Abstract
Kaposi sarcoma-associated herpesvirus (KSHV) encodes a viral interleukin 6 (vIL6) that mimics many activities of human IL6 (hIL6). Both vIL6 and hIL6 play important roles in stimulating the proliferation of tumors caused by KSHV. Here, we provide evidence that a miRNA pathway is involved in regulation of vIL6 and hIL6 expression through binding sites in their open reading frames (ORF). We show a direct repression of vIL6 by hsa-miR-1293 and hIL6 by hsa-miR-608. The repression of vIL6 by miR-1293 was reversed by disruption of the vIL6 miR-1293 seed match through the introduction of point mutations. In addition, expression of vIL6 or hIL6 in KSHV-infected cells could be enhanced by transfection of the respective miRNA inhibitors. In situ hybridization of human lymph node sections revealed that miR-1293 is primarily expressed in the germinal center, but is deficient in the mantle zone of lymph nodes where the expression of vIL6 is often found in patients with KSHV-associated multicentric Castleman’s disease, providing evidence of an anatomic correlation. Together, our study indicates that IL6 expression can be regulated by miRNA interactions in its ORF and provides evidence for the role of these interactions in the pathogenesis of KSHV-associated diseases.
Keywords: Viral IL6, IL6, miRNA, Kaposi sarcoma-associated herpesvirus, germinal center, gene expression, post-transcriptional regulation
Introduction
IL6 plays a critical role in cell proliferation, inflammatory response, and oncogenesis[1–3]. IL6 expression is regulated by NF-κB and is influenced by a variety of external stimuli. Various RNA elements in the 3′ untranslated region (UTR) of IL6 mRNA contribute to IL6 mRNA degradation and regulate IL6 expression. These include an AU-rich element (ARE) that interacts with AU-rich binding factor 1 (AUF1)[4] and tristetraprolin[5], and a non-ARE element that interacts with Zc3h12a endonuclease[6].
miRNAs are non-coding regulatory RNAs in size of 21–25 nucleotides and modulate gene expression by base-pairing with complementary nucleotide sequences (seed matches) in the 5′ and 3′ UTRs of target mRNAs[7]. miRNA-mediated gene suppression, in an RNA-induced silencing complex (RISC), is the result of inhibiting translation and causing mRNA decay [7]. There is evidence that let-7[8] and miR-26[9] miRNA can interact with their correspondent seed matches in the 3′ UTR of human IL6 (hIL6) and regulate hIL6 expression. Other short-lived cytokine mRNAs are also subject to miRNA regulation, including miR-16-1 repression[10] and miR-369-3 increase of TNFα expression[11], and miR-155 indirect repression of IL4, IL5, and IL10 expression in activated T cells[12].
Kaposi sarcoma-associated herpesvirus (KSHV) is a lymphotropic tumor virus and is the causal agent for Kaposi sarcoma (KS) [13], primary effusion lymphoma (PEL)[14], and a form of multicentric Castleman’s disease (MCD)[15]. KSHV encodes a number of mimics of human genes, including a viral IL6 (vIL6) with about 24.8 % amino acid sequence identity and 49.7% sequence similarity to hIL6[16]. Unlike hIL6, vIL6 binds to the signal transducer protein, gp130, without the need to bind to the hIL6 receptor-α. Nonetheless, it induces similar cellular effects to hIL6[16–21]. IL6 promotes B cell and KS spindle cell proliferation, with the increased expression of vIL6 and hIL6 being important in the pathogenesis of KS, PEL, and KSHV-associated MCD[17,18,21,22] and is associated with severe KSHV-associated inflammatory cytokine syndromes in patients co-infected with HIV and KSHV[23]. vIL6 expression has been ascribed to transcriptional activation of the vIL6 gene by KSHV ORF50 [24] and by IFN-α [22]. KSHV ORF50 also transactivates the hIL6 gene[25]. Although generally considered a viral lytic gene[26], vIL6 is detectable at a low level in uninduced PEL cells without expression of other viral lytic genes [20,27,28].
