Abstract
Telomeres terminate in 3´ single stranded G-overhangs that function in telomere end protection and telomerase action. An accurate measurement of overhang length is challenging due to the presence of many kilobases of double-stranded telomere DNA. Here, a simple method is described that utilizes duplex specific nuclease (DSN) to digest all genomic DNA including telomeres, leaving the single-stranded overhangs intact. The telomere single strand G-rich overhang length can then be determined by Southern blot based assays.
Keywords: Telomere, G-overhang, duplex specific nuclease (DSN), genomic DNA, Southern blot
1. Introduction
Mammalian telomeric DNA consists of repetitive hexamers of TTAGGG, ending in a single-stranded G-overhang at the 3´ ends of the chromosome(1). Telomeric overhangs are thought to be involved in the formation of T-loops, in which the single-stranded overhang invades the double-stranded telomere, forming a so called D-loop(2). This structure is thought to hide the chromosome ends from being recognized as DNA double-stranded breaks. Telomeric overhangs play an essential role in the protection of telomeres from end-to-end fusions, abnormal recombination, degradation(3), and serve as the substrate of telomerase in vivo.
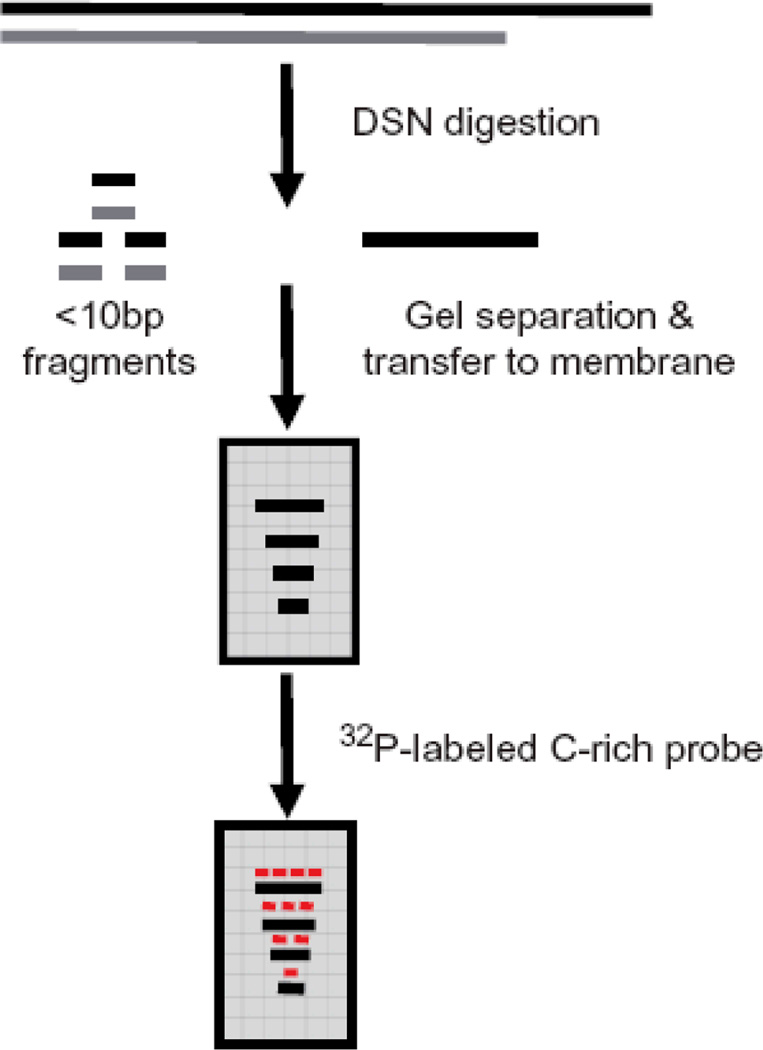
As G-overhangs perform many critical functions, it is important to have an assay that can accurately determine the length of telomeric overhangs. Current overhang assays fall into two groups: relative analysis and absolute length measurements. With relative analysis such as the non-denaturing hybridization(4) and HPA methods(5), the relative strength of overhang signals with respect to total telomeric DNA is obtained. In comparison, the absolute length measurements such as primer extension-nick translation (PENT)(6), telomeric-oligonucleotide ligation assay (T-OLA)(7) and electron microscopy methods(1), provide an actual overhang length but fail to detect very short overhangs. In this chapter, we present details of a new method for the direct measurement of overhang length that utilizes Kamchatka crab duplex specific nuclease (DSN). DSN is a newly characterized endonuclease that is highly specific for double-stranded (ds) DNA and is practically inactive on single-stranded (ss) DNA(8). DSN is able to digest double stranded DNA into <10bp fragments while leaving the single-stranded telomeric overhangs intact(9). These single-stranded overhangs can then be used for size determination on Southern blots upon hybridization with a telomere-specific probe (Figure 1). This procedure has been successfully applied to analyze overhang lengths as short as 12nt in human cells(9).
