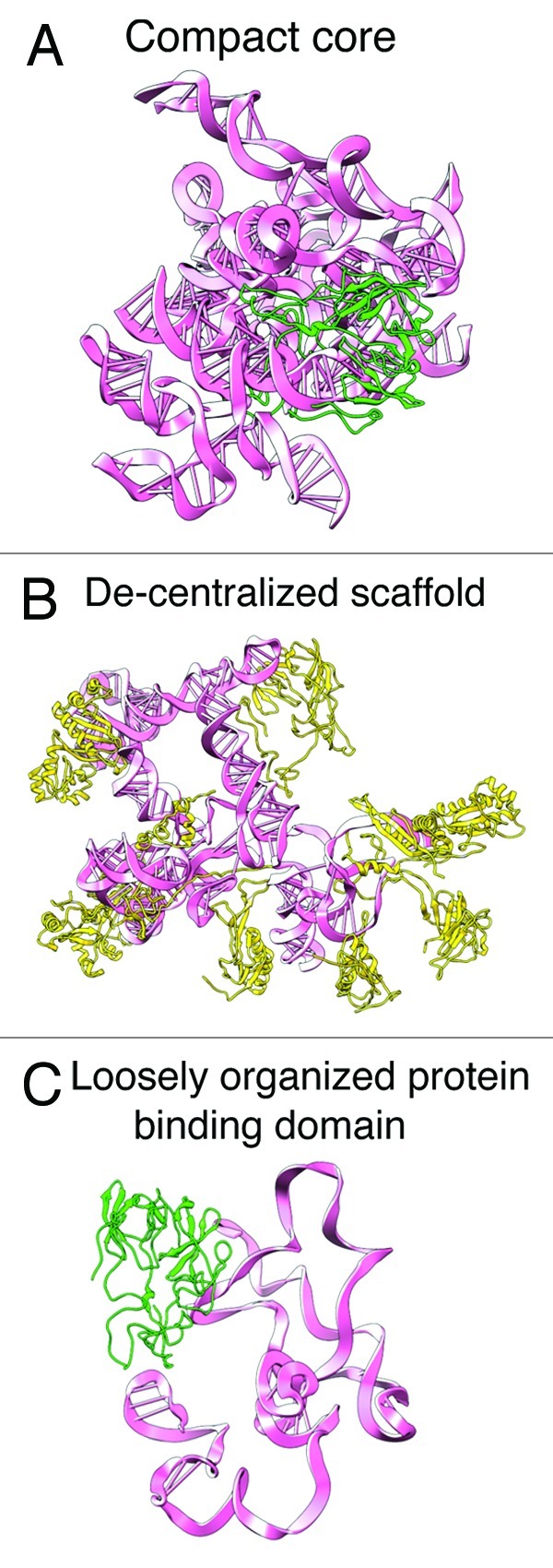
Figure 3. Possibilities for lncRNA three-dimensional architecture. These homology models represent concepts for possible lncRNA 3D structures. (A) lncRNA (pink) contains a compact tertiary core. The lncRNA may have a main protein (green) binding site, responsible for binding various protein factors. (B) De-centralized scaffold. In this scenario, the lncRNA does not have a compact core. The lncRNA may have several protein (yellow) binding sites. (C) Loosely organized protein binding domain with regions of unstructured RNA. The lncRNA may contain several long stretches of disordered single stranded RNA.
Table 1. Possibilities for structural configurations of lncRNAs.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| Core secondary structure? |
+ |
+ |
+ |
+ |
+ |
- |
- |
- |
| Binding domain secondary structure? |
+ |
+ |
+ |
- |
- |
+ |
+ |
- |
| |
|
|
|
|
|
|
|
|
| Core tertiary structure? |
+ |
+ |
- |
+ |
- |
- |
- |
- |
| Binding domain tertiary structure? | + | - | - | - | - | + | - | - |
Columns 1–8 represent different RNA structural configurations. Column 1 represents a highly structured configuration similar to the ribosome. Column 8 represents an unstructured RNA. Columns 2–7 represent various intermediate cases. Columns 6–8 represent a decentralized structural configuration. Our recent study demonstrates that the entire SRA lncRNA has well-organized secondary structure, corresponding to columns 1–3, depending on the degree of tertiary structure. All cases may also include single-stranded regions that organized upon protein binding.
