Abstract
Identifying genes that influence behavioral responses to alcohol is critical for understanding the molecular basis of alcoholism and ultimately developing therapeutic interventions for the disease. Using an integrated approach that combined the power of the Drosophila, C. elegans and mouse model systems with bioinformatics analyses, we established a novel, conserved role for Chloride Intracellular Channels (CLICs) in alcohol-related behavior. CLIC proteins might have several biochemical functions including intracellular chloride channel activity, modulation of TGF-β signaling, and regulation of ryanodine receptors and A-kinase anchoring proteins. We initially identified vertebrate Clic4 as a candidate ethanol-responsive gene via bioinformatic analysis of data from published microarray studies of mouse and human ethanol-related genes. We confirmed that Clic4 expression was increased by ethanol treatment in mouse prefrontal cortex and also uncovered a correlation between basal expression of Clic4 in prefrontal cortex and the locomotor activating and sedating properties of ethanol across the BXD mouse genetic reference panel. Furthermore, we found that disruption of the sole Clic Drosophila orthologue significantly blunted sensitivity to alcohol in flies, that mutations in two C. elegans Clic orthologues, exc-4 and exl-1, altered behavioral responses to acute ethanol in worms, and that viral-mediated overexpression of Clic4 in mouse brain decreased the sedating properties of ethanol. Together, our studies demonstrate key roles for Clic genes in behavioral responses to acute alcohol in Drosophila, C. elegans and mice.
Keywords: alcohol, invertebrate, vertebrate, sensitivity, tolerance, genetics
Introduction
Alcohol abuse has broad negative effects on human health (Dhhs, 2000). Despite these health consequences, few effective treatments are available and currently there are few genes with established roles in human alcohol abuse (Bierut et al., 2010, Edenberg et al., 2010, Johnson et al., 2006, Kendler et al., 2011, Kumar et al., 2009, Lind et al., 2010, Treutlein et al., 2009). A more comprehensive understanding of the genes that influence behavioral responses to alcohol is critical for meaningfully predicting the potential of an individual to abuse the drug and for developing new therapeutic strategies aimed at novel molecular targets.
Short-term exposure to moderate doses of alcohol causes sedation in humans, mice, fruit flies (Drosophila melanogaster) and nematode worms (Caenorhabditis elegans), whereas repeated or longer-term exposure to ethanol leads to tolerance in these same species (Crabbe et al., 2006, Davies et al., 2004, Davies et al., 2003, Heberlein, 2000, Kapfhamer et al., 2008, Scholz et al., 2000). Given the conservation in behavioral responses to ethanol, animal models have been used to investigate genetic pathways that influence behavioral responses to the drug. Forward genetic strategies have identified genes important for ethanol-related behaviors in flies (Scholz et al., 2005), worms (Davies et al., 2003) and, more recently, mice (Kapfhamer et al., 2008). Reverse genetic approaches in flies and mice have also been used to examine the influence of genes predicted to play roles in behavioral responses to ethanol (Crabbe et al., 2006, Rodan & Rothenfluh, 2010). Interestingly, flies harboring putative alleles of ethanol responsive genes (Kong et al., 2009, Morozova et al., 2006) or genes that are differentially expressed in flies artificially selected for ethanol sensitivity (Morozova et al., 2007) exhibit altered ethanol-related behaviors. Thus, ethanol-responsive genes (genes that change expression in response to ethanol) or genes with expression levels that correlate with ethanol phenotypes are excellent candidate loci for influencing behavioral responses to the drug. Understandably, the vast majority of published studies have investigated the role of a gene or pathway of interest in a single species only, although there are several notable exceptions (Corl et al., 2009, Kapfhamer et al., 2008, Lasek et al., 2011a, Lasek et al., 2011b, Schumann et al., 2011). The general lack of cross-species studies, however, leaves unresolved whether many genes that influence ethanol-related behavior in one species have effects in others.
We took a two-step approach to identify novel genes with roles in alcohol behavior. First, we developed a list of candidate genes by analyzing ethanol-responsive loci in several microarray studies in mice and humans (Kerns et al., 2005, Liu et al., 2006, Mayfield et al., 2002, Mulligan et al., 2006). Second, we investigated the consequences of genetic manipulation of several of these genes on acute behavioral responses to alcohol in Drosophila, C. elegans and mice. Here, we report our studies on multiple members of the Chloride Intracellular Channel (Clic) family of genes and their effects on alcohol related behavior. CLICs are typically small proteins with single GST-CLIC domains at the N- and C-termini. CLICs have several proposed biochemical functions including intracellular chloride channel activity (Ashley, 2003), modulation of TGF-β or bone morphogenic protein (BMP) signaling (Shukla et al., 2009), and regulation of ryanodine receptors (Jalilian et al., 2008), 14-3-3 proteins (Suginta et al., 2001) and A-kinase anchoring proteins (Shanks et al., 2002). The Clic gene family is represented in mammals, flies and worms by six (Clic1–Clic6), one (Clic) and two (exc-4 and exl-1) loci, respectively. Our studies establish that members of the Clic gene family are key modulators of ethanol behavior in flies, worms and mice.
Materials and Methods
Mouse husbandry
Mice were maintained in a room under controlled temperature (23±1°C) with 12 h light/dark cycles and free access to standard chow (Harlan Teklad #7912, Madison, WI) and water. Cages (4 mice/cage until viral injections and then single housing thereafter) and bedding (Harlan Sani-chips, #7090A, Harlan, Teklad, Madison, WI) were changed weekly. All experimental manipulations or behavioral testing were done between 0900 and 1200 h. Procedures were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University and followed the NIH Guide for the Care and Use of Laboratory Animals. All mice were DBA/2J males from Jackson Laboratories (Bar Harbor, Maine) purchased at 10–13 weeks of age. All animals were allowed to habituate to the animal facility for at least 2 weeks prior to experimental procedures.
Recombinant adeno-associated virus over-expression of Clic4 in mice
Helper-free adeno-associated serotype 2 was used throughout this study. Mouse Clic4 cDNA was generated by PCR and cloned into the plasmid pAAV-IRES-hrGFP (Stratagene, La Jolla, CA) in frame with the FLAG epitope tag. FLAG-tagged pAAV-CLIC4-GFP was verified through sequencing and by green fluorescent protein (GFP) expression in HEK-293 cells via Arrest-in transfection (Open Biosystems, Huntsville, AL). Additional details are provided as Supplementary Material.