Figure 1.
Strategy of DSN assay to determine the length of telomeric overhangs.
2. Materials
2.1 Cell harvest and DNA isolation
1×PBS, pH 7.4, made from 10×PBS (Gibco)
DNeasy Blood & Tissue Kit (Qiagen, Inc)
RNase A (100mg/ml, Qiagen, Inc)
Ethyl Alcohol (100%)
Millipore water with 5mM Tris-HCl, pH 8.0
2.2 DSN digestion
Exonuclease I, 20U/µl (Epicentre Biotechnologies)
Duplex Specific Nuclease DSN (EVROGEN, Moscow, Russia), prepared as 0.2U/µl following instructions provided by manufacture and stored at −20°C.
10×DSN buffer provided by the manufacture containing 500mM Tris-HCl, pH 8.0; 50mM MgCl2; 10mM DTT
0.5M EDTA, pH 8.0
37 °C water bath or PCR machine
2.3 Electrophoresis on alkaline agarose gel and transfer to membrane
10M NaOH and 0.5M EDTA
2×loading dye: 100mM NaOH; 2mM EDTA; 5% Ficoll (type 400); 0.05% bromophenol blue (Bio-Rad, Inc)
Alkali electrophoresis buffer: 50mM NaOH; 1mM EDTA
UltraPure™ Agarose (Invitrogen, Inc)
Telomeric DNA markers: synthesized 36, 54 and 96-mer oligonucleotides of repetitive telomere sequence were used as low MW DNA markers; high MW telomeric markers were made as described by Chai et al(10). (see Note 1)
Amersham Hybond™-XL (GE Healthcare, Inc)
10×SSC (transfer buffer), made from 20×SSC: 3M NaCl; 0.3M sodium citrate, pH 7.0
3MM Whatman Paper
2.4 Making a high specific activity telomere C-rich probe
10 pmol/µl GTU4 oligonucleotide dissolved in TE buffer: 5′-GGGUUAGGGUUAGGGUUAGGGAAA-3′
100 pmol/µl T3C3+9 oligonucleotide (T3C3 is complementary to the 3′ end of GTU4 above, and +9 refers to 9nt of telomeric repeats) dissolved in TE buffer: 5′-TTTCCCTAACCCTAA-3′
1 M NaCl
10× buffer M: 100 mM Tris·Cl (pH 7.5); 100 mM MgCl2; 500 mM NaCl; 10 mM dithioerythritol
2 M Tris·Cl, pH 7.4 to 7.6
10 mg/ml BSA (Ambion, Inc)
1.25 mM dAdT: 1.25 mM each of dATP and dTTP
[α-32P]dCTP (3000 Ci/mmol) (PerkinElmer)
5U/µl Klenow large fragment of E. coli DNA polymerase I (NEB)
1 U/µl uracil deglycosylase (UDG) (Invitrogen)
PCR machine
2.5 Hybridization and washing buffer
Hybridization buffer: 6×SSC; 5×Denhardt’s solution; 0.5%(w/v) SDS
Washing buffer 1: 2×SSC and 0.1% SDS
Washing buffer 2: 0.5×SSC and 0.1% SDS
UVC 500 Crosslinker (GE Healthcare, Inc)
Hybridization oven (Hybridiser HB-1D, Techne, Inc)
2.6 Image capture and data analysis
Typhoon™ (GE Healthcare)
Phosphor screen (Molecular Dynamics)
Software: ImageQuant 5.2 and IQTools 3.0
3. Methods
3.1 Cell harvest and DNA purification (see Note 2)
Harvest cells by trypsinizing and spinning at 500g for 5min, wash the cell pellet once with 1×PBS and resuspend the cells in 1×PBS (5×106 cells/200µl).
