Abstract
Embryonic patterning relies upon an exquisitely timed program of gene regulation. While the regulation of this process via the action of transcription factor networks is well understood, new lines of study have highlighted the importance of a concurrently regulated program of chromatin remodeling during development. Chromatin remodeling refers to the manipulation of the chromatin architecture through rearrangement, repositioning, or restructuring of nucleosomes to either favor or hinder the expression of associated genes. While the role of chromatin remodeling pathways during tumor development and cancer progression are beginning to be clarified, the roles of these pathways in the course of tissue specification, morphogenesis and patterning remains relatively unknown. Further, relatively little is understood as to the mechanism whereby developmentally critical transcription factors coordinate with chromatin remodeling factors to optimize target gene loci for gene expression. Such a mechanism might involve direct transcription factor/chromatin remodeling factor interactions, or could likely be mediated via an unknown intermediary. Our group has identified the relatively unknown protein Akirin as a putative member of this latter group: a secondary cofactor that serves as an interface between a developmentally critical transcription factor and the chromatin remodeling machinery. This role for the Akirin protein suggests a novel regulatory mode for regulating gene expression during development.
Keywords: Akirin, Twist, SWI/SNF, chromatin, transcription, muscle, Drosophila
Akirin: A Conserved Transcription Cofactor
Akirin is a highly conserved 201-residue protein with orthologs identified throughout metazoa including Drosophila, mice, teleosts, and humans.1 The function of the Akirin protein has been enigmatic; despite its high degree of conservation, it remains almost completely devoid of known protein domains, defined catalytic activity, or demonstrable ability to bind DNA.1-3 Nevertheless, Akirin appears to be a crucial regulator of gene expression in nearly every context examined.1-5 Akirin was first identified as a critical positive regulator of NFκB-dependent gene expression in the Drosophila innate immunity pathway.3 Akirin was shown to function in parallel with or downstream of NFκB transcription factor, Relish, for gene activation, despite no detected physical association between Akirin and Relish. Importantly, Akirin appeared to have conserved function in both the Drosophila and mouse immunity pathways, reinforcing the notion that the role of the Akirin protein has been conserved throughout evolution.
Other studies have established that Akirin has a promyogenic role during muscle regeneration in mice.4,5 Akirin1, the mouse homolog of Drosophila akirin, is highly expressed in activated satellite myoblasts during muscle regeneration.5 akirin1 promotes muscle growth presumably through activation of transcription of regulators of the myogenic pathway and is negatively regulated by myostatin signaling.5 Under differentiation conditions, expression of akirin1 mRNA preceded that of myoD by 12 h. Inhibition of akirin1 expression by siRNA led to decreased expression of myogenic regulatory factors (MRFs) such as MyoD, with a corresponding decrease in myoblast differentiation and myotube formation.4 Despite this correlation, the mechanism by which Akirin1 promotes transcription of myoD, as well as downstream targets of the mammalian myogenic pathway, remains unknown.
Akirin Mediates Gene Activation Through Interactions Between Transcription Factors and Chromatin Remodeling Complexes
A number of recent studies are increasingly demonstrating that an organized system of chromatin modification or remodeling plays an important role during embryonic development in a variety of organisms and contexts.6-9 Recent work by our group has determined that akirin is a critical transcriptional co-factor, with important roles in the embryonic development of Drosophila.2 During Drosophila embryogenesis, Akirin interacts with and is critical for proper expression of genes that are regulated by the bHLH transcription factor, Twist. Twist, a key regulator of Drosophila mesodermal development, has numerous roles in the establishment, patterning, and development of the skeletal musculature.10 Correspondingly akirin mutants display a range of phenotypes indicative of defective muscle patterning, including missing muscles, improperly attached muscles, and duplicated muscles. Accordingly, Akirin is required for proper expression of Dmef2, a Twist-regulated myogenic differentiation gene, as Dmef2 transcript levels are highly reduced in akirin mutant embryos.2 This finding was highly reminiscent of previous studies in mammalian myoblasts that observed decreased levels of myoD following akirin1 knockdown by siRNA.4 We further determined that Akirin mediates a physical and functional link with core subunits of the Drosophila SWI/SNF-class Brahma chromatin remodeling complex (BRM).2 In both whole-genome polytene chromosome assays and chromatin immunoprecipitation experiments, Akirin was observed to immunocolocalize with core BRM subunits such as Moira, Snr1, and Brahma. The interactions between Twist and Akirin, and Akirin and core BRM subunits suggested a mechanism whereby Akirin fosters the recruitment or stabilization of BRM chromatin remodeling complexes at Twist-regulated genes.2
In addition to Twist, Akirin is further predicted to interact with a variety of other transcription factors: whole genome yeast 2-hybrid analysis,11 as well as overexpression studies, have suggested interactions between Akirin and Charlatan, a zinc-finger transcription factor involved in development of the embryonic and adult peripheral nervous system,12 and the Drosophila GATA-2 homolog pannier, which is involved in not only PNS development, but also cardiovascular, lymph gland and nephrocyte development.13-15 This variety of transcription factors, contexts, and gene expression pathways suggests a robust and diverse array of Akirin-dependent outputs.