In this report, we examined the function of cellular miRNAs in the regulation of vIL6 and hIL6 production. We found that vIL6 and hIL6 are regulated by two separate miRNAs, each of which binds to a seed match residing in the open reading frame (ORF) of the respective IL6. We also found that vIL6 is expressed primarily in a region of lymph nodes in which there is little expression of the vIL6-suppressive miRNA.
Material and methods
Cells
HEK293 and HeLa cells were grown in Dulbecco’s modified Eagle’s medium with 10% FBS. RKO and RKOdicer- cells[29] were grown in McCoy’s 5A medium with 10% FBS. JSC-1 (KSHV+/EBV+)[30] and BCBL-1 cells (KSHV+)[31] were cultured in RPMI 1640 containing 10% FBS. Doxycycline-inducible TREx BCBL1-Rta and vector control cell lines were cultured as described [32]. A HEK293 cell line stably harboring a wild-type KSHV genome (Bac36 wt)[33] was maintained in DMEM medium with 10% FBS and hygromycin at 150 ng/ml.
Plasmids
To create a KSHV vIL6 (K2) ORF expression vector pNP4 (Figure S1), KSHV K2 (nt 17875 to 17264 of the KSHV genome [NCBI accession number U75698]) was amplified by PCR from JSC-1 cell DNA with a primer pair of oNP34 and oNP35 (See all primer sequences in Table S1), digested with NotI and BamHI, and cloned into a p3xFLAG-CMV-14 vector. To create a miR-re vIL6 (pJGK10) vector, C17423A, C17424G, C17425A, and C17427G mutations were introduced into vIL6 gene by overlapping PCR with the paired primers oNP34 and oNP35 on a mixture of two separate PCR products: the first PCR product was amplified from pNP4 by a primer pair of oJGK32 and oNP34, and the second PCR by a primer pair of oJGK33 and oNP35. The overlapped PCR product was digested with NotI and BamHI and cloned into a p3xFLAG-CMV-14 vector. To express a 3x FLAG-tagged hIL6 (pJGK6, Figure S1), the human IL6 ORF (GenBank accession number NM_000600) from the SC125236 plasmid (Origene, Rockville, MD) was amplified by PCR with a primer pair of oJGK24 and oJGK25. The PCR product was then cloned into the p3xFLAG-CMV-14 vector. The HA/FLAG-hAgo2 plasmid[34] was purchased from Addgene (Cambridge, MA). All plasmids were confirmed by sequencing.
Transient cotransfection
Cells were cotransfected with the indicated plasmids in each figure using Lipofectamine 2000 for analyses 24 h after transfection. siRNA duplexes for Ago2 and miR-1293 were transfected with Lipofectamine 2000. Pre-miR-NC#1 served as a negative control, Pre-miR-608, Pre-miR-1293 and Pre-miR-214 were purchased from Ambion (Austin, TX). Cells were transfected with Pre-miRs using Ambion siPORT-NeoFX and followed by vIL6 or hIL6 vector 48 h after Pre-miR transfection.
Anti-miRs [Peptide nucleic acids (PNAs)-based miRNA inhibitors] for hsa-miR-1293, hsa-miR-608, hsa-miR-214 and a negative control (NC), were purchased from Panagene Co. (Daejeon, South Korea). Cell transfection with each anti-miR was performed according to manufacturer’s instruction by direct addition of the testing anti-miR to culture medium. Cell lysates were prepared and analyzed 3 days after transfection.
RNA-protein pulldown assay
RNA oligomers labeled with biotin at the 5′ end (Fig. 2D) were immobilized on NeutrAvidin beads. The cell lysates were applied to RNA oligomer-immobilized beads in 1x binding buffer [20 mM Tris-HCl (pH7.5), 200 mM NaCl, 6 mM EDTA, 5 mM potassium fluoride, 5 mM β-glycerophosphate, and 1 tablet of protease inhibitors (Roche, Nutley, NJ)/10ml]. Pulldown was conducted at 4°C overnight with rotation. Proteins on the beads were washed 3 times with 1x binding buffer and dissolved in 2X SDS sample buffer for Western blot analysis.