Behavioral analyses in mice
Three weeks after viral stereotactic injections, AAV-CLIC4 (Clic4 over-expressing) and AAV-IRES (control) injected mice were evaluated for a sequential battery of behavioral responses to acute ethanol administration. Mice were habituated to injections with saline in their home cage for 2 days prior to behavioral studies. All animals were allowed a 1-hour acclimation period to the behavioral room prior to testing. Locomotor activity was measured immediately following injection with either saline or ethanol (2.0 g/kg) by photobeam breaks during a 10-minute session in locomotor activity chambers (Med-Associates, model ENV-510; St. Albans, VT) in ventilated sound attenuating boxes (ENV-022MD-27). Two weeks later, anxiety testing was done using the light-dark transition model (Crawley & Goodwin, 1980). Mice were injected i.p. with saline or ethanol (1.8 g/kg) and placed in the light compartment of the light-dark box (Med-Associates ENV-510 with black plexiglass inserts to divide into two 13.5 cm × 27 cm × 20 cm compartments. Light compartment was illuminated with 100 mA incandescent bulbs. Timing was initiated when animals entered the dark compartment and locomotor activity/position monitored for 10 minutes. Results were expressed as percent of time or locomotor activity in the light vs. dark. One week after anxiety testing, loss-of-righting reflex (LORR) assays were done by injecting mice i.p. with 3.8 g/kg of ethanol and hypnotic effects of ethanol were observed by inverting mice onto their backs in V-shaped troughs (Linsenbardt et al., 2009). For LORR testing, all animals were injected with ethanol (n=28 each for AAV-IRES and AAV-CLIC4). The duration of LORR was initiated when the animal lost the ability to right itself onto its paws and terminated when the animal recovered its righting reflex by righting itself three times in 30 seconds. Behavioral measurements were discarded when ethanol leaked from the injection site and/or the genital area or when the animal did not lose the righting reflex. Two animals each for AAV-IRES and AAV-CLIC4 groups was excluded for these reasons (final n=26 each). All animals were subjected to supervised randomization between each phase of the behavioral battery so that both groups being tested for each behavior had identical prior experimental exposure.
Detection of in vivo Clic4 expression by immunohistochemistry and qRT-PCR
Two weeks following the conclusion of behavioral studies (~9 weeks after stereotactic injections) Clic4 expression from AAV-CLIC4 injected animals was detected in vivo by immunohistochemistry using antisera against the FLAG epitope tag. Ethanol regulation of endogenous Clic4 mRNA in mouse brain was determined through quantitative real-time PCR (qRT-PCR) to confirm prior microarray studies (Kerns et al., 2005). Additional details are provided as Supplementary Material.
Bioinformatics analyses of Clic4 gene expression in BXD mice
A Clic4 co-expression network was initially generated using Pearson product-moment correlation coefficients with basal gene expression data from medial PFC across 27 BXD recombinant inbred mice, as well as C57BL/6J and DBA/2J progenitors (n = 29). This was done within the GeneNetwork resource for genetic analysis of genomic and phenotypic traits (www.genenetwork.org). To address possible false positives due to multiple comparisons, p-values were corrected using the q-value false-discovery rate method (Qian & Huang, 2005) within the R statistical framework (http://www.bioconductor.org/packages/2.4/bioc/html/qvalue.html), and resulting q-values were filtered for q<0.05, resulting in a total of 1015 probe sets. Ingenuity Pathway Analysis (www.ingenuity.com) was utilized to assess the biological significance and relationships between these genes, based on current scientific literature, using the Pearson correlation coefficients (positive and negative) as signals for individual genes (Figure S7).
Drosophila husbandry, genetics and qRT-PCR
Flies were grown at 20°C (Clic homozygous mutants) or 25°C (all other genotypes) and 55% relative humidity on a standard sugar:yeast:cornmeal:agar medium (10%:2%:3.3%:1% w/v) supplemented with 0.2% Tegosept (Sigma Chemical Co., St. Louis, MO, USA) and active dry yeast under a 12-h light-dark cycle. P[EPgy2]ClicEY04209 (i.e. ClicEY04209) and P[w[+mC]=lacW]Clic[G0472] (i.e. ClicG0472) were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN, USA). To assess Clic expression, qRT-PCR studies were performed using standard methods as described (Jones et al., 2009). Additional details are provided as Supplementary Material.
Drosophila ethanol-related behaviors and internal ethanol measurements
For all fly behavioral studies, groups of 25 adult flies (2–5 days-old, grown in parallel) were collected under brief CO2 anesthesia, transferred to fresh food vials, and housed overnight at 25°C and 55% relative humidity. Each group of 25 flies constituted n=1 (7,425 total flies were used for the reported behavioral studies). Sensitivity and rapid tolerance to ethanol (provided as a vapor) were determined in flies using ethanol Rapid Iterative Negative Geotaxis (eRING) assays at 25°C and 55–65% relative humidity as described (Bhandari et al., 2009). Internal ethanol concentrations in flies exposed to ethanol in eRING assays were determined as described previously (Bhandari et al., 2009). Additional details are provided as Supplementary Material.
C. elegans genetics and ethanol-related behaviors
C. elegans were cultured according to Brenner (Brenner, 1974). Strains used in this study (N2 control and Clic mutant strains exc-4 (rh133) and exl-1(ok857)) were obtained from the C. elegans Genetics Center, which is funded by NIH-NCRR. The double mutant exc-4(rh133);exl-1(ok857) was generated by standard genetic crosses, using the recessive excretory canal phenotype that is associated with exc-4(rh133) to detect that mutation and a set of PCR primers to detect the exl-1(ok857) deletion mutation (5'-GTGCAATCTCGTCAGGACCAGGC-3', 5'-ATGCGTTACGATGCCCCGACAC-3'). Ethanol response assays were carried out as previously described (Davies et al., 2004, Davies et al., 2003) except additional time points were measured and ImagePro Plus was used for object tracking. Additional details are provided as Supplementary Materials.
Internal ethanol measurements in C. elegans
Young-adult age-matched worms, reared at 20°C, were placed on unseeded plates containing 0 mM or 400 mM ethanol (prepared as for ethanol response assays) for 5, 10 or 30 minutes. Ethanol-exposed worms were frozen at −80°C and homogenized in H2O. Internal ethanol was determined in the homogenates using a commercially available alcohol reagent (Pointe Scientific) and by calculating the volume of worms. Additional details are provided as Supplementary Materials.
Statistical Analyses
To determine statistical significance (p≤0.05) in studies comparing two or more groups, t tests (Prism, GraphPad, San Diego, CA, USA), one- and two-way ANOVA (JMP, SAS Institute, Cary, NC) followed by Bonferroni multiple comparison tests were performed as appropriate. Data from qRT-PCR studies in Figure 2B were analyzed by one-sample t tests (Prism) to individually compare Clic mRNA levels in mutants and revertants to control (100%).