Using Akirin to Link Transcriptional Activation to Chromatin Remodeling
A general mechanism for establishing tissue-, lineage-, and target gene-specific regulation of BRM activity in Drosophila remains poorly understood. In mammals, targeting of tissue-specific SWI/SNF complex activity is often achieved through restricted expression of SWI/SNF complex subunits, differential complex subunit composition, and/or correspondent subunit exchange/replacement.16-18 As an example, human neuronal BAF complexes have been identified that incorporate neuron-specific isoforms of the BAF55 core subunit. This neuronal BAF complex is a critical regulator of the postmitotic neuronal differentiation process.19 Additionally, isoforms of the BAF60 core subunit are each preferentially expressed and incorporated into tissue-specific BAF complexes: loss of BAF60c, for example, leads to defects in cardiac and skeletal myogenesis in mice.20 However, subunit replacement is thus far not an analogous mechanism for generating tissue specificity of BRM complex activity in Drosophila. While the Drosophila genome has only one homolog for each BRM complex subunit, vertebrates often have multiple homologs of each BAF subunit.17 Also, unlike vertebrate and yeast SWI/SNF complexes, all Drosophila cells contain roughly equal numbers of BAP (Osa) and PBAP (Polybromo/BAP170, BAP200 and SAYP) complexes, and a preference of one complex over another in a particular lineage or tissue has not been identified.18
Understandably, the targeting of BRM complex activity to specific gene loci is an area of great interest, and direct interactions of BRM complex subunits with particular transcription factors and coactivators have been identified in Drosophila and other species. For example, the Drosophila BAP-specific subunit Osa directly interacts with Zeste21 as well as with the transcription cofactor Chip.22 In humans PBAF subunits have been shown to bind Serum response factor23 and retinoic acid receptor targets.24 Finally, the Baf60c core subunit has been identified recently as a direct participant in myogenic gene expression.25 In response to differentiation-specific cues, the BAF60c subunit interacts specifically with MyoD transcription factor, and this interaction promotes binding of MyoD to its target genes. This BAF60c-associated binding then marks the locus for recruitment of core SWI/SNF subunits to myogenic genes and promotes gene expression.25
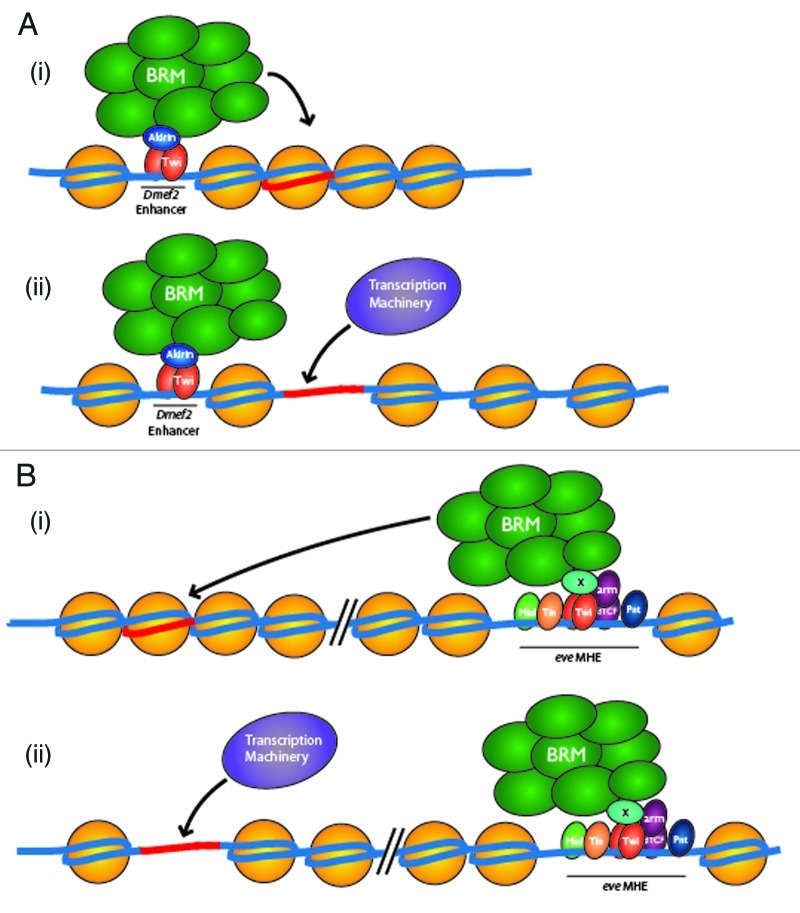
Genetic and chromatin immunoprecipitation data from our study demonstrated that Akirin interacts with BRM core subunits, including Brahma, Moira, Bap60, and Snr1. Nevertheless, Akirin is not a BRM subunit itself but rather a BRM-interacting accessory protein.2 This interaction between Akirin and BRM complex core subunits, together with the apparent selective requirement of Akirin for optimal expression from some, but not all, Twist-regulated genes establishes an intriguing possibility that Akirin might provide a means to provide another level of target specificity for BRM-mediated chromatin remodeling activity at various Twist-regulated loci (Fig. 1). The embryonic distribution of the Akirin protein is widespread, appearing restricted to the nucleus, but found in all tissue layers and derivatives.2 This distribution of Akirin throughout the entire embryo suggests that specificity of Akirin function is determined not by restriction of Akirin localization, but rather by the associated transcription factor with which it interacts. Through this paradigm, the specificity of BRM activity would therefore be achieved through the tissue-specific Akirin-interacting transcription factor. By binding to tissue specific transcription factors such as Twist and promoting their association with the BRM complex, Akirin would serve as a mechanism to confer tissue and/or gene specific information to BRM chromatin remodeling complexes.
