Abstract
Verticillium wilt is a major threat to alfalfa (Medicago sativa) and many other crops. The model legume Medicago truncatula was used as a host for studying resistance and susceptibility to Verticillium albo-atrum. In addition to presenting well-established genetic resources, this wild plant species enables to investigate biodiversity of the response to the pathogen and putative crosstalk between disease and symbiosis. Symptom scoring after root inoculation and modelling of disease curves allowed assessing susceptibility levels in recombinant lines of three crosses between susceptible and resistant lines, in a core collection of 32 lines, and in mutants affected in symbiosis with rhizobia. A GFP-expressing V. albo-atrum strain was used to study colonization of susceptible plants. Symptoms and colonization pattern in infected M. truncatula plants were typical of Verticillium wilt. Three distinct major quantitative trait loci were identified using a multicross, multisite design, suggesting that simple genetic mechanisms appear to control Verticillium wilt resistance in M. truncatula lines A17 and DZA45.5. The disease functional parameters varied largely in lines of the core collection. This biodiversity with regard to disease response encourages the development of association genetics and ecological approaches. Several mutants of the resistant line, impaired in different steps of rhizobial symbiosis, were affected in their response to V. albo-atrum, which suggests that mechanisms involved in the establishment of symbiosis or disease might have some common regulatory control points.
Key words: biodiversity, Medicago truncatula, nodulation mutants, partial resistance, quantitative trait loci, root disease, vascular wilt, Verticillium albo-atrum.
Introduction
Vascular wilt diseases are major constraints for plant production. They are characterized by the colonization of the plant’s xylem vessels by the pathogen and are mainly caused by soil-borne microorganisms, which might be bacteria or fungi, such as Fusarium or Verticillium (Agrios, 2005). The most important plant pathogenic Verticillium species are Verticillium dahliae and Verticillium albo-atrum, infecting more than 200 plant species and destroying high amounts of crops worldwide (Fradin and Thomma, 2006; Klosterman et al., 2009). The colonization of xylem vessels by spores and hyphae, together with gum formation by the plant cells, induce clogging of the vessels, stop water flow, and result in the typical wilting symptoms. At the end of the infection cycle, the fungus grows outside of the stele and colonizes other tissues (Agrios, 2005). Owing to survival structures, which remain viable for many years in the soil, and the protected localization in infected plants, Verticillium wilt is difficult to control. So far the most efficient way is by breeding resistant or tolerant varieties. Examples of polygenic and monogenic dominant resistance have been described for several plant species and resistance loci or genes have been identified. A well-studied example is the Ve locus in tomato which confers resistance against race 1 of V. dahliae and V. albo-atrum (Fradin and Thomma, 2006). The tomato Ve locus has been cloned and shown to contain two genes encoding receptor-like proteins with extracellular leucine-rich repeat (LRR) domains, Ve1 and Ve2, which are each able to confer resistance to susceptible potato (Kawchuk et al., 2001). In Arabidopsis, the tomato Ve1 gene has been reported to confer resistance to V. dahliae and V. albo-atrum (Fradin et al., 2011). Studies on the interaction between lettuce and V. dahliae indicated that a single dominant gene with homology to tomato Ve1 and Ve2 controls resistance against race 1 of the fungus in this species (Hayes et al., 2011). Similarly, Ve homologues are proposed to be involved in tolerance to V. dahliae in cotton and mint (Vining and Davis, 2009; Zhang et al., 2011). The V. dahliae effector Ave1, recognized by tomato VE1, has been cloned recently (de Jonge et al., 2012).
Because of the high protein content of their seeds and their capacity to fix atmospheric nitrogen due to a symbiosis with rhizobia, legume plants have an essential role in human and animal nutrition and in sustainable agriculture. Alfalfa (Medicago sativa) yields in Europe are strongly reduced by the impact of Verticillium wilt caused by V. albo-atrum. In alfalfa, monogenic and quantitative resistance to Verticillium wilt has been reported (Pennypacker, 2000; Acharya and Huang, 2003). Antimicrobial compounds such as phytoalexins and saponins have been suggested to be involved, as well as deposit of electron-dense material in xylem vessels (Acharya and Huang, 2003). Although tolerant cultivars are available (Molinéro-Demilly et al., 2007), the molecular mechanisms underlying resistance are still not understood. This lack of data stresses the need for a model system to investigate Verticillium wilt response in legume plants.
Because of its autotetraploid and outcrossing nature, genetic studies on disease resistance in alfalfa are difficult. The closely related wild species Medicago truncatula has been established during the last 15 years as a model for legume plants, particularly attractive for the study of plant–microbe interactions (Samac and Graham, 2007; Rose, 2008; Young and Uvardi, 2009). It is diploid, self-fertile, has a short generation cycle, a small genome size, and shows high synteny with cultivated legume crops. Hence, knowledge obtained by studying M. truncatula can be transferred to other legumes. The understanding of the molecular mechanisms involved in symbiosis with rhizobia and mycorrhiza has greatly advanced thanks to studies with this plant, and common and specific symbiotic pathways for these symbioses have been identified (Gough and Cullimore, 2011).
In addition to the tremendous amount of data on symbiotic interactions, pathogenic interactions have also been described. Resistance genes and loci against various pathogens and pests, such as Colletotrichum trifolii, Erysiphe pisi, Aphanomyces euteiches, Ralstonia solanacearum, and pea aphid have been identified in M. truncatula (Vailleau et al., 2007; Yang et al., 2007; Ameline-Torregrosa et al., 2008; Djébali et al., 2009; Pilet-Nayel et al., 2009; Stewart et al., 2009). Furthermore, the RCT1 gene responsible for resistance against C. trifolii race 1 in M. truncatula was able to confer resistance against several races of the fungus in alfalfa (Yang et al., 2008b). Yet, compared to other plant families, genetic mechanisms of disease resistance in legumes, particularly for root diseases, are less understood. They may present particular characteristics due to their putative interactions with symbiotic processes (Mithöfer, 2002), which are absent in non-legume plants. Such putative interaction with symbiosis may lead to novel recommendations in terms of plant breeding for root disease tolerance in legumes.
In the present work, the model plant M. truncatula is used to study Verticillium wilt in legumes. The infection was characterized and colonization of the host plant was studied with a green fluorescent protein (GFP)-expressing strain. Resistance and susceptibility levels of different M. truncatula lines were quantified through disease symptoms modelling. Loci responsible for resistance were identified in three crosses, and a collection of accessions was used to analyse biodiversity of the interaction. In order to address the question of crosstalk between disease response and symbiosis, various nodulation mutants in line A17 were inoculated with the fungus and symptom development was analysed.
Materials and methods
Plants
M. truncatula Gaertn. seeds were obtained from plants grown in the greenhouse. The lines, except Jemalong-A17, and mutants are derived from natural populations of different geographical origins (http://www1.montpellier.inra.fr/BRC-MTR/mauguio/mauguio.php). Scarification and germination of seeds, as well as culture in peat substrate (Jiffy pots, Jiffy, France), were performed as described by Vailleau et al. (2007). For microscopy, plants were grown in a miniaturized hydroponic culture system on N-containing Farhäeus medium (Farhäeus, 1957). In vitro plants were grown on solid Farhaeus medium as described by Vailleau et al. (2007). Plants were grown in a growth chamber with 16h light (170 µmol m–2 s–1) at 25 °C and 8h dark at 23 °C.
Fungal isolates
V. albo-atrum V31-2 was grown on PDA medium at 24 °C in the dark. Spore suspensions were obtained by flooding 2-week-old cultures in Petri dishes with sterile water. The spore concentration was determined with a Malassez haematocytometer. Wild type and GFP-expressing strains were maintained on PDA plates at 14 °C or stored as spore suspensions in 25% glycerol at –80 °C.
Genetic transformation of V. albo-atrum
V. albo-atrum V31-2 was transformed with the GFP gene by Agrobacterium tumefaciens-mediated transformation as described by Eynck et al. (2007), using A. tumefaciens AGL-1 containing the binary vector pBin-GFP-hph. This vector carries the GFP gene under the control of the constitutive glyceraldehyde-3-phosphate dehydrogenase promoter of Aspergillus nidulans and a hygromycin B resistance marker (O’Connell et al., 2004). Equal volumes of fresh bacterial and spore suspensions were mixed and plated onto sterile cellophane sheets placed on co-cultivation medium containing 200 µM acetosyringone. After 2 days of incubation at 21 °C in the dark, the fungal and bacterial culture growing on the cellophane sheets was resuspended in 3ml of sterile 0.9% NaCl containing 200 µM cefotaxime, and the suspension was plated on selective Czapek Dox medium containing 200 µM cefotaxime and 50 µg.ml–1 hygromycin B for selection of transformants. Colonies of the fungus appeared after 5 days of incubation at 21°C and putative transformants were transferred to fresh Czapek Dox medium containing 50 µg.ml–1 hygromycin B. Monosporic strains were obtained after sporulation of fluorescent isolates on this medium. Their stability was tested by four generations of culture on PDA without selection pressure, then culture on PDA with 50 µg.ml–1 hygromycin B and assessment of their fluorescence under the microscope.