Figure 2. Transposon insertions cause partial loss of function in Drosophila Clic.
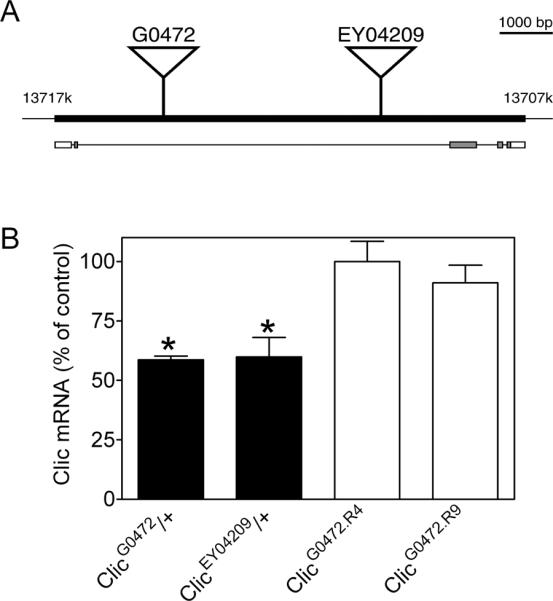
(A) The Clic locus and transposon insertions. Transcription of Clic is from left to right. The Clic transcription unit is represented by the filled rectangle with nucleotide positions indicated above (coordinates from FlyBase annotation release 5.33). Exons are represented by rectangles below the transcription unit with protein coding sequences and untranslated regions depicted as grey and open rectangles, respectively. Introns are represented as a line and transposons as triangles. Scale bar (upper right) is 1000 bp. Schematic adapted from FlyBase. (B) Whole-body Clic mRNA expression in transposon lines. Expression of Clic mRNA in flies heterozygous for the G0472 and the EY04209 transposons was reduced relative to w1118 controls.
Results
Expression of Mammalian Clic4 and Acute Ethanol Behaviors
To identify novel candidate genes involved in alcohol responses, we used gene set overlap analysis within the Ontological Discovery Environment (Baker et al., 2009) and the Ethanol-Related Gene Resource (Guo et al., 2009) to rank data from multiple microarray studies of ethanol-related genes in mouse (Kerns et al., 2005, Mulligan et al., 2006) and human alcoholic brain (Liu et al., 2006, Mayfield et al., 2002). The most highly ranked gene from our analyses, Chloride Intracellular Channel 4 (Clic4), was represented across six independent microarray, linkage and association studies. For example, our prior microarray studies showed that Clic4 had lower basal expression but greater ethanol-responsive expression in prefrontal cortex (PFC) of DBA/2J mice compared to C57BL/6 mice (Supplemental Figure S1; two-way ANOVA; treatment, F(1,12)=7.28, p=0.027; genotype, F(1,12)=8.082, p=0.022; interaction, F(1,12)=11.158, p=0.010, n=3); also see (Kerns et al., 2005)). These two mouse strains differ widely in terms of multiple behavioral responses to acute or chronic ethanol, as well as ethanol consumption (Metten et al., 1998). We also found that Clic4 was in other published datasets for genes associated with ethanol drinking preference in mice (Mulligan et al., 2006) and genes with altered expression in frontal cortex of alcoholics (Mayfield et al., 2002). Furthermore, Clic4 is located within a confirmed quantitative trait locus (QTL) for ethanol drinking behavior on distal mouse chromosome 4 (Tarantino et al., 1998) and we also found that ethanol treatment (4 g/kg × 4 h) increased expression of Clic4 in PFC of DBA/2J mice (Figure 1A; *, two-sample t-test; t(6)=3.091, p=0.027, n=4 for EtOH and 3 for Saline), validating our prior microarray data (Kerns et al., 2005).
Figure 1. Ethanol-responsive and basal expression of Clic4 in mouse PFC.
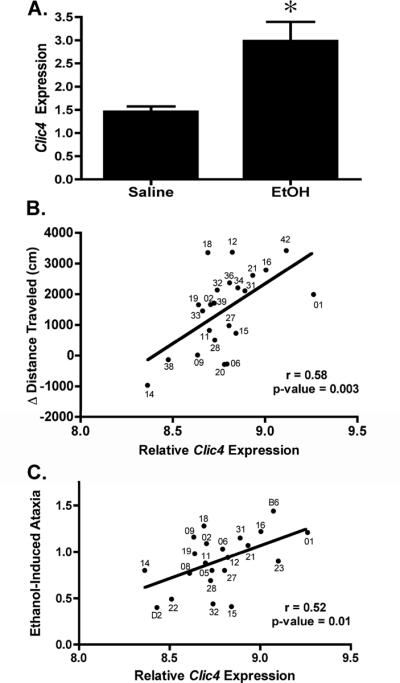
(A) qRT-PCR analysis of basal (Saline) and ethanol-responsive (EtOH, 4 g/kg, 4 hr) Clic4 expression in DBA2/2J mice. Expression of Clic4 is elevated after ethanol treatment, validating prior microarray results (Kerns et al., 2005) . (B and C) Pearson correlation of Clic4 basal expression in PFC (x-axis) of BXD recombinant inbred lines (numbered points) with ethanol-induced locomotor activity (GeneNetwork trait ID 11962 (Philip et al., 2010)) (B) and initial sensitivity to ethanol-induced rotarod ataxia following first of five injections (onset of ataxia brain ethanol threshold, mg ethanol/g brain – GeneNetwork ID 10144 (Gallaher et al., 1996)) (C). Scattergrams were generated in GeneNetwork (www.genenetwork.org) using the VCU PFC saline database (Wolen and Miles, unpublished). B6 and D2 strains were not tested in experiments shown in panel B
As an initial characterization of the role for Clic4 in ethanol responses in mammals, we used genomic studies across the BXD genetic panel of mice to determine whether basal expression of Clic4 in PFC correlated with ethanol-related behaviors in the GeneNetwork web resource (www.genenetwork.org). Through this analysis we found that Clic4 PFC basal expression correlated positively with low dose (2 g/kg) ethanol-induced locomotor activation (Figure 1B) and inversely with high dose (4 g/kg) ethanol-induced ataxia (Figure 1C) in mice. Our analysis of data from human and rodent studies thus suggests that Clic4 or related genes might be important determinants of acute level of response to ethanol across a range of species.
