Abstract
Recently, genome-wide association studies have identified and validated genetic variations associated with urinary bladder cancer (UBC). However, it is still unknown whether the high-risk alleles of several SNPs interact with one another, leading to an even higher disease risk. Additionally, there is no information available on how the UBC risk due to these SNPs compare to the risk of cigarette smoking and to occupational exposure to urinary bladder carcinogens, and whether the same or different SNP combinations are relevant in smokers and non-smokers. To address these questions, we analyzed the genotypes of six SNPs, previously found to be associated with UBC, together with the GSTM1 deletion, in 1,595 UBC cases and 1,760 controls, stratified for smoking habits. We identified the strongest interactions of different orders and tested the stability of their effect by bootstrapping. We found that different SNP combinations were relevant in smokers and non-smokers. In smokers, polymorphisms involved in detoxification of cigarette smoke carcinogens were most relevant (GSTM1, rs11892031), in contrast to those in non-smokers with MYC and APOBEC3A near polymorphisms (rs9642880, rs1014971) being the most influential. Stable combinations of up to three high-risk alleles resulted in higher odds ratios (OR) than the individual SNPs, although the interaction effect was less than additive. The highest stable combination effects resulted in an OR of about 2.0, which is still lower than the ORs of cigarette smoking (here, current smokers' OR: 3.28) and comparable to occupational carcinogen exposure risks which, depending on the workplace, show mostly ORs up to 2.0.
Introduction
Urinary bladder cancer (UBC) is the ninth most common cancer worldwide [1]. The strongest known risk factors include cigarette smoking, occupational exposure to urinary bladder carcinogens, and male gender. It is well established that a deletion variant of the detoxifying phase II metabolizing enzyme glutathione S-transferase M1 (GSTM1), in addition to N-acetyltransferase 2 (NAT2) slow acetylation are associated with increased urinary bladder cancer risk [2]–[6]. Recently, further genetic variants have been identified and validated in several genome-wide association studies [7]–[12] and were extended to occupational exposure [13]–[15].
The recently discovered SNPs and the corresponding genes have already been comprehensively discussed [1]. Briefly, rs1014971 maps to a non-genic region of chromosome 22q13.1 [9] close to CBX6 and APOBEC3A. Chromobox homolog 7 (CBX7) positively regulates E-cadherin expression by interacting with histone deacetylase 2 [16]. This possibly explains why loss of CBX7 expression is associated with a highly malignant phenotype of carcinomas. Overexpression of APOBEC3 genes may lead to genetic instability [17]. Rs11892031 is located on chromosome 2q37 in an intronic region of the UDP-glucuronosyltransferase 1A (UGT1A) locus. UGT1A is a phase II metabolizing enzyme that catalyzes the glucuronidation and elimination of numerous xenobiotics [18], [19]. Rs1495741 (on chromosome 8p22) is known as a tagging SNP of N-acetyltransferase 2 (NAT2) that distinguishes between fast and slow acetylators [20], [21]. Compared to fast acetylators, slow acetylators have an increased bladder cancer risk, probably because of their decreased ability to efficiently detoxify aromatic amines. Rs710521[A] on chromosome 3q28 close to TP63 is associated with urinary bladder cancer risk [7], [14]. TP63 shows strong homology to the tumour suppressor P53 [22,23; review: 1]. Rs8102137 on 19q12 maps to Cyclin E (CCNE1) which controls cell cycle progression at the G1/S transition [24; review: 1]. Rs9642889, 30 kb upstream of the MYC gene on chromosome 8q24.21, confers susceptibility to bladder cancer and influences expression of MYC [7], [13]. The well-known proto oncogene MYC is involved in the control of proliferation and cell cycle progression [25]. Deletion of the detoxifying phase II enzyme glutathione S-transferase M1 (GSTM1) on chromosome 1q13.3 leads to a decreased detoxification of numerous xenobiotics, including polycyclic aromatic hydrocarbons that are known bladder carcinogens [13], [26]. Although the association of each of these SNPs with urinary bladder cancer risk has been validated and confirmed in several independent cohorts, it is still not known if there is an interaction among the high-risk alleles, and if their influence differs between smokers and non-smokers. Therefore, we determined the most influential genetic variants (rs1014971, rs11892031, rs1495741, rs710521, rs8102137, rs9642880, and GSTM1) in 1,595 bladder cancer cases and 1,760 controls. We performed interaction analyses addressing the following questions: Are there specific and stable SNP interactions resulting in higher odds ratios than individual SNPs? If so, are these SNP combinations identical or distinct between smokers and non-smokers? Finally, how high is the combined genetic (SNP-based) risk compared to that of cigarette smoking and occupational exposure? We report that specific SNP combinations show a higher UBC risk than individual SNPs, where distinct SNP combinations confer susceptibility in smokers and non-smokers. These risks are, however, still small when compared to that of cigarette smoking.
Materials and Methods
Ethics Statement
The sample collection by the Leibniz Research Centre for Working Environment and Human Factors (IfADo) was approved by the ethics commission of the Leibniz Research Centre for Working Environment and Human Factors (Ethikkommission des Leibniz-Instituts für Arbeitsforschung an der TU Dortmund) and the institutional review board of the Leibniz Research Centre for Working Environment and Human Factors (Wissenschaftlicher Beirat des Leibniz-Instituts für Arbeitsforschung an der TU Dortmund). All participants provided their written informed consent.
Patients
To investigate whether there is a combined effect of SNPs associated with UBC, a total of 1,595 UBC cases of European descent and 1,760 controls of European descent from four case-control series collected by the Leibniz Research Centre for Working Environment and Human Factors (IfADo) were genotyped at the glutathione S-transferase M1 (GSTM1) and six SNPs (rs1014971, rs11892031, rs1495741, rs710521, rs8102137, rs9642880) previously identified in genome-wide association studies to be associated with UBC [7], [9].
This data set comprised confirmed urinary bladder cancer cases and controls without malignant disease from the Department of Urology, Semmelweis University, Budapest, Hungary (“Hungary”; 246 cases and 78 controls), the Department of Urology, Paul Gerhardt Foundation, Lutherstadt Wittenberg, Germany (“East Germany”; 218 cases and 213 controls), the “West Germany – Ongoing” case-control series conducted at five hospitals (in total, 646 cases and 525 controls), and the “West Germany – Industrial” burdened case-control series (in total, 485 cases –111 UBC cases from the Department of Urology, Klinikum Dortmund, Germany, and 374 UBC cases surveyed for recognition of an occupational disease – and 944 controls). Information on profession obtained by questionnaire was available for the “East Germany” case-control series only (information on profession: 216 cases and 211 controls) [27], [28]. Detailed descriptions of these four case-control series can be found in [15].
Patients’ characteristics, such as distribution of gender, age at diagnosis for cases and age at examination for controls, as well as numbers of cases and controls in the individual case-control series, are summarized in Tables S1, S2, and S3. 101 cases and 37 controls with unknown smoking habits were excluded from the interaction analysis in the study groups, leading to a total of 1,494 cases and 1,723 controls that were finally considered to determine the impact of SNP combinations on the UBC risk.
Polymorphisms
Isolation of genomic DNA of leucocytes was performed according to standard procedures. Genotypes of the SNPs rs1014971, rs11892031, rs1495741, rs710521, rs8102137, and rs9642880 were detected via TaqMan® Assay. Details of the SNPs are given in Appendix S1 and Table S4.
The homozygous GSTM1 deletion was detected by the amplification of the GSTM1 DNA sequence segment with 218 base pairs by means of PCR [29], [30]. After gel-electrophoresis using ethidium bromide, the DNA product was detected using UV light. This method helped determine whether at least one copy of the GSTM1 gene was present or totally missing.
Statistical Analysis
Cigarette smoking was defined as non-smokers, former smokers, i.e. smokers that quit smoking at least one year before diagnosis (cases) or examination (controls), and current smokers. Former and current smokers were pooled together as “ever smokers”. Analyses were performed stratified for non-smokers, former smokers and current smokers as well as for ever smokers. Analyses on the combined ever smokers groups reflect the past exposure to bladder carcinogens accounting for the latency time of bladder cancer of several decades. Age was defined as “age at diagnosis” for the cases and “age at examination” for the control persons.
Deviations from Hardy-Weinberg equilibrium (HWE) were checked in each study group and separately for cases and controls using χ2 tests (for the results, see Table S5). Associations of polymorphisms and smoking habits with UBC were evaluated applying χ2 tests, odds ratios (OR), and 95% confidence intervals (95% CI). Moreover, ORs and 95% CIs adjusted for age, gender, smoking habits, and study site were estimated using logistic regression.