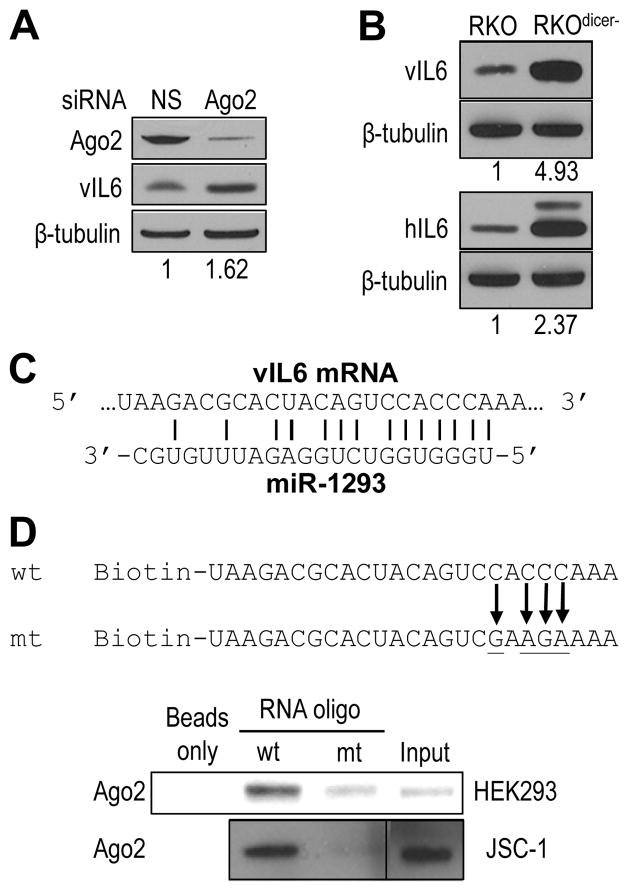
Figure 2.
Involvement of the miRNA pathway in regulation of IL6 expression. (A) Reduction of endogenous Ago2 expression by siRNA promotes vIL6 production in HEK293 cells. The cell lysates were immunoblotted 24 h after vIL6 expression vector transfection of HEK293 cells pretreated with Ago2 siRNA (40 nM) for 24 h. (B) Increased expression of vIL6 and hIL6 in Dicer-deficient RKO cells. Both wt RKO and Dicer-knockout RKO cells (RKOdicer-) were transfected with a vIL6 (top) or hIL6 (bottom) expression vector and were immunoblotted for vIL6 or hIL6 expression 24 h after transfection. The upper band of hIL6 is N-glycosylated hIL6[64]. The numbers below each panel of blots in (A–B) indicate a relative level of IL6 expression after being normalized to β-tubulin. (C) A putative human miR-1293 seed match in the coding region of vIL6 mRNA. (D) The miR-1293 seed match of vIL6 mRNA interacts with Ago2. Biotinylated RNA oligomers containing a vIL6 wild-type (wt) miR-1293 seed match or a mutant (mt) seed match with mutations underlined and by arrows were used for RNA pulldown and immunoblot assays with a fractionated cytoplasmic extract from HEK293 cells ectopically expressing HA-Ago2 (top) or from JSC-1 cells. The proteins in the pulldowns were blotted with anti-Ago2 (JSC-1) or anti-HA (HEK293 for Ago2) antibodies.
Western blot analysis
Protein samples from the pulldowns or prepared by lysis of cells in 1x RIPA buffer were dissolved in 2x SDS sample buffer and were resolved by electrophoresis. Western blot analyses were conducted with the following antibodies: monoclonal or polyclonal anti-vIL6 [35,36], polyclonal anti-Ago2 (Upstate, Waltham, MA), monoclonal anti-β-tubulin (Sigma), and monoclonal anti-FLAG (M2, Sigma), together with corresponding horseradish peroxidase-conjugated secondary antibodies (Sigma). The signal on blots was detected with a West Pico or Femto chemiluminescence substrate (Thermo Scientific).