Functional analysis of Clic in Drosophila ethanol behavior
We used Drosophila to directly test the hypothesis that Clic genes are important for acute ethanol behavior. We confirmed two independent transposon insertions (G0472 and EY04209, Figure 2A) in the sole Clic locus in flies and subsequently backcrossed both elements into w1118 (our standard laboratory stock) to control for genetic background effects on ethanol behavior. Both transposon insertions were homozygous female and hemizygous male lethal at 25°C. Female flies heterozygous for G0472 and EY04209, however, were fertile, appeared morphologically normal and seemed generally healthy. qRT-PCR analysis showed that Clic mRNA in G0472/+ and EY04209/+ flies was reduced by approximately 40% compared to w1118 control (Figure 2B; *, one sample t tests; t(2)=27.01 and t(2)=4.95 respectively, p<0.05, n=3). G0472 and EY04209 are therefore partial loss of function mutations in Drosophila Clic.
To assess ethanol behavior in Clic mutant flies, we used an ethanol Rapid Iterative Negative Geotaxis (eRING) assay developed in one of our component laboratories (Bhandari et al., 2009). Female ClicG0472/+ and ClicEY04209/+ mutants maintained their negative geotaxis behavior longer than did the w1118 female controls during exposure to ethanol (Supplemental Figure S2; two-way ANOVA; time, F(7,216)=249.75, p<0.0001; genotype, F(2,216)=40.36, p<0.0001; n=5), suggesting that Clic mutants have reduced sensitivity to the sedative effects of ethanol. Data from several experiments confirmed the blunted ethanol sensitivity in female ClicG0472/+ and ClicEY04209/+ flies (Figure 3A and B, respectively; *, t tests; ClicG0472/+, t(26)=6.31, p<0.0001; ClicEY04209/+, t(28)=6.87, p<0.0001, n=13–15). Additionally, ClicG0472 and ClicEY04209 hemizygous males and homozygous females (reared at 20°C to improve viability) had diminished ethanol sensitivity (Supplemental Figure S3; two-sample t tests; ClicG0472 males, t(6)=3.55, p=0.012; ClicG0472 females, t(6)=9.39, p<0.0001; ClicEY04209 males, t(7)=5.62, p=0.0008; ClicEY04209 females, t(8)=13.04, p<0.0001, n=3–5). (Note that data in Figure 3 and Supplemental Figure S3 are T50 values that reflect the time required for ethanol to inhibit negative geotaxis by 50%. Higher values indicate blunted ethanol sensitivity; i.e. higher doses of ethanol are required to produce the same level of response.) In contrast to the effect of Clic mutations on ethanol sensitivity, we did not observe a significant alteration in rapid ethanol tolerance in these animals (Supplemental Figure S4; two-sample t tests; ClicG0472/+, t(16)=0.777, p=0.448; ClicEY04209/+, t(17)=1.86, p=0.080; n=9–10). Clic might therefore be an important determinant of ethanol sensitivity in Drosophila without having major effects on rapid ethanol tolerance in this species.
Figure 3. Ethanol sensitivity in Drosophila Clic transposon mutants and revertants.
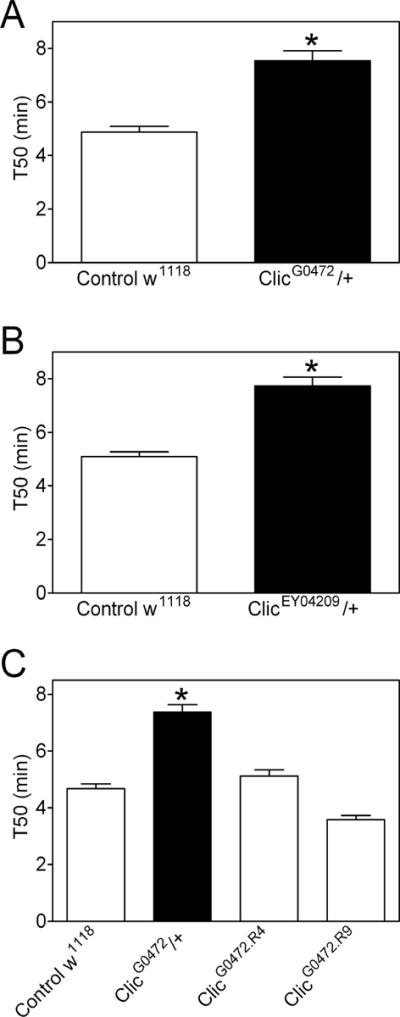
Ethanol sensitivity represented as T50 values in ClicG0472/+ and ClicEY04209/+ (black bars, A and B, respectively) and Control w1118 flies (open bars). Clic mutants had significantly higher T50 values than controls. (C) Ethanol sensitivity (T50 values) in ClicG0472 heterozygous transposon mutants and revertants. ClicG0472/+ flies (black bar) had higher T50 values than Control w1118 flies or revertants (ClicG0472.R4 and ClicG0472.R9) (white bars).
Although the ClicG0472 and ClicEY04209 alleles were independently derived and backcrossed for several generations to normalize the genetic background, it was formally possible that the transposon insertions in Clic were not responsible for the blunted ethanol sensitivity in flies. To address this issue, we generated two revertants (ClicG0472.R4 and ClicG0472.R9) of the G0472 chromosome through precise excision of the transposon. Expression of Clic returned to normal in the two revertants (Figure 2B; one sample t tests; ClicG0472R.4, t(2)=0.0, p=1.00, n.s.; ClicG0472.R9, t(2)=1.22, p=0.348, n.s.; n=3) as did the ethanol sensitivity phenotype of ClicG0472 flies (Figure 3C; one-way ANOVA; genotype, F(3,44)=69.93, p<0.0001; *Bonferroni multiple comparison, Control w1118 t(22)=9.40, p<0.05; ClicG0472R.4 t(22)=7.68, p<0.05; ClicG0472R.9 t(25)=14.29, p<0.05; n=10–15). These data confirm that the G0472 transposon insertion disrupted Clic expression and blunted ethanol sensitivity.