The ORs of the individual polymorphisms, and combinations of these polymorphisms in the total cohort as well as in subgroups defined by the smoking status of the subjects, were determined by considering the dominant and recessive effects of the SNPs. For each interaction of p polymorphisms (p = 2, …, 7), the ten combinations showing the OR with the lowest p-values were identified in each of the subgroups. To check whether it is appropriate to compute p-values for higher-order SNP interactions based on a χ2 distribution with one degree of freedom, we also determined permutation p-values and compared these with the parametric p-values. Additionally, a bootstrap strategy was used to investigate the stability of the ORs of the SNP combinations of different sizes in the subgroups. To achieve this, 500 bootstrap samples were drawn from the respective subgroup and counted to determine how often the top 10 SNP combinations from the original analysis appeared among the top 10, top 20, and top 50 SNP combinations (of the same number of SNPs) from the analyses of the corresponding 500 bootstrap samples.
To test whether the OR of a certain SNP combination differs between the ever smokers and the non-smokers, logistic regression models were fitted containing parameters for the respective SNP combination, smoking status, and the interaction between these two factors. The standard test for the interaction parameter in this logistic regression model was used to test whether the ORs differ significantly between smokers and non-smokers. Details on this and other statistical analyses can be found in Appendix S2.
Population attributable risks (PAR) indicating the proportion of cases that could be attributed to a certain risk factor, and combined PARs for two or more independent risk factors were calculated according to [31]. The PARs of the individual polymorphism were calculated based on adjusted and unadjusted ORs. Combined PARs were determined based on the adjusted ORs of the homozygous and heterozygous vs. the reference genotypes of each SNP. ORs were adjusted for age, gender, smoking habits, study site (in case of combined study groups) and all measured polymorphisms but rs11892031, as this SNP has a rather protective effect in about 16% of the population of European descent [32]. All four study groups were used to determine the PAR due to smoking habits and genetic risk factors in the present study, whereas the PAR for certain professions was based on the “East Germany” case-control series only.
For an overview of UBC risk factors from the literature, we performed an extensive literature search using PubMed. We included the relevant papers on UBC causes in populations of European descent. If possible, we used the given adjusted ORs to determine the PAR from published studies. Otherwise, unadjusted ORs or ORs calculated from the published frequencies were used. Estimation of ORs of combined genetic risk factors was done for varying frequencies assuming a PAR of 30%.
Results
Analysis of ORs of SNP Combinations
Currently, it is unknown whether genetic variants associated with increased UBC risk interact with one another resulting in higher odds ratios (OR) for combinations than for individual SNPs. Therefore, we analyzed the ORs from combinations of up to seven polymorphisms that were previously found to be individually associated with UBC [2], [7], [9], [20]. The ORs as well as the corresponding 95% confidence intervals (95% CI) and p-values for the individual SNPs, determined in the analysis of our total study group and subgroups defined by the smoking habits, are summarized in Table S6.
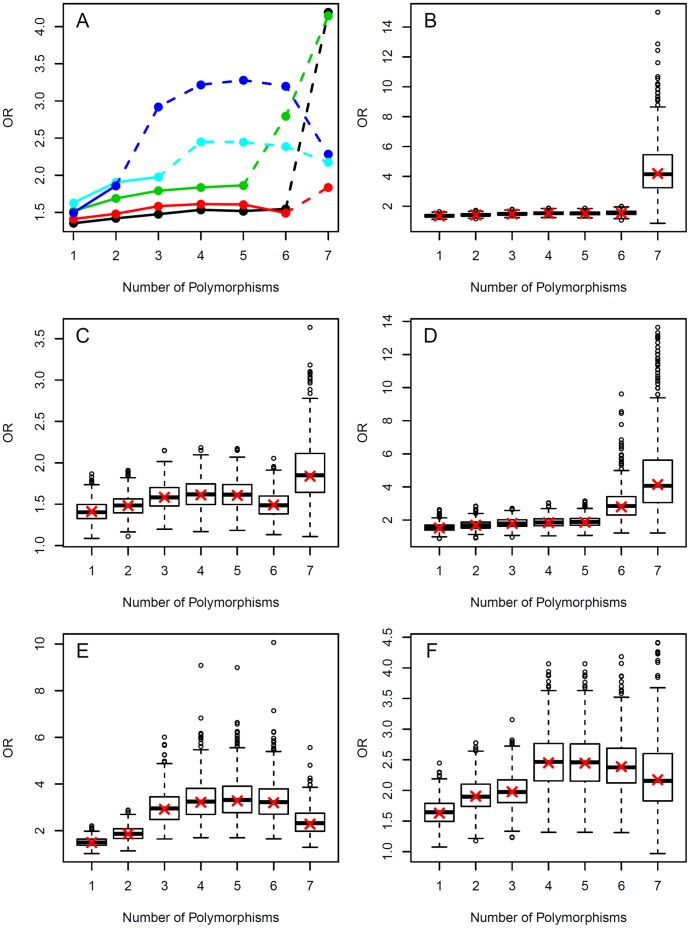
Analyzing the SNP combinations, the ORs of the optimal SNP combinations, in general, increased with the numbers of combined SNPs (Figure 1A). However, case numbers of the high-risk alleles decreased rapidly when several SNPs were combined, thus leading to relatively high variability of the odds ratios in the bootstrap sample (Figure 1B–F). Here the variation typically increased with decreasing number of subjects. In contrast to the ORs, the Wald statistics corresponding to the ORs increased from individual SNPs to combinations of three polymorphisms. However, no further increase was observed (Figure S1), which is again due to high variances and small sample sizes.
Figure 1. Optimal odds ratios for combinations of one to seven polymorphisms.
For the computation of the optimal odds ratios (OR), all possible combinations of one to seven of the polymorphisms rs1014971, rs9642880, rs710521, rs8102137, rs11892031, rs1495741 and GSTM1 were considered. (A) Profile plots for the odds ratios in the total group (black line) and the subgroups of ever smokers (red line), current smokers (green), former smokers (blue) and non-smokers (cyan). The lines were included for clarity of information and not to suggest a continuous development. Dashed lines indicate when number of cases and/or number of controls fall below 100. In these situations, the corresponding odds ratios should be interpreted with caution. (B)–(F): For the optimal combinations shown in (A), box plots of odds ratios computed in 500 bootstrap samples from (B) the total group, (C) the ever smokers, (D) the current smokers, (E) the former smokers and (F) the non-smokers. In twelve of the bootstrap samples (all but one in the analyses of the seven-way interactions in the total and the smoker group), the odds ratios were larger than 15. For a better presentation, these odds ratios are not displayed in the corresponding box plots. The crosses mark the odds ratios of the optimal combinations in the original analysis. The corresponding plots of the test statistics are shown in Figure S1.
In Tables 1, 2, 3 and 4, the ORs with 95% CIs and the p-values of the ten combinations of two and three polymorphisms with the smallest p-values found in the analysis of the ever smokers and the non-smokers are shown. The ORs of the top ten individual effects as well as the top ten two-way and three-way interactions in the total group and in the smoker subgroups are presented in Tables S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19 S20 and S21. Additionally, we summarized how often the seven polymorphisms occur in the top ten two-way and three-way interactions in the different subgroups (Table 5).
Table 1. Top ten two-way interactions found in the analysis of the ever smokers.
| SNP combination | OR (95% CI) | P-value |
| rs11892031 [A/A] × GSTM1 null | 1.48 (1.25–1.76) | 0.0024 |
| rs8102137 [C/T, T/T] × GSTM1 null | 1.51 (1.25–1.82) | 0.0040 |
| rs710521 [A/A, A/G] × GSTM1 null | 1.46 (1.22–1.73) | 0.0062 |
| rs710521 [A/A, A/G] × GSTM1 present | 0.69 (0.58–0.83) | 0.0105 |
| rs9642880 [G/G, G/T] × GSTM1 present | 0.69 (0.57–0.82) | 0.0113 |
| rs11892031 [A/A, A/C] × GSTM1 present | 0.70 (0.59–0.84) | 0.0185 |
| rs11892031 [A/A, A/C] × GSTM1 null | 1.42 (1.19–1.69) | 0.0204 |
| rs1014971 [C/C, C/T] × GSTM1 present | 0.71 (0.60–0.84) | 0.0303 |
| rs1495741 [A/A, A/G] × GSTM1 null | 1.40 (1.18–1.66) | 0.0398 |
| rs1014971 [C/C, C/T] × GSTM1 null | 1.38 (1.16–1.64) | 0.0703 |
The top ten of the 288 possible two-way interactions comprised of the six SNPs and GSTM1 as well as their odds ratios (OR) with 95% confidence intervals (CI) are listed in order of their p-values, where the p-values were adjusted for multiple comparisons by the Bonferroni correction.
