Abstract
Induced pluripotent stem cells (iPSCs) have the potential for creating patient-specific regenerative medicine therapies, but the links between pluripotency and tumorigenicity raise important safety concerns. More specifically, the methods employed for the production of iPSCs and oncogenic foci (OF), a form of in vitro produced tumor cells, are surprisingly similar, raising potential concerns about iPSCs. To test the hypotheses that iPSCs and OF are related cell types and, more broadly, that the induction of pluripotency and tumorigenicity are related processes, we produced iPSCs and OF in parallel from common parental fibroblasts. When we compared the transcriptomes of these iPSCs and OF to their parental fibroblasts, similar transcriptional changes were observed in both iPSCs and OF. A significant number of genes repressed during the iPSC formation were also repressed in OF, including a large cohort of differentiation-associated genes. iPSCs and OF shared a limited number of genes that were upregulated relative to parental fibroblasts, but gene ontology analysis pointed toward monosaccharide metabolism as upregulated in both iPSCs and OF. iPSCs and OF were distinct in that only iPSCs activated a host of pluripotency-related genes, while OF activated cellular damage and specific metabolic pathways. We reprogrammed oncogenic foci (ROF) to produce iPSC-like cells, a process dependent on Nanog. However, the ROF had reduced differentiation potential compared to iPSC, suggesting that oncogenic transformation leads to cellular changes that impair complete reprogramming. Taken together, these findings support a model in which OF and iPSCs are related, yet distinct cell types, and in which induced pluripotency and induced tumorigenesis are similar processes.
Introduction
Differentiated cells can be reprogrammed into induced pluripotent stem cells (iPSCs) that share many properties with embryonic stem cells (ESCs) [1–6,26]. Many elements of ESC gene expression are invoked during iPSC formation, including both activation and repression of specific genes, but interestingly there are important differences as well [7], arguing that iPSCs are their own unique subtype of pluripotent cells. Both iPSCs and ESCs can readily form teratomas in xenograft assays, which are tumors consisting of tissues that include derivatives of all 3 germ layers. However, the tumorigenic properties of iPSCs have not yet been investigated to the same degree as ESCs.
Many of the genes involved in producing iPSCs are either known or suspected oncogenes including, most notably, Myc and Klf4. However, Nanog, Sox2, and Oct4 (also known as Pou5f1) are linked to tumorigenesis as well ([8–10] and reviewed in ref. [11]). The degree to which iPSCs can form malignant tumors remains mostly undefined as are the mechanisms involved when tumor formation has been shown to occur [12]. ESCs themselves not only form teratoma, but also can more rarely form malignant tumors [13].
An ESC-like module of gene expression, regulated by Myc, is to some extent operative in many cancer cells and correlates with poor prognosis [14,15]. Myc also maintains ESC self-renewal and pluripotency [16], at least in part by blocking their differentiation through regulation of histone modifications and specific miRNAs [17,18]. Both Myc and N-Myc also bind and, together with their cofactor Miz-1, repress differentiation-associated genes, including Hox genes in human ESC [19], and Myc repression of differentiation genes has also been noted as important in iPSCs as well [20]. In the context of human neuroblastoma, N-Myc also directly binds and regulates expression of pluripotency-related genes, including KLF2, KLF4, LIF, and LIN28B [21]. These findings have led us to hypothesize that a process analogous to that which leads to the genesis of many cancers may also be involved in reprogramming to pluripotency. A greater understanding of the relationship between pluripotency and tumorigenicity will be instrumental in the development of regenerative therapies as well as the use of ESCs and iPSCs for modeling human diseases. iPSCs have been used as a platform to model a number of human diseases, including most recently Parkinson's disease [22]. However, if iPSCs are inherently similar to tumor cells, this may confound some of these model systems of disease states, particularly those having no link to tumorigenesis.
Also of concern is the compelling similarity that we first noted between the methods used for iPSC production and those used to produce so-called oncogenic foci (OF) [11]. OF formation assays have been used to study oncogenes for decades [23]. For both the production of iPSCs and OF, fibroblasts are transduced with viruses encoding a variety of genes, most often suspected or known oncogenes, a process that ultimately leads to the formation of colonies with a variety of morphologies. OF are in essence colonies of transformed cells that develop into tumors when injected into immunodeficient mice. OF produced from fibroblasts form fibrosarcoma [23], malignant cancers made up of immature proliferating fibroblasts. Myc not only induces OF formation on its own [24], but can also enhance production of OF by other oncogenes, such as Ras [23,25]. Exogenous Myc, while not essential for iPSC formation, greatly enhances the efficiency of iPSC formation when combined with other iPSC inducing genes [6,26,27]. Lowering p53 levels also enhances both iPSC formation efficiency [28–33] and the formation of OF [34]. The addition of SV40 large T antigen, a well-known oncogene that suppresses normal p53 function [35], also increases the efficiency of iPSC formation [36]. In addition, immortalization appears to eliminate a roadblock to iPSC formation [33]. Thus, there are numerous methodological similarities between OF and iPSC production, but the relationship between stem and tumor cells created in vitro, (iPSC and OF, respectively), is still largely unknown.
Here we examined iPSCs and OF produced from common parental fibroblasts to test the hypothesis that they are related cell types. To this end, we conducted gene expression microarray studies to compare the transcriptomes of parental mouse embryonic fibroblasts (MEFs) to those of iPSCs and OF derived from them. We found substantial overlap in gene expression changes that occur in iPSCs and OF relative to parental MEFs. The specific group of differentiation-associated genes that we found to be downregulated during iPSC production was quite similar to those downregulated during OF formation, and the overlapping downregulated genes were highly related to those found by Sridharan et al. [20] as down in iPSCs. We also found that both cell types commonly upregulated the glycolysis pathway, although through different genes in each case. We also examined 2 other published microarray data sets generated from iPSCs that were derived either by using different cocktails of reprogramming factors [37] or different starting cells [38] to determine the global validity of our comparisons and found very similar results, suggesting that the similarity of iPSCs and OF is independent of both Myc and fibroblastic parental cells. There were some key differences, however, between iPSCs and OF. For example, iPSCs exhibited a cluster of 17 activated pluripotency genes absent from OF, while OF uniquely expressed cell damage and specific metabolic programs. Interestingly, OF were able to be reprogrammed in a Nanog-dependent manner into iPSC-like cells based on morphology and gene expression, although their differentiation was less robust. Our findings indicate that both OF and iPSC switch to a glycolytic metabolic state, a finding that is supported in iPSCs by a recent metabolomics study [39]. Our findings also show that cell-type-specific differentiation is commonly downregulated during both the reprogramming and transformation processes. Together, these results support a model in which iPSCs and OF are related cell types as well as more broadly arguing that the processes of tumorigenesis and induced pluripotency are related.
Methods
MEF isolation
MEFs were isolated from E14.5 CF-1 embryos. The isolation protocol was as described in Ref. [40].
OF, iPSC, and reprogrammed oncogenic foci derivation and in vitro differentiation
iPSC and OF were derived using retroviral transduction of pMXs based plasmids by methods similar to what was previously described [2]. At 7 days post-transduction, cells transduced with Oct4, Sox2, Klf4, and Myc (OSKM) were replated on irradiated MEF feeder cells. Media was replaced with the mouse embryonic stem cells (mESC) medium [2] on cells from all conditions the following day and was subsequently changed daily. Starting at day 21 post-transduction, colonies from the OSKM condition were selected based on having colony morphology similar to mESC. Selected colonies were manually picked up and cultured using mESC conditions. Reprogrammed oncogenic foci (ROF) derivation was similar, but cells were transduced with Nanog and the complementary factors. Also starting at day 21 post-transduction, colonies from all other transduced cells were manually picked for the first passage, and then enzymatically passaged as needed. Knoepfler Lab and Millipore MEFs (MEF1 and MEF2, respectively), are derived from CF1 mouse strains, passage 2 and 4, respectively. To derive embryoid bodies (EBs), mESC, ROF, and iPSC were harvested by trypsinization and seeded into low-binding 9-cm tissue culture dishes (Thermo Scientific; part no. 145401) in the mESC medium without leukemia inhibitory factor. After 7 days in suspension culture, EBs were transferred to gelatin-coated coverslips and allowed to differentiate for another 7 days.
Expression microarrays
RNA was isolated from MEF, OF, iPSC, ROF, and mESC using the RNeasy Plus Mini Kit (Qiagen). RNA was submitted to the UC Davis Genome Center Expression Analysis Core for hybridization and scanning of MouseWG-6 beadchip microarrays (Illumina; v2). Microarray data were normalized by quantile normalization and analyzed using GenomeStudio (Illumina; v2010.1). Venn diagrams and lists of overlapping genes were generated using VENNY [41]. P values for the degree of overlap between gene sets were calculated using the Chi-squared test.