In vitro transcription/translation assay
DNA templates for in vitro transcription of full-length vIL6 (pNP4) or miR-re vIL6 (pJGK10) were amplified from pNP4 or pJGK10 by PCR using a primer pair of T7 chimeric oJGK46 and oJGK47 oligomer dT(T30/2 stop codons/vector sequence, which provides a poly-A tail in mRNA.
In vitro translation of wt vIL6 and miR-re vIL6 in the presence of an miRNA duplex and HA-tagged human Ago2 (HA-hAgo2) protein was performed by using Promega (Madison, WI) rabbit reticulocyte lysate (RRL) as described [37]. Firefly luciferase (FL) mRNA served as an internal control.
ELISA
hIL6 levels in the culture supernatant of KSHV-infected cells was determined by an IL6 Single-Analyte ELISArray kit (SABiosciences, Frederick, MD).
Immunohistochemistry and Immunofluorescence
Immunohistochemical staining of MCD lymph node sections was performed as described [38] by using an anti-vIL6 antibody [36].
Stable Bac36 cells grown on coverslips and transfected with anti-miRs were fixed with 4% paraformaldehyde in PBS for 15 min. The cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min and blocked with 2% BSA in PBST (PBS containing 0.05% Tween 20) for 1 h at 37°C. Anti-vIL6 antibody [36] was applied to cells for 1 h at 37 °C followed by three washes in PBST, 10 min each. The cells were incubated with a corresponding secondary antibody conjugated with Alexa Fluor 546 for 1 h at 37°C. Confocal fluorescence optical slices, 2.0 μm in thickness, were acquired using a Zeiss LSM510 META (Carl Zeiss MicroImaging, Inc., Thornwood, NY) microscope equipped with a 20x plan-apochromat (N.A. 0.8) objective lens.
In situ hybridization (ISH)
ISH was performed based on a published protocol [39]. DIG-labeled LNA miRCURY probes for detection of miR-155 (5′ DIG-labeling) and miR-1293 (5′ and 3′ double DIG labeling) were purchased from Exiqon (Woburn, MA). Lymph node sections from patients with KSHV-associated MCD or HIV-associated follicular hyperplasia approved by the NIH Office of Human Subjects Research were deparaffinized with xylene for 5 min twice and hydrated with ethanol dilutions (100%, 70%, 30% and DEPC water) for 2 min each (twice for each step). After washing twice in PBS, 5 min each, the sections were deproteinated with proteinase K (10 ug/ml) at 37°C for 5 min, fixed for 10 min in 4% paraformaldehyde, rinsed twice in PBS, prehybridized for 1 h in 1X hybridization buffer in an ENZO ISH AP Detection Kit (ENZO, Farmingdale, NY) at 37°C, and hybridized with a probe (500 nM) at 37 °C for 16 h in a humidified chamber. After hybridization, the slides were washed 2 times, 5 min each, in ISH wash reagent at 4 °C, blocked for 30 min in antibody blocking buffer, and incubated for 1 h at 37 °C with anti-DIG-AP Fab fragments (1:100 in blocking buffer) (Roche). After washed for 1 min in SignaSure Wash buffer (ENZO), the slides were incubated with NBT/BCIP reaction mixture until color development, washed three times in PBST, 5 min each, counterstained with FastRed nuclear staining reagent, rinsed with tap water, dehydrated, and mounted for microscopy. Brightfield images were acquired using a AxioVision software (v. 4.6) controlling a Zeiss axiovert 200M microscope equipped with 10x plan-apochromat (N.A. 0.45) air and 63x plan-apochromat (N.A. 1.4) oil objective lenses and an Axiocam MRc5 color CCD camera (Carl Zeiss MicroImaging Inc.).
qRT-PCR
First-strand cDNA was synthesized from 100 ng of total RNA using random hexamers and SuperScript II RT. The qPCR was conducted using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). A primer pair of oJGK51 and oJGK52 was used for vIL6 amplification and a primer pair of oJGK24 and oJGK25 for hIL6 amplification. For GAPDH amplification, a primer pair of oZMZ269 and oZMZ270[40] was used.