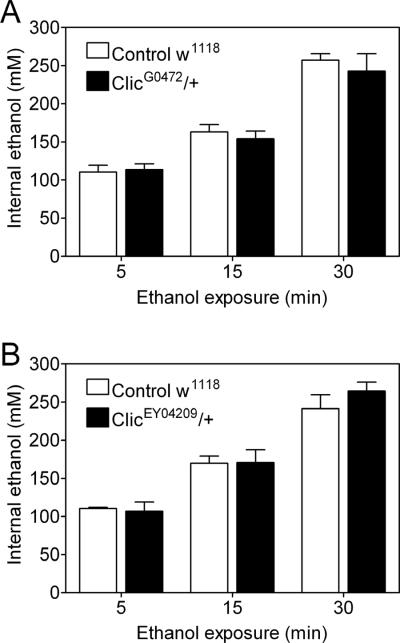
To address the possibility that blunted ethanol sensitivity in Drosophila Clic mutants could be secondary to altered ethanol uptake or metabolism, we determined internal ethanol concentrations in flies exposed to ethanol in eRING assays (Bhandari et al., 2009). Duration of ethanol exposure had a significant effect on internal ethanol concentrations as expected (individual two-way ANOVAs; Figure 4A, F(2,12)=115.69, p<0.0001; Figure 4B, F(2,12)=84.39, p<0.0001; n=3), but there was no effect of Clic genotype on internal ethanol concentrations (Figure 4A, F(1,12)=0.417, p=0.529; Figure 4B, F(1,12)=0.283, p=0.603) or an interaction between genotype and duration of ethanol exposure (Figure 4A, F(2,12)=0.340, p=0.569; Figure 4B, F(2,12)=0.888, p=0.362). Clic mutants, therefore, have a blunted level of response to acute ethanol exposure that is not related to a significant change in tissue concentrations of the drug.
Figure 4. Internal ethanol concentrations in Drosophila Clic mutants.
ClicG0472/+ (A) and ClicEY04209/+ (B) were exposed to ethanol vapor for the indicated durations in eRING assays in parallel with Control w1118 flies. Internal ethanol concentrations were determined as described in Materials and Methods
It seemed possible that partial loss of function in Clic could, in principle, lead to a global improvement in negative geotaxis behavior that manifested as blunted sensitivity to ethanol in eRING assays. We addressed this issue by carefully examining two key components of negative geotaxis, latency to initiate the behavior and climbing speed, in control and Clic mutants in the absence of ethanol. Climbing speed in negative geotaxis assays was indistinguishable in ClicG04072/+, ClicEY04209/+ and control w1118 flies (Supplemental Figure S5A and C; two-sample t tests; ClicG04072/+, t(28)=1.66, p=0.107; ClicEY04209/+, t(28)=0.891, p=0.381; n=14–15). Although climbing latency was decreased in ClicG0472/+ (Supplemental Figure S5B; two-sample t test; t(26)=3.40, p=0.002, n=13–14), this effect was not seen in ClicEY04209/+ (Supplemental Figure S5D; two-sample t test; t(27)=1.26, p=0.218; n=14). We conclude that partial loss of function in Clic does not consistently alter negative geotaxis in flies and, therefore, that the blunted ethanol sensitivity in Clic mutants must be independent of global improvements in negative geotaxis or climbing behavior.
Functional Analysis of Clic orthologues in C. elegans
To determine if Clic genes act exclusively in initial sensitivity or if they also influence the development of acute functional tolerance to ethanol, we extended our genetic analysis to another invertebrate model, the nematode C. elegans. Worms have two Clic genes, exc-4 and exl-1 (Berry et al., 2003). We assessed the responses to acute ethanol using an established behavioral assay in which worms were exposed to the drug on agar plates and locomotor speed was assessed as a function of exposure time (Davies et al., 2004, Davies et al., 2003, Kapfhamer et al., 2008). In this assay, locomotor speed in N2 control worms is increasingly depressed in the presence of ethanol over the first 15 minutes of drug exposure (indicating initial sensitivity). Thereafter, locomotor speed increases despite the continued presence of ethanol (indicating the development of acute functional tolerance to ethanol) (Davies et al., 2004). Repeated measures two-way ANOVA indicated significant effects of time (F(10,120)=14.66, p=0.0244) and genotype (F(3,120)=11.06, p=0.0009) on locomotor behavior in the presence of ethanol with no interaction between the factors (F(10,120)=1.78, p=0.177). Worms harboring exc-4(rh133), a null allele (Berry et al., 2003), had diminished ethanol sensitivity during the first few minutes of drug exposure (Figure 5A, Supplemental Figure S6A; #, Bonferroni multiple comparison, t(7)=2.91–4.92, p<0.05) and a trend toward blunted acute tolerance to ethanol at later time-points (Figure 5A). In contrast, worms carrying the exl-1(ok857) mutation, also likely a null allele (Berry & Hobert, 2006), had wild-type initial sensitivity to ethanol followed by significantly enhanced acute functional ethanol tolerance (Figure 5A; †, Bonferroni multiple comparison; t(7)=2.91–4.92, p<0.05). Importantly, while internal ethanol concentrations increased with the duration of ethanol exposure as expected, neither mutation significantly altered internal tissue concentrations of ethanol determined at key time-points of behavioral testing as compared to N2 control animals (Figure 5B; two-way ANOVA; duration, (F(2,60)=29.19, p<0.0001); genotype (F(3,60)=3.07, p=0.0439; interaction between duration and genotype, (F(3,60)=0.1143, p=0.951, n.s.; Bonferroni multiple comparisons between N2 and other genotypes, t(11)=0.0824–1.94, p>0.05, n.s., n=6). The behavioral consequences of exc-4 and exl-1 mutations are therefore likely to be related to changes in the pharmacodynamic responses to ethanol as opposed to altered pharmacokinetics of the drug. These studies indicate that both Clic orthologues in C. elegans influence ethanol-related behavior. The effects of mutations in exc-4 and exl-1 on initial sensitivity and acute tolerance in worms, however, are wholly distinct and suggest that different Clic genes could play distinct roles in ethanol-related behavior.
Figure 5. Ethanol sensitivity and acute functional tolerance in C. elegans with mutations in Clic orthologues.
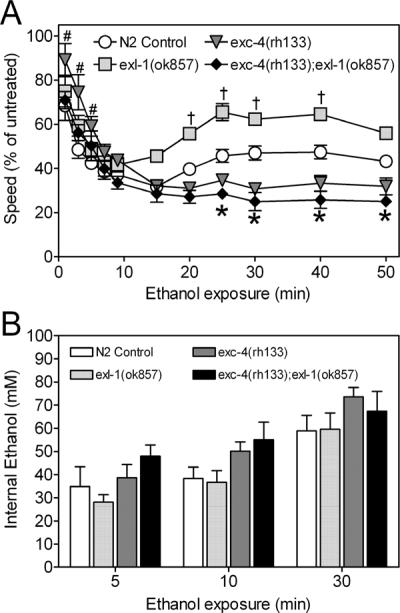
(A) Effect of ethanol exposure on relative locomotor speed (percent of untreated animals) in N2 control (open circles) and Clic mutants (exl-1(ok857), light grey squares; exc-4(rh133), dark grey triangles; exc-4(rh133);exl-1(ok857), black diamonds). Data are from 4 independent experiments where 10 worms per genotype contributed to an average speed for a population. (B) Internal ethanol concentrations in N2 control and Clic mutants were determined as described in Materials and Methods
To address the possibility that the Clic genes exc-4 and exl-1 might work in concert, we also assessed the behavioral response to ethanol in exc-4;exl-1 double mutants. Interestingly, initial sensitivity in the double mutants was essentially normal (similar to exl-1 mutants), whereas acute tolerance was significantly suppressed in these animals (similar to exc-4 mutants) (Figure 5A). Since internal ethanol concentrations were not significantly altered in exc-4(rh133);exl-1(ok857) double mutants compared to wild-type N2 controls (Figure 5B), we tentatively conclude that the effect of loss of exc-4 function on initial sensitivity requires normal expression of exl-1 and conversely that the effect of loss of exl-1 function on acute tolerance requires normal expression of exc-4.