Figure 1. Proposed model of action for Akirin during regulation of Twist-dependent genes. (A) Following binding of the Twist transcription factor (Twi) with its target enhancer, Akirin likely mediates an interaction between Twist and the Brahma chromatin remodeling (BRM) (i). This interaction between Twist and BRM would then serve to remodel the local chromatin environment, allowing the general transcription machinery access for expression of Dmef2 (ORF and promoter element indicated in red). (B) Conversely, BRM is located at the even-skipped MHE enhancer in an Akirin-independent manner, either via direct interaction with Twist, interaction with an as-yet-unidentified factor (”X”), or other transcription factors known to bind the MHE element (i). Presumably, this localization of BRM to the MHE element is required to remodel the local chromatin environment for optimal eve expression (ii).
Intriguingly, not all Twist-regulated genes exhibited this Akirin dependence (Fig. 1): while Akirin was largely absent from the Twist-regulated even-skipped MHE enhancer, Moira occupancy was detected at this element.2 One possibility to explain the selective requirement for Akirin at some, but not all Twist-regulated enhancers would be that the two enhancers have distinct chromatin landscapes. Another could be the presence of other transcription factor binding sites at the even-skipped enhancer that make Akirin recruitment/stabilization of BRM redundant. At present, the nature of the chromatin landscape at each of these enhancers over the course of embryonic development remains unclear and something that we are currently exploring. Together, our model (Fig. 1) would suggest that Akirin would work with Twist to improve the local chromatin environment by linking BRM complex activity to Twist-regulated enhancers. This potential role of Akirin as a “chromatin sensor” is an area of considerable interest at present.
Applicability of the Akirin-Twist-BRM Interaction Paradigm to Other Contexts
It remains to be determined if our hypothesized role of Akirin as an interface between specific transcription factors and the SWI/SNF complexes can be extrapolated to other gene expression contexts in other organisms, such as the above described innate immunity pathways and mammalian myogenic gene expression. The high degree of primary sequence conservation, together with the ubiquity of akirin orthologs in higher metazoans, would certainly suggest that Akirin plays a key role in gene regulation across multiple possible contexts. As described above, Akirin/Relish interactions and Akirin-mediated regulation of NFκB gene expression likely occurs via an unknown intermediary.3 Our previous work2 would suggest that Akirin may be interacting with the BRM complex to promote gene activation. To date, a direct association of SWI/SNF with NFκB factors has not been demonstrated. SWI/SNF does not directly interact with NFκB to activate HIV-1 templates in vitro, but its activity is critical for downstream gene expression in this process.26 It remains an intriguing possibility that Akirin would mediate such an interaction between Relish and BRM complexes. This association would presumably lead to chromatin remodeling that produces an environment favorable for NFκB gene expression. In the case of myogenic gene expression, our study2 would suggest that in mammals Akirin1 might mediate an interaction between MRFs and SWI/SNF complexes to promote expression of myogenic genes during muscle patterning and/or regeneration. For added complexity, it has been recently postulated that differing SWI/SNF subcomplexes participate in expression of different myogenic genes.25,27 If true, it is unknown if Akirin1 would exhibit a preference for interaction with a particular composition of SWI/SNF complex in regenerating muscle.