Isolate genomic DNA following the DNeasy kit instructions with a slight modification: instead of adding RNase A before lysing the cells, add it afterwards and incubate at RT for at least 10min.
Re-precipitate the DNA by adding 2 volumes of 100% ethanol and spinning down.
Wash DNA pellet twice with 70% ethanol.
Let DNA pellet dry (not too dry) at room temperature. It will take 10min to 1h depending on the size of the pellet.
Resuspend the DNA pellet in deionized water (Millipore) containing 10mM Tris-HCl, pH 8.0.
Incubate DNA in a 37°C water bath overnight to allow genomic DNA to completely dissolve in solution, and to disrupt any potential overhang structures.
Measure DNA concentration. (The ratio of 260/280 must be between 1.8–1.9)
3.2 DSN digestion of genomic DNA
- Prepare samples for DSN digestion:
Genomic DNA(5 µg) χ µl (dependent on DNA concentration) 10×DSN buffer 2 µl H2O 17- χ µl DSN(0.2U/ µl) 1 µl Total 20 µl As a control, add 10 U (0.5 µl) ExoI to genomic DNA instead of DSN in the above reaction solution and incubate at 37°C for 1 h to digest 3' overhangs. Then add DSN.
Perform DSN digestion at 37°C for 2h (see Note 3).
Stop reaction by adding 0.5µl 0.5M EDTA (pH 8.0).
3.3 Electrophoresis of telomeric overhangs on alkaline agarose gels and DNA transfer to membranes (see Note 4)
Add the correct amount of agarose to a measured quantity of boiling Millipore water to make a 1.2% gel.
Cool the solution to ~50°C, add 10M NaOH and 0.5M EDTA to a final concentration of 50mM and 1mM respectively.
Make the gel. After it sets add sufficient alkaline electrophoresis buffer to cover the gel.
Add the same volume (20µl) of 2×loading dye to each sample.
Load the samples; telomeric DNA markers are loaded on both sides of the gel, perform electrophoresis in a cold room (4°C) at low voltage (1–2V/cm) until the dye has migrated approximately 6–8cm.