TaqMan miRNA assays (Applied Biosystems, Foster City, CA) were used in accordance with manufacturer’s protocol to detect endogenous miR-1293 and miR-608 from total cell small RNA (10 or 100 ng ) prepared with Ambion miRNA isolation kit. The CT values of qRT-PCR data from 3 repeats were analyzed by the 2-ΔΔCT method [41] and plotted to against a standard linear equation generated by qRT-PCR on double-stranded synthetic miR-1293 or miR-608 as described for lin-4 miRNA [42]. After dividing by a factor of total small RNA per cell, the calculated values were averaged as a copy number (mean + SD) per cell.
Results
miRNA pathway is involved in regulation of vIL6 and hIL6 expression by its effect on vIL6 and hIL6 ORFs
Since hIL6 expression can be regulated by several miRNAs through their interaction with the 3′ UTR (~ 430 nts in length) of hIL6 mRNA[8,9], we considered the possibility that KSHV-encoded vIL6 could be also regulated by miRNAs. However, vIL6 has a short (<70 nts) 3′ UTR region and there are no conserved seed matches with the miRNAs that bind to the 3′ UTR of hIL6. Expanding the scope of our search, we attempted to identify putative miRNA binding sites in both ORFs. By searching a miRNA database (http://microrna.sanger.ac.uk), we identified a miR-214 seed match and a miR-1293 seed match in a sequence conserved between the ORFs of vIL6 and hIL6 (Figure 1). To investigate whether miRNA is involved in regulating IL6 expression, we used RNAi to knock down the expression of Ago2 [43] in HEK293 cells and then transfected the cells with a vIL6 ORF expression vector containing only the coding region. We found that the expression of vIL6 protein increased in the Ago2-deficient cells (Figure 2A). This was confirmed in RKOdicer- cells, which have global reduction of mature miRNAs[29]. When a vIL6 or hIL6 ORF expression vector was transfected into wild-type (wt) RKO or RKOdicer- cells, a substantial increase of vIL6 and hIL6 expression was observed in RKOdicer- cells over wt RKO cells (Figure 2B), indicating that miRNA regulates the expression of both vIL6 and hIL6 through its effect on the corresponding ORF. We then focused on miR-1293 regulation of vIL6 because the identified miR-1293 seed match in vIL6 is a binding site for viral ORF57 [37] and this seed match in hIL6 overlaps with miR-608. We then examined the binding of Ago2 to the predicted miR-1293 seed match in vIL6 (Figure 2C) as Ago2 is a major component of RISC which associates with targeted mRNA via a miRNA seed match[44]. RNA-protein pulldown assays in the presence of HEK293 or JSC-1 cell lysates were performed with a biotin-conjugated vIL6 RNA oligomer with a wt or mt miR-1293 seed match. As expected, Ago2 was found to bind to wt vIL6 RNA, but only weakly to the mt vIL6 RNA (Figure 2D), providing evidence that an efficient Ago2 binding to the miR-1293 seed match region of vIL6 is mediated by miR-1293.
Figure 1.
Conserved miRNA seed matches in the coding regions of both hIL6 (NCBI accession number NM_000600) and vIL6 mRNAs (NCBI accession number U75698). * indicates conserved sequences between hIL6 and vIL6. The box indicates the conserved region that has the same seed match for both miR-608 and miR-1293 in hIL6 and a miR-1293 seed match in vIL6.