Possibilities for structure-based mechanisms of lncRNAs
Although many more proteins have been studied in mechanistic detail relative to RNAs, a diverse portfolio of RNA mechanisms has emerged, based on either sequence, secondary or tertiary organization of RNA molecules, as well as combinations of these mechanisms. In sequence-based mechanisms, such as RNA interference by siRNAs and RNA silencing by miRNAs, the RNA plays a very minor structural role. Here, the role of RNA is mainly to add sequence specificity to the process, allowing the RISC complex to find its target and trigger a largely protein-based regulation mechanism.73,74
Over the past decade, a new mechanism of regulation has emerged, which is almost entirely based on RNA secondary structure.75–80 In riboswitch RNA systems, two secondary structures compete with each other to control termination of transcription (some riboswitch RNAs also control translation by sequestering the start codon). Here, one sequence in the 5′-UTR of the mRNA codes for two different secondary structures. The presence or absence of a metabolite selects one of the two structures, switching gene expression on or off. For example, in the case of the SAM-I riboswitch, the presence of a metabolite (SAM) causes the RNA to fold into a compact aptamer, favoring formation of the transcriptional terminator helix, turning gene expression of SAM synthetase off. In the absence of the metabolite, a second, alternative helix is formed, preventing formation of the transcriptional terminator helix, turning gene expression on. Riboswitches are more ‘secondary structure specific’ than ‘sequence specific’, often mandating stochastic context-free grammar algorithms for searches, as opposed more conventional BLAST-like searches. Interestingly, artificial riboswitch-like systems were first designed and produced in the lab and only later discovered in bacteria.81
RNA mechanisms based on tertiary structure are often allosteric and may be described by ‘induced-fit’ or ‘conformational selection’.78,82,83 In induced-fit, an event, such as protein- or ligand-binding, triggers a large conformational change. In conformational selection, the system is often frustrated between two conformations. A protein- or ligand-binding event shifts the equilibrium to one of the two conformations. In the case of the ribosome, many conformational fluctuations occur simultaneously and at different time scales. Protein binding or GTP hydrolysis events act to synchronize the fluctuations, shifting the equilibrium to the next basin in the energy landscape, allowing the ribosome to progress through the elongation cycle.84–87
Time scales and order of events