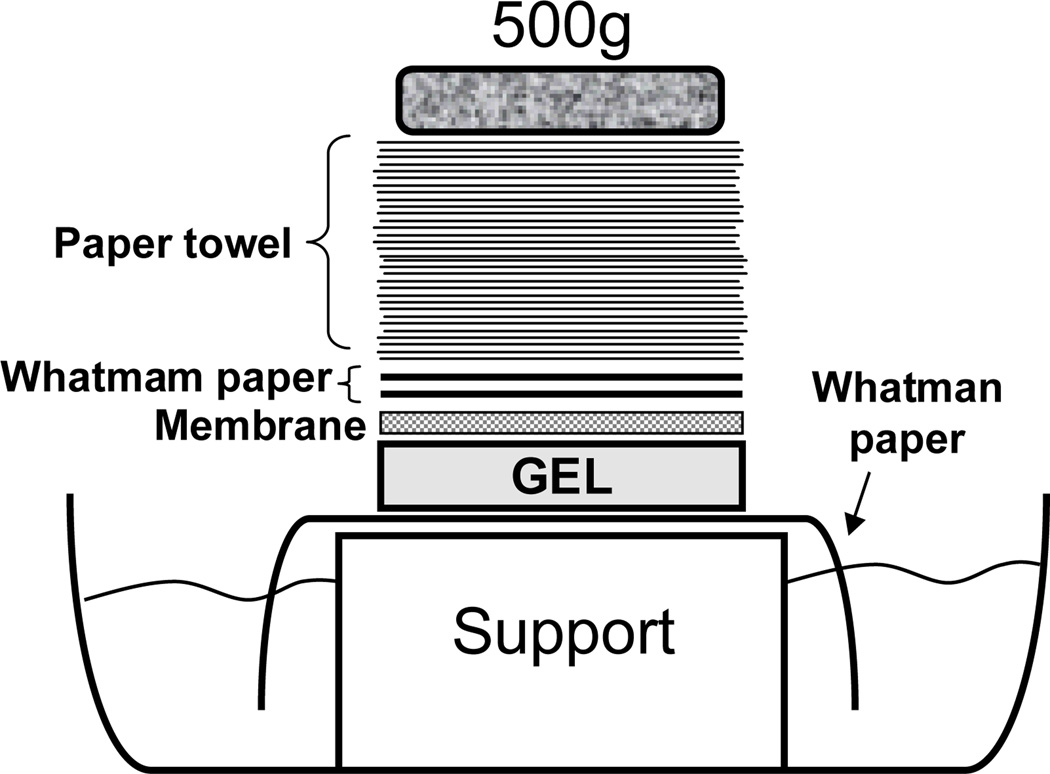
Set up a blotting tray for the capillary transfer as shown in Fig 2.
Carefully place the gel on 10×SSC presoaked Whatman paper with both of its sides draped in transfer buffer and remove all bubbles in between gel and paper.
Cut a piece of Hybond™-XL membrane and two pieces of 3MM Whatman paper slightly larger than gel, wet the membrane in dH2O and then soak it in transfer buffer until ready, wet Whatman filter paper in transfer buffer.
Cut four strips of SaranWrap™ to cover around the edge of the gel.
Carefully overlay membrane on top of the gel, roll away all bubble between gel and membrane.
Lay wet Whatman filter paper, one at a time, on the top of the membrane, making sure no bubbles are introduced.
Place a stack of paper towels (5–10cm high, cut to appreciate size) over the Whatman filter paper. Put glass plate on top of paper towels and apply a weight (such as a 2 inch book) on top.
Allow capillary transfer to occur overnight.
Remove the membrane from the gel and let it dry in air for 1h.
Fix DNA on the membrane by UV cross-linking at 70 mj/cm2 in a commercial UV crosslinker.
Figure 2.
Illustration of upward capillary transfer.
3.4 Making a high specific activity telomere C-rich probe (see Note 5)
- Make 1.7pmol/µl annealed oligonucleotide template by mixing the following:
10 pmol/µl GTU4 oligonucleotide 3.4 µl 100 pmol/µl T3C3+9 oligonucleotide 15.6 µl 1 µl 1M NaCl 1 µl Total: 20 µl Incubate for 1 min at 99°C, 15 min at 37°C, and 15 min at room temperature, the annealed template is stored at −20°C.
- Make 8× modified buffer M, by mixing the following:
10× buffer M 500 µl 2 M Tris·Cl, pH 7.4 to 7.6 100 µl 10 mg/ml BSA 25 µl Total 625 µl - Prepare the reaction mixture:
8× buffer M 3.125 µl Annealed template oligo 1 µl 1.25 mM dAdT (final 50 µM) 1 µl dH2O 13.875 µl [α-32P]dCTP 5 µl Klenow large fragment 1 µl Total 25 µl Incubate the reaction mixture at room temperature for 30min and then 95°C for 5min.