Clic4 influences ethanol sensitivity in mice
The gene expression-correlation studies in Figure 1, combined with the functional genetic analysis of Clic orthologues in Drosophila (Figures 2–4) and C. elegans (Figure 5), suggested that Clic4 might influence acute behavioral sensitivity to ethanol in mammals. To directly test this possibility, we constructed AAV2 viral vectors to over-express CLIC4 protein in specific regions of mouse brain. As expected, stereotactic injection of AAV2/Clic4 virus in medial prefrontal cortex (including cingulate and secondary motor cortex) in DBA/2J mice increased Clic4 transcript abundance (not shown) and increased expression of epitope-tagged CLIC4 protein (Figure 6A) compared to control animals injected with empty vector (Figure 6B). AAV2-mediated overexpression of Clic4 in DBA/2J mice also decreased the sensitivity to high-dose ethanol (3.8 g/kg) loss-of-righting reflex (LORR) (Figure 6C; *, two-sample t-test, t(49)=2.49, p=0.016, n=26) without changing the anxiolytic or locomotor-activating responses to lower doses of ethanol (1.8 g/kg, not shown). The decreased LORR time with Clic4 overexpression is consistent with results of Fig. 1C showing that higher Clic4 expression correlates with higher blood ethanol levels being required for initiation of rotarod ataxia (decreased sensitivity). These results indicate that the expression level of Clic4 can influence the level of response to high-doses of ethanol in DBA/2J mice, consistent with a role for CLIC proteins in behavioral responses to acute ethanol in mammals.
Figure 6. Altered loss of righting reflex (LORR) in mice with AAV2 viral vector-mediated expression of Clic4 in brain.
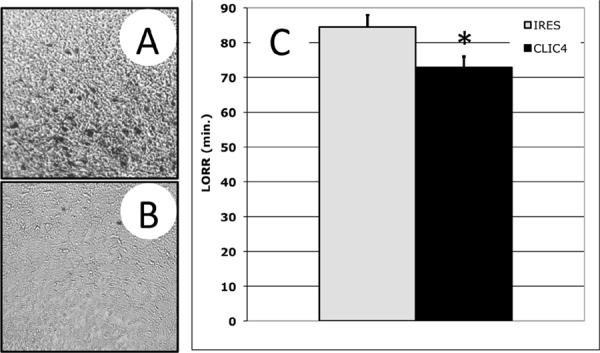
AAV2 vectors expressing a Clic4-FLAG fusion protein (AAV-CLIC4; panel A) or empty vector (AAV-IRES; panel B) were stereotactically injected into male DBA/2J mouse PFC. Panels A and B show immunohistochemistry results for FLAG epitope primary antibody staining. Immunohistochemistry was done 2 weeks after the last behavioral studies (~9 weeks after viral injections). Panel C shows that Clic4 over-expressing animals (black) had a shorter duration of LORR following 3.8 g/kg IP of ethanol compared to control (grey). Behavioral testing for LORR was done ~7 weeks after viral injections.
Bioinformatics analysis for Clic4 functional mechanisms in mice
The Clic family of genes has been implicated in a number of different biological processes or molecular functions (Maeda et al., 2008, Shukla et al., 2009, Singh & Ashley, 2007, Suh et al., 2004, Ulmasov et al., 2009). Many of these functions, however, remain largely unconfirmed or poorly characterized. As a non-biased analysis of possible mechanisms for Clic4 action on ethanol responses in mouse brain PFC, where we observed altered LORR after Clic4 over-expression, we performed a genetic correlation analysis using the GeneNetwork web resource (www.genenetwork.org) to identify genes with expression tightly coupled with basal Clic4 expression in PFC across the BXD genetic panel of mice. Clic4 expression was strongly correlated with a large set of genes in PFC (Supplemental Table S1). This gene set was strikingly over-represented (p< 10−15) for a network of genes involved in RNA processing as determined via Ingenuity Pathway Analysis (Supplementary Figure S7 and Supplemental Table S2). This result raises the possibility that Clic4 might influence ethanol sensitivity in mice by functioning within a network of genes that impinge on RNA processing and trafficking.
Discussion
Using animal models to identify genes and genetic pathways that influence ethanol-related behavior holds tremendous promise for providing mechanistic insight into the basic biology of alcohol abuse. Studies in animal models could lay the foundation for developing novel drug targets for treating alcohol abuse and for estimating risk of abusing alcohol in human populations. Starting with ethanol-responsive gene sets and other ethanol-related data from studies in mammals, we selected mammalian Clic4 as a candidate gene in ethanol behavior. Subsequent studies in multiple animal models directly linked the Clic family of genes to acute behavioral responses to alcohol. Partial loss of function in the sole Clic orthologue in Drosophila blunted the locomotor sedating effects of ethanol. In C. elegans, loss of function in the Clic orthologue exc-4 blunted ethanol sensitivity during the first few minutes of drug exposure, whereas loss of function in another Clic orthologue, exl-1, enhanced the development of acute functional tolerance to alcohol. Additionally, viral-mediated brain-specific overexpression of mammalian Clic4 decreased the sedating effects of high dose ethanol in mice. The ethanol behavior phenotypes were unrelated to altered uptake/metabolism of the drug in worms or flies, indicating that CLIC proteins influence the pharmacodynamic properties of ethanol in these species. We did not perform pharmacokinetic studies on ethanol in the virally injected mice, but we have not observed altered ethanol metabolism in other AAV gene delivery studies on PFC (Miles and Meng, unpublished data). Although we cannot totally exclude the possibility that expression of Clic4 in a relatively small population of PFC neurons in adult animals would lead to peripheral differences in ethanol metabolism (as might occur with a traditional gene knockout approach), we believe it highly unlikely. Collectively, our data establish that several members of the Clic gene family influence ethanol-related behaviors in multiple species. We are currently investigating whether variance in Clic genes might be associated with human responses to alcohol or alcohol abuse.