Reverse transcriptase-polymerase chain reaction and quantitative RT-polymerase chain reaction for marker genes
First strand cDNA synthesis was carried out using the SuperScript III First Strand Synthesis Supermix kit (Invitrogen) with oligo(dT)20 primers. GoTaq (Promega) was used to perform conventional reverse transcriptase-polymerase chain reaction (RT-PCR). ABsolute Blue QPCR Master Mix (Thermo) was used to perform SYBR green-based quantitative PCR (qPCR). Fold changes were normalized to PPIA and analyzed by the ΔΔCt method. Sequences of primers used for the analysis of marker genes are in Supplementary Table ST4 (Supplementary Data are available online at www.liebertpub.com/scd).
Alkaline phosphatase and immunofluorescence staining
Cells were fixed with 4% paraformaldehyde. Alkaline phosphatase (AP) staining was performed using the Vector Blue Alkaline Phosphatase Substrate Kit (Vector Laboratories) according to the manufacturer's instructions. For immunofluorescence, cells were stained with primary anti-SSEA1 (Chemicon; MAB4301), anti-Nanog (Abcam; ab80892), anti-β-tubulin (Covance; PRB-435P), anti-α-Fetoprotein (R&D systems; MAB1368), or anti-α-smooth muscle actin (Abcam; ab5694) antibodies. Primary antibodies were visualized with AlexaFluor® 488–conjugated anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG or AlexaFluor® 555–conjugated anti-mouse IgM. Coverslips were mounted using Vectashield with 4′,6-diamidino-2-phenylindole (Vector Laboratories).
Western blotting
Equivalent levels of protein from each cell type were separated on 6%–12% Bis-Tris gel (Invitrogen protocol). The protein was then transferred onto polyvinylidene fluoride membrane and blocked in 5% milk for an hour. Membranes were then incubated with a primary antibody overnight at 4°C. Primary antibody concentrations and catalog numbers are listed below. All secondary antibodies were applied for 1 h at room temperature.
c-Myc: Santa Cruz Biotechnology rabbit polyclonal—sc-764; lot K2008. 1:400 dilution.
N-Myc: Abcam mouse monoclonal—ab16898; lot GR46870. 1:100 dilution.
Sox2: Abcam rabbit polyclonal—ab15830. 1:166 dilution.
Nanog: Abcam rabbit polyclonal—ab80892; lot GR40243-9. 1:300 dilution.
Oct4: Abcam rabbit polyclonal—ab19857; lot GR54542-1. 1:800 dilution.
p57Kip2: Abcam rabbit polyclonal—ab4058; lot 750614. 1:1,000 dilution.
p53: Abcam mouse monoclonal—ab26; lot GR68134-1. 1:200 dilution.
B-actin: Sigma mouse monoclonal—A 1978; lot 047K4768. 1:8,000 dilution.
Tumor formation assays
OF, iPSC, ROF, and mESC were harvested by trypsinization and brought to a concentration of 107 cells/mL in the mESC medium with 30% Matrigel. One hundred microliter suspensions (106 cells) were injected subcutaneously into the left flank of NOD/SCID IL2RG mice (n=4 for all cell types) [42]. Tumors were harvested as they appeared (4–8 weeks), fixed with formaldehyde, sectioned, and stained with hematoxylin and eosin.
Results
Production of iPSC and OF with identical genetic backgrounds
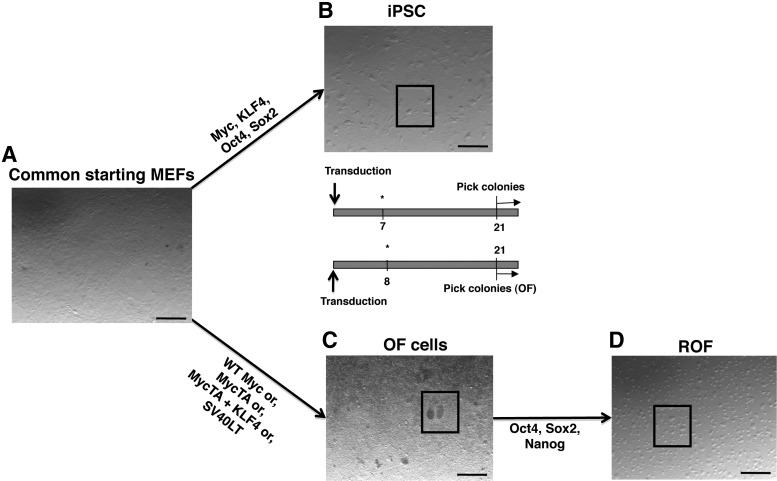
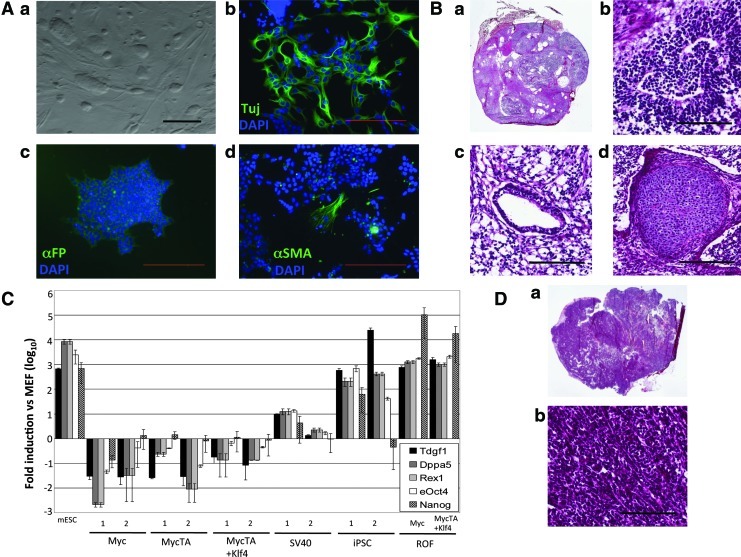
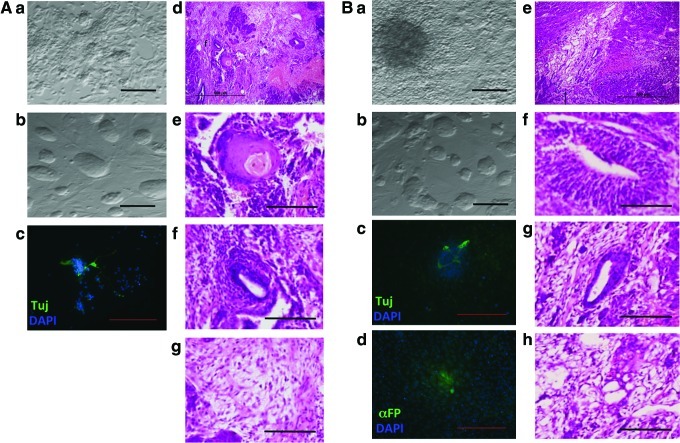
To analyze the relationship between iPSC and OF, we produced both in parallel using the same MEFs (MEF1) as starting parental cells (Fig. 1). We transduced MEF1 cells with Myc, Klf4, Oct4, and Sox2 (MKOS) to produce iPS1. We generated iPS2 by transduction of the MKOS cocktail into commercially available MEFs (Millipore; MEF2) of an identical genetic background to MEF1 [2]. Colonies with typical mESC morphology were picked and propagated (Fig. 1B, see boxed area for an example of such colonies and Supplementary Fig. S1) to establish clonal lines. In parallel, we produced OF through transduction of MEF1 with different combinations of retrovirus encoding the following: (1) wild-type Myc (WT), (2) the stabilized mutant MycT58A (MycTA), (3) MycTA and Klf4 together (MycTA+Klf4), or (4) SV40 Large T antigen (SV40) (Fig. 1). In each case, individual OF were picked and propagated (see boxed area in Fig. 1C for examples of OF colonies). Biological replicates of clonal OF lines were produced in two separate transductions and one clone from each was selected for further analysis. We also transduced MEFs with Klf4 alone, but no OF were produced (not shown). Thus, we produced (or in the case of MEF2, obtained) MEFs, OF, and iPSC all with identical genetic backgrounds. For both OF and iPSC, transgene expression was verified (Supplementary Fig. S2). iPSCs were initially screened by colony morphology [Fig. 2A(a) and Supplementary Fig. S1]. Differentiation potential of the iPSC was examined through in vitro EB formation resulting in tripotent differentiation producing mesoderm, endoderm, and ectoderm derivatives [Fig. 2A(b–d) and Supplementary Fig. S3], teratoma formation (Fig. 2B), and endogenous stem cell-related gene expression profiling (Fig. 2C). OF produced sarcoma upon xenograft (Fig. 2D).