Inoculation and symptom scoring
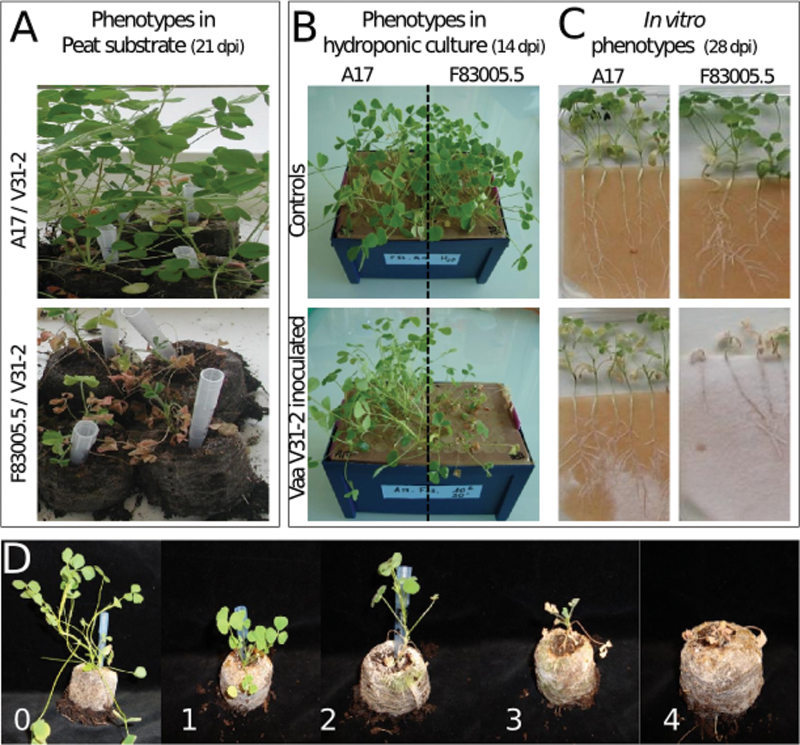
Inoculation experiments were performed at three sites using the same standard protocol: Ecolab at Auzeville, R2n at Druelle, and Barenbrug Tourneur at Mas Grenier. Suspensions of conidia were obtained as described above, and the concentration was adjusted to 106 spores ml–1 for all inoculations. Ten-day-old plants (first pair of leaves stage) grown in peat substrate were inoculated after cutting 1cm of the bottom of the Jiffy pots which were then dipped in the conidial suspension during 30 minutes before being transferred to trays containing moist soil. For microscopy studies, inoculation was performed with 10-day-old plants grown in hydroponic conditions by cutting 1cm of their roots and dipping them in conidial suspension for 30min before transferring them back to the nutritive solution. For in vitro experiments, 4-day-old plants grown on solid Farhaeus medium were inoculated with 100 µl of conidia solution after cutting 1cm of the root. In some assays, 3–4-week-old plants grown in Jiffy pots were inoculated with conidial suspension sprayed on cut aerial parts as described by Molinéro-Demilly et al. (2007). All inoculated plants and respective controls treated with water were incubated in a growth chamber at 20 °C with 16h photoperiod. Symptoms were rated on a scale from 0 to 4, similar to that described for alfalfa (http://www.naaic.org/stdtests/verticil.htm, Fig. 1D) and the proportion of dead plants (PDP) was determined at the end of the experiment.
Fig. 1.
Verticillium wilt symptoms in Medicago truncatula. (A–C) Phenotypes of the two most contrasted lines of M. truncatula (Jemalong-A17 and F83005.5) after root inoculation with a conidia suspension of Verticillium albo-atrum V31-2 (106 spores ml–1) in peat substrate (A), hydroponic system (B), and in vitro culture (C). (D) Symptom scale for disease index scoring. Scores from 0 to 4 correspond to characteristic stages of disease development.
The area under the disease progress curve (AUDPC, Shaner and Finney, 1977) was computed based on the disease scores from 0 to 4, using the ‘agricolae’ package of the R system (R Core Team, 2012). Model for disease progress empirical curves was based on a logistic model. The curves were fitted to disease index data based on the overall methodology described in Gilligan (1990). Model fitting was done using the ‘nlme’ package for the R system (Pinheiro et al., 2012). Full models had fixed components for line and when required, treatment and repetition effects and had random components for plants within line. When needed to improve model fit, autocorrelation among residuals was specified. Models were fitted using maximum-likelihood method, and analysis of variance (ANOVA) allowed testing for significance of effects. BLUE and BLUP of effects were obtained using the most parsimonious model for a given experimental design. The modelling allowed the description of the disease progress in terms of three functional parameters: (i) maximum disease index (MDI) at the end of the experiment; (ii) time to reach 50% MDI; and (iii) time to proceed from 50% MDI to (50% MDI)/(1 + e–1) (c.75% MDI).
Microscopic observations
Roots were rinsed briefly in water and 1-cm long fragments were embedded in 5% low melting point agarose. Longitudinal and radial sections of 80 and 45 µm, respectively, were prepared on a vibratom (VT 1000S, Leica, Germany) and mounted on a glass slide with distilled water. Confocal images were acquired with a spectral confocal laser scanning system (SP2 SE, Leica) equipped with an upright microscope (DM 6000, Leica). Observations were made using ×10 (HC PL Fluotar, N.A. 0.3) and ×40 (HCX PL APO, N.A. 0.8) dry and water immersion objectives, respectively. The 488-nm ray line of an argon laser was used to detect the GFP fluorescence emission collected in the range between 490 and 540nm.
Genetic maps
Previously described low-density genetic maps of A17 × DZA315.16 (LR4; Julier et al., 2007) and A17 × F83005.5 (LR5; Arraouadi et al., 2012) recombinant inbred line (RIL) populations were improved by adding simple sequence repeat markers based on the M. truncatula genome sequence assembly (Young et al., 2011, http://www.medicagohapmap.org/?genome) and unigene set of the Medicago Gene Index at DFCI (http://compbio.dfci.harvard.edu/). PCR conditions, gel electrophoresis, and genotype scoring were done as previously reported (Julier et al., 2007). The genetic map of the F83005.5 × DZA45.5 cross (LR3) is described in Hamon et al. (2010).
Quantitative trait locus analysis
Response to V. albo-atrum was evaluated in three different RIL populations and in different sites and/or years. For each RIL, 8–12 plants were inoculated as described above and scored regularly during 4 weeks. A total of 173 RILs of the A17 × F83005.5 (LR5) population in the F8 and/or F9 generations were evaluated in two different sites (ENSAT and Barenbrug-Tourneur), and AUDPC, disease curve parameters, and PDP were computed. Similarly, 137 RILs of the A17 × DZA315.16 (LR4) population in the F8 and/or F9 generations were evaluated in three sites (ENSAT, Barenbrug-Tourneur, and R2n) and 178 RILs of the F83005.5 × DZA45.5 (LR3; Hamon et al., 2010) RIL populations were evaluated at R2n in two independent experiments and mean PDP were determined.
Quantitative trait loci (QTLs) for resistance to V. albo-atrum were detected by Multiple QTL Mapping (MQM) (Jansen, 1993; Jansen and Stam, 1994) using the ‘qtl’ package (Broman et al., 2003; Arends et al., 2010) of the R system. AUDPC, PDP, and disease parameters were used as variables for QTL detection. Empirical threshold values for the logarithm of odd scores (LODs) were determined by computing 5000 permutations (Churchill and Doerge, 1994). Depending on the phenotype and the cross, the critical LOD score to indicate QTL significance ranges from 2.5 to 3.7. Heritabilities were computed using variance-components methods, by equating mean squares to their expectations and using WLS method for variance estimations (Kearsey and Pooni, 1996). All computations were done using the R system.
Results
V. albo-atrum is a pathogen of M. truncatula
To assess if the model legume M. truncatula is also a host plant for the alfalfa pathogen V. albo-atrum, the alfalfa isolate V31-2 was inoculated on six M. truncatula genotypes (Jemalong A17, A20, DZA45.5, DZA315.16, F83005.5, and TN1.11) which are parental lines of mapped RIL populations.
Young plants cultivated in peat substrate were root inoculated and compatible and incompatible interactions were identified: pronounced wilting symptoms and death of the plants were observed in some lines such as F83005.5 and DZA315.16 whereas other genotypes like A17 and DZA45.5 seemed to be highly resistant.
The two most contrasting lines A17 and F83005.5 (Fig. 1A) were also root inoculated in hydroponic and in vitro culture conditions. Again, plants developed typical disease symptoms and the same differential behaviour (Fig. 1B and 1C, respectively).