Table 2. Top ten two-way interactions found in the analysis of the non-smokers.
| SNP combination | OR (95% CI) | P-value |
| rs9642880 [G/T, T/T] × rs1014971 [C/C] | 1.91 (1.44–2.51) | 0.0015 |
| rs9642880 [G/G, G/T] × rs1014971 [C/T, T/T] | 0.56 (0.43–0.74) | 0.0112 |
| rs710521 [A/A, A/G] × rs1014971 [C/C] | 1.68 (1.28–2.20) | 0.0458 |
| rs1014971 [C/C] × rs1495741 [A/A, A/G] | 1.66 (1.27–2.16) | 0.0524 |
| rs1014971 [C/C] × rs11892031 [A/A, A/C] | 1.65 (1.27–2.16) | 0.0564 |
| rs1014971 [C/T, T/T] × rs8102137 [C/C, C/T] | 0.61 (0.46–0.79) | 0.0640 |
| rs1014971 [C/C] × rs11892031 [A/A] | 1.65 (1.26–2.15) | 0.0761 |
| rs9642880 [T/T] × rs710521 [A/A, A/G] | 1.75 (1.29–2.37) | 0.0827 |
| rs1014971 [C/T, T/T] × rs1495741 [A/A, A/G] | 0.62 (0.47–0.81) | 0.1051 |
| rs1014971 [C/C] × GSTM1 null | 1.73 (1.28–2.35) | 0.1111 |
The top ten of the 288 possible two-way interactions comprised of the six SNPs and GSTM1 as well as their odds ratios (OR) with 95% confidence intervals (CI) are listed in order of their p-values, where the p-values were adjusted for multiple comparisons by the Bonferroni correction.
Table 3. Top ten three-way interactions found in the analysis of the ever smokers.
| SNP combination | OR (95% CI) | P-value |
| rs8102137 [C/T, T/T] × rs11892031 [A/A] × GSTM1 null | 1.58 (1.30–1.92) | 0.0059 |
| rs710521 [A/A, A/G] × rs11892031 [A/A] × GSTM1 null | 1.51 (1.26–1.80) | 0.0080 |
| rs710521 [A/A, A/G] × rs8102137 [C/T, T/T] × GSTM1 null | 1.55 (1.28–1.88) | 0.0135 |
| rs9642880 [G/G, G/T] × rs710521 [A/A, A/G] × GSTM1 present | 0.66 (0.55–0.79) | 0.0171 |
| rs8102137 [C/T, T/T] × rs1495741 [A/A, A/G] × GSTM1 null | 1.52 (1.26–1.84) | 0.0248 |
| rs8102137 [C/T, T/T] × rs11892031 [A/A, A/C] × GSTM1 null | 1.50 (1.25–1.81) | 0.0315 |
| rs1014971 [C/C, C/T] × rs11892031 [A/A] × GSTM1 null | 1.46 (1.22–1.74) | 0.0419 |
| rs9642880 [G/G, G/T] × rs11892031 [A/A, A/C] × GSTM1 present | 0.68 (0.57–0.82) | 0.0520 |
| rs710521 [A/A, A/G] × rs11892031 [A/A, A/C] × GSTM1 null | 1.45 (1.22–1.72) | 0.0552 |
| rs710521 [A/A, A/G] × rs1014971 [C/C, C/T] × GSTM1 present | 0.69 (0.57–0.82) | 0.0582 |
The top ten of the 1,760 possible three-way interactions comprised of the six SNPs and GSTM1, as well as their odds ratios (OR) with 95% confidence intervals (CI) are listed in order of their p-values, where the p-values were adjusted for multiple comparisons by the Bonferroni correction.
Table 4. Top ten three-way interactions found in the analysis of the non-smokers.
| SNP combination | OR (95% CI) | P-value |
| rs9642880 [G/T, T/T] × rs710521 [A/A, A/G] × rs1014971 [C/C] | 1.98 (1.49–2.63) | 0.0044 |
| rs9642880 [G/T, T/T] × rs1014971 [C/C] × rs1495741 [A/A, A/G] | 1.95 (1.47–2.58) | 0.0054 |
| rs9642880 [G/T, T/T] × rs1014971 [C/C] × rs11892031 [A/A, A/C] | 1.93 (1.46–2.55) | 0.0061 |
| rs9642880 [G/T, T/T] × rs1014971 [C/C] × GSTM1 null | 2.21 (1.58–3.10) | 0.0070 |
| rs9642880 [G/G, G/T] × rs1014971 [C/T, T/T] × rs8102137 [C/C, C/T] | 0.54 (0.40–0.71) | 0.0318 |
| rs9642880 [G/G, G/T] × rs710521 [A/A, A/G] × rs1014971 [C/T, T/T] | 0.54 (0.40–0.71) | 0.0325 |
| rs9642880 [G/G, G/T] × rs1014971 [C/T, T/T] × rs1495741 [A/A, A/G] | 0.56 (0.42–0.74) | 0.0735 |
| rs710521 [A/A, A/G] × rs1014971 [C/C] × GSTM1 null | 1.93 (1.41–2.64) | 0.0773 |
| rs9642880 [G/T, T/T] × rs1014971 [C/C] × rs11892031 [A/A] | 1.80 (1.35–2.40) | 0.0954 |
| rs710521 [A/A, A/G] × rs1014971 [C/C] × rs11892031 [A/A] | 1.74 (1.33–2.29) | 0.1142 |
The top ten of the 1,760 possible three-way interactions comprised of the six SNPs and GSTM1, as well as their odds ratios (OR) with 95% confidence intervals (CI) are listed in order of their p-values, where the p-values were adjusted for multiple comparisons by the Bonferroni correction.
Table 5. Number of times the considered polymorphisms appear in the ten top two- and three-way interactions when analyzing the different smoker groups.
| Polymorphism | Total | Ever | Current | Former | Never |
| GSTM1 | 10 (9) | 10 (10) | 10 (10) | 7 (4) | 2 (1) |
| rs11892031 | 6 (3) | 6 (3) | 4 (3) | 3 (2) | 3 (2) |
| rs710521 | 5 (4) | 5 (2) | 4 (2) | 5 (3) | 4 (2) |
| rs9642880 | 5 (3) | 2 (1) | 3 (1) | 8 (7) | 8 (3) |
| rs8102137 | 3 (1) | 4 (1) | 2 (1) | 5 (2) | 1 (1) |
| rs1495741 | 1 (0) | 1 (1) | 0 (0) | 2 (1) | 2 (2) |
| rs1014971 | 0 (0) | 2 (2) | 7 (3) | 0 (1) | 10 (9) |
Numbers in brackets are from the analysis of the two-way interactions. Numbers outside the brackets are from the analysis of the three-way interactions. The corresponding groupwise top ten two-way interactions are listed in Tables S12, S13, S14, S15 and S16, and the top ten three-way interactions are presented in Tables S17, S18, S19, S20 and S21.
Appropriateness of Parametric p-values
Since the p-values were determined using a χ2 distribution with one degree of freedom, we examined the suitability of employing such parametric p-values for combinations of several SNPs by comparing these p-values with the corresponding permutation-based p-values. In addition, we computed both the mean and the variance of the test statistics determined in the 100,000 permutations used in the derivation of the latter p-values. The results of these computations are displayed in the supporting information. Figures S2, S3 and S4 indicate that the χ2 approximation worked well for most of the combinations of two or three SNPs, and in particular, for the respective top ten combinations. However, the χ2 approximation became worse as the number of SNPs forming an interaction increased. Surprisingly, the most extreme differences in p-values for the combinations of two SNPs were larger than the ones for, for example, three-way interactions. This, however, was only relevant for a few combinations.
Stability of the Estimated ORs
The above results, together with the relatively small case numbers in the subgroups of current, former and non-smoker for combinations of more than three SNPs, led us to focus on the interaction of two and three polymorphisms when we analyzed the stability of the ranks of the SNP combinations in the bootstrap samples (Tables S12, S13, S14, S15, S16 and Tables S17, S18, S19, S20 and S21, respectively). The ranks were very stable considering the individual variables coding for the polymorphisms (Tables S7, S8, S9, S10 and S11). In addition, the top two-way interactions occurred among the top ten interactions in a large majority of the bootstrap samples (Tables S12, S13, S14, S15 and S16). However, the instability of the ranks increased with the number of polymorphisms forming a combination (for example, the ranks for three-way SNP combinations in Tables S17, S18, S19, S20 and S21).