FIG. 1.
Parallel generation and analysis of iPSCs and OF from common parental fibroblasts. (A) Common parental MEFs. (B) Typical iPSC colonies before picking (boxed). (C) Typical OF colonies before picking (boxed). (D) Typical ROF colonies (boxed) (scale bars=500 μm). iPSCs, induced pluripotent stem cells; OF, oncogenic foci; MEFs, mouse embryonic fibroblasts; ROF, reprogrammed oncogenic foci.
FIG. 2.
Characterization of iPSC and OF. (A) (a) Phase-contrast image of typical iPSC colonies (scale bar=100 μm). (b–d) Immunofluorescence (IF) staining images of: (b) Tuj (ectoderm), (c) αFP (endoderm), (d) αSMA (mesoderm) from embryoid body differentiation (scale bars=200 μm). (B) (a) Hematoxylin and Eosin stained section of whole teratoma produced by iPS1. Higher magnification images of tissues within teratoma, including (scale bars=100 μm) (b) ectoderm, (c) endoderm, and (d) mesoderm. (C) qPCR analysis of stem cell markers in all conditions, normalized to Ppia. (D) (a) Hematoxylin and Eosin stained section of whole sarcoma and (b) higher magnification image of sarcoma tissue (scale bar=100 μm). αFP, α-fetoprotein; αSMA, smooth muscle actin; qPCR, quantitative polymerase chain reaction. Color images available online at www.liebertpub.com/scd
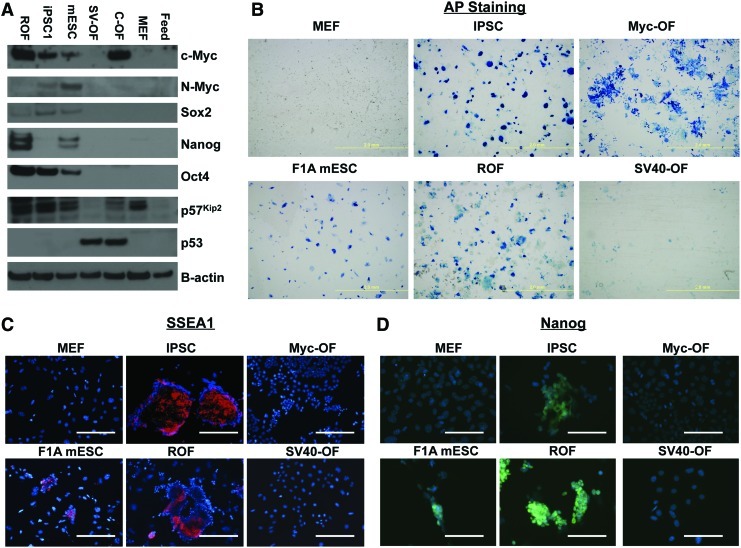
Expression of a panel of stem cell-related genes was also assessed by immunoblotting for their protein products (Fig. 3A). As expected, we found elevated levels of OCT4 and SOX2 in pluripotent cells, but not parental cells, feeder cells, or OF. There was a noticeable lack of Nanog at the RNA level in iPS2, though it was very weakly detected by western blot (WB) and much more strongly by immunofluorescence in iPS1. Interestingly, OF were found to have much higher levels of p53 than any other cell type. In addition, p57Kip2, a negative cell cycle regulator and possible tumor suppressor, was found at levels similar to that in the parental cells in all, but the 2 OF lines, which had significantly lower levels (Fig. 3). Thus, there was an inverse relationship between p53 and p57, and OF were the only cell type that had high p53 and low p57.
FIG. 3.
Immunoblotting for the indicated proteins in iPSC and OF. (A) Western blots of indicated protein markers in representative cells. (B) Alkaline phosphatase staining in representative cells (scale bar=2 mm). (C, D) Immunofluorescence images of indicated cells costained with DAPI (blue) and SSEA1 [red (C)] or Nanog [green (D)] (scale bars=200 μm). DAPI, 4′,6-diamidino-2-phenylindole. Color images available online at www.liebertpub.com/scd
iPSCs were further validated by AP activity (Fig. 3B), as well as by immunoreactivity for SSEA1 (Fig. 3C) and Nanog (Fig. 3D). Interestingly, both Myc OF and SV40 OF were also positive for AP which, aside from its use as a marker for stemness, has also been described as a poor prognostic marker in tumors of mesodermal origin, including sarcoma [43]. Biological replicates were created and analyzed for iPSCs (iPS1 and iPS2) and OF (infection 1 and infection 2) cell lines. Control mESCs were tested for pluripotency-related gene expression and were also found to produce teratoma (not shown).
OF and iPSC produce distinct tumor types in xenograft assays
OF cells produced by MycTA+Klf4 were injected (1×106, n=4) into immunodeficient mice, which were observed over time for tumor formation. Starting at 4 weeks postinjection of cells, tumors formed at all 4 injection sites and were found to be histologically consistent with sarcoma in all cases (Fig. 2D). When 1×106 iPS1 and iPS2 cells were injected (n=4), different tumor types were observed. iPS1 produced teratoma (4/4 injections; Fig. 2B), while iPS2 produced malignant teratoma-bearing immature neuroepithelial components (4/4 injections, not shown). While both types of teratoma (iPS1 benign and iPS2 malignant) contained ecto, endo, and mesoderm derivatives and validated pluripotency in both iPSC lines; the malignant teratoma contained a higher proportion of undifferentiated cells. These findings demonstrate that different iPSC lines can have distinct tumorigenic properties while appearing almost identical morphologically and in terms of pluripotency-related gene expression.
iPSC are related to OF in terms of global gene expression
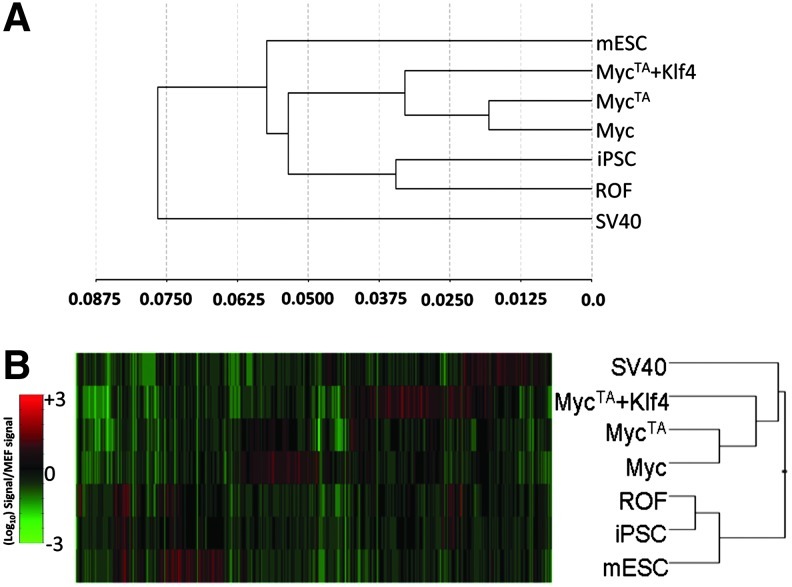
To examine global gene expression profiles of OF versus iPSC, we conducted expression microarray studies on RNA that was isolated from iPSC, OF, and control mESC. When total gene expression profiles were organized into a family cluster tree, 2 main branches were evident. SV40 OF constituted one unique branch, while the other branch was divided into 3 sub-branches. mESC formed its own sub-branch, while the remaining OF and iPSC each formed their own sub-branches. Surprisingly, iPSCs clustered more closely to the OF than to mESC (Fig. 4A). Thus, the global gene expression profile of iPSC is more similar to OF than to mESC. The 3 distinct types of OF formed with Myc, MycTA, and MycTA+ Klf4 were similar in global expression, while SV40 OF diverged (Fig. 4A). Not surprisingly, the presence of Klf4 led to some divergence in the Myc-containing OF (Myc/MycTA vs. MycTA+Klf4).
FIG. 4.