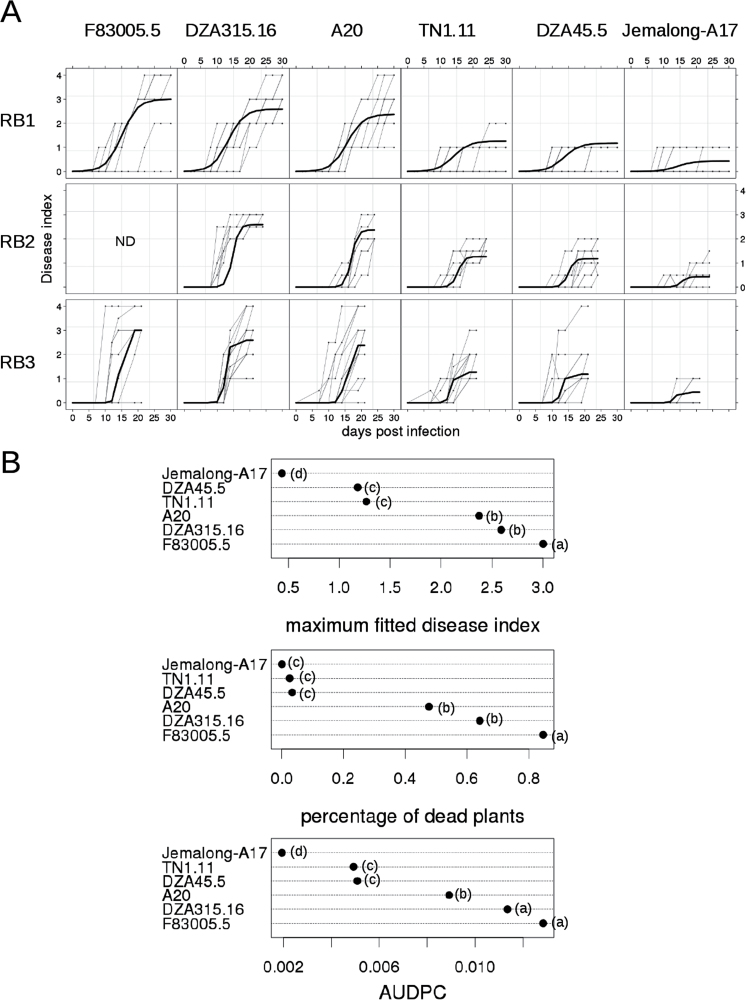
To quantify the degree of susceptibility, symptom development in the six parental lines was regularly scored on a scale from 0 to 4 (Fig. 1D). AUDPC and PDP at the end of the experiment were computed. Both disease parameters showed significant differences among the six parental lines (ANOVA P-values <2.10–16). The symptom curves were also modelled with a non-linear mixed model which allows characterizing the disease development more precisely with three functional parameters: the MDI which measures the severity of the disease, the time to reach 50% MDI which describes the onset of disease, and the time to proceed from 50 to 75% of the MDI which represents the rate of disease progression (Gilligan, 1990) (Fig. 2A). MDI was the only parameter of this model which showed significant differences among the six lines (ANOVA P-value <0.0001). As assessed by MDI and AUDPC values, line F83005.5 was the most susceptible one, followed by DZA315.16; A20 was intermediate, and DZA45.5, TN1.11, and A17 were resistant, with A17 being highly resistant (Fig. 2). When considering PDP, DZA45.5, TN1.11, and A17 all appeared highly resistant (Fig. 2B). Thus PDP seems to be less discriminating than AUDPC or MDI to describe disease resistance levels among genotypes. The same six lines were also spray inoculated on wounded aerial parts, as described for resistance screening in M. sativa (Molinéro-Demilly et al., 2007), and displayed similar phenotypes as in the root inoculation assay (Supplementary Fig. S1, available at JXB online). Taken together, the results showed that M. truncatula is a host plant for V. albo-atrum and that sources of resistance exist in the germplasm of this model plant.
Fig. 2.
Evolution of wilting symptoms and disease evaluation parameters of six Medicago truncatula lines with different levels of quantitative resistance. (A) Fitted logistic curves for disease kinetics for six genotypes (F83005.5, DZA315.16, A20, DZA45.5, TN1.11, Jemalong-A17) in three independent experiments (RB1, RB2, and RB3), following root inoculation in peat substrate with Verticillium albo-atrum V31-2 at 106 spores ml–1. Wilting symptoms were scored for 20–30 days after inoculation, on a scale from 0 to 4. Raw disease curves were modelled with a non-linear mixed model. Lines are ordered by decreasing susceptibility from left to right. (B) Distribution of the disease evaluation parameters after root inoculation among the six lines. Lines with the same letter belong to the same group of means, defined by Newman-Keuls test. AUDPC, area under disease progress curve. Data were obtained in three independent experiments performed at INP-ENSAT and R2n sites with 8–10 plants per line per experiment.
V. albo-atrum colonizes the host plant through xylem vessels
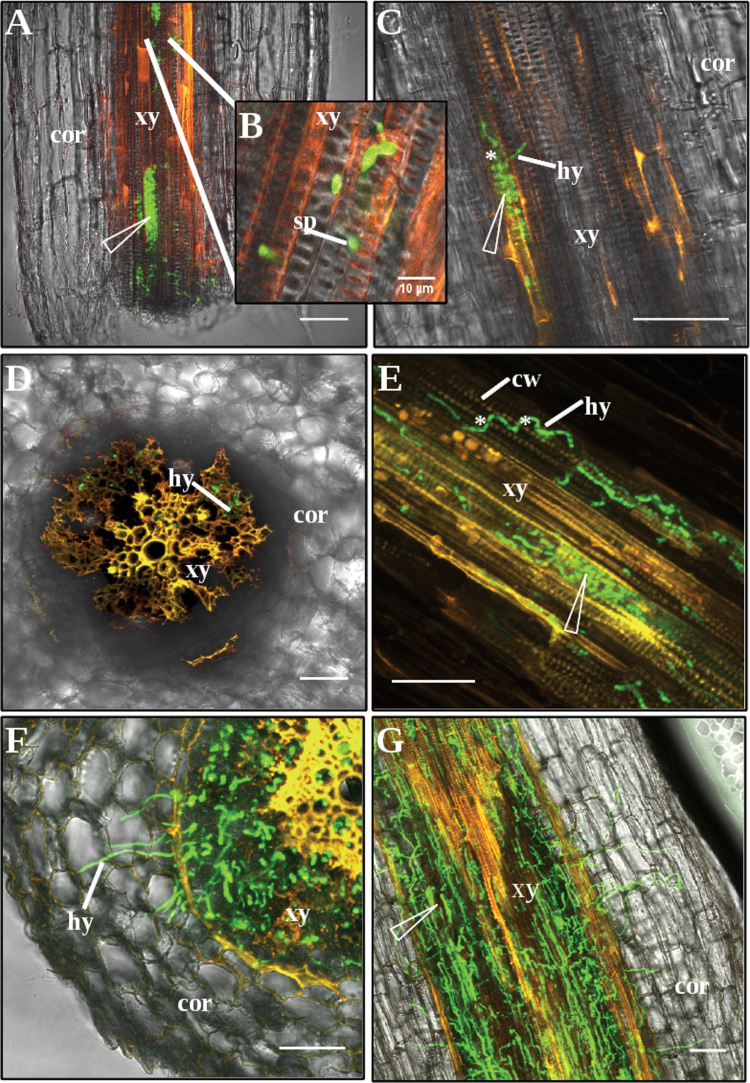
To study colonization by the pathogen, V. albo-atrum V31-2 was transformed, via A. tumefaciens, with the GFP reporter gene following a protocol described for V. dahliae (Eynck et al., 2007). Four monosporic fluorescent strains were obtained. Their growth rate on PDA medium and their aggressiveness on the susceptible M. truncatula line F83005.5 were similar to the wild type, and they were also unable to cause disease in the resistant line A17 (Supplementary Fig. S2). V. albo-atrum A1b2, which has high fluorescence, was then used for root inoculation in hydroponic cultures of the susceptible line F83005.5 and for microscopic observations. Roots were harvested at various times after inoculation, and sections were observed by confocal laser scanning microscopy. Conidia were rapidly absorbed into the xylem vessels by the transpiration stream as seen at 2 hours post inoculation (Fig. 3A). It was observed that their movement was stopped by the plaques which could then be crossed by germinating hyphae (Fig. 3B). Germination occurred as soon as after 3 hours. Hyphae then grew inside the vessels along the root axis but were also able to penetrate into adjacent xylem vessels (Fig. 3C). Sporulation was observed in xylem vessels after several days (data not shown). After 12 days, hyphae were detected in the hypocotyl, but the fungal growth was still restricted to the central cylinder. At later stages, when plants were severely diseased (i.e. symptom score >3.5 after 21 days), a massive production of mycelium was observed and hyphae colonized stele parenchyma, penetrated through the endodermis, and colonized the root cortex (Fig. 3F and 3G).
Fig. 3.
Root colonization of Medicago truncatula F83005.5 (susceptible line) by a GFP-expressing strain of Verticillium albo-atrum, observed by confocal laser scanning microscopy. (A) At 2 hours post inoculation (HPI), Conidia are seen in xylem vessels and accumulate at the end of a tracheid (arrow). (B) Conidia penetrating with its germination hypha through the plaque at the end of a xylem vessel (enlargement of A). (C) At 24 HPI, germinating conidia and hyphae. (D) At 7 days post inoculation (DPI), fungal hyphae in the central cylinder; no hyphae detected in the cortex. (E) At 7 DPI, proliferation of mycelium in xylem vessels and penetration into adjacent vessels (asterisk). (F and G) At 21 DPI, proliferation of mycelium in vascular (F) and cortical (G) tissues. Bars = 50 µm, and 10 µm in B. Longitudinal sections in A, B, C, E, G; cross-sections in D and F. cor = cortex; cw = cell wall; hy = hypha; sp = conidia; xy = xylem elements.