Differences in relevant SNP interactions between smokers and non-smokers
Interestingly, different SNP combinations were obtained for non-smokers and smokers. The optimal three-way SNP combinations (resulting in maximal odds ratios) for non-smokers consisted of (i) rs1014971, (ii) rs9642880, and (iii) one of the three SNPs: rs11892031, rs1495741, or rs710521 (Tables 2 and 4 as well as Table 5). In contrast, the optimal combinations for the current smokers were composed of GSTM1, rs1014971, and one of the three SNPs: rs11892031, rs710521, and rs9642880 (Table 5 as well as Tables S14 and S19). A similar result was obtained for the ever smokers in which, however, rs1014971 was only rarely present in the top SNP combinations (Table 5 as well as Tables 1 and 3). This SNP also did not appear in any of the top ten three-way interactions in the former smokers (Table 4 as well as Tables S15 and S20). Interestingly, the former smokers showed a mixed SNP pattern of smokers and non-smokers, including GSTM1 (the top “smoker SNP”), rs9642880 (the second-best scoring “non-smoker SNP”), rs710521 (present in both the smoker and non-smoker SNP combination), as well as rs8102137 (the least or second least important SNP when considering the three-way interactions in non-smokers and current smokers, respectively).
Comparison with Published Results
Considering the genetic risks due to single well-known and novel polymorphisms, ORs range between null and 1.34 in the present study in accordance with the published results from case-controls studies, meta-analyses and GWAS that did not exceed 1.81 (Table 6). Particularly, UBC risks attributed to GSTM1 and NAT2 show a remarkable variation in the literature ranging from 1.28 to 1.70 in case of GSTM1, and no considerable effect to mild risks of 1.43 due to slow NAT2 genotypes not stratified by smoking habits. In terms of relevance for the populations – depending on relative risks and frequency of the risk factors – a considerable fraction of the UBC cases can be attributed to overall genetic risks (30%) or single polymorphisms, in particular GSTM1 with population attributable risks (PAR) ranging from 13% to 26% (Table 6).
Table 6. Population attributable risks and odds ratios due to genetic factors.
| Present study | Published | |||
| Genetic Factors | PAR | OR | PAR | OR |
| All | – | – | 30% [47], [51] | 1.04–1.81 [1], [9], [31], [48] |
| GSTM1 | 13%a | 1.28 | 14–26% [47], [48], [54], [55] | 1.28–1.70 [47], [48], [54], [55] |
| NAT2 | 1%b | 1.02b | 8.2%c [54] | 1.04–1.43 [47], [48], [54], [56] |
| “wimp” SNPs | 33%d | 1.02–1.34e | – | 1.11–1.81 [1], [9], [32] |
| Top 3-way interaction | 16% | 1.48 | – | – |
Population attributable risks (PARs) and odds ratios (ORs) were calculated from the data of the present study and summarized from previously published studies for different genetic factors. Numbers in brackets refer to the publications in which the PARs and ORs were published.
Adjusted for age, gender, smoking habits, all measured SNPs and study site; crude PAR/OR: 16%/1.39; adjusted for age and gender: 16%/1.37; adjusted for all measured SNPs: 15%/1.36.
Adjusted for age, gender, smoking habits, all measured SNPs and study site; crude PAR/OR: 5%/1.09; adjusted for age and gender: 3%/1.05; adjusted for all measured SNPs: 5%/1.10.
Combined PAR, individual SNP OR and PAR adjusted for age, gender, smoking habits and all measured SNPs.
Range of individual SNP OR adjusted for age, gender, smoking habits, all measured SNPs and study site depending on the mode of inheritance.
Comparison of Interaction Effects with Occupational and Environmental Risk
The situation is less clear for risks due to occupational exposure to bladder carcinogens. The risk depends strongly on the population under investigation and time of recruitment, both of which reflects the structure of the local industry and changes in exposure (Table 7). Estimates of overall PARs range from 2–5% for women and 7–10% for men [33], [34] to 20–26% [35]–[37] for highly industrialized areas. Strongly increased risks due to exposure to bladder carcinogens, in particular β-naphthylamine, 4-aminobiphenyl and 4-chloro-o-toluidine, can be found in old studies on highly exposed workers whereas clearly and moderately increased risks are still present but do not exceed ORs of two [38]. Determination of PARs for single professions is hampered by their different frequencies in different regions, though common occupations as painters or hairdressers contribute to 0.2–0.9% of the UBC cases.
Table 7. Population attributable risks and odds ratios for different occupational exposures.
| Present study | Published | ||||
| Increased risk | Occupation/Exposure | PAR | OR | PAR | OR |
| All | – | – | 20–26% [35]–[37] | – | |
| M: 7–10% [33], [34] | |||||
| F: 2–5% [33], [34] | |||||
| Moderately | Painter | 0.89% | 1.38 | 0.7% [33], [34] | 1.17–1.98 [36], [38], [57]–[59] a |
| Hairdresser | – | – | 0.2% [33], [34] | 1.23–2.10 [36], [38], [63] | |
| Coal miner | 2.81% | 1.47 | – | 1.31–2.40 [38], [58], [64], [65] | |
| Clearly | Aluminium Workerb | – | – | – | 1.50–2.34 [36], [66] |
| Rubber Industry | 2.80% | 1.76 | – | 1.29–1.30 [36], [38] | |
| Roofer and Slater | – | – | – | 1.70 [36] | |
| Strongly | Benzidine/β-Naphthylamine | – | – | – | 1.60 [37] |
| Benzidinec | – | – | – | 30–75 [67], [68] | |
| β-Naphthylaminec | – | – | – | 5–200 [68] | |
| 4-Aminobiphenylc | – | – | – | 11%d [68] | |
| 4-Chloro-o-toluidine | – | – | – | 38–90 [68] | |
Population attributable risks (PARs) and odds ratios (ORs) were calculated from the data of the present study and summarized from previously published studies for different occupations and occupational exposures, partly stratified by gender (M: Male, F: Female). Numbers in brackets refer to the publications in which the PARs and ORs were published.
More exactly, Aluminium Workers (Soderberg Processing).
Results from historical studies.
Prevalence in exposed workers.
Most UBC cases can clearly be attributed to cigarette smoking (Table 8; present study PAR: 46%; other studies PAR: 50–56%). While current smokers have an approximately 3-fold risk (present study OR = 3.28, other studies OR = 2.77–4.95) of developing UBC – increasing with amount and time – the UBC risk of former smokers decreases to an OR of about two (present study OR = 2.12, other studies OR = 1.74–2.34). Both subgroups contribute almost equally to the UBC cases in the present study (former smokers PAR = 29%, current smokers PAR = 30%), whereas in published studies estimates of the PAR range from 28–40% for former smokers to 39% in current smokers. Interestingly, among men more UBC cases are attributable to smoking (former 41%, current 55%, ever 66%) than among women (former 17%, current 32%, ever 30%).
Table 8. Population attributable risks and odds ratios in the different smoker groups.
| Present study | Published | |||
| Smokinghabits | PAR | OR | PAR | OR |
| Former smokers | 30%a | 2.12a | 28–40% [48], [53] | 1.74–2.34 [48], [53], [71] |
| M: 41% [69] | M: 2.74 [69] | |||
| F: 17% [70] | F: 1.42 [70] | |||
| Current smokers | 29%b | 3.28b | 39% [48], [53] | 2.77–4.95 [48], [53], [71] |
| M: 55% [69] | M: 4.72 [69] | |||
| F: 32% [70] | F: 1.89 [70] | |||
| Ever smokers | 46%c | 2.47c | 50–56% [48], [52], [53] | 2.61–2.89 [48], [53] |
| M: 66% [69] | M: 3.65 [69] | |||
| F: 30% [70] | F: 1.69 [70] | |||
Population attributable risks (PARs) and odds ratios (ORs) were calculated from the data of the present study and summarized from previously published studies for the different smoker groups, partly stratified by gender (M: Male, F: Female), where non-smokers were used as a reference group having no additional risk. Numbers in brackets refer to the publications in which the PARs and ORs were published.
Adjusted for age and gender; crude PAR/OR: 39%/2.65; adjusted for age, gender, SNPs: 30%/2.15.
Adjusted for age and gender; crude PAR/OR: 29%/3.21; adjusted for age, gender, SNPs: 28%/3.17.
Adjusted for age and gender; crude PAR/OR: 51%/2.83; adjusted for age, gender, SNPs: 46%/2.47.
Discussion
Comparison of the Results from Analyzing Non-smokers and Smokers
The distinct SNP patterns for smokers and non-smokers found in our analysis are remarkable, since the genes closest to the top scoring “smoker variants” are involved in the detoxification of carcinogens in cigarette smoke, whereas the top scoring “non-smoker SNPs” are associated with cell cycle control and DNA stability. The deletion variant of GSTM1, the polymorphism found in our analysis to be the most important in smokers, results in loss of activity of the phase II metabolizing enzyme glutathione S-transferase M1, which is involved in detoxification of numerous polycyclic aromatic hydrocarbons [39], [40]. The second scoring “smoker variant” rs11892031 is located closest to the UGT1A cluster [9]. UDP-glucuronosyltransferase is also a phase II metabolizing enzyme responsible for the conjugation and detoxification of several urinary bladder carcinogens present in cigarette smoke [27], [41]–[45].