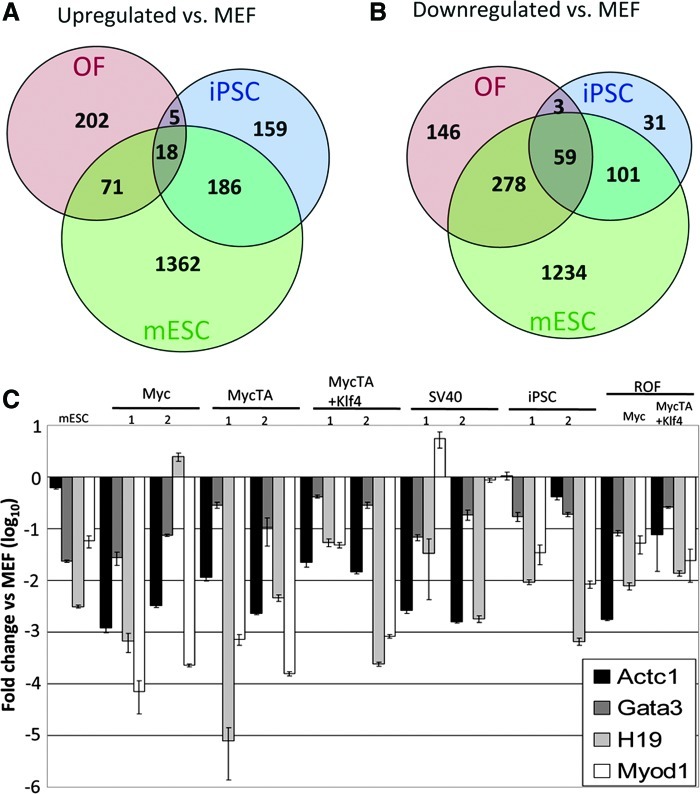
Microarray data analysis of relationships between conditions. (A) Dendrogram based on similarities of global gene expression patterns. (B) Heatmap and dendrogram based on genes up- or downregulated 5-fold or greater versus MEFs.
To further analyze the groups of genes that were most highly up- or downregulated, we generated a list of 642 genes that were a minimum of 5-fold up- or downregulated as compared to parental MEFs (Supplementary Table ST1). We organized the profiles of the different conditions into a family cluster tree and heat map to visualize groups of genes that were similarly regulated in the different conditions (Fig. 4B). When taking into account only the genes with expression that was most highly changed upon generation of either OF or iPSC, 2 main branches were evident. One was comprised of all OF conditions, and the other contained mESCs and iPSCs. SV40 OF were again the most divergent OF, followed by the MycTA+Klf4 OF. The Myc and MycTA OF formed their own sub-branch and, as expected, were the 2 most closely related of the OF. The branch that contained mESC and iPSC was separate from the OF branch indicating particularly strong similarity between iPSC and mESC in genes that are highly up- or downregulated.
OF and iPSCs exhibit a high degree of overlap with mESCs in genes with altered expression versus parental MEFs
Genes were divided into those whose expression was up- or downregulated 1.5-fold or more in both iPSC conditions as compared to parental MEFs. Similarly, to obtain a general OF profile and remove some of the bias toward only Myc-regulated genes, genes that were commonly up- or downregulated in to all OF conditions versus parental MEFs were divided (gene lists in Supplementary Tables ST2 and ST3). Both gene lists were compared for overlap with each other and with mESCs (Fig. 5A, B). Both iPSC and OF individually had a high level of overlap with mESC in both up- and downregulated genes. Surprisingly, most genes that were commonly up- or downregulated within OF and iPSCs were also similarly expressed in mESCs as compared to MEFs. Further validating the identity of the iPSC, genes downregulated in both iPS1 and iPS2 relative to MEFs were similar to those genes with lower abundance in mESCs relative to MEFs (Supplementary Fig. S4). A significant portion of the upregulated genes in iPSCs was also upregulated in mESCs relative to MEFs as well (Supplementary Fig. S4). A similarly high proportion of genes (nearly 30%, P<0.0001) repressed during iPS1 formation were also repressed during OF formation. About 8% (P<0.0001) of repressed genes in iPS2 were also repressed in OF (Supplementary Fig. S5).
FIG. 5.
Gene expression similarities between iPSC and OF. (A) Venn diagram of genes upregulated 1.5-fold in OF (genes commonly upregulated in all OF), iPSC, and mESC. (B) Venn diagram of genes downregulated 1.5-fold in OF (genes commonly downregulated in all OF), iPSC, and mESC. (C) qPCR of differentiation genes in all conditions. mESC. mESC, mouse embryonic stem cells. Color images available online at www.liebertpub.com/scd
Among the common expression patterns between iPSCs and ESCs were high levels of expression of pluripotency-associated genes relative to both MEFs and OF, including many factors associated specifically with iPSC induction (Oct4, Lin28, Sox2, Nanog, Klf4, Utf1, and Fbxo15). Neither Myc alone nor MycTA and Klf4 together significantly induced pluripotency genes during OF formation, suggesting they do not act alone to induce these genes during the iPSC formation. In addition, DAVID bioinformatics software [44,45], which generates clusters of genes based on gene ontology (GO), showed that iPSCs uniquely expressed genes that fell into ontological clusters involved in sugar transport, regulation of cell growth, various metabolic processes (such as that for coenzymes, glutathione, nicotinamide, and alkaloids), and histone H2b as well as general chromatin assembly (Table 1, light gray and Supplementary Tables ST4 and ST5). Collectively, these clusters suggest that pluripotency-related transcription factors, as well as cell cycling, chromatin, and metabolism are uniquely activated during and may promote induced pluripotency.
Table 1.
Ontological Clusters Formed by Genes Commonly Upregulated in Induced Pluripotent Stem Cells, Oncogenic Foci
| Ontological clusters of upregulated genes | P value |
|---|---|
| Carbohydrate catabolic process | 9.92×10−04 |
| Glucose metabolic process | 2.08×10−03 |
| Cellular carbohydrate catabolic process | 2.44×10−03 |
| Lysosome | 2.73×10−03 |
| Sphingolipid metabolic process | 2.75×10−03 |
| Lytic vacuole | 2.81×10−03 |
| Membrane lipid metabolic process | 3.09×10−03 |
| Organophosphate metabolic process | 6.40×10−03 |
| Glucose catabolic process | 1.29×10−02 |
| Monosaccharide metabolic process | 2.20×10−02 |
| Positive regulation of inflammatory response | 2.12×10−02 |
| Response to wounding | 4.58×10−02 |
| Histone core | 1.07×10−05 |
| Nucleosome core | 1.82×10−05 |
| Stem cell maintenance | 2.92×10−03 |
| Stem cell development | 3.33×10−03 |
| Sugar/inositol transporter | 7.46×10−03 |
| Monosaccharide biosynthetic process | 7.83×10−03 |
| Alcohol biosynthetic process | 1.28×10−02 |
| Glutathione metabolic process | 3.98×10−02 |
| Nicotinamide metabolic process | 3.98×10−02 |
| Embryonic organ development | 4.17×10−02 |
| Monosaccharide metabolic process | 4.17×10−02 |
Genes common in iPS1 and iPS2 (in dark gray), OF (Myc, MycTA, MycTA+Klf4, and SV40, in light gray).
Bolded entries denote clusters shared between conditions that arise from specific genes that are not commonly regulated; i.e., overlapping clusters from non-overlapping genes.
About 10% of upregulated genes in iPS1 and 3% of upregulated genes in iPS2 were also activated in OF compared to the starting MEFs (Supplementary Fig. S5). This represents a statistically significant degree of concordance between iPS and OF, but is less than the amount of overlap seen between iPSCs and ESCs. When data sets were divided in this way into up- and downregulated genes versus MEFs, iPSCs therefore appeared more similar to mESCs than to OF. However, taken together, these data support the notion that iPSCs are not simply analogous to mESCs and have significant similarities to OF as well.
To validate and analyze the microarray data further, we employed qRT-PCR to quantify levels of gene expression of a group of known markers of pluripotent cells, including endogenous Oct4 (eOct4), Tdgf1 (also known as Cripto), Rex1, and Nanog by comparing levels in OF and iPSCs to levels in their parental MEFs (Fig. 2C). While expression of most of these markers of pluripotency was largely absent in OF cells produced using Myc alone or in combination with other factors, we observed expression of very low levels of Nanog in some of these OF. All of these markers of pluripotency were, to some degree, expressed in the SV40 OF, although expression was lower than in pluripotent cells, which exhibited robust expression (Fig. 2C). Thus, the general pattern was that OF could be distinguished from ESC or iPSC by the lack of strong pluripotency gene expression.