Taken together, the colonization pattern of V. albo-atrum in M. truncatula was typical for other described vascular wilt pathogens (Agrios, 2005; Fradin and Thomma, 2006) and confirmed that V. albo-atrum is a true pathogen of M. truncatula.
Evidence for diverse quantitative genetic mechanisms controlling resistance to V. albo-atrum in M. truncatula
Resistance parameters segregate in different RIL populations
To search for loci controlling the resistance to V. albo-atrum, RILs derived from crosses involving the most contrasting parental lines were analysed. Disease symptoms were screened within three RIL populations, namely 178 RILs of the DZA45.5 × F83005.5 (LR3) population, 137 RILs of the A17 × DZA315.16 (LR4) population, and 173 RILs of the A17 × F83005.5 (LR5) population.
The PDP at the end of the experiment was evaluated in different sites for all three populations (21–30 days post inoculation, depending on the site). For LR4 and LR5 populations, foliar wilting symptoms were also scored regularly and AUDPC and disease functional parameters were computed (Supplementary Fig. S3). For these two crosses, three independent experiments were performed in at least two different sites. LR3 population was analysed in two independent blocks at one site and mean PDP values were determined. ANOVA showed a significant effect of the trial (block or site) for most crosses and variables (Table 1). Yet, the high positive Spearman’s correlations obtained on genotype ranks for different repetitions showed that relative susceptibility levels of the RILs were reproducible over the different environments.
Table 1.
Variance analysis for several disease evaluation parameters in three RIL populations: proportion of dead plants at the end of the experiment, disease functional parameters, and AUDPC Significance levels are indicated for each factor. For LR3 population, only proportion of dead plants at the end of the experiment is available. [R] and [S] indicate resistant and susceptible parental lines, respectively. AUDPC, area under the disease progress curve; NS, not significant at α = 1%. Percentage of dead plants was tested by generalized linear model.
| Cross | Source of variation | df | Percentage of dead plants | Disease curve modelling | AUDPC | ||
|---|---|---|---|---|---|---|---|
| Maximum disease index | Time to 50% disease (days) | Rate of disease progression (days) | |||||
| LR3: F83005.5 [S] × DZA45.5 [R] | Block | 1 | NS | ||||
| Genotype | 212 | <2.10–16 | |||||
| LR4: A17 [R] × DZA315.16 [S] | Site | 2 | 2.10–9 | <2.10–16 | <2.10–16 | NS | <2.10–16 |
| Genotype | 158 | <2.10–16 | 4.10–10 | NS | NS | 4.10–9 | |
| LR5: A17 [R] × F83005.5 [S] | Site | 1 | NS | <2.10–16 | <2.10–16 | NS | 3.10–16 |
| Genotype | 159 | <2.10–16 | 1.10–5 | NS | NS | 1.10–5 | |
Among the four parameters describing the disease kinetics in LR4 and LR5 RILs, only MDI and AUDPC values were significantly different between genotypes (Table 1). ANOVA also showed significant genetic variation (P < 2.10–16) for the PDP at the end of the experiment in the three studied RIL populations. As expected, highly significant positive correlations were observed between these three parameters (Pearson’s correlation coefficient R > 0.95 in each population).
For each of these traits, some RILs showed lower or higher mean values compared to parental means, suggesting transgressions in phenotypes (Supplementary Fig. S4). Broad sense heritabilities, that describe the part of total phenotypic variation due to genetic variation, range from 31 to 33% for AUDPC and MDI values and can reach up to 65% for the PDP in LR5 population (Table 2). Taken together, the existence of transgressive lines and moderate to high heritabilities for all phenotypes confirm that the populations and traits are suited to QTL mapping through integration of RILs genotyping and phenotyping data.
Table 2.
Characteristics for the quantitative trait loci of resistance to Verticillium albo-atrum detected in LR3, LR4, and LR5 recombinant inbred line populations [R] and [S] indicate resistant and susceptible parental lines, respectively. h2, broad sense heritability; (H), narrow sense heritability; LOD, logarithm of odd score; QTL, quantitative trait locus.
| Cross | Trait | Trait heritability h2 (H) | Chromosome | Nearest marker | Peak LOD score | Peak (cM) | 1–LOD support interval (cM) | QTL effect | Percentage of phenotypic variance | Percentage of additive variance |
|---|---|---|---|---|---|---|---|---|---|---|
| LR3: F83005.5 [S] × DZA45.5 [R] | Percentage of dead plants | 0.32 (0.19) | 2 | EM4.250 | 6.0 | 196 | 193–197 | –0.095 | 22.0 | 36 |
| 6 | mtic343 | 3.6 | 142 | 133–154 | –0.067 | 18 | ||||
| LR4: A17 [R] × DZA315.16 [S] | Maximum disease index | 0.33 (0.20) | 7 | mtic432 | 11.1 | 60 | 55–70 | 0.41 | 32.2 | 92 |
| AUDPC | 0.31 (0.18) | 7 | mtic273 | 12.3 | 58 | 55–70 | 0.0022 | 32.9 | 83 | |
| Percentage of dead plants | 0.46 (0.29) | 7 | mtic82 | 5.7 | 76 | 65–78 | 0.0907 | 20.6 | 32 | |
| LR5: A17 [R] × F83005.5 [S] | Maximum disease index | 0.32 (0.19) | 7 | MTE85 | 22.6 | 58 | 54–61 | 0.5093 | 39.4 | 99 |
| AUDPC | 0.32 (0.19) | 7 | MTE85 | 26.1 | 58 | 54–61 | 0.0028 | 42.5 | 99 | |
| Percentage of dead plants | 0.65 (0.48) | 7 | MTE85 | 14.9 | 65 | 56–68 | 0.1973 | 29.3 | 48 |
Two distinct combinations of QTLs on chromosome 7 and chromosomes 2 and 6 govern resistance to Verticillium.
To facilitate precise QTL localization, marker densities were increased for LR4 (Julier et al., 2007) and LR5 (Arraouadi et al., 2012) genetic maps. Additional simple sequence repeats were thus genotyped in these two RIL populations. The LR4 map now comprises 370 loci, evenly distributed over each linkage group (Supplementary Fig. S5A). It spans a total of 910 cM with an average distance between markers of 3 cM. The LR5 map comprises 158 loci and spans a total of 780 cM with an average distance between markers of 5 cM (Supplementary Fig. S5B). The linkage groups of the genetic maps presented here are longer than in previously published maps. This shows that detection of crossing-over events was more accurate by increasing the number of mapped simple sequence repeats.
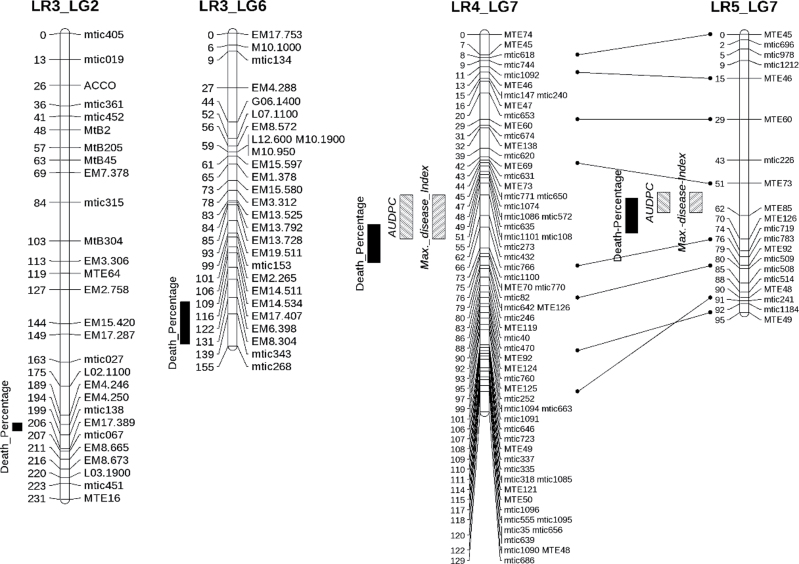
The RIL populations LR4 and LR5, both derived from crosses involving the resistant Jemalong-A17 line as a parent, were first analysed for response to V. albo-atrum. A single QTL, located on linkage group 7, was detected for both populations and with each of the three disease parameters (Table 2). The 1-LOD support intervals of the QTL for each trait ranged from 7 to 15 cM and were overlapping. This suggests that, for both populations, the same genomic region between markers MTE73 and MTE126, is involved in the genetic control of Verticillium wilt (Fig. 4). This single major QTL, named MtVa1, was detected with data obtained in different sites for both populations (Supplementary Fig. S6). The MtVa1 QTL showed high LOD scores (up to 26 for AUDPC in LR5) and accounted for large effects, as it explained from 20.6% of the phenotype for the PDP in LR4 to 42.5% of phenotypic variation for AUDPC parameter in LR5. The occurrence of one single QTL suggests also that the two susceptible lines F83005.5 and DZA315.16 do not contain additional resistance loci with strong effects.