In contrast, the two top scoring “non-smoker SNPs” are not involved in carcinogen detoxification. Rs1014971 is located approximately 25 kb centromeric of APOBEC3A, which deaminates cytosine to uracil, thereby playing a role in endogenous mutagenesis [1], [9]. The second, rs9642880 is known to influence the expression of the proto oncogene MYC, which controls transcription of numerous genes involved in proliferation [7], [13]. This scenario suggests that control factors of proliferation and DNA integrity are critical for susceptibility to bladder cancer in non-smokers. In contrast, enzymes detoxifying cigarette smoke carcinogens seem to be of highest relevance in smokers.
Another striking observation is that the three SNPs forming the optimal three-way SNP combination in non-smokers, i.e. rs9642880[G/T, T/T] x rs710521[A/A, A/G] x rs1014971[C/C], differ from the three polymorphisms composing the optimal three-way interaction in ever smokers, i.e. rs8102137[C/T, T/T] x rs11892031[A/A] x GSTM1 null. Moreover, the optimal three-SNP combination in non-smokers results in an OR of 1.98 (95% Cl: 1.49–2.63) that is significantly higher (p-value: 1.78×10−4) than the OR of this combination in the ever smokers (OR: 1.03, 95% CI: 0.86–1.24). Conversely, the optimal three-SNP combination in ever smokers exhibits an OR of 1.58 (95% Cl: 1.30–1.92), which is substantially, but not significantly (p-value: 0.143) higher than the OR of this three-SNP combination in non-smokers (OR: 1.21; 95% CI: 0.90–1.64). However, cigarette smoking is already associated with an OR of 3.28 (95% Cl: 2.67–4.03) when current smokers are compared to non-smokers in our study population (Table 9). This high OR suggests that under conditions of continuous exposure to cigarette smoke carcinogens, the contribution of the “non-smoker SNPs” with their relatively small influence on cell cycle and DNA integrity control, is of minor relevance.
Table 9. Distribution of smoking habits and UBC risk in the present case-control study.
| Smoking Habit (n Ca/n Co) | Cases | Controls | OR (95% CI) | OR adj (95% CI adj) |
| Non-smokers (321/752) | 21% | 44% | (reference) | (reference) |
| Former smokers (742/656) | 50% | 38% | 2.65 (2.24–3.31) | 2.12 (1.78–2.53) |
| Current smokers (431/315) | 29% | 18% | 3.21 (2.64–3.90) | 3.28 (2.67–4.03) |
| Ever smokers (1173/971) | 79% | 56% | 2.83 (2.42–3.31) | 2.47 (2.10–2.90) |
For each of the smoker subgroups containing nCa cases and nCo controls, the odds ratios (OR) and their corresponding 95% confidence intervals (95% CI) were computed, both not adjusted and adjusted for age and gender. The latter odds ratios are abbreviated by OR adj.
Comparison with Published Results
To study the consistency of this observation, we re-visited the data of the genome-wide association study on UBC of Rothman et al. [9] who validated rs9642880 and rs710521 in 3,532 UBC cases and 5,120 controls, and confirmed the impact of the GSTM1 deletion in 2,480 cases and 3,222 controls. Assuming a multiplicative model, they also obtained higher ORs for non-smokers compared to ever smokers for rs9642880 (1.24 for non-smokers versus 1.16 for smokers) and rs11892031 (1.49 versus 1.31). The higher OR for rs9642880 contradicts the study of Kiemeney et al. [7] who reported no association of rs9642880 with smoking habits. Also, the findings of a higher OR for rs11892031 in non-smokers is in contrast to Tang et al. [32] who found a higher risk in ever smokers (OR = 1.28) than in non-smokers (OR = 1.23) based on a subset of study groups from Rothman et al. (GWAS stage 1 [9]). However, no difference was found for rs710521 (1.13 vs. 1.14) in accordance with the discovery GWAS [7], and an opposite trend was shown for rs1014971 (1.11 vs. 1.16) and the NAT2 tagging SNP rs1495741 (1.00 vs. 1.18) in accordance with the assumed higher risk of slow acetylators in smokers. Therefore, the difference should still be interpreted with caution until independent confirmatory data are available.
The association among the GSTM1 null genotype, smoking habits and bladder cancer has been controversial since the first study by Bell et al. in 1993 [2]. In their study, smokers had an OR of 1.8 and non-smokers an OR of 1.3, indicating higher risks in smokers due to the lack of GSTM1. However, recent meta analyses and large or pooled studies found no or only weak evidence for an association between GSTM1 and smoking habits [9], [46]–[48], whereas Rothman et al. [9] reported an even higher OR for non-smokers than for ever smokers (1.71 vs. 1.47). In this context, it should be mentioned that our study groups present a higher proportion of occupationally exposed bladder cancer cases. This may be particularly important for GSTM1. For example, it was shown that bladder cancer patients with occupational histories in coal, iron, and steel industries, i.e. exposure to polycyclic aromatic hydrocarbons, presented with high percentages of GSTM1 null genotypes [49]. Decades after the closure of these industries, the GSTM1 genotypes were equal in both cases and controls (GSTM1 null: 52%) [50].
Gain of Considering SNP Interactions
We have shown that SNP combinations result in less than additive ORs compared to the influence of the individual SNPs. For example, the ORs of the “non-smoker SNPs” rs1014971 and rs9642880 are 1.63 = 1/0.61 and 1.48, respectively, in non-smokers (for all ORs of individual SNPs, see Table S6). In comparison, the combination of both SNPs results in an OR of 1.91 in this subgroup (Table S16), which is larger than the individual effects, but smaller than 1.63+1.48 = 3.11. Adding a third SNP to the rs1014971 × rs9642880 combination results in an increase of only 0.07 (Table S21). The less than additive effect is not surprising considering the relatively high frequencies of the high-risk alleles (rs1014971 [C/C]: 40%; rs9642880 [G/T, T/T]: 71%; rs710521 [A/A, A/G]: 93%) in non-smoking controls and their overlap between individual SNPs (two-way interaction: 27%; three-way interaction 24%). Therefore, it seems unlikely that the addition of further “low impact” or “wimp SNPs” [1] would lead to a relevant increase in the combined ORs in populations of European descent.
Analysis of Population Attributable Risks
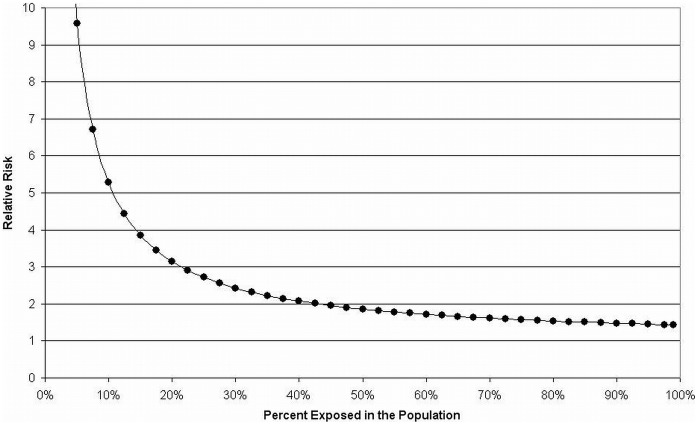
Altogether, it is estimated that up to 30% of bladder cancer cases can be explained by genetic risk factors [47], [51] (see also Table 6), whereas about half of all UBC cases are caused by cigarette smoking [48], [52], [53] (see also Table 8). Estimates of the population attributable risk (PAR) for occupations vary widely, ranging from 7.1% in men and 1.9% in women [34] to 20–26% in both genders [35]–[37] (see also Table 7). The PAR – as a measure of the proportion of cases that could be explained by a certain risk factor – depends on and increases with both the frequency of the risk factor in the population and the relative risk (which is often approximated by the OR). Thus, assuming that the PAR of the genetic risk factors is limited to about 30% in the general population, the OR of the frequent combinations of these polymorphisms must be limited to modest ORs of about two (Figure 2). For instance, a PAR of 30% results from a risk factor present in 40% of the population and a relative risk of 2.1, whereas a risk factor present in 10% of the population requires a relative risk of 5.3 to obtain the same PAR. However, in subgroups different impacts of the genetic risk factors can be observed, not only in terms of relevance of single SNPs and their combinations, but also with respect to their combined attributable risks. In our study, combined PARs for the “wimp SNPs” range from 28% in ever smokers to 43% in non-smokers and also reflect the different impact of genetic risk factors in subpopulations with higher or lower exposure to bladder carcinogens from tobacco smoke.
Figure 2. Relative risks and frequency of risk factors assuming a PAR of 30%.