Gene expression changes unique to OF point to cell damage and specific metabolic pathways as important during oncogenic transformation
Some ontological clusters of genes that had no clear links to pluripotency were identified as uniquely upregulated in OF, but not iPSCs, as compared to MEFs. These clusters included response to wounding, lysosome, positive regulation of inflammatory response, and some specific types of metabolism (Table 1, light gray). The cell damage and inflammatory response ontological clusters elevated in the OF suggest that oncogenic transformation may be identified by the cell as a type of injury.
iPSC and OF exhibit strongly overlapping patterns of downregulation of differentiation-associated gene expression compared to parental fibroblasts
When the ontology of genes repressed in both iPSC and OF was analyzed and intercompared, some interesting patterns emerged (Fig. 5B; Table 2). DAVID analysis of the 62 genes commonly repressed in both OF and iPSC produced a list of significant ontological clusters almost exclusively related to differentiation with the only exception being 2 similar clusters related to phospholipid metabolism (Table 2, white). This list was also subjected to STRING analysis (Supplementary Fig. S6) [46], which highlights functional protein associations, further validating some of the clusters inferred by ontological analysis. A subset of differentiation-associated genes that were downregulated in both OF and iPSCs, including Actc1, Gata3, H19, and Myod1, was validated by qRT-PCR (Fig. 5C), though we were not able to observe MYOD protein by WB in any of the cell lines (not shown). Interestingly, when we performed DAVID analysis on lists of genes repressed in either iPSC or OF, we observed a subset of clusters that were shared between these cell types (Table 2, bold). These shared clusters were also mainly involved in differentiation, but also included clusters related to positive regulation of gene transcription. The genes in the clusters related to transcription mainly encoded differentiation-associated transcription factors, including Myog in OF and Gata6, Gli2, Sox9 in iPSCs. The observation that OF and iPSC up- or downregulated distinct genes that nonetheless comprised common ontological clusters (Tables 1 and 2, common clusters in bold) suggests that, in some cases, cells may take different paths to reach common phenotypic end points, in this case, suppression of differentiation.
Table 2.
Ontological Clusters Formed by Genes Commonly Downregulated in Induced Pluripotent Stem Cells, Oncogenic Foci, and Both Induced Pluripotent Stem Cells and Oncogenic Foci
| Ontological clusters of downregulated genes | P value |
|---|---|
| Positive regulation of transcription | 1.50×10−04 |
| Positive regulation of gene expression | 1.86×10−04 |
| Positive regulation of cell differentiation | 2.78×10−04 |
| Positive regulation of nitrogen compound metabolic process | 3.40×10−04 |
| Epithelium development | 4.77×10−04 |
| Heart development | 9.98×10−04 |
| Positive regulation of transcription from RNA polymerase II promoter | 2.40×10−03 |
| Skeletal system development | 3.44×10−03 |
| Chordate embryonic development | 5.83×10−03 |
| Regulation of cell proliferation | 6.26×10−03 |
| Blood vessel development | 8.41×10−03 |
| Vasculature development | 9.28×10−03 |
| Vasculature development | 9.28×10−03 |
| Neural crest cell development | 1.25×10−02 |
| Mesenchymal cell development | 2.44×10−02 |
| Skeletal system development | 5.75×10−04 |
| Ossification | 2.12×10−03 |
| Bone development | 2.88×10−03 |
| Osteoblast differentiation | 5.77×10−03 |
| Sarcolemma | 6.78×10−03 |
| Glycerophospholipid metabolic process | 1.92×10−02 |
| Branching morphogenesis of a tube | 2.13×10−02 |
| Myofibril | 2.39×10−02 |
| Tube development | 2.58×10−02 |
| Morphogenesis of a branching structure | 3.68×10−02 |
| Glycerolipid metabolic process | 3.90×10−02 |
| Angiogenesis | 4.12×10−02 |
| Skeletal muscle tissue development | 1.67×10−07 |
| Muscle cell differentiation | 2.20×10−06 |
| Heart development | 1.12×10−05 |
| Regulation of cell growth | 9.62×10−05 |
| Regulation of apoptosis | 1.13×10−03 |
| Regulation of programmed cell death | 1.31×10−03 |
| Positive regulation of neuron apoptosis | 3.06×10−03 |
| Positive regulation of programmed cell death | 6.14×10−03 |
| Positive regulation of transcription from RNA polymerase II promoter | 1.09×10−02 |
| Skeletal system development | 1.45×10−02 |
| Blood vessel development | 1.50×10−02 |
| Vasculature development | 1.72×10−02 |
| Positive regulation of transcription | 3.68×10−02 |
| Positive regulation of gene expression | 4.39×10−02 |
| Induction of apoptosis | 4.79×10−02 |
Genes common in iPS1 and iPS2 (in dark gray), OF (Myc, MycTA, MycTA+Klf4, and SV40, in light gray), and both iPSC and OF (in white).
Bolded entries denote clusters shared between conditions that arise from specific genes that are not commonly regulated; i.e., overlapping clusters from non-overlapping genes.
iPSC, induced pluripotent stem cells; OF, oncogenic foci.
Common transcriptome changes in OF and iPSCs are at least, in part, Myc-independent and are not specific only to fibroblastic parental cells
Many of our inducing conditions share one or more factors, mainly Myc. To determine whether our comparisons were biased toward a Myc-controlled transcriptional program, we analyzed a published dataset in which only Oct4, Sox2, and Esrrb were used to produce iPSCs (OSE iPSC) without Myc from MEFs [37]. We generated lists of genes 1.5-fold or more downregulated in the OSE iPSC as compared to their starting MEFs and compared them to our gene expression data from mESCs and OF (Supplementary Fig. S7). This comparison yielded 103 shared downregulated genes in both OSE iPSCs and OF. The overlapping genes set encompassed many of the same genes found in our initial analysis, including the Actc1 gene. DAVID analysis of the 96 downregulated genes shared by OSE iPSCs with both mESCs and OF yielded almost exclusively differentiation-associated GO clusters, in particular, those related to muscle/skeletal system development (Supplementary Fig. S7A). These findings support the notion that the transcriptional changes shared between OF and iPSCs that we observed are not solely attributable to Myc.
To ascertain the potential influence of parental cell type on loss of differentiation-associated gene expression during the iPSC formation, we also examined a data set published on iPSCs derived using bone marrow mononuclear cells (BM iPSCs) from adult mice as starting material [38]. We generated lists of genes at least 1.5-fold downregulated in BM iPSCs as compared to their starting cells, and then compared that list of genes to those genes downregulated in our OF and mESCs compared to parental MEFs (Supplementary Fig. S7B). DAVID analysis of the overlapping genes between OF and BM iPSCs again yielded GO clusters related to differentiation. This time, however, the GO clusters of downregulated differentiation genes were mostly hematopoietic specific, such as hemoglobin and oxygen transport (Supplementary Fig. S7B). These findings suggest that differentiation-associated gene downregulation during the iPSC formation is a common event that shares similarities to processes that occur during OF formation, but the specific differentiation-associated genes that are downregulated depend on the starting parental cell type.
Cellular metabolism-associated gene expression is strongly upregulated in iPSCs and OF
When the ontology of genes upregulated in iPSCs and OF versus MEFs was analyzed, the most common clusters involved cellular metabolism in both cases (Table 1). There were no significant ontological clusters observed when DAVID analysis was performed specifically on the set of identically overlapping genes upregulated in both iPSCs and OF. However, when comparing clusters of genes uniquely upregulated in either iPSCs or OF, one cluster was found to be similar: monosaccharide metabolic process (Table 1, bold). Upon closer examination of the lists of unique genes leading to the common GO cluster, we found that both iPSC and OF upregulated 2 of the main regulatory enzymes in the glycolysis pathway, hexokinase-2 (HK2) in iPSC and the muscle isoform of phosphofructokinase (PFKM) in OF (Supplementary Table ST4). These data suggest that activation of monosaccharide metabolism is an important element of transformation of fibroblasts into both iPSCs and OF, although expression of different gene constituents of this metabolic pathway was different in each cell type. While there were no significant ontological clusters related to the set of overlapping genes between both iPSC lines and OF, there were some functionally related genes when we made a list of genes upregulated in either iPS1 or iPS2 in common with OF and subjected it to STRING analysis (Supplementary Fig. S8). Among the groups of interconnected nodes was a group, including the metabolism-related genes Tpi1, Pfkl, Rpia, and others.
Again, we wanted to study the dependence of gene expression changes on the specific reprogramming cocktail and cell type of origin. We generated lists of genes that were upregulated at least 1.5-fold in OSE iPSCs and BM iPSCs compared to their respective cells of origin. When we compared the lists generated from OSE iPSCs and BM iPSCs separately to mESCs and OF, DAVID analysis uncovered metabolism as a whole to be upregulated in both OSE iPSCs and BM iPSCs. GO clusters of OSE iPSCs yielded mainly those related to mitochondria (Supplementary Fig. S9A). Monosaccharide metabolism was found in the GO clusters of overlap between BM iPSCs and OF (Supplementary Fig. S9B). GO clusters related to nucleotide binding were upregulated in both OSE iPSCs and BM iPSCs suggesting an increased utilization of ATP in both cell types. These results indicate that a more active metabolism is common to OF and iPSCs regardless of their specific reprogramming mixture, and further, that this aspect is not dependent on the cell type of origin. See Supplementary Figs. S10 and S11 for Venn diagram comparisons of OSE iPSCs and BM iPSCs, respectively, to iPSCs generated in this study.