Fig. 4.
Position of quantitative trait loci (QTL) for Verticillium albo-atrum resistance in Medicago truncatula assessed through maximum fitted disease index and AUDPC in LR4 and LR5 populations and proportion of dead plants in LR3, LR4, and LR5 populations. Bars at the left of considered linkage groups indicate 1 logarithm of odd score support for QTL interval.
In order to study resistance in a parent different from Jemalong-A17, the cross F83005.5 (susceptible) × DZA45.5 (resistant) (LR3) was used. QTL analysis of PDP in this mapping population evidences one major locus on chromosome 2 and another minor locus on chromosome 6 (Table 2). These QTLs, called MtVa2 and MtVa3, had respective LOD scores of 6.0 and 3.6, and together accounted for 22% of the resistance phenotype and covered confidence intervals of 4 and 21 cM.
At each of the three loci detected by analysis of the different RIL populations, the allele from the resistant parent (A17 or DZA45.5 depending on the concerned RIL populations) was associated with lower susceptibility level computed as AUDPC, MDI, or PDP (Table 2).
V. albo-atrum resistance QTLs do not co-localize with identified Ve homologues
To check if Ve gene homologues are localized in the region underlying the QTLs, a blastp analysis on the M. truncatula genome was performed using the peptide sequence of tomato VE1 (GeneBank accession AAK58681.2). Eight homologous sequences were detected which were further analysed by multiple alignment and establishment of a phylogenetic tree using clustal w version 2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Supplementary Fig. S7A). Among them, seven are localized on chromosome 4 and one on chromosome 5. Hence these homologous sequences do not co-localize with the major QTLs of partial resistance to V. albo-atrum in this study’s pathosystem.
The four most homologous candidates (Medtr4g017280.1, Medtr4g017350.1 Medtr4g017370.1, and Medtr5g046350.1) were compared with the complete open reading frame amino-acid sequences of Ve genes described to be involved in resistance to Verticillium wilt, such as Ve1 and Ve2 from tomato, StVe from Solanum torvum, GbVe from cotton, as well as MsVe1 and MlVe1 from Mentha spicata and Mentha latifolia respectively (Supplementary Fig. S7B). The comparison shows a high diversity in the peptide sequence of the validated Ve proteins with identities ranging from 42 to 96%. The four M. truncatula homologues share from 69 to 91% identity. The highest homology between the candidates and validated Ve proteins was 46% identity for three Mt genes with Ve1. The four candidates have similar protein structures as Ve, such as LRRs, signal peptides, and transmembrane domains (Supplementary Fig. S7C).
M. truncatula presents a wide biodiversity in its response to V. albo-atrum
The QTL analysis revealed that different resistance sources exist within the species. To gain a broader view of this finding, the natural biodiversity in response to V. albo-atrum infection was analysed by phenotypic evaluation of a M. truncatula core collection of 32 lines (CC32), representing the genetic variability within this Mediterranean natural weed (Ronfort et al., 2006). ANOVA indicated statistically significant differences in terms of disease onset and severity in the responses of these lines (Fig. 5A, Table 3, and Supplementary Table S1). The response to V. albo-atrum of the CC32 lines appeared to be independent of the M. truncatula subpopulation structure defined with neutral markers by Ronfort et al. (2006). This result (Fig. 5B), was confirmed through ANOVA, which showed that P-values for the subpopulations effect were non-significant for MDI and for disease onset (P = 0.26 and P = 0.12, respectively).
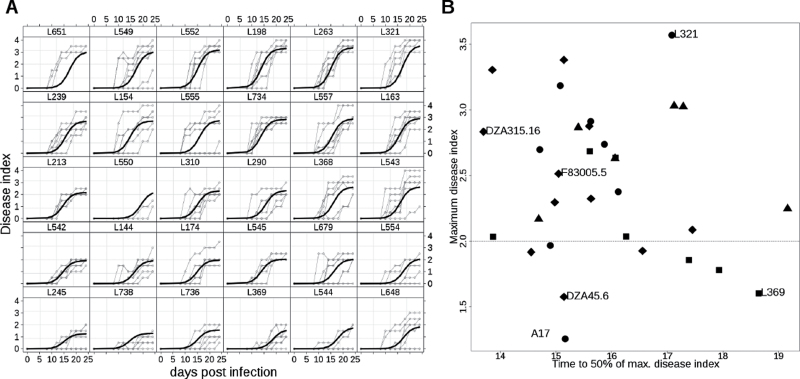
Fig. 5.
Diversity in the response of Medicago truncatula to Verticillium albo-atrum within the Core Collection 32. (A) Fitted logistic curves for disease kinetics; M. truncatula lines are ordered from top left to bottom right by increasing maximum disease index, from the resistant line Jemalong-A17 (L738) to the most susceptible line L321; similar results were obtained in two independent experiments. (B) Quantitative resistance levels as evaluated through maximum fitted disease index and time to reach 50% of maximum disease index, adjusted over two biological repeats. Each of the four putative M. truncatula sub-populations (Ronfort et al. 2006) is identified by a distinct symbol (1=●, 2=■, 3=◆, 4=▲). DZA45.6 is a sister line of DZA45.5.
Table 3.
Disease functional parameters in the core collection 32 of Medicago truncatula. Lines are ordered in increasing maximum disease index parameter (i.e. from most resistant to most susceptible. Plants were grown in peat substrate and inoculated by a conidial suspension of Verticillium albo-atrum (106 spores ml–1). Disease symptom scoring was done in two independent experiments. Populations are noted according to Ronfort et al. (2006). CC32, core collection 32.
| CC32 line code | Usual name | Maximum disease index | Time to 50% disease (days) | Geographical origin | Population |
|---|---|---|---|---|---|
| L245 | SA014161 | 1.26 | 15.17 | Jordan | 1 |
| L738 | Jemalong A17 | 1.29 | 14.51 | Australia | |
| L736 | DZA045.6 | 1.58 | 15.14 | Algeria | 3 |
| L369 | PRT180-A | 1.60 | 18.64 | Portugal | 2 |
| L544 | ESP105-L | 1.78 | 17.93 | Spain | 2 |
| L648 | SALSES 42B | 1.86 | 17.39 | France | 2 |
| L542 | DZA233.4 | 1.92 | 14.55 | Algeria | 3 |
| L144 | SA014163 | 1.93 | 16.56 | Jordan | 3 |
| L174 | SA028064 | 1.97 | 14.90 | Cyprus | 1 |
| L545 | ESP 158-A | 2.03 | 13.87 | Spain | 2 |
| L679 | F66017 | 2.04 | 16.26 | France | 2 |
| L554 | F20089-B | 2.09 | 17.45 | France | 3 |
| L213 | SA027882 | 2.17 | 14.69 | Morocco | 4 |
| L550 | F11013-3 | 2.25 | 19.16 | France | 4 |
| L310 | SA009944 | 2.30 | 14.98 | Tunisia | 3 |
| L290 | SA009119 | 2.32 | 15.63 | Turkey | 3 |
| L337 | GRC 042-1 | 2.38 | 16.12 | Greece | 1 |
| L530 | F83005-5 | 2.51 | 15.05 | France | 3 |
| L368 | DZA 012-J | 2.63 | 16.06 | Algeria | 4 |
| L543 | DZA 327-7 | 2.64 | 16.07 | Algeria | 2 |
| L239 | SA026063 | 2.69 | 15.61 | Morocco | 2 |
| L154 | SA024714 | 2.70 | 14.71 | Italy | 1 |
| L555 | GRC 020-B | 2.74 | 15.87 | Greece | 1 |
| L734 | DZA315.16 | 2.83 | 13.70 | Algeria | 3 |
| L049 | SA009707 | 2.87 | 15.40 | Tunisia | 4 |
| L557 | GRC 064-B | 2.88 | 15.60 | Greece | 3 |
| L163 | SA022322 | 2.91 | 15.62 | Syria | 1 |
| L651 | SALSES 71B | 3.02 | 17.29 | France | 4 |
| L549 | F11005-E | 3.03 | 17.12 | France | 4 |
| L552 | F20047-A | 3.18 | 15.08 | France | 1 |
| L198 | SA009048 | 3.30 | 13.86 | Libya | 3 |
| L263 | SA003116 | 3.38 | 15.14 | Israel | 3 |
| L321 | SA003780 | 3.57 | 17.08 | Italy | 1 |
Phenotypic screening of the CC32 also showed that the speed of disease onset was independent of the symptom severity at the end of the experiment (Fig. 5B). Thus, classes of typical behaviour could be identified. Among highly susceptible lines, a group represented by DZA315.16, presented fast appearance of symptoms whereas another group, typified by L321, showed a slower evolution of the disease. The same variability of response kinetics was observed in the most resistant lines, with some lines such as A17 that showed weak symptoms even early after inoculation and some genotypes such as L369 for which weak symptoms developed late. These results suggest the existence of diverse genetic systems controlling the resistance to Verticillium within M. truncatula species.
This evaluation is leading to the identification of even more contrasted lines than the parents used in the present study, notably more susceptible ones which exhibit fast and intensely evolving wilting symptoms. In the future, it could be interesting to study the progenies of new crosses involving the most contrasted lines; such populations are currently under development.