In addition to the three-dimensional structure of lncRNAs, the order of events and kinetics of these systems is essential for mechanistic understanding. For example, crystallographic structures have been solved for many ribosome complexes; however, the mechanism of ribosome translocation is still not understood. Rapid kinetics studies88,89 define the overall order of events. Single-molecule studies help elucidate the mechanism for transitions between states.90 A fusion of structural and kinetic information is required to unlock mechanism. Interestingly, the overall order of events can often be obtained before high-resolution crystallographic structures are available.
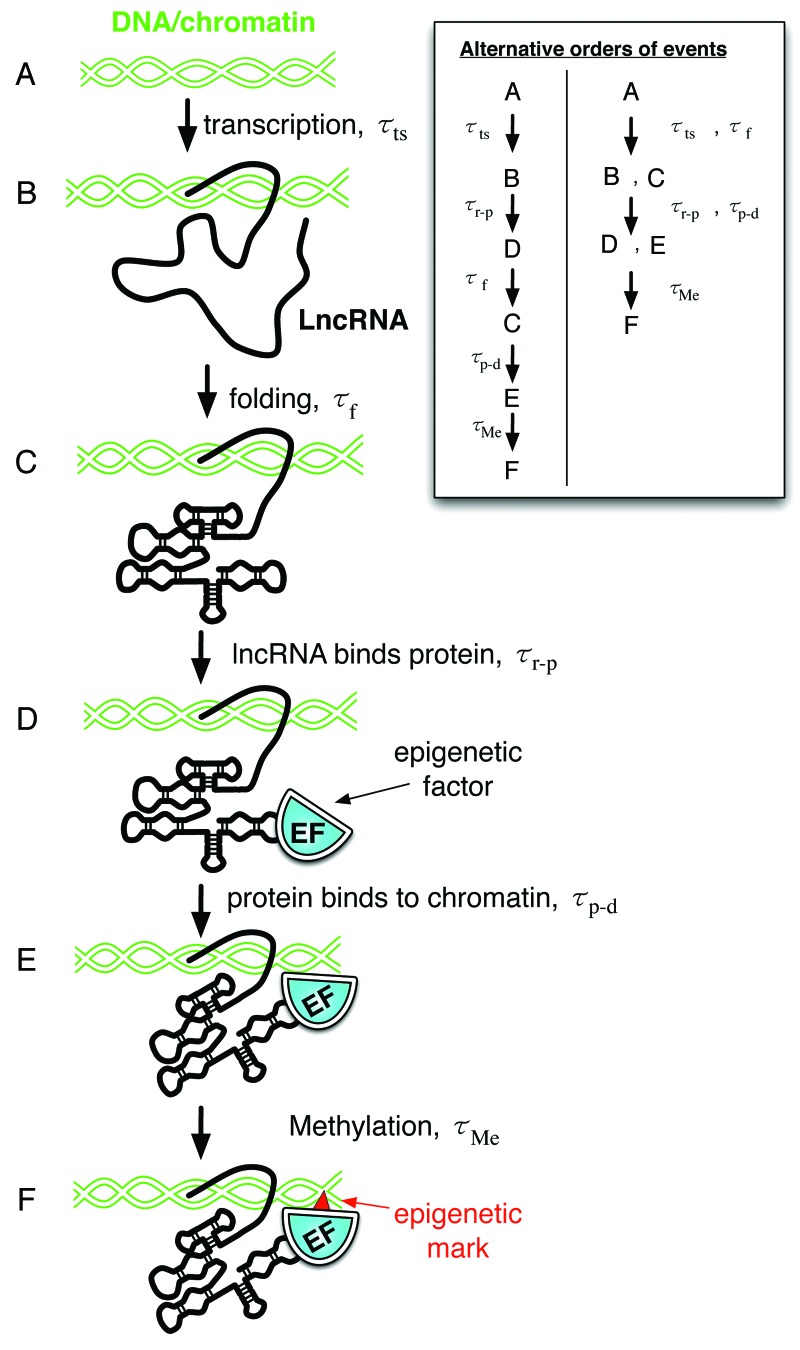
To illustrate potential time scales involved in lncRNA mechanism, we consider the lncRNA DBE-T, a key component of the epigenetic switch associated with Facioscapulohumeral muscular dystrophy (FSHD).37 This lncRNA is a cis-acting tether that recruits the epigenetic factors D4Z4 and Ash1L to the D4Z4 binding element (DBE) on chromatin, driving histone methylation and 4q35 gene transcription. One possible order of events may be (Fig. 4): (A) transcription, (B) lncRNA folding, (C) epigenetic protein binding to the lncRNA, (D) epigenetic protein binding to the chromatin, and (F) action of the epigenetic protein (e.g., histone methylation). Each of these steps will have its own time scale. Identifying the rate-limiting step will yield significant insight into the mechanism of lncRNA action. Other scenarios are also possible, involving different orders of events and different combinations of steps. For other classes of lncRNAs, entirely different events may occur.
Figure 4. An example of potentially relevant time scales for lncRNA activity: DBE-T lncRNA. DBE-T is a cis-acting tether than recruits epigenetic factors to chromatin. Steps af each have an associated timescale, t. (A) Initial state. (B) Transcription of DBE-T. (C) DBE-T folding. (D) The epigenetic factor binds to the lncRNA. (E) The epigenetic factor binds to chromatin. (F) The epigenetic factor marks the chromatin (e.g., histone methylation).
Conclusions
The structural biology of lncRNAs presents a brave new RNA world, where many fundamental questions have not been addressed. With the thousands of new lncRNAs recently discovered in disparate areas of biology, it is likely that a zoo of distinct structural architectures and structural mechanisms will be revealed. These may be sequence-based, secondary structure based, tertiary structure based, or some unusual combination of these. The diverse range of lncRNA structures will be accompanied by a corresponding array of kinetic mechanisms. As RNA molecules are notoriously difficult to crystallize, it may be useful to first apply alternative strategies to gain three-dimensional information about lncRNAs. Ultimately, the identification of common structural features and structure/function relationships will help us understand the role of lncRNAs in development and disease. As with many established therapeutic strategies, mechanistic understanding will help lay the foundation for development of lncRNA-based therapy.