Add 0.5µl of 1 U/µl UDG and incubate for 10 min at 37°C and then 10 min at 95°C, the probe is ready to use.
3.5 Hybridizing, washing, exposing on PhosphoImager screens and imaging
Prehybridize the membrane at 42°C for at least 30min using hybridization buffer.
Add 5–10µl high specific activity telomere C-rich probe to hybridization buffer to a concentration of 1µl/ml.
Hybridize at 42°C overnight.
After the hybridization, wash the membrane by incubating twice, 15min each, in washing buffer 1, followed by washing buffer 2 for 15min at 42°C.
Remove the membrane from the last wash, drain and wrap in SaranWrap, expose to PhosphoImager screens for an appropriate time (unusually 2–3h is enough to get a good signal, but overnight exposure is recommended to obtain better images).
After exposure, scan PhosphoImager screen on Typhoon.
3.6 Analysis of data to obtain mean length of the telomeric overhang
Divide each lane into 100–300 intervals from the highest MW to the lowest using ImageQuant 5.2.
- Determine the MW for each interval by fitting the molecular weight (Y) and migration distance (X) of each marker to one phase exponential decay to generate a standard curve using the following function:
-
i.Y= (Y0 − Plateau)×exp (−KX) + Plateau,
-
i.
Use the ExoI treated sample as a background and subtract its signal from that of the untreated sample at each measured size interval.
- Calculate the average overhang length as the weighted mean using the following formula:
-
ii.Mean overhang length= Σ(ODi)/Σ(ODi/Li),
-
ii.
Footnotes
Telomeric DNA markers can be replaced by commercial DNA markers with 5´-ends labeled by 32P.
DSN is highly sensitive to ionic strength, and the presence of salt results in a dramatic decrease in catalytic activity. Therefore, it is important to avoid any unnecessary salt input. In order to get rid of any potential contaminations from the purification process, we re-precipitate the genomic DNA, wash with 70% ethanol twice and dissolve in low-salt solution. If DNA is from other sources, the same protocol is highly recommended.
The maximum activity of DSN is 65°C, but this will melt small DNA fragments well before the limit size of <10bp is reached.
Overhangs vary in size and give a smear. This smear is more compact and easier to see using 1.2% agarose gels compared to 8% polyacrylamide gels, but either can be used.
This primer extension protocol is designed for synthesis of super-sensitive probes that have 6 32P nucleotides incorporation (For details, see ref.11 (11)). Using regular 32P end-labeled C-rich probes may not be sensitive enough to detect telomeric overhangs from 5µg genomic DNA. The use of dU and uracil deglycosylase to remove the G-strand template maximizes the hybridization of the single stranded C-rich probe to the G-rich overhangs.
References
- 1.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 4.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara H, Kusunoki M, Yamanaka Y, Matsumura S, Ide T. G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat Methods. 2005;2:829–831. doi: 10.1038/nmeth797. [DOI] [PubMed] [Google Scholar]
- 6.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 7.Cimino-Reale G, Pascale E, Battiloro E, Starace G, Verna R, D'Ambrosio E. The length of telomeric G-rich strand 3'-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 2001;29:E35. doi: 10.1093/nar/29.7.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002;12:1935–1942. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Hoshiyama H, Shay JW, Wright WE. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008;36:e14. doi: 10.1093/nar/gkm1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai W, Shay JW, Wright WE. Human telomeres maintain their overhang length at senescence. Mol Cell Biol. 2005;25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbert BS, Shay JW, Wright WE. Analysis of telomeres and telomerase. Curr Protoc Cell Biol. 2003;Chapter 18(Unit 18):16. doi: 10.1002/0471143030.cb1806s20. [DOI] [PubMed] [Google Scholar]