Relative risks are calculated depending on the frequency of the risk factor in the population assuming a population attributable risk (PAR) of 30%, corresponding to the supposed PAR of genetic risk factors for UBC. Given a PAR of 30%, the relative risk does not fall below 1.43 if the frequency of the risk factor is present in almost the entire population.
Conclusion
In conclusion, we have shown that different types of genetic variants confer different susceptibility to smokers and non-smokers. In addition, the present work fuels the debate regarding the degree to which genetic disposition or environmental exposure contributes to carcinogenesis. Whereas the odds ratio of cigarette smoking is approximately 3.5 for current smokers in most studies, the combined high-risk alleles of the SNPs recently discovered in genome-wide association studies add up to ORs of approximately 2.0. Therefore, the environmental factors seem to have a higher impact on the UBC risk than genetic disposition based on the SNPs derived from recent genome-wide association studies.
Supporting Information
Test statistics for the optimal combinations consisting of one to seven SNPs in the bootstrap samples. Data are shown for (A) the total group, (B) the ever smokers, (C) the current smokers, (D) the former smokers, and (E) the non-smokers. For each of the optimal combinations from in Figure 1A, box plots of the test statistics in the 500 bootstrap samples drawn from the respective subgroups are displayed. The plots correspond to the odds ratios shown in Figure 1B–F. The crosses mark the test statistics for the respective optimal combinations.
(TIFF)
Mean test statistic over 100,000 permutations of the case-control status. For the top 100 SNP combinations of each size and in each subgroup, the means over the Wald statistics in 100,000 permutations of the case-control status were computed. The subgroup-wise distributions of these means are shown as box plots, and the subgroup-wise means of the top 10 combinations are marked by red crosses. For a better representation, six outliers (with means smaller than 0.9) were removed from the box plots for the two-way interactions. For reference, the minimum and maximum of the sample means from 100 samples consisting of 100,000 random draws from a χ2-distribution with 1 degree of freedom are marked by dashed blue lines. If the χ2-approximation is reasonable, the mean test statistic over the 100,000 permutations should be approximately 1, i.e. close to the solid blue lines marking the mean of the χ2-distribution with 1 degree of freedom.
(TIFF)
Variance of the test statistic over 100,000 permutations of the case-control status. In addition to the mean test statistic displayed in Figure S2, the variances of the test statistics for the top 100 interactions in the different subgroups were computed. The subgroup-wise distributions of these variances are shown as box plots, and the variances of the top ten combinations in the subgroups are marked by red crosses. For a better representation, six outliers (with variances smaller than 1.3) were removed from the box plots for the two-way interactions. For reference, the minimum and maximum of the sample variances from 100 samples consisting of 100,000 random draws from a χ2-distribution with 1 degree of freedom are marked by dashed blue lines. If the χ2-approximation is reasonable, the variance of the test statistic over the 100,000 permutations should be approximately 2, i.e. close to the solid blue lines marking the variance of the χ2-distribution with 1 degree of freedom.
(TIFF)
Differences between parametric and permutation-based p-values. Box plots of the differences between the parametric p-values of the top 100 SNP combinations of each size from the analysis of each subgroup and the corresponding p-values computed based on 100,000 permutations of the case-control status. The differences of the respective top ten SNPs are additionally marked by red crosses. Ideally, this difference is zero (which is marked by a dashed blue line). The six outliers removed from Figures S2 and S3 (with means smaller than 0.9 and variances smaller than 1.3) were also removed before constructing the box plots.
(TIFF)
Distribution of gender in the study groups.
(DOC)
Distribution of age at diagnosis (cases) or examination (controls) in the study groups.
(DOC)
Frequency of non-smokers, former smokers and current smokers in the study groups.
(DOC)
Chromosomal and data base information on the six analyzed SNPs.
(DOC)
Testing for Hardy-Weinberg equilibrium.
(DOC)
Maximum odds ratios of the seven polymorphisms in the subgroups.
(DOCX)
Stability of the ranks of the top ten individual effects in the total study group.
(DOC)
Stability of the ranks of the top ten individual effects in the ever smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the current smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the former smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the non-smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the total study group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the ever smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the current smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the former smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the non-smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the total study group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the ever smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the current smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the former smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the non-smoker group.
(DOC)
Details on the polymorphisms.
(DOCX)
Details on the statistical analysis.
(DOCX)
Acknowledgments
The authors thank Ms. Kirsten Liesenhoff-Henze, Ms. Marion Page, and Ms. Claudia Schulte-Dahmann for excellent technical support. We also wish to acknowledge the contribution of our collaborating partners from the Department of Urology, St.-Josefs-Hospital, Dortmund, Germany; Department of Urology and Department of Surgery, Klinikum Dortmund gGmbH, Dortmund, Germany; Department of Urology, Lukaskrankenhaus Neuss, Germany; Department of Urology, Heinrich Heine University Düsseldorf, Germany; Department of Urology, Johannes Gutenberg University Mainz, Germany; Department of Urology, Paul Gerhardt Foundation, Lutherstadt Wittenberg, Germany; Institute for Occupational, Social and Environmental Medicine, Castrop-Rauxel, Germany; Department of Anaesthesia and Critical Care, St. Vincenz Hospital, Menden, Germany; Institute and Outpatient Clinic of Occupational, Social and Environmental Medicine (IPASUM), University of Erlangen-Nuremberg, Erlangen, Germany; Department of Urology, Semmelweis University Budapest, Budapest, Hungary; Practice for Urology, St. Augustin, Germany; Institute for General Medicine, University Hospital of Essen, Essen, Germany.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (Project C4 of the SFB 876 “Providing Information by Resource-Constrained Data Analysis” to KI and grant SCHW 1508/3-1 to HS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Golka K, Selinski S, Lehmann ML, Blaszkewicz M, Marchan R, et al. (2011) Genetic variants in urinary bladder cancer: collective power of the “wimp SNPs”. Arch Toxicol 85: 539–554. [DOI] [PubMed] [Google Scholar]
- 2. Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW (1993) Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst 85: 1159–1164. [DOI] [PubMed] [Google Scholar]
- 3. Cartwright RA, Glashan RW, Rogers HJ, Ahmad RA, Barham-Hall D, et al. (1984) Role of N-acetyltransferase phenotypes in bladder carcinogenesis: a pharmacogenetic epidemiological approach to bladder cancer. Lancet 2: 842–845. [DOI] [PubMed] [Google Scholar]
- 4. Golka K, Prior V, Blaszkewicz M, Cascorbi I, Schöps W, et al. (1996) Occupational history and genetic N-acetyltransferase polymorphism in urothelial cancer patients of Leverkusen, Germany. Scand J Work Environ Health 22: 332–338. [DOI] [PubMed] [Google Scholar]
- 5. Kempkes M, Golka K, Reich S, Reckwitz T, Bolt HM (1996) Glutathione S-transferase GSTM1 and GSTT1 null genotypes as potential risk factors for urothelial cancer of the bladder. Arch Toxicol 71: 123–126. [DOI] [PubMed] [Google Scholar]
- 6. Hengstler JG, Arand M, Herrero ME, Oesch F (1998) Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res 154: 47–85. [DOI] [PubMed] [Google Scholar]
- 7. Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, et al. (2008) Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 40: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kiemeney LA, Sulem P, Besenbacher S, Vermeulen SH, Sigurdsson A, et al. (2010) A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet 42: 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, et al. (2010) A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet 42: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, et al. (2009) Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet 41: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rafnar T, Vermeulen SH, Sulem P, Thorleifsson G, Aben KK, et al. (2011) European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum Mol Genet 20: 4268–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, et al. (2009) Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet 41, 991–995. Erratum in: Nat Genet 41: 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Golka K, Hermes M, Selinski S, Blaszkewicz M, Bolt HM, et al. (2009) Susceptibility to urinary bladder cancer: relevance of rs9642880[T], GSTM1 0/0 and occupational exposure. Pharmacogenet Genomics 19: 903–906. [DOI] [PubMed] [Google Scholar]