Response to Verticillium interacts with nodulation pathways
As a legume plant, M. truncatula has the ability to interact with both root symbiotic or pathogenic microbes. As a preliminary survey to identify putative crosstalks in signalling pathways involved in plant defence and nodulation, ten mutants of M. truncatula known to be affected in distinct steps of the rhizobia symbiosis were inoculated with V. albo-atrum (for review, see Kouchi et al., 2010). NFP and DMI2 membrane receptors perceive the Nod factors released by the soil bacterium Sinorhizobium meliloti and initiate the signal cascade involving proteins such as DMI1, DMI3, and NSP1 to elicit the gene expression programme leading to infection and nodulation. HCL, LIN, and RPG play roles in the rhizobia infection and SUNN and SICKLE participate to the autoregulation of the symbiotic process. These mutants are in the genetic background of the resistant line Jemalong-A17.
The hypernodulation mutants sickle and sunn showed altered but opposed responses to V. albo-atrum (Table 4 and Supplementary Fig. S8). As assessed by all three parameters of the disease curve, sunn was significantly more susceptible than the wild type, with increased wilting symptoms and faster disease development. In contrast, sickle seemed to be more resistant than the wild type as evidenced by the clearly slower disease progression and the subsignificant reduction in symptom severity. The nfp mutant appeared to be more susceptible to V. albo-atrum than the wild type, as revealed by the slight increase in MDI. Besides these three mutants that showed highly altered disease phenotypes, three other ones, namely dmi1, lin, and nsp1, also showed some increase in their resistance level as revealed by the significant reduction in the disease progression rate.
Table 4.
Comparison of disease functional parameters of Medicago truncatula nodulation mutants Mutants are in the Jemalong-A17 (resistant) genetic background. Plants were grown in peat substrate and inoculated by a conidial suspension of V. albo-atrum (106 spores ml–1). Disease symptom scoring was done in three independent experiments. P-values refer to test for difference between the parameter of the mutant line compared to that of the wild reference Jemalong-A17. Bold indicates significant (P < 0.05) or subsignificant (P < 0.10) differences compared to the reference line Jemalong-A17. [R] and [S] indicate resistant and susceptible lines, respectively.
| Line | Maximum disease index | Time to 50% disease (days) | Rate of disease progression (days) | |||
|---|---|---|---|---|---|---|
| Value | P | Value | P | Value | P | |
| Mutants | ||||||
| nfp | 1.3 | 0.051 | 24.6 | 0.210 | 2.7 | 0.293 |
| dmi2 | 1.1 | 0.233 | 19.1 | 0.144 | 2.3 | 0.810 |
| dmi1 | 1.0 | 0.387 | 19.7 | 0.311 | 3.8 | 0.001 |
| dmi3 | 1.1 | 0.176 | 21.2 | 0.754 | 1.9 | 0.459 |
| nsp1 | 0.9 | 0.640 | 24.2 | 0.244 | 3.4 | 0.021 |
| hcl | 0.6 | 0.465 | 21.3 | 0.787 | 2.4 | 0.505 |
| lin | 0.9 | 0.680 | 22.1 | 0.865 | 2.9 | 0.064 |
| rpg | 1.0 | 0.293 | 24.2 | 0.264 | 1.6 | 0.219 |
| skl | 0.4 | 0.092 | 24.8 | 0.210 | 3.2 | 0.013 |
| sunn | 1.3 | 0.042 | 18.3 | 0.065 | 0.8 | 0.000 |
| References | ||||||
| A17 [R] | 0.8 | – | 21.8 | – | 2.2 | – |
| F8300.5 [S] | 2.8 | 0.000 | 16.7 | 0.004 | 2.0 | 0.636 |
Discussion
In order to study Verticillium wilt in legumes, a new pathosystem involving the model legume M. truncatula and an alfalfa isolate of V. albo-atrum was established in this study. Susceptible and resistant lines were identified with different inoculation methods that reproducibly established lines A17 and DZA45.5 as resistant and lines F83005.5 and DZA315.16 as susceptible. The colonization of susceptible roots as observed with a GFP-expressing V. albo-atrum strain was limited to the vascular system until the final stage of disease when the fungus invaded the whole plant tissue. This is in agreement with colonization patterns reported in other host plants (Fradin and Thomma, 2006) and confirms M. truncatula as a true host plant for V. albo-atrum. The tools available for this model plant allowed the study of the genetic basis of resistance to V. albo-atrum.
Analysis of three RIL populations led to the identification of three QTLs for resistance in two resistant parental lines. These loci are located on linkage group 7 for A17 and on linkage groups 2 and 6 for DZA45.5, respectively. The analysis revealed thus that only a small number of major QTLs are involved in resistance to V. albo-atrum in M. truncatula, suggesting a simple genetic control. Model fitting of disease progression curves in RIL populations allowed this study to show that these resistance traits are more characterized by lower symptoms severity at the end of the experiment, rather than delay in disease onset or lower disease progression rate. AUDPC combines all the three functional parameters and may be less powerful in discriminating between resistance QTLs for disease progression and resistance QTLs for symptoms severity. These two types of resistance traits probably involve distinct genetic mechanisms.
Experiments in three different sites with two RIL populations using A17 as the resistant parent led to the identification of the same major QTL on linkage group 7. Thus, while environmental variations significantly affect the RILs’ responses to Verticillium (Table 1), they do not modify the genetic control for resistance. Such a stability in QTL effects towards environments and resistance parameters has already been reported in several pathosystems (Buerstmayr et al., 2009; Hamon et al., 2011; Lu et al., 2012). A survey of the biodiversity of the response to V. albo-atrum infection revealed a large variety of response, in terms of disease onset and severity, suggesting that different genetic systems may exist within the species. Response to Verticillium wilt does not appear to be correlated with population structure based on neutral markers (Ronfort et al., 2006) which represents the ancient history of the species. This suggests that resistance to Verticillium is being selected within environments. In their native environment, M. truncatula populations may encounter different Verticillium strains, against which distinct resistance mechanisms may have evolved. Depending on the QTL, the resistance towards V. albo-atrum V31-2 may or may not be affected by environmental conditions as exemplified by the range of additive variance explained by this QTL (36% for the LR4 and 99% for LR5 RILs populations). Thus, whether or not the resistance mechanisms evolved in the natural populations are influenced by environmental variations remains to be studied. A more in-depth assessment of this hypothesis may be promoted by mapping QTLs in additional RIL populations derived from crosses between lines with strong contrasts in the response to Verticillium. Additionally, putative correlation of the resistance with geographical origin of the accessions may be detected using a wider collection of genotypes and comparing with eco-geographical covariates.
Genetic studies of resistance or tolerance against Verticillium wilt have been reported for several plant species (Schaible et al., 1951; Diwan et al., 1999; Bae et al., 2008; Yang et al., 2008a). The best-characterized example is monogenic resistance against V. albo-atrum and V. dahliae in tomato, where the resistance gene Ve1 is effective against race 1 of both species (Diwan et al., 1999; Fradin et al., 2009). Homologues of the Ve gene have been reported in other species as well (Fradin and Thomma, 2006). In M. truncatula, four Ve homologues, exhibiting 41–46 % identity at the protein level with Ve genes from other plant species were detected. As described for tomato Ve1 and Ve2 which occur in tandem (Fradin et al., 2009), most of the putative M. truncatula Ve homologues were localized near each other. Although these genes are on chromosomes 4 and 5 and do not co-localize with the resistance QTLs identified in the present study, it cannot be excluded that another homologue exists in the unsequenced regions underlying part of the QTLs. Although 94% of the gene space of M. truncatula genome is now sequenced (Young et al., 2011), information is still lacking for some regions. Taking into account the large confidence interval of the MtVa1 QTL covering at least 5 Mbp and the existence of sequence gaps, identification of putative candidate genes predicted in this region was not possible.
In Arabidopsis, overlaps between plant development and susceptibility to Verticillium wilt have been described. Notably, early flowering was reported to be correlated to susceptibility to the Brassicaceae-infecting Verticillium longisporum (Veronese et al., 2003). QTLs involved in colonization by V. longisporum were reported to be localized in the vicinity of loci for the regulation of flowering such as constans and flowering locus (Häffner et al., 2010). A Constans-like gene on chromosome 7 has been proposed recently to control flowering date in M. truncatula (Pierre et al., 2011). This gene localizes 25 cM south of the QTL involved in resistance to V. albo-atrum, indicating that M. truncatula resistance does not seem to interfere with regulation of flowering, under the experimental conditions.