Cellular transformation establishes roadblocks to differentiation that are not readily reversed during reprogramming
Because of the similarities in overall gene expression between OF and iPSCs as well as previous data showing that immortalization catalyzes induced pluripotency [32], we examined whether OF could be reprogrammed into iPSCs. OF produced with either Myc, or MycTA+Klf4 were transduced with complementary factors to complete the MKOS mixture, but no iPSC-like colonies were observed (data not shown). Only upon the further addition of Nanog to the reprogramming mixture were iPSC-like colonies produced, which we termed ROF (Fig. 6 and Supplementary Fig. S2). These colonies could be propagated and they retained their ESC-like appearance for many weeks, suggesting they might have been reprogrammed into iPSC-like cells. Expression of markers of pluripotency in ROF was validated by qRT-PCR (Fig. 2C), WB (Fig. 3A), and immunostaining (Fig. 3B–D). The ROF also gained an iPSC-like gene expression profile, clustering closely with iPSC in the transcriptome analysis (Fig. 4). When the ROF were injected into immunodeficient mice, tumors were formed in 100% of mice within 6–8 weeks. The tumors were found to contain derivatives of all 3 germ layers, confirming pluripotency in the ROF (Fig. 6). However, these tumors grew rapidly and were mainly composed of undifferentiated cells, indicating the tumors were malignant teratomas. This finding was further substantiated by the inability of the ROF to fully differentiate through in vitro EB differentiation (Fig. 6). Immunofluorescence confirmed ectodermal differentiation in both ROF conditions and endodermal differentiation only in the ROF made from MycTA+Klf4 OF (Fig. 6). Differentiated cells staining positive for α-smooth muscle actin (mesodermal differentiation) were not detected in either ROF condition in 2 independent experiments. These data, taken together, indicate that oncogenic transformation is not necessarily incompatible with aspects of pluripotency, but can lead to cellular changes that inhibit differentiation.
FIG. 6.
Reprogramming of OF into ROF. (A) Characterization of ROF from Myc OF (a) Phase-contrast image of Myc OF (scale bar=100 μm), (b) phase-contrast image of Myc ROF colonies (scale bar=100 μm), (c) anti-Tuj (ectoderm) immuno fluorescent (IF) stain of differentiated Myc ROF (scale bar=200 μm), (d) hematoxylin and eosin stained section of Myc ROF teratoma showing various tissues (scale bar=500 μm) as well as higher magnification images (scale bars=100 μm) of (e) ectoderm, (f) endoderm, and (g) mesoderm within Myc ROF teratoma. (B) Characterization of ROF from MycTA+Klf4 OF. (a) Phase-contrast image of MycTA+Klf4 OF (scale bar=100 μm), (b) phase-contrast image of MycTA+Klf4 ROF colonies (scale bar=100 μm), (c) anti-Tuj (ectoderm) IF stain of differentiated MycTA+Klf4 ROF (scale bar=200 μm), (d) anti-αFP (endoderm) IF stain of differentiated MycTA+Klf4 ROF (scale bar=200 μm), and (e) hematoxylin and eosin stained section of MycTA+Klf4 ROF teratoma showing various tissues (scale bar=500 μm). Higher magnification images (scale bars=100 μm) of (f) ectoderm, (g) endoderm, and (h) mesoderm within MycTA+Klf4 ROF teratoma. Color images available online at www.liebertpub.com/scd
Discussion
One of the earliest assays used by tumor biologists to measure the potential tumorigenicity of genes and by which many of the first oncogenes were identified was the oncogenic focus formation assay. From a historical perspective, it is interesting that the first method to induce pluripotency and produce iPSCs bears such striking resemblance in methodology to the oncogenic focus formation assay. Here we found that the similarities extend beyond the methods. OF cells and iPSCs, while distinct, are nonetheless surprisingly similar cell types in terms of their transcriptomes.
Our findings suggest that within the shared gene expression programming between OF and iPSCs are the factors that make iPSCs tumorigenic. Because, for the most part, these gene expression signatures are also shared with ESCs, they may be of particular importance for understanding the pathogenesis of teratoma. Importantly, these putative tumorigenicity-related factors appear to have little or nothing to do with core pluripotency factors. iPSCs have been shown to be similar to, yet distinct from ESCs in terms of gene expression [7]. In further support of the findings of Chin et al., [7] we only observed about 10% overlap between genes uniquely upregulated in iPS1 and iPS2 relative to MEFs and the published ESC-like core of genes [14]. Interestingly there was ∼10% overlap between OF-specific genes relative to MEFs and the ESC-like core further supporting the notion that tumorigenic cells, though different, share properties with ESCs.
The lack of induction of pluripotency gene expression in OF by the combination of MycTA and Klf4 supports the idea that the contributions of Myc and Klf4 to the iPSC formation are mostly independent of the core pluripotency machinery. Rather, Myc and Klf4 appear to predominantly act to enhance iPSC formation by suppressing the expression of differentiation-associated genes and by activating cellular metabolic programs. Although Klf4 overexpression made OF more like mESCs, surprisingly, Klf4 also made OF less like iPSCs when not combined with Oct4, Sox2, and Nanog. Klf4 is known to associate with and co-occupy promoters of genes in a complex with Oct4 and Sox2 during reprogramming, but its expression is not limited to ESCs [47,48]. These data indicate that the effects of Klf4 on gene expression are not strictly stem cell-specific.
There was a slight discrepancy between the qRT-PCR and WB data (Figs. 2C and 3A respectively) in the SV40 cells as it pertains to their expression of markers of pluripotency. qRT-PCR is a very sensitive assay in which very low levels of expression can be quantified. Indeed, while the SV40 cells express RNAs for pluripotency markers above the level of MEFs, their expression of those genes is far less than in pluripotent cells. Since there is much less of these RNAs in the SV40 cells it may lead to much less protein being made. It is also well known that there are many factors that influence the regulation of protein levels that are independent of RNA levels. It is likely that the SV40 cells express these proteins at a level lower than the detection limit of the WB assay. It is also possible that there is some regulatory mechanism in nonpluripotent cells that is still active in the SV40 cells that leads to the degradation of some of those proteins.
In the case of Myc, there is existing evidence that it may function at least, in part, during iPSC formation by directly binding and regulating differentiation promoting target genes [19,20], while the role of Klf4, particularly on chromatin, is less clear. Myc may also drive the formation of both OF and iPSCs through promoting a highly metabolically active state. However, it may stimulate cellular reprogramming and, to some extent, tumorigenicity as well through, as yet, unknown pluripotency-related genes. Exogenous Myc has been used, to our knowledge, in all, but one of the studies aimed at exploring Myc function in reprogramming. That one study highlights the importance of endogenous Myc during reprogramming [49]. Yamanaka and coworkers found that L-Myc lacked the ability to transform cells, but enhanced iPSC reprogramming efficiency [50]. Myc maintains the undifferentiated state of mESCs at least, in part, through regulating miRNA expression and through its influence on chromatin [16–18]. The regulation of chromatin by Myc is surprisingly widespread in both normal stem and tumor cells for a basic-helix-loop-helix-zipper transcription factor [51–54]. Myc may function in this more global manner on chromatin during the formation of both iPSCs and OF, a possibility being addressed by functional genomics studies on iPSCs.
Given the similarities between OF and iPSCs, it was notable that OF could be reprogrammed into iPSC-like cells (ROF), but had a somewhat reduced differentiation capacity. Why might cancer cells be able to be induced to express a pluripotent gene expression profile, but continue to be at least partially resistant to differentiation? One possible explanation is the heterogeneous culture conditions that arise during reprogramming. It was published recently that normal ESCs, when cocultured with transformed ESCs, acquired a neoplastic phenotype through cell-to-cell contact that led to a loss of the ability of normal ESCs to fully differentiate [55]. Since only a small portion of cells is fully reprogrammed during iPSC formation, the surrounding cells may provide signals that lead to a diminished differentiation capacity of the newly reprogrammed cells. Alternatively, Myc overexpression in the absence of Oct4 and Sox2 may induce cell injury and DNA damage [56] that renders cells incompetent for normal differentiation and leaves them with impaired pluripotency. Pre-existing Myc overexpression could also sensitize the cells to subsequent transgene (e.g., Oct4) overexpression leading to apoptosis. It is possible as well that exogenous Myc, alone in the absence of coexpression of other reprogramming factors, leads to unrestrained Myc activity that irreversibly oncogenically transforms the cells leading to partially impaired differentiation, a hallmark of many tumors. Finally, elevated Myc in the absence of other reprogramming factors may lead to global chromatin changes in cells that allow for induction of pluripotency, but have permanently silenced genes that are essential for complete differentiation. In the presence of Oct4 and Sox2, Myc function on chromatin may be more specific and constrained, potentially through co-occupancy of some of the same target genes [20]. There is precedent for some of these possibilities, as multiple groups have now reported genetic and epigenetic abnormalities attributed to iPSC formation [57].