- 14. Lehmann ML, Selinski S, Blaszkewicz M, Orlich M, Ovsiannikov D, et al. (2010) Rs710521[A] on chromosome 3q28 close to TP63 is associated with increased urinary bladder cancer risk. Arch Toxicol 84: 967–978. [DOI] [PubMed] [Google Scholar]
- 15. Selinski S, Lehmann ML, Blaszkewicz M, Ovsiannikov D, Moormann O, et al. (2012) Rs11892031[A] on chromosome 2q37 in an intronic region of the UGT1A locus is associated with urinary bladder cancer risk. Arch Toxicol 86: 1369–1378. [DOI] [PubMed] [Google Scholar]
- 16. Federico A, Pallante P, Bianco M, Ferraro A, Esposito F, et al. (2009) Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein. Cancer Res 69: 7079–7087. [DOI] [PubMed] [Google Scholar]
- 17. Vartanian J-P, Guetard D, Henry M, Wain-Hobson S (2008) Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320: 230–233. [DOI] [PubMed] [Google Scholar]
- 18. Hengstler JG, Utesch D, Steinberg P, Platt KL, Diener B, et al. (2000) Cryopreserved primary hepatocytes as a constantly available in vitro model for the evaluation of human and animal drug metabolism and enzyme induction. Drug Metab Rev 32: 81–118. [DOI] [PubMed] [Google Scholar]
- 19. Strassburg CP, Lankisch TO, Manns MP, Ehmer U (2008) Family 1 uridine-5-diphosphate glucuronosyltransferases (UGT1A): from Gilbert’s syndrome to genetic organization and variability. Arch Toxicol 82: 415–433. [DOI] [PubMed] [Google Scholar]
- 20. García-Closas M, Hein DW, Silverman D, Malats N, Yeager M, et al. (2011) A single nucleotide polymorphism tags variation in the arylamine N-acetyltransferase 2 phenotype in populations of European background. Pharmacogenet Genomics 21: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Selinski S, Blaszkewicz M, Lehmann ML, Ovsiannikov D, Moormann O, et al. (2011) Genotyping NAT2 with only two SNPs (rs1041983 and rs1801280) outperforms the tagging SNP rs1495741 and is equivalent to the conventional 7-SNP NAT2 genotype. Pharmacogenet Genomics 21: 673–678. [DOI] [PubMed] [Google Scholar]
- 22. Lefkimmiatis K, Caratozzolo MF, Merlo P, D'Erchia AM, Navarro B, et al. (2009) p73 and p63 sustain cellular growth by transcriptional activation of cell cycle progression genes. Cancer Res 69: 8563–8571. [DOI] [PubMed] [Google Scholar]
- 23. Sayan BS, Sayan AE, Yang AL, Aqeilan RI, Candi E, et al. (2007) Cleavage of the transactivation-inhibitory domain of p63 by caspases enhances apoptosis. Proc Natl Acad Sci USA 104: 10871–10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, et al. (1991) Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66: 1217–1228. [DOI] [PubMed] [Google Scholar]
- 25. Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, et al. (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451. [DOI] [PubMed] [Google Scholar]
- 26. Bolt HM, Thier R (2006) Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab 7: 613–628. [DOI] [PubMed] [Google Scholar]
- 27. Zimmermann A, Blaszkewicz M, Roth G, Seidel T, Dietrich H, et al. (2008) UDP-glucuronosyltransferase 2B7 C802T (His268Tyr) polymorphism in bladder cancer cases. J Toxicol Environ Health A 71: 911–914. [DOI] [PubMed] [Google Scholar]
- 28. Golka K, Abreu-Villaca Y, Anbari Attar R, Angeli-Greaves M, Aslam M, et al. (2012) Bladder cancer documentation of causes: multilingual questionnaire “Bladder Cancer Doc”. Front Biosci E4: 2709–2722. [DOI] [PubMed] [Google Scholar]
- 29. Arand M, Mühlbauer R, Hengstler J, Jäger E, Fuchs J, et al. (1996) A multiplex polymerase chain reaction protocol for the simultaneous analysis of the glutathione S-transferase GSTM1 and GSTT1 polymorphisms. Anal Biochem 236: 184–186. [DOI] [PubMed] [Google Scholar]
- 30.Krause G, Müller M, Lewalter J, Schulz T (2004) Glutathione S-transferase T1 and M1 (GSTT1, GSTM1) (genotyping). In: Angerer J, Müller, M, editors. Analyses of hazardous substances in biological materials, Vol 9, Special issue: Marker of susceptibility. Weinheim, Germany: Wiley-VCH Verlag GmbH. 183–210.
- 31. Steenland K, Armstrong B (2006) An overview of methods for calculating the burden of disease due to specific risk factors. Epidemiology 17: 512–519. [DOI] [PubMed] [Google Scholar]
- 32. Tang W, Fu YP, Figueroa JD, Malats N, Garcia-Closas M, et al. (2012) Mapping of the UGT1A locus identifies an uncommon coding variant that affects mRNA expression and protects from bladder cancer. Hum Mol Genet 21: 1918–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doll R, Peto R (1981) The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 66: 1191–1308. [PubMed] [Google Scholar]
- 34. Rushton L, Bagga S, Bevan R, Brown TP, Cherrie JW, et al. (2010) Occupation and cancer in Britain. Br J Cancer 102: 1428–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delclos GL, Lerner SP (2008) Occupational risk factors. Scand J Urol Nephrol 42 (Suppl 218)58–63. [DOI] [PubMed] [Google Scholar]
- 36. Silverman DT, Levin LI, Hoover RN, Hartge P (1989) Occupational risks of bladder cancer in the United States: I. White men. J Natl Cancer Inst 81: 1472–1480. [DOI] [PubMed] [Google Scholar]
- 37. Vineis P, Pirastu R (1997) Aromatic amines and cancer. Cancer Causes Control 8: 346–355. [DOI] [PubMed] [Google Scholar]
- 38. Reulen RC, Kellen E, Buntinx F, Brinkman M, Zeegers MP (2008) A meta-analysis on the association between bladder cancer and occupation. Scand J Urol Nephrol 42 (Suppl 218)64–78. [DOI] [PubMed] [Google Scholar]
- 39. Ketterer B, Harris JM, Talaska G, Meyer DJ, Pemble SE, et al. (1992) The human glutathione S-transferase supergene family, its polymorphism, and its effects on susceptibility to lung cancer. Environ Health Perspect 98: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee KH, Lee J, Ha M, Choi JW, Cho SH, et al. (2002) Influence of polymorphism of GSTM1 gene on association between glycophorin a mutant frequency and urinary PAH metabolites in incineration workers. J Toxicol Environ Health A 65: 355–363. [DOI] [PubMed] [Google Scholar]
- 41. Carreón T, LeMasters GK, Ruder AM, Schulte PA (2006) The genetic and environmental factors involved in benzidine metabolism and bladder carcinogenesis in exposed workers. Front Biosci 11: 2889–2902. [DOI] [PubMed] [Google Scholar]
- 42. Giuliani L, Ciotti M, Stoppacciaro A, Pasquini A, Silvestri I, et al. (2005) UDP-glucuronosyltransferases 1A expression in human urinary bladder and colon cancer by immunohistochemistry. Oncol Rep 13: 185–191. [PubMed] [Google Scholar]
- 43. Giuliani L, Gazzaniga P, Caporuscio F, Ciotti M, Frati L, Aglianò AM (2001) Can down-regulation of UDP-glucuronosyltransferases in the urinary bladder tissue impact the risk of chemical carcinogenesis? Int J Cancer 91: 141–143. [DOI] [PubMed] [Google Scholar]
- 44. Lin GF, Guo WC, Chen JG, Qin YQ, Golka K, et al. (2005) An association of UDP-glucuronosyltransferase 2B7 C802T (His268Tyr) polymorphism with bladder cancer in benzidine-exposed workers in China. Toxicol Sci 85: 502–506. [DOI] [PubMed] [Google Scholar]
- 45. Zenser TV, Lakshmi VM, Hsu FF, Davis BB (2002) Metabolism of N-acetylbenzidine and initiation of bladder cancer. Mutat Res 506–507: 29–40. [DOI] [PubMed] [Google Scholar]
- 46. Engel LS, Taioli E, Pfeiffer R, Garcia-Closas M, Marcus PM, et al. (2002) Pooled analysis and meta-analysis of glutathione S-transferase M1 and bladder cancer: a HuGE review. Am J Epidemiol 156: 95–109. [DOI] [PubMed] [Google Scholar]
- 47. García-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, et al. (2005) NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 366: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moore LE, Baris DR, Figueroa JD, Garcia-Closas M, Karagas MR, et al. (2011) GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis 32: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Golka K, Reckwitz T, Kempkes M, Cascorbi II, Blaskewicz M, et al. (1997) N-Acetyltransferase 2 (NAT2) and glutathione S-transferase µ (GSTM1) in bladder-cancer patients in a highly industrialized area. Int J Occup Environ Health 3: 105–110. [DOI] [PubMed] [Google Scholar]
- 50. Ovsiannikov D, Selinski S, Lehmann ML, Blaszkewicz M, Moormann O, et al. (2012) Polymorphic enzymes, urinary bladder cancer risk, and structural change in the local industry. J Toxicol Environ Health A 75: 557–565. [DOI] [PubMed] [Google Scholar]
- 51. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. (2000) Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78–85. [DOI] [PubMed] [Google Scholar]
- 52. Yu MC, Skipper PL, Tannenbaum SR, Chan KK, Ross RK (2002) Arylamine exposures and bladder cancer risk. Mutat Res 506–507: 21–28. [DOI] [PubMed] [Google Scholar]
- 53. Puente D, Hartge P, Greiser E, Cantor KP, King WD, et al. (2006) A pooled analysis of bladder cancer case-control studies evaluating smoking in men and women. Cancer Causes Control 17: 71–79. [DOI] [PubMed] [Google Scholar]
- 54. Vineis P (2002) The relationship between polymorphisms of xenobiotic metabolizing enzymes and susceptibility to cancer. Toxicology 181–182: 457–462. [DOI] [PubMed] [Google Scholar]
- 55. Zhang R, Xu G, Chen W, Zhang W (2011) Genetic polymorphisms of glutathione S-transferase M1 and bladder cancer risk: a meta-analysis of 26 studies. Mol Biol Rep 38: 2491–2497. [DOI] [PubMed] [Google Scholar]
- 56. Vineis P, Marinelli D, Autrup H, Brockmoller J, Cascorbi I, et al. (2001) Current smoking, occupation, N-acetyltransferase-2 and bladder cancer: a pooled analysis of genotype-based studies. Cancer Epidemiol Biomarkers Prev 10: 1249–1252. [PubMed] [Google Scholar]
- 57.Bolm-Audorff U, Jöckel KH, Kilguss B, Pohlabeln H, Siepenkothen T (1993). Bösartige Tumoren der ableitenden Harnwege und Risiken am Arbeitsplatz. Scientific report Fb 697 from the report series of the Federal Institute for Occupational Safety and Health, Dortmund, Germany. Bremerhaven, Germany: Wissenschaftsverlag NW. In German.