M. truncatula is now well established as a model for the study of plant–pathogen interactions, involving bacteria, fungi, oomycetes, nematodes, and aphids (Torregrosa et al., 2004; Vailleau et al., 2007; Ameline-Torregrosa et al., 2008; Dhandaydham et al., 2008; Djébali et al., 2009; Stewart et al., 2009; Uppalapati et al., 2009; Ramirez-Suero et al., 2010). The lines A17 and F83005.5 are often compared for pathogenic interactions and used for comparative and/or genetic approaches to study the interaction with C. trifolii (Ameline-Torregrosa et al., 2008), A. euteiches (Djébali et al., 2009), R. solanacearum (Vailleau et al., 2007), and Fusarium oxysporum (Ramirez-Suero et al., 2010). Genetic studies using RIL populations derived from a cross between lines A17 and F83005.5 revealed different quantitative mechanisms of resistance to R. solanacearum, A. euteiches, and C. trifolii, which always rely on a small number of loci. Two major QTLs against A. euteiches were reported in overlapping genomic regions of LG3 in two different RIL populations, LR5 (Djébali et al., 2009) and LR3 (Pilet-Nayel et al., 2009). These populations were also used by this study which, in contrast, led to reveal that different QTLs control resistance in the parents Jemalong-A17 and DZA45.5. So far, only one disease resistance gene has been cloned in M. truncatula. RCT1 controls resistance against C. trifolii in line A17 and codes for a protein of the TIR-NBS-LRR type. Introduction of RCT1 from M. truncatula into alfalfa conferred broad resistance against the anthracnose fungus (Yang et al., 2008b), confirming the utility of model plants for knowledge transfer to cultivated crops.
Compared to the model plant Arabidopsis thaliana, M. truncatula allows studying crosstalk between pathways involved in symbiotic and pathogenic interactions. Such crosstalk has been demonstrated by Penmetsa et al. (2008) by using sickle, one of the numerous nodulation mutants in M. truncatula. This mutant was first identified by its hypernodulating phenotype and the Sickle gene was shown to be an orthologue of the A. thaliana Ein2 gene involved in ethylene signalling. It was also found more susceptible in its interactions with the root pathogens Rhizoctonia solani and Phytophthora medicaginis. Four out of the ten tested nodulation mutants, skl, dmi1, lin, and nsp1, showed putatively enhanced resistance to V. albo-atrum as shown by reduced disease progression. However, the high resistance of the wild-type A17 makes enhanced resistance levels difficult to detect. In order to confirm these observations, a screening with V. albo-atrum strains for which line A17 is susceptible will be performed.
In this study, sickle was slightly more resistant than the wild type, indicating that the ethylene signalling pathway could be necessary for Verticillium wilt development. The Sickle gene (Mtr7g101410, corresponding to TC176805; Penmetsa et al., 2008) is located near locus mtic337, approximately 50 cM south of the MtVa1 QTL peak on chromosome 7. Data from the literature indicate that ethylene signalling is involved in the response to Verticillium wilt, but its role seems to be complex since different mutants in the ethylene pathway have different disease phenotypes (Veronese et al., 2003; Johansson et al., 2006; Pantelides et al., 2010).
The second hypernodulation mutant, sunn, was also shown to be modified in response to V. albo-atrum in this study, but the phenotype was opposite to that of skl (i.e. the mutant was less resistant than the wild type). The sunn mutant is altered in a CLAVATA1-like LRR receptor-like kinase (RLK) (Schnabel et al., 2005). The Sunn gene (Medtr4g070970, which corresponds to accession AY769943) is located on linkage group 4. Besides hypernodulation, the mutant also shows increased mycorrhization (Sagan et al., 1995). Although the role of LRR-RLK in plant disease resistance is well established now, this is the first time that the sunn mutant has been shown to be modified in response to a pathogen.
The nfp mutant, which is defective in a putative Nod factor receptor, is also affected in its resistance to V. albo-atrum. Since the backbone of this factor is a chitin oligomer, a modified phenotype might suggest involvement of PAMP perception such as chitin-derived compounds of fungal walls.
As far as is known, this study is the first where a collection of nodulation mutants was assessed for its response to a pathogen. Several mutants, affected in different steps of the nodulation process, show altered response to Verticillium wilt. The results emphasize the existence of crosstalks between signalling pathways in symbiosis and disease, and the need for detailed studies of Verticillium wilts and other root pathogens in legume plants. Such crosstalks with symbiosis should be considered in breeding for legume tolerance for root diseases, in order to avoid putative trade off between nodulation and response to root diseases. It also raises questions concerning hopes to transfer rhizobial symbioses to non-legumes, as it may endanger the tolerance mechanisms to root disease in these crops.
Future work aims at cloning the MtVa1 resistance gene by fine mapping the major QTL. However, remaining gaps in the sequences under this QTL render this objective a long-term goal relying on additional sequencing efforts in M. truncatula. Transcriptomic studies have now been initiated to describe the molecular response of the resistant A17 and the susceptible F83005.5 lines. The gene expression profiles might also reveal candidate resistance genes if regulated at the transcriptional level. As more M. truncatula genotypes and Verticillium sp. strains are evaluated, there will be a focus on the discovery of new resistance loci through an enlargement of the analysis of biodiversity and subsequent exhaustive genome-wide genetics association studies. This may give insights towards adaptation to root diseases in Mediterranean legumes.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Comparisons of the quantitative resistance levels of six Medicago truncatula lines depending on inoculation method with Verticillium albo-atrum.
Supplementary Fig. S2. Analysis of the growth and aggressiveness of four monosporic isolates of Verticillium albo-atrum expressing the green fluorescent protein gene in comparison to the wild-type V31-2.
Supplementary Fig. S3. Example of model fitting for wilting symptoms in 113 LR4 recombinant inbred lines at site R2N (A) and 158 LR5 recombinant inbred lines at site ENSAT (B).
Supplementary Fig. S4. Distribution of disease evaluation parameters in LR3, LR4, and LR5 recombinant inbred lines upon infection with Verticillium albo-atrum V31-2.
Supplementary Fig. S5. Dense genetic maps for A17 × DZA315.16 (LR4, A) and A17 × F83005.5 (LR5, B) recombinant inbred line populations of Medicago truncatula.
Supplementary Fig. S6. Whole-genome scans for quantitative trait loci detection for Verticillium resistance assessed through maximum disease index and area under disease progress curve parameters in LR4 and LR5 populations for each site.
Supplementary Fig. S7. Identification of four Medicago truncatula paralogues putatively orthologous to Verticillium resistance genes.
Supplementary Fig. S8. Response to Verticillium albo-atrum inoculation of Medicago truncatula nodulation mutants.
Supplementary Table S1. Variance analysis for functional disease parameters in the core collection 32, including Jemalong-A17.
Acknowledgements
MT was supported by a doctoral grant from Mayotte department, and AN by a doctoral grant from the French Government. The authors thank S. Penverne (FR3450 Toulouse) for training at the confocal microscope, and V. Olive, S. Amatya, S. Suwal, and R. Oustrières who helped with the inoculations and phenotyping experiments, as well as J. Faivre d’Arcier for help with genetic transformation of V. albo-atrum. The authors also thank D. Cook (University of California at Davis, USA) for kindly providing M. truncatula mutant lines. Part of this work was financed by a Contrat de Branche by the French Ministry of Agriculture (coordinated by BJ).