Finally in human cancers, loss of SWI/SNF function via silencing or loss of the BRG1 or BRM1 subunits is emerging as a new mechanism for the promotion of cancer progression, leading to malignant growth (reviewed in28). Additionally, Twist has been identified as an essential control factor for metastasis.29 Our identification of Akirin as a positive cofactor for Twist-dependent gene expression and its link to BRM complex activity presents a new regulatory mechanism of Twist transcription factor activity. It remains to be determined whether Akirin also has a similarly critical role during the process of tumor biogenesis and could present a new target for therapeutics aimed at reversing or inhibiting this process.
Conclusion
Akirin represents a new class of transcription cofactor: a relatively small protein, without catalytic or DNA-binding capability, that links the activities of transcription factors with those of chromatin remodeling complexes to influence gene expression in a context-dependent manner. As such, Akirin represents another potential regulatory facet for control of gene expression. Future studies, aimed at the molecular mechanism of Akirin action, as well as the identification of new Akirin-dependent gene expression pathways will undoubtedly shed further light on this already fascinating yet enigmatic cofactor.
Acknowledgments
The authors wish to thank K. Dobi for essential comments on this manuscript. This work was supported by a Kennesaw State University Mentor/Protege Award 06FY201301 (S.J.N.) and by NIH grant GM56989 and GM78318 (M.K.B.).
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/22907
References
- 1.Macqueen DJ, Johnston IA. Evolution of the multifaceted eukaryotic akirin gene family. BMC Evol Biol. 2009;9:34. doi: 10.1186/1471-2148-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowak SJ, Aihara H, Gonzalez K, Nibu Y, Baylies MK. Akirin links twist-regulated transcription with the Brahma chromatin remodeling complex during embryogenesis. PLoS Genet. 2012;8:e1002547. doi: 10.1371/journal.pgen.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goto A, Matsushita K, Gesellchen V, El Chamy L, Kuttenkeuler D, Takeuchi O, et al. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat Immunol. 2008;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salerno MS, Dyer K, Bracegirdle J, Platt L, Thomas M, Siriett V, et al. Akirin1 (Mighty), a novel promyogenic factor regulates muscle regeneration and cell chemotaxis. Exp Cell Res. 2009;315:2012–21. doi: 10.1016/j.yexcr.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Marshall A, Salerno MS, Thomas M, Davies T, Berry C, Dyer K, et al. Mighty is a novel promyogenic factor in skeletal myogenesis. Exp Cell Res. 2008;314:1013–29. doi: 10.1016/j.yexcr.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A. 2011;108:12758–63. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci U S A. 2011;108:2282–7. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas M, Langley B, Berry C, Sharma M, Kirk S, Bass J, et al. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem. 2000;275:40235–43. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 9.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–56. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 10.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–4. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 11.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 12.Escudero LM, Caminero E, Schulze KL, Bellen HJ, Modolell J. Charlatan, a Zn-finger transcription factor, establishes a novel level of regulation of the proneural achaete/scute genes of Drosophila. Development. 2005;132:1211–22. doi: 10.1242/dev.01691. [DOI] [PubMed] [Google Scholar]
- 13.García-García MJ, Ramain P, Simpson P, Modolell J. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development. 1999;126:3523–32. doi: 10.1242/dev.126.16.3523. [DOI] [PubMed] [Google Scholar]
- 14.Peña-Rangel MT, Rodriguez I, Riesgo-Escovar JR. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics. 2002;160:1035–50. doi: 10.1093/genetics/160.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet. 2004;36:1019–23. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- 16.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 17.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 18.Carrera I, Treisman JE. Message in a nucleus: signaling to the transcriptional machinery. Curr Opin Genet Dev. 2008;18:397–403. doi: 10.1016/j.gde.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 21.Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 2000;14:1058–71. [PMC free article] [PubMed] [Google Scholar]
- 22.Heitzler P, Vanolst L, Biryukova I, Ramain P. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev. 2003;17:591–6. doi: 10.1101/gad.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Azhar G, Zhong Y, Wei JY. Zipzap/p200 is a novel zinc finger protein contributing to cardiac gene regulation. Biochem Biophys Res Commun. 2006;346:794–801. doi: 10.1016/j.bbrc.2006.05.211. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–16. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, et al. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2012;31:301–16. doi: 10.1038/emboj.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–89. doi: 10.1016/S1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 27.Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it’s time to exchange! Exp Cell Res. 2010;316:3073–80. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–30. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]