- 58.Golka K, Bandel T, Reckwitz T, Urfer W, Bolt HM, et al.. (1999) Occupational risk factors for bladder carcinoma. A case control study. Urologe A 38: 358–363. In German. [DOI] [PubMed]
- 59. Guha N, Steenland NK, Merletti F, Altieri A, Cogliano V, Straif K (2010) Bladder cancer risk in painters: a meta-analysis. Occup Environ Med 67: 568–573. [DOI] [PubMed] [Google Scholar]
- 60. Golka K, Bandel T, Schlaefke S, Reich SE, Reckwitz T, et al. (1998) Urothelial cancer of the bladder in an area of former coal, iron, and steel industries in Germany: a case-control study. Int J Occup Environ Health 4: 79–84. [DOI] [PubMed] [Google Scholar]
- 61.Golka K, Goebell PJ, Rettenmeier AW (2007) Bladder cancer: etiology and prevention. Dtsch Arztebl 104: A 719–723.
- 62. Myslak ZW, Bolt HM, Brockmann W (1991) Tumors of the urinary bladder in painters: a case-control study. Am J Ind Med 19: 705–713. [DOI] [PubMed] [Google Scholar]
- 63. Colt JS, Baris D, Stewart P, Schned AR, Heaney JA, et al. (2004) Occupation and bladder cancer risk in a population-based case-control study in New Hampshire. Cancer Causes Control 15: 759–769. [DOI] [PubMed] [Google Scholar]
- 64. Cordier S, Clavel J, Limasset JC, Boccon-Gibod L, Le Moual N, et al. (1993) Occupational risks of bladder cancer in France: a multicentre case-control study. Int J Epidemiol 22: 403–411. [DOI] [PubMed] [Google Scholar]
- 65. Schifflers E, Jamart J, Renard V (1987) Tobacco and occupation as risk factors in bladder cancer: a case-control study in southern Belgium. Int J Cancer 39: 287–292. [DOI] [PubMed] [Google Scholar]
- 66. Thériault G, Tremblay C, Cordier S, Gingras S (1984) Bladder cancer in the aluminium industry. Lancet 1: 947–950. [DOI] [PubMed] [Google Scholar]
- 67. Golka K, Prior V, Blaszkewicz M, Bolt HM (2002) The enhanced bladder cancer susceptibility of NAT2 slow acetylators towards aromatic amines: a review considering ethnic differences. Toxicol Lett 128: 229–241. [DOI] [PubMed] [Google Scholar]
- 68. Golka K, Wiese A, Assennato G, Bolt HM (2004) Occupational exposure and urological cancer. World J Urol 21: 382–391. [DOI] [PubMed] [Google Scholar]
- 69. Brennan P, Bogillot O, Cordier S, Greiser E, Schill W, et al. (2000) Cigarette smoking and bladder cancer in men: a pooled analysis of 11 case-control studies. Int J Cancer 86: 289–294. [DOI] [PubMed] [Google Scholar]
- 70. Brennan P, Bogillot O, Greiser E, Chang-Claude J, Wahrendorf J, et al. (2001) The contribution of cigarette smoking to bladder cancer in women (pooled European data). Cancer Causes Control 12: 411–417. [DOI] [PubMed] [Google Scholar]
- 71. Gandini S, Botteri E, Iodice S, Boniol M, Lowenfels AB, et al. (2008) Tobacco smoking and cancer: a meta-analysis. Int J Cancer 122: 155–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Test statistics for the optimal combinations consisting of one to seven SNPs in the bootstrap samples. Data are shown for (A) the total group, (B) the ever smokers, (C) the current smokers, (D) the former smokers, and (E) the non-smokers. For each of the optimal combinations from in Figure 1A, box plots of the test statistics in the 500 bootstrap samples drawn from the respective subgroups are displayed. The plots correspond to the odds ratios shown in Figure 1B–F. The crosses mark the test statistics for the respective optimal combinations.
(TIFF)
Mean test statistic over 100,000 permutations of the case-control status. For the top 100 SNP combinations of each size and in each subgroup, the means over the Wald statistics in 100,000 permutations of the case-control status were computed. The subgroup-wise distributions of these means are shown as box plots, and the subgroup-wise means of the top 10 combinations are marked by red crosses. For a better representation, six outliers (with means smaller than 0.9) were removed from the box plots for the two-way interactions. For reference, the minimum and maximum of the sample means from 100 samples consisting of 100,000 random draws from a χ2-distribution with 1 degree of freedom are marked by dashed blue lines. If the χ2-approximation is reasonable, the mean test statistic over the 100,000 permutations should be approximately 1, i.e. close to the solid blue lines marking the mean of the χ2-distribution with 1 degree of freedom.
(TIFF)
Variance of the test statistic over 100,000 permutations of the case-control status. In addition to the mean test statistic displayed in Figure S2, the variances of the test statistics for the top 100 interactions in the different subgroups were computed. The subgroup-wise distributions of these variances are shown as box plots, and the variances of the top ten combinations in the subgroups are marked by red crosses. For a better representation, six outliers (with variances smaller than 1.3) were removed from the box plots for the two-way interactions. For reference, the minimum and maximum of the sample variances from 100 samples consisting of 100,000 random draws from a χ2-distribution with 1 degree of freedom are marked by dashed blue lines. If the χ2-approximation is reasonable, the variance of the test statistic over the 100,000 permutations should be approximately 2, i.e. close to the solid blue lines marking the variance of the χ2-distribution with 1 degree of freedom.
(TIFF)
Differences between parametric and permutation-based p-values. Box plots of the differences between the parametric p-values of the top 100 SNP combinations of each size from the analysis of each subgroup and the corresponding p-values computed based on 100,000 permutations of the case-control status. The differences of the respective top ten SNPs are additionally marked by red crosses. Ideally, this difference is zero (which is marked by a dashed blue line). The six outliers removed from Figures S2 and S3 (with means smaller than 0.9 and variances smaller than 1.3) were also removed before constructing the box plots.
(TIFF)
Distribution of gender in the study groups.
(DOC)
Distribution of age at diagnosis (cases) or examination (controls) in the study groups.
(DOC)
Frequency of non-smokers, former smokers and current smokers in the study groups.
(DOC)
Chromosomal and data base information on the six analyzed SNPs.
(DOC)
Testing for Hardy-Weinberg equilibrium.
(DOC)
Maximum odds ratios of the seven polymorphisms in the subgroups.
(DOCX)
Stability of the ranks of the top ten individual effects in the total study group.
(DOC)
Stability of the ranks of the top ten individual effects in the ever smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the current smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the former smoker group.
(DOC)
Stability of the ranks of the top ten individual effects in the non-smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the total study group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the ever smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the current smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the former smoker group.
(DOC)
Stability of the ranks of the top ten two-way interactions in the non-smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the total study group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the ever smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the current smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the former smoker group.
(DOC)
Stability of the ranks of the top ten three-way interactions in the non-smoker group.
(DOC)
Details on the polymorphisms.
(DOCX)
Details on the statistical analysis.
(DOCX)