We focused our Clic genetic studies reported here on the level of response to acute ethanol exposure because there is an intriguing literature indicating that level of response correlates with alcohol abuse or intake. Men with low level of response to acute ethanol are more likely to develop alcohol dependence than those with high level of response (Schuckit, 1994, Schuckit & Smith, 1996). A similar inverse relationship between acute ethanol sensitivity and ethanol drinking behavior is also often seen in several studies with gene-targeted mice. For example, mice with knockout of the A2A adenosine receptor (Naassila et al., 2002), neuropeptide Y (Thiele et al., 1998) or the protein kinase A regulatory subunit RIIβ (Thiele et al., 2000) exhibit decreased sensitivity to ethanol and increased ethanol consumption. Conversely, protein kinase-C epsilon knockout mice have increased sensitivity to the effects of ethanol and decreased ethanol drinking (Hodge et al., 1999). Not all gene-targeted mouse studies, however, show this inverse relationship. In an extensive review of the subject, Crabbe et al. found that 19 out of 48 (40%) genes studied showed opposite effects on ethanol consumption vs. acute sedation, but 12% showed increases in both traits and another 48% were equivocal (Crabbe et al., 2006). These studies thus indicate that acute level of response to ethanol often, but not always, has predictive value for identifying molecular events relevant to ethanol consumption including long-term ethanol drinking behavior in humans. The studies on Clic in this report document a novel gene affecting the acute level of response to ethanol across multiple species, and thus could have important implications for understanding the mechanisms that affect ethanol consumption.
All genetic manipulations of Clic that we investigated led to altered ethanol-response behavior. The observed phenotypes between or even within a species, however, were not identical. Although we currently do not understand the range of phenotypes observed in animals with altered Clic expression, one possible explanation is that CLIC proteins could be direct targets of ethanol, but different Clic genes might be expressed in different cells (e.g. excitatory versus inhibitory neurons) leading to varying effects on ethanol responses. The C. elegans CLIC protein EXL-1 is expressed in PVD and CAN neurons in addition to neurons that have not been identified, while EXC-4, the other CLIC protein in worms, is expressed in a distinct but somewhat overlapping pattern in the nervous system (Berry et al., 2003, Berry & Hobert, 2006). Additionally, while both EXL-1 and EXC-4 are expressed in the intestine, EXL-1 is expressed in body muscle and coloemocytes whereas EXC-4 is localized to the excretory canal cell, hypodermis, sheath cells, rectal gland and vulval cells (Berry & Hobert, 2006). Although the connections between these various cells or tissues and ethanol behavior in worms are not immediately obvious, the distinct patterns of EXL-1 and EXC-4 expression could contribute to the different ethanol-related behaviors observed in exl-1 and exc-4 mutants. Similarly, it is possible that Clic4 over-expression in regions of the mouse brain other than the PFC could produce varying effects on LORR or even other ethanol behavioral phenotypes.
Although there is evidence that Clic genes encode intracellular chloride channels (Ashley, 2003), this channel activity is somewhat controversial and several other functions have been suggested. Vertebrate CLIC4 or other CLIC proteins bind and likely regulate the function of ryanodine receptors (Jalilian et al., 2008), 14-3-3 proteins (Suginta et al., 2001) and A-kinase anchoring proteins (Shanks et al., 2002) Additionally, CLIC4 interacts with Smad proteins and is consequently a mediator of TGF-β or bone morphogenic protein (BMP) signaling (Shukla et al., 2009). The relevance of these biochemical functions for CLIC4 or other CLIC proteins in behavioral responses to ethanol is unknown currently. Our bioinformatics studies on gene networks associated with Clic4 basal expression in mouse PFC (Supplementary Figure S7), however, provide correlative evidence that Clic4 might modulate RNA processing and trafficking. This possibility is particularly interesting since CLIC4 is a mediator of TGF-β signaling and this pathway is known to modulate RNA processing via Smad protein interaction with RNA binding sites on miRNA (Davis et al., 2008). Thus, CLIC4 might regulate TGF-β/Smad-mediated modulation of RNA processing and therefore protein expression, which could ultimately influence neuronal responses to ethanol. While such a role for CLIC proteins in modulating ethanol-related behavior is speculative at this time, our approach of using multiple model organisms will allow us to investigate this and other possible mechanisms of CLIC proteins in behavioral responses to ethanol.
Supplementary Material
Acknowledgements
The authors thank the Bloomington Drosophila Stock Center and the C. elegans Genetics Center for contribution of genetic reagents. The authors thank Devin Rhodenizer, Lauren Thomas, Gina Blackwell, and Ryan Friedberg for technical assistance, Dr. Thomas Green for helpful discussions in development of the Clic4-AAV2 vector, and other members of the Virginia Commonwealth University Alcohol Research Center for insightful comments on the project. The authors thank Soichi Tanda and Mark Berryman (Ohio University) for sharing unpublished information. This study was supported by the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, via the following: P20 AA017828 (M Miles, J Bettinger, A Davies, M Grotewiel), R01 AA016842 (A Davies, J Bettinger), R01 AA016837 (J Bettinger, A Davies), U01 AA016662, U01 AA016667 (M Miles) and F31 AA018615 (S Farris).