The requirement for Nanog in addition to the other reprogramming factors to reprogram the OF is notable. It suggests that the molecular basis for the increased difficulty in inducing pluripotency in the OF cells may be due to events that occurred while they were originally produced or subsequently grown as OF and was not a product of the ROF reprogramming event. However, another theory is that the transgenes were not efficiently silenced, which rendered the ROF unable to differentiate. Evidence for this theory includes the observation that ROF expressed Nanog at higher levels than that seen in iPSCs or ESCs (Fig. 3). Whatever the mechanism, which remains an open question, the fact that ROF were not able to differentiate as efficiently as iPSCs suggest that faithful reprogramming of cancer cells may require a different approach from that used in reprogramming noncancerous somatic cells, a notion supported by the reprogramming of melanoma cells using only miRNAs [58]. To date, a number of cancer cell types have been reprogrammed into iPS-like cells [59], including colorectal, pancreatic, hepatocellular carcinoma, and embryonal carcinoma [60]. It is probable that each specific type of cancer may require customized methods for reprogramming. Our data also suggests that the function of Myc alone is fundamentally distinct from its activity in the presence of Klf4, Oct4, and Sox2, which has important implications for tumorigenesis and induced pluripotency.
Despite the similarities between iPSCs and OF, key differences stood out. The OF form sarcoma, while the iPSCs form either benign or malignant teratoma. At the molecular level, both cell types exhibit gene expression signatures consistent with highly active cellular metabolism relative to parental MEFs. However, transcriptional profiling indicated that OF likely have an even more highly active metabolic state and, interestingly, this included the activation of protein metabolism-related genes that was not as apparent in iPSCs. OF also exhibited activation of ontological clusters of genes related to cell damage and an immune response, indicating that oncogenic transformation is sensed by the cell as damage and/or induces actual cell damage. The most striking molecular feature distinguishing iPSCs from OF was the module of induced pluripotency-related factor expression in iPSCs, including nearly all the factors shown to facilitate iPSC formation. The absence of robust induction of these genes in OF suggests they do not play a role in the maintenance of tumors, at least in the context here of sarcoma. The most strongly induced pluripotency genes in iPSCs were Oct4 and Dppa5, induced ∼50 and 15-fold, respectively. The potential role of Dppa5 in the iPSC formation remains unclear at this point. Dnmt3b and Dnmt3l were also induced specifically during the iPSC formation, implicating specific DNA methylation changes in the process. The observation of slightly enriched Mdm2 expression and the absence of the detectable p53 protein in iPSCs are intriguing given the links between the loss of the p53 pathway function and induced pluripotency [28–33]. To our knowledge, our study is the first to suggest a potential role for Mdm2 in reprogramming. It is also interesting that expression of the tumor suppressor Rb as well as Cdkn1a (p21) were enriched in iPSCs over MEFs and OF, suggesting substantial complexity in the changes that occur during iPSC formation.
We provide evidence that a subset of the expression changes that are common to both OF and iPSCs are cell-type specific. An inherent aspect of expression changes in cells that have been reprogrammed either into pluripotent cells or OF is the loss of the cells' differentiation signature. In this case, our choice of using identical starting parental cells, while having the benefit of creating a system with genetically identical cells, in addition, introduced a potential bias into our comparison. This possibility was further examined by examining the gene expression profile of BM iPSCs [38]. Indeed, there was a decrease in expression of differentiation-associated genes in BM iPSCs that are associated with the parental bone marrow cells that are not shared with fibroblasts. This finding supports the notion that downregulation of differentiation-associated genes is a common event during both iPSC and OF formation, but that the nature of the specific genes that are downregulated in each case will depend largely on the starting cell type.
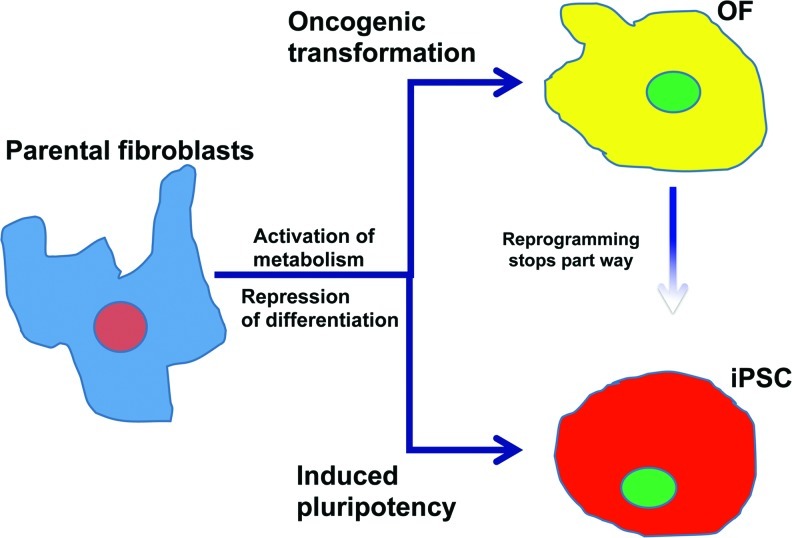
Taken together, our findings support a working model for the mechanisms by which differentiated cells may acquire either pluripotency or oncogenic transformation (Fig. 7). A common initiating event may be repression of differentiation-associated gene expression modules, while at the same time switching to a more glycolytic metabolic state. The cellular path then diverges depending on the genes that are being expressed. Notably, others have successfully reprogrammed other cancer cell lines [60] and our ROF were highly similar, but not identical to iPSCs. In this model, oncogenic transformation may actually be a special case of cellular reprogramming.
FIG. 7.
Diagram of similarities between reprogramming and transformation of fibroblasts. In our model comparing cellular reprogramming and transformation, normal fibroblasts first acquire changes that lead to a downregulation of the cell's differentiation machinery and a concomitant upregulation of glycolysis and other metabolic pathways. Only then do the pathways diverge into either an oncogenic or pluripotent phenotype depending on other factors, such as pluripotency genes. Finally, pluripotent cells can acquire traits that render them oncogenic, while we did not observe the reverse process (OF acquiring full pluripotency). Color images available online at www.liebertpub.com/scd
The identification of iPSC and OF as related cell types and the description of specific molecular pathways that they share, lays the foundation for further exploration of the mechanism by which iPSCs form teratoma and other malignant tumors, at least in the context of fibroblasts as starting cells. In that way, our studies add to the growing knowledge base that will aid in the discovery of novel methods to make iPSCs in a more efficient and less tumorigenic manner. One novel type of target in this regard is cellular metabolism. Studies of cancer-associated metabolic reprogramming are yielding many surprising results. There are currently several clinical trials testing anticancer drugs that target cellular metabolism [61], one of which is specifically targeting HK2, a gene we found to be uniquely upregulated in our iPSC. Our findings along with others suggest that targeting specific metabolic pathways may enhance the iPSC formation, while modulating others may improve safety. Further study, including functional genomics and metabolomics approaches will more clearly elucidate the relationship between induced pluripotency and tumorigenicity.