References
- Acharya SN, Huang AJ. 2003. Breeding alfalfa for resistance to verticillium wilt: a sound strategy. In: Huang HC, Acharya SN, eds, Advances in plant disease management. Trivandrum, India: Research Signpost, pp 345–371 [Google Scholar]
- Agrios GN. 2005. Plant pathology, 5thedn. USA, UK: Elsevier Academic Press; [Google Scholar]
- Ameline-Torregrosa C, Cazaux M, Danesh D, et al. 2008. Genetic dissection of resistance to anthracnose and powdery mildew in Medicago truncatula . Molecular Plant–Microbe Interactions. 21, 61–69 [DOI] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, Broman KW. 2010. R/qtl: high-throughput multiple QTL mapping. Bioinformatics. 26, 2990–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraouadi S, Badri M, Abdelly C, Huguet T, Aouani ME. 2012. QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula recombinant inbred lines. Genomics. 99, 118–125 [DOI] [PubMed] [Google Scholar]
- Bae J, Halterman DA, Jansky SH. 2008. Development of a molecular marker associated with Verticillium wilt resistance in diploid interspecific potato hybrids. Molecular Breeding. 22, 61–69 [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19, 889 –890 [DOI] [PubMed] [Google Scholar]
- Buerstmayr H, Bant T, Anderson JA. 2009. QTLmapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breeding. 128, 1–26 [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics. 138, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, van Esse PH, Maruthachalam K, et al. 2012. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proceedings of the National Academy of Sciences, USA. 109, 5110–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhandaydham M, Charles L, Zhu H, Starr JL, Huguet T, Cook DR, Prosperi JM, Opperman C. 2008. Characterisation of root-knot nematode resistance in Medicago truncatula . Journal of Nematology. 40, 46–54 [PMC free article] [PubMed] [Google Scholar]
- Diwan N, Fluhr R, Eshed Y, Zamir D, Tanksley SD. 1999. Mapping of Ve in tomato: a gene conferring resistance to the broad-spectrum pathogen Verticillium dahliae race 1. Theoretical and Applied Genetics. 98, 315–319 [Google Scholar]
- Djébali N, Jauneau A, Ameline-Torregrosa C, et al. 2009. Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Molecular Plant–Microbe Interactions. 22, 1043–1055 [DOI] [PubMed] [Google Scholar]
- Eynck C, Koopmann B, Grunewaldt-Stoecker G, Karlovsky P, von Tiedemann A. 2007. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. European Journal of Plant Pathology. 118, 259–274 [Google Scholar]
- Farhäeus G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass technique. Journal of General Microbiology. 16, 374–381 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MHAJ, Thomma BPHJ. 2011. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis . Plant Physiology. 156, 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Bart PHJ, Thomma BPHJ. 2006. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum . Molecular Plant Pathology. 7, 71–86 [DOI] [PubMed] [Google Scholar]
- Fradin EF, Zhang Z, Ayala JCJ, Castroverde CDM, Nazar RN, Robb J, Liu CM, Thomma B PHJ. 2009. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1 . Plant Physiology. 150, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan CA. 1990. Comparison of disease progress curves. New Phytologist. 115, 223–242 [DOI] [PubMed] [Google Scholar]
- Gough C, Cullimore J. 2011. Lipo-chitooligosaccharide signaling in endosymbiotic plant–microbe interactions. Molecular Plant–Microbe Interactions. 24, 867–878 [DOI] [PubMed] [Google Scholar]
- Häffner E, Karlovsky P, Diederichsen E. 2010. Genetic and environmental control of the Verticillium syndrome in Arabidopsis thaliana . BMC Plant Biology. 10, 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon C, Baranger A, Miteul H, et al. 2010. A complex genetic network involving a broad-spectrum locus and strain-specific loci controls resistance to different pathotypes of Aphanomyces euteiches in Medicago truncatula . Theoretical and Applied Genetics. 120, 955–970 [DOI] [PubMed] [Google Scholar]
- Hamon C, Baranger A, Coyne CJ, et al. 2011. New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in multiple French and American environments. Theoretical and Applied Genetics. 123, 261–281 [DOI] [PubMed] [Google Scholar]
- Hayes RJ, McHale LK, Vallad GE, Truco MJ, Michelmore RW, Klosterman SJ, Maruthachalam K, Subbarao KV. 2011. The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. Theoretical and Applied Genetics. 123, 509–517 [DOI] [PubMed] [Google Scholar]
- Jansen RC. 1993. Interval mapping of multiple quantitative trait loci. Genetics. 135, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P. 1994. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 136, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Staal J, Dixelius C. 2006. Early responses in the Arabidopsis-Verticillium longisporum pathosystem are dependent on NDR1, JA- and ET-associated signals via cytosolic NPR1 and RFO1. Molecular Plant–Microbe Interactions. 19, 958–969 [DOI] [PubMed] [Google Scholar]
- Julier B, Huguet T, Chardon F, Ayadi R, Pierre JB, Prosperi JM, Barre P, Huyghe C. 2007. Identification of quantitative trait loci influencing aerial morphogenesis in the model legume Medicago truncatula . Theoretical and Applied Genetics. 114, 1391–1406 [DOI] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, et al. 2001. Tomato Ve disease resistance genes encode cell surface-like receptors. Proceedings of the National Academy of Sciences, USA. 98, 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey MJ, Pooni HS. 1996. The genetical analysis of quantitative traits. London, UK: Chapman and Hall; [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. 2009. Diversity, pathogenicity, and management of Verticillium species. Annual Review of Phytopathology. 47, 39–62 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. 2010. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiology. 51, 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Bjørnstad Å, Ren Y, Asad MA, Xia X, Chen X, Ji F, Shi J, Lillemo M. 2012. Partial resistance to powdery mildew in German spring wheat ‘Naxos’ is based on multiple genes with stable effects in diverse environments. Theoretical and Applied Genetics. 125, 297–309 [DOI] [PubMed] [Google Scholar]
- Mithöfer A. 2002. Suppression of plant defence in rhizobia–legume symbiosis. Trends in Plant Science. 7, 440–444 [DOI] [PubMed] [Google Scholar]
- Molinéro-Demilly V, Montegano B, Julier B, Giroult C, Baudouin P, Chosson JF, Bayle B, Noël D, Guénard M, Gensollen V. 2007. Resistance to Verticillium albo-atrum in Lucerne (Medicago sativa L.) to distinguish between varieties. Euphytica. 153, 227–232 [Google Scholar]
- O’Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerré-Tugayé MT, Dumas B. 2004. A novel Arabidopsis–Colletotrichum pathosystem for the molecular dissection of plant–fungal interactions. Molecular Plant–Microbe Interactions. 17, 272–282 [DOI] [PubMed] [Google Scholar]
- Pantelides IS, Tjamos SE, Paplomatas EJ. 2010. Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae . Molecular Plant Pathology. 11, 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Uribe P, Anderson J, et al. 2008. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. The Plant Journal. 55, 580–595 [DOI] [PubMed] [Google Scholar]
- Pennypacker BW. 2000. Differential impact of carbon assimilation on the expression of quantitative and qualitative resistance in alfalfa (Medicago sativa). Physiological and Molecular Plant Pathology. 57, 87–93 [Google Scholar]
- Pierre JB, Bogard M, Herrmann D, Huyghe C, Julier B. 2011. A CONSTANS-like gene candidate that could explain most of the genetic variation for flowering date in Medicago truncatula . Molecular Breeding. 28, 25–35 [Google Scholar]
- Pilet-Nayel ML, Prospéri JM, Hamon C, et al. 2009. AER1, a major gene conferring resistance to Aphanomyces euteiches in Medicago truncatula . Phytopathology. 99, 203–208 [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. 2012. nlme: linear and nonlinear mixed effects models. R package version 3. 1–104 [Google Scholar]
- Ramirez-Suero M, Khanshour A, Martinez Y, Rickauer M. 2010. A study on the susceptibility of the model legume Medicago truncatula to the soil-borne pathogen Fusarium oxysporum . European Journal of Plant Pathology. 126, 517–530 [Google Scholar]
- R Core Team 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; [Google Scholar]
- Ronfort J, Bataillon T, Santoni S, Delalande M, David JL, Prosperi JM. 2006. Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula . BMC Plant Biology. 6, 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ. 2008. Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Functional Plant Biology. 35, 253–264 [DOI] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. 1995. Selection of nodulation and mycorrhizal mutants in the model plant Medicago trucatula (Gaertn.) after gamma ray mutagenesis. Plant Science. 111, 63–71 [Google Scholar]
- Samac DA, Graham MA. 2007. Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiology. 144, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible L, Cannon OS, Waddoups V. 1951. Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology. 41, 986–990 [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology. 58, 809–822 [DOI] [PubMed] [Google Scholar]
- Shaner G, Finney RE. 1977. The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology. 67, 1051–1056 [Google Scholar]
- Stewart SA, Hodge S, Ismail N, Mansfield JW, Feys BJ, Prospéri JM, Huguet T, Ben C, Gentzbittel L, Powel G. 2009. The RAP1 gene confers effective, race-specific resistance to the pea aphid in Medicago truncatula independent of the hypersensitive reaction. Molecular Plant–Microbe Interactions. 22, 1645–1655 [DOI] [PubMed] [Google Scholar]
- Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prosperi JM, Esquerre-Tugaye MT, Dumas B, Jacquet C. 2004. Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii . Molecular Plant–Microbe Interactions. 17, 909–920 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Marek SM, Lee HK, Nakashima J, Tang Y, Sledge MK, Dixon RA, Mysore KS. 2009. Global gene expression profiling during Medicago truncatula– Phymatotrichopsis omnivora interaction reveals a role for jasmonic acid, ethylene, and the flavonoid pathway in disease development. Molecular Plant–Microbe Interactions. 22, 7–17 [DOI] [PubMed] [Google Scholar]
- Vailleau F, Sartorel E, Jardinaud MF, Chardon F, Genin S, Huguet T, Gentzbittel L, Petitprez M. 2007. Characterisation of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula . Molecular Plant–Microbe Interactions. 20, 159–167 [DOI] [PubMed] [Google Scholar]
- Veronese P, Narasimhan ML, Stevenson RA, Zhu JK, Weller SC, Subbarao KV, Bressan RA. 2003. Identification of a locus controlling verticillium disease symptom response in Arabidopsis thaliana . The Plant Journal. 35, 574–587 [DOI] [PubMed] [Google Scholar]
- Vining K, Davis T. 2009. Isolation of a Ve homolog, mVe1, and its relationship to verticillium wilt resistance in Mentha longifolia (L.) Huds. Molecular Genetics and Genomics. 282, 173–184 [DOI] [PubMed] [Google Scholar]
- Yang S, Gao M, Deshpande S, Lin S, Roe BA, Zhu H. 2007. Genetic and physical localization of an anthracnose resistance gene in Medicago truncatula . Theoretical and Applied Genetics. 116, 45–52 [DOI] [PubMed] [Google Scholar]
- Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ. 2008a. QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Science. 174, 290–298 [Google Scholar]
- Yang S, Gao M, Xu C, Gao J, Deshpande S, Lin S, Roe BA, Zhu H. 2008b. Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proceedings of the National Academy of Sciences, USA. 105, 12164–12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 480, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M. 2009. Translating Medicago truncatula genomics to crop legumes. Current Opinion in Plant Biology. 12, 193–201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Yang S, Chi J, Zhang G, Ma Z. 2011. Cloning and characterisation of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana . Plant Cell Reports. 30, 2085–2096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.