References
- Ashley RH. Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins (Review) Molecular membrane biology. 2003;20:1–11. doi: 10.1080/09687680210042746. [DOI] [PubMed] [Google Scholar]
- Baker EJ, Jay JJ, Philip VM, Zhang Y, Li Z, Kirova R, Langston MA, Chesler EJ. Ontological Discovery Environment: a system for integrating gene-phenotype associations. Genomics. 2009;94:377–387. doi: 10.1016/j.ygeno.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KL, Bulow HE, Hall DH, Hobert O. A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science (New York, N.Y. 2003;302:2134–2137. doi: 10.1126/science.1087667. [DOI] [PubMed] [Google Scholar]
- Berry KL, Hobert O. Mapping functional domains of chloride intracellular channel (CLIC) proteins in vivo. Journal of molecular biology. 2006;359:1316–1333. doi: 10.1016/j.jmb.2006.04.046. [DOI] [PubMed] [Google Scholar]
- Bhandari P, Bettinger JC, Davies AG, Kendler K, Grotewiel M. An assay for evoked locomotor behavior in Drosophila reveals a role for integrins in ethanol sensitivity and rapid tolerance. Alcohol Clin Exp Res. 2009;33:1794–1805. doi: 10.1111/j.1530-0277.2009.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crawley J, Goodwin FK. Preliminary report of a simple animal behavior model for the anxiolytic effects of benzodiazepines. Pharmacol Biochem Behav. 1980;13:167–170. doi: 10.1016/0091-3057(80)90067-2. [DOI] [PubMed] [Google Scholar]
- Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DHHS 10th Special Report to the U.S. Congress on Alcohol and Health: Highlights from Current Research. 2000 [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher EJ, Jones GE, Belknap JK, Crabbe JC. Identification of genetic markers for initial sensitivity and rapid toleranceto ethanol-induced ataxia using quantitative trait locus analysis in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1996;277:604–612. [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z. ERGR: An ethanol-related gene resource. Nucleic Acids Res. 2009;37:D840–845. doi: 10.1093/nar/gkn816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U. Genetics of alcohol-induced behaviors in Drosophila. Alcohol Res Health. 2000;24:185–188. [PMC free article] [PubMed] [Google Scholar]
- Hermens WT, ter Brake O, Dijkhuizen PA, Sonnemans MA, Grimm D, Kleinschmidt JA, Verhaagen J. Purification of recombinant adeno-associated virus by iodixanol gradient ultracentrifugation allows rapid and reproducible preparation of vector stocks for gene transfer in the nervous system. Hum Gene Ther. 1999;10:1885–1891. doi: 10.1089/10430349950017563. [DOI] [PubMed] [Google Scholar]
- Hodge CW, Mehmert KK, Kelley SP, McMahon T, Haywood A, Olive MF, Wang D, Sanchez-Perez AM, Messing RO. Supersensitivity to allosteric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat Neurosci. 1999;2:997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- Jalilian C, Gallant EM, Board PG, Dulhunty AF. Redox potential and the response of cardiac ryanodine receptors to CLIC-2, a member of the glutathione S-transferase structural family. Antioxidants & redox signaling. 2008;10:1675–1686. doi: 10.1089/ars.2007.1994. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Gargano JW, Rhodenizer D, Martin I, Bhandari P, Grotewiel M. A forward genetic screen in Drosophila implicates insulin signaling in age-related locomotor impairment. Experimental gerontology. 2009;44:532–540. doi: 10.1016/j.exger.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, McIntire SL. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 2008;7:669–676. doi: 10.1111/j.1601-183X.2008.00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide Association Analysis of Symptoms of Alcohol Dependence in the Molecular Genetics of Schizophrenia (MGS2) Control Sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-Regulated Genes That Contribute to Ethanol Sensitivity and Rapid Tolerance in Drosophila. Alcohol Clin Exp Res. 2009;34:302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Porcu P, Werner DF, Matthews DB, Diaz-Granados JL, Helfand RS, Morrow AL. The role of GABA(A) receptors in the acute and chronic effects of ethanol: a decade of progress. Psychopharmacology. 2009;205:529–564. doi: 10.1007/s00213-009-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011a;35:1600–1606. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, Brush G, Xue L, Robertson M, Moore MS, Vranizan K, Morris SW, Schuckit MA, White RL, Heberlein U. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS ONE. 2011b;6:e22636. doi: 10.1371/journal.pone.0022636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, Martin NG, Willemsen G, de Geus EJ, Vogelzangs N, Penninx BW, Whitfield JB, Montgomery GW, Boomsma DI, Madden PA. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenbardt DN, Moore EM, Gross CD, Goldfarb KJ, Blackman LC, Boehm SL., 2nd Sensitivity and tolerance to the hypnotic and ataxic effects of ethanol in adolescent and adult C57BL/6J and DBA/2J mice. Alcoholism, clinical and experimental research. 2009;33:464–476. doi: 10.1111/j.1530-0277.2008.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Maeda K, Haraguchi M, Kuramasu A, Sato T, Ariake K, Sakagami H, Kondo H, Yanai K, Fukunaga K, Yanagisawa T, Sukegawa J. CLIC4 interacts with histamine H3 receptor and enhances the receptor cell surface expression. Biochem Biophys Res Commun. 2008;369:603–608. doi: 10.1016/j.bbrc.2008.02.071. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome biology. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome biology. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naassila M, Ledent C, Daoust M. Low ethanol sensitivity and increased ethanol consumption in mice lacking adenosine A2A receptors. J Neurosci. 2002;22:10487–10493. doi: 10.1523/JNEUROSCI.22-23-10487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes, brain, and behavior. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian HR, Huang S. Comparison of false discovery rate methods in identifying genes with differential expression. Genomics. 2005;86:495–503. doi: 10.1016/j.ygeno.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Experimental gerontology. 2008;43:739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan AR, Rothenfluh A. The genetics of behavioral alcohol responses in Drosophila. International review of neurobiology. 2010;91:25–51. doi: 10.1016/S0074-7742(10)91002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O'Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RA, Larocca MC, Berryman M, Edwards JC, Urushidani T, Navarre J, Goldenring JR. AKAP350 at the Golgi apparatus. II. Association of AKAP350 with a novel chloride intracellular channel (CLIC) family member. The Journal of biological chemistry. 2002;277:40973–40980. doi: 10.1074/jbc.M112277200. [DOI] [PubMed] [Google Scholar]
- Shukla A, Malik M, Cataisson C, Ho Y, Friesen T, Suh KS, Yuspa SH. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nature cell biology. 2009;11:777–784. doi: 10.1038/ncb1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Ashley RH. CLIC4 (p64H1) and its putative transmembrane domain form poorly selective, redox-regulated ion channels. Mol Membr Biol. 2007;24:41–52. doi: 10.1080/09687860600927907. [DOI] [PubMed] [Google Scholar]
- Suginta W, Karoulias N, Aitken A, Ashley RH. Chloride intracellular channel protein CLIC4 (p64H1) binds directly to brain dynamin I in a complex containing actin, tubulin and 14-3-3 isoforms. The Biochemical journal. 2001;359:55–64. doi: 10.1042/0264-6021:3590055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KS, Mutoh M, Nagashima K, Fernandez-Salas E, Edwards LE, Hayes DD, Crutchley JM, Marin KG, Dumont RA, Levy JM, Cheng C, Garfield S, Yuspa SH. The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis. J Biol Chem. 2004;279:4632–4641. doi: 10.1074/jbc.M311632200. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res. 1998;22:1099–1105. [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Willis B, Stadler J, Reynolds JG, Bernstein IL, McKnight GS. High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J Neurosci. 2000;20:RC75. doi: 10.1523/JNEUROSCI.20-10-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Archives of general psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov B, Bruno J, Gordon N, Hartnett ME, Edwards JC. Chloride intracellular channel protein-4 functions in angiogenesis by supporting acidification of vacuoles along the intracellular tubulogenic pathway. Am J Pathol. 2009;174:1084–1096. doi: 10.2353/ajpath.2009.080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.