Acknowledgments
This effort is supported by the LANL LDRD program.
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/22592
References
- 1.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–49. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barsotti AM, Prives C. Noncoding RNAs: the missing “linc” in p53-mediated repression. Cell. 2010;142:358–60. doi: 10.1016/j.cell.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 5.Valadkhan S, Nilsen TW. Reprogramming of the non-coding transcriptome during brain development. J Biol. 2010;9:5. doi: 10.1186/jbiol197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28:445–53. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 10.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–90. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Anthony PC, Perez CF, García-García C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci USA. 2012;109:1485–9. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shcherbakova I, Brenowitz M. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nat Protoc. 2008;3:288–302. doi: 10.1038/nprot.2007.533. [DOI] [PubMed] [Google Scholar]
- 13.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–98. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 18.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 19.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–23. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 21.Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, et al. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011;12:1280–5. doi: 10.1038/embor.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr., et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korostowski L, Raval A, Breuer G, Engel N. Enhancer-driven chromatin interactions during development promote escape from silencing by a long non-coding RNA. Epigenetics Chromatin. 2011;4:21. doi: 10.1186/1756-8935-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 25.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziats MN, Rennert OM. (2012). Aberrant Expression of Long Noncoding RNAs in Autistic Brain. Journal of molecular neuroscience: MN [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sati S, Ghosh S, Jain V, Scaria V, Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 33.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–93. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–31. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sclavi B, Woodson S, Sullivan M, Chance M, Brenowitz M. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 1998;295:379–402. doi: 10.1016/S0076-6879(98)95050-9. [DOI] [PubMed] [Google Scholar]
- 41.Appelt K, Dijk J, Reinhardt R, Sanhuesa S, White SW, Wilson KS, et al. The crystallization of ribosomal proteins from the 50 S subunit of the Escherichia coli and Bacillus stearothermophilus ribosome. J Biol Chem. 1981;256:11787–90. [PubMed] [Google Scholar]
- 42.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 43.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 47.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 52.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–66. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beniaminov A, Westhof E, Krol A. Distinctive structures between chimpanzee and human in a brain noncoding RNA. RNA. 2008;14:1270–5. doi: 10.1261/rna.1054608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 57.Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–51. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holbrook SR. RNA structure: the long and the short of it. Curr Opin Struct Biol. 2005;15:302–8. doi: 10.1016/j.sbi.2005.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–45. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Chan RT, Robart AR, Rajashankar KR, Pyle AM, Toor N. Crystal structure of a group II intron in the pre-catalytic state. Nat Struct Mol Biol. 2012;19:555–7. doi: 10.1038/nsmb.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–8. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 64.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA. 2004;101:10024–9. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–9. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–15. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–44. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 70.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–7. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng R, Dye BT, Pérez I, Barnard DC, Thompson AB, Patton JG. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8:1334–47. doi: 10.1017/S1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–9. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 74.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breaker RR. Complex riboswitches. Science. 2008;319:1795–7. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 76.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–6. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 77.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011;39:2416–31. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–97. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J. 2009;96:L7–9. doi: 10.1016/j.bpj.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayes RL, Noel JK, Mohanty U, Whitford PC, Hennelly SP, Onuchic JN, et al. Magnesium fluctuations modulate RNA dynamics in the SAM-I riboswitch. J Am Chem Soc. 2012;134:12043–53. doi: 10.1021/ja301454u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–9. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vaiana AC, Sanbonmatsu KY. Stochastic gating and drug-ribosome interactions. J Mol Biol. 2009;386:648–61. doi: 10.1016/j.jmb.2008.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88:1053–9. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 84.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–77. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–81. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanbonmatsu KY. Computational studies of molecular machines: the ribosome. Curr Opin Struct Biol. 2012;22:168–74. doi: 10.1016/j.sbi.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whitford PC, Onuchic JN, Sanbonmatsu KY. Connecting energy landscapes with experimental rates for aminoacyl-tRNA accommodation in the ribosome. J Am Chem Soc. 2010;132:13170–1. doi: 10.1021/ja1061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem. 2001;70:415–35. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 89.Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem Sci. 2001;26:124–30. doi: 10.1016/S0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 90.Blanchard SC. Single-molecule observations of ribosome function. Curr Opin Struct Biol. 2009;19:103–9. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]