Supplementary Material
Acknowledgments
The authors want to thank Rebecca Cotterman for reading the manuscript, Jan Nolta and Jeannine McGee for assistance with the teratoma/tumor formation assays, and Katie Bell and Sandy Borowsky for aid in analysis of tumor types. They also thank Charlie Nicolet and the UC Davis Expression Analysis Core, Dawei Lin of the UCD Bioinformatics Core, and Neil Willits of the UCD Statistics department. This work was supported by the CIRM grant RN2-00922 and the NIH grant 5R01GM100782-01.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M. Meissner A. Foreman R. Brambrink T. Ku M. Hochedlinger K. Bernstein BE. Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:260–262. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka S. Takahashi K. [Induction of pluripotent stem cells from mouse fibroblast cultures] Tanpakushitsu Kakusan Koso. 2006;51:2346–2351. [PubMed] [Google Scholar]
- 5.Park IH. Zhao R. West JA. Yabuuchi A. Huo H. Ince TA. Lerou PH. Lensch MW. Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M. Koyanagi M. Tanabe K. Takahashi K. Ichisaka T. Aoi T. Mochiduki Y. Takizawa N. Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Chin MH. Mason MJ. Xie W. Volinia S. Singer M. Peterson C. Ambartsumyan G. Aimiuwu O. Richter L, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiou SH. Yu CC. Huang CY. Lin SC. Liu CJ. Tsai TH. Chou SH. Chien CS. Ku HH. Lo JF. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 9.Liu A. Cheng L. Du J. Peng Y. Allan RW. Wei L. Li J. Cao D. Diagnostic utility of novel stem cell markers SALL4, OCT4, NANOG, SOX2, UTF1, and TCL1 in primary mediastinal germ cell tumors. Am J Surg Pathol. 2010;34:697–706. doi: 10.1097/PAS.0b013e3181db84aa. [DOI] [PubMed] [Google Scholar]
- 10.Meng HM. Zheng P. Wang XY. Liu C. Sui HM. Wu SJ. Zhou J. Ding YQ. Li JM. Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:295–302. doi: 10.4161/cbt.9.4.10666. [DOI] [PubMed] [Google Scholar]
- 11.Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miura K. Okada Y. Aoi T. Okada A. Takahashi K. Okita K. Nakagawa M. Koyanagi M. Tanabe K, et al. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 13.Blum B. Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Cancer Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- 14.Wong DJ. Liu H. Ridky TW. Cassarino D. Segal E. Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben-Porath I. Thomson MW. Carey VJ. Ge R. Bell GW. Regev A. Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varlakhanova NV. Cotterman RF. de Vries WN. Morgan J. Donahue LR. Murray S. Knowles BB. Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin CH. Lin C. Tanaka H. Fero ML. Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS One. 2009;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CH. Jackson AL. Guo J. Linsley PS. Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varlakhanova N. Cotterman R. Bradnam K. Korf I. Knoepfler PS. Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics Chromatin. 2011;4:20. doi: 10.1186/1756-8935-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sridharan R. Tchieu J. Mason MJ. Yachechko R. Kuoy E. Horvath S. Zhou Q. Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotterman R. Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One. 2009;4:e5799. doi: 10.1371/journal.pone.0005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devine MJ. Ryten M. Vodicka P. Thomson AJ. Burdon T. Houlden H. Cavaleri F. Nagano M. Drummond NJ, et al. Parkinson's disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat Commun. 2011;2:440. doi: 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yancopoulos GD. Nisen PD. Tesfaye A. Kohl NE. Goldfarb MP. Alt FW. N-myc can cooperate with ras to transform normal cells in culture. Proc Natl Acad Sci U S A. 1985;82:5455–5459. doi: 10.1073/pnas.82.16.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keath EJ. Caimi PG. Cole MD. Fiboblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984;39:339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- 25.Breitman ML. Vigne R. Vogt PK. Growth-related alterations induced in chick embryo fibroblasts by src-gene deletion mutants of the Schmidt-Ruppin strain of Rous sarcoma virus. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1047–1055. doi: 10.1101/sqb.1980.044.01.113. [DOI] [PubMed] [Google Scholar]
- 26.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 27.Aoi T. Yae K. Nakagawa M. Ichisaka T. Okita K. Takahashi K. Chiba T. Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y. Yin X. Qin H. Zhu F. Liu H. Yang W. Zhang Q. Xiang C. Hou P, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Hong H. Takahashi K. Ichisaka T. Aoi T. Kanagawa O. Nakagawa M. Okita K. Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura T. Suzuki J. Wang YV. Menendez S. Morera LB. Raya A. Wahl GM. Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marion RM. Strati K. Li H. Murga M. Blanco R. Ortega S. Fernandez-Capetillo O. Serrano M. Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utikal J. Polo JM. Stadtfeld M. Maherali N. Kulalert W. Walsh RM. Khalil A. Rheinwald JG. Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. Collado M. Villasante A. Strati K. Ortega S. Canamero M. Blasco MA. Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinds P. Finlay C. Levine AJ. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol. 1989;63:739–746. doi: 10.1128/jvi.63.2.739-746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohaly G. Korf K. Dehde S. Dornreiter I. Simian virus 40 activates ATR-Delta p53 signaling to override cell cycle and DNA replication control. J Virol. 2010;84:10727–10747. doi: 10.1128/JVI.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou BK. Mali P. Huang X. Ye Z. Dowey SN. Resar LM. Zou C. Zhang YA. Tong J. Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng B. Jiang J. Kraus P. Ng JH. Heng JC. Chan YS. Yaw LP. Zhang W. Loh YH, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 38.Kunisato A. Wakatsuki M. Kodama Y. Shinba H. Ishida I. Nagao K. Generation of induced pluripotent stem cells by efficient reprogramming of adult bone marrow cells. Stem Cells Dev. 2010;19:229–238. doi: 10.1089/scd.2009.0149. [DOI] [PubMed] [Google Scholar]
- 39.Panopoulos AD. Yanes O. Ruiz S. Kida YS. Diep D. Tautenhahn R. Herrerias A. Batchelder EM. Plongthongkum N, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodnar MS. Meneses JJ. Rodriguez RT. Firpo MT. Propagation and maintenance of undifferentiated human embryonic stem cells. Stem Cells Dev. 2004;13:243–253. doi: 10.1089/154732804323099172. [DOI] [PubMed] [Google Scholar]
- 41.Oliveros JC. VENNY an interactive tool for comparing lists with Venn Diagrams. 2007. http://bioinfogp.cnb.csic.es/tools/venny/index.html. [Oct 11;2012 ]. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 42.Ito M. Hiramatsu H. Kobayashi K. Suzue K. Kawahata M. Hioki K. Ueyama Y. Koyanagi Y. Sugamura K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 43.Thorpe WP. Reilly JJ. Rosenberg SA. Prognostic significance of alkaline phosphatase measurements in patients with osteogenic sarcoma receiving chemotherapy. Cancer. 1979;43:2178–2181. doi: 10.1002/1097-0142(197906)43:6<2178::aid-cncr2820430603>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Huang da W. Sherman BT. Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W. Sherman BT. Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Jensen LJ. Kuhn M. Stark M. Chaffron S. Creevey C. Muller J. Doerks T. Julien P. Roth A, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J. Chen T. Liu X. Jiang J. Li J. Li D. Liu XS. Li W. Kang J. Pei G. More synergetic cooperation of Yamanaka factors in induced pluripotent stem cells than in embryonic stem cells. Cell Res. 2009;19:1127–1138. doi: 10.1038/cr.2009.106. [DOI] [PubMed] [Google Scholar]
- 48.Wei Z. Yang Y. Zhang P. Andrianakos R. Hasegawa K. Lyu J. Chen X. Bai G. Liu C. Pera M. Lu W. Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells. 2009;27:2969–2978. doi: 10.1002/stem.231. [DOI] [PubMed] [Google Scholar]
- 49.Smith KN. Singh AM. Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakagawa M. Takizawa N. Narita M. Ichisaka T. Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cotterman R. Jin VX. Krig SR. Lemen JM. Wey A. Farnham PJ. Knoepfler PS. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654–9662. doi: 10.1158/0008-5472.CAN-08-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knoepfler PS. Zhang XY. Cheng PF. Gafken PR. McMahon SB. Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guccione E. Martinato F. Finocchiaro G. Luzi L. Tizzoni L. Dall’ Olio V. Zardo G. Nervi C. Bernard L. Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 54.Martinato F. Cesaroni M. Amati B. Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Werbowetski-Ogilvie TE. Schnerch A. Rampalli S. Mills CE. Lee JB. Hong SH. Levadoux-Martin M. Bhatia M. Evidence for the transmission of neoplastic properties from transformed to normal human stem cells. Oncogene. 2011;30:4632–4644. doi: 10.1038/onc.2011.175. [DOI] [PubMed] [Google Scholar]
- 56.Felsher DW. Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pera MF. Stem cells: the dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- 58.Lin SL. Chang DC. Chang-Lin S. Lin CH. Wu DT. Chen DT. Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyoshi N. Ishii H. Nagai K. Hoshino H. Mimori K. Tanaka F. Nagano H. Sekimoto M. Doki Y. Mori M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2009;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun C. Liu YK. Induced pluripotent cancer cells: progress and application. J Cancer Res Clin Oncol. 2011;137:1–8. doi: 10.1007/s00432-010-0955-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheong H. Lu C. Lindsten T. Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nature Biotechnology. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.