Suppression of vIL6 expression by miR-1293 and hIL6 expression by miR-608, with each binding to the corresponding IL6 ORF
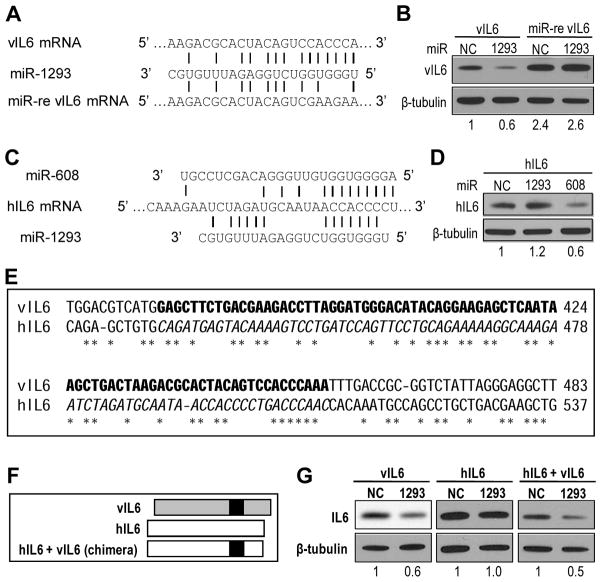
The role of miR-1293 in vIL6 expression was examined by a miR-1293-resistant vIL6 (miR-re vIL6) with mutations in the seed match within vIL6 ORF (Figure 3A). The expression efficiency of this miR-re vIL6 in HEK293 cells in the presence of ectopic miR-1293 or a miR-NC (a non-specific miRNA control) was compared to the expression of a wt vIL6 by transfection. We found that miR-re vIL6 showed increased expression relative to its wt counterpart (2.4 vs 1) in the presence of ectopic miR-NC and ectopic miR-1293 inhibited the expression of wt vIL6, but not miR-re vIL6 (Figure 3B). These data indicate that miR-1293 suppression of vIL6 is miRNA seed-match-specific, demonstrating that vIL6 is a direct target of both endogenous and exogenous miR-1293.
Figure 3.
Inhibition of vIL6 and hIL6 expression by miRNAs. (A) Generation of a miRNA-resistant vIL6 (miR-re vIL6) by introduction of point mutations into the miR-1293 seed match. (B) Repression of vIL6 expression by ectopic miR-1293 in HEK293 cells. The cells were pretreated with 10 nM Pre-miR-1293 or a negative control (NC) Pre-miR-NC for 48 h before transfection with an indicated vIL6-FLAG expression vector. Cell lysates prepared 24 h after transfection were blotted for vIL6 expression by using an anti-FLAG antibody. (C) Two putative, overlapped miRNA seed matches in the ORF of hIL6 mRNA corresponding to the miR-1293 responsive region of vIL6 mRNA. (D) Repression of hIL6 expression by miR-608, but not by miR-1293. HeLa cells pretreated for 48 h with 10 nM Pre-miR-608, Pre-miR-1293 or Pre-miR-NC were transfected with an hIL6-FLAG expression vector for additional 24 h before Western blotting for hIL6 expression using an anti-FLAG antibody. The numbers below panels B and D indicate a relative level of vIL6 and hIL6 in each sample after being normalized to β-tubulin. (E) The IL6 sequences for swapping between hIL6 and vIl6. The vIL6 sequences indicated by the black box in (F) for swapping are bolded to replace the corresponding hIL6 sequence in Italic. The conserved nts between vIL6 and hIL6 are marked with *. (F) Diagram of the strategy for swapping the vIL6 region (black box) into hIL6. (G) The vIL6 region responsive to miR-1293 functions in hIL6 in the response to miR-1293-mediated repression. HEK293 cells transfected with Pre-miR-NC or Pre-miR-1293 for 48 h were transfected again with the indicated IL6-FLAG expression vector for additional 24 h before being immunoblotted with anti-FLAG M2 and β-tubulin antibodies. The numbers below each sample set indicate a relative level of hIL6 in each sample in a representative gel after being normalized to β-tubulin.
As hIL6 expression was greatly increased in RKOdicer- cells (Figure 2B) and the ORF region of hIL6 mRNA also contains overlapped miR-608 and miR-1293 seed matches in the corresponding region of vIL6 (Figure 3C), the effects of both miR-608 and miR-1293 on hIL6 expression were examined in transient cotransfection of HEK293 cells. We found that only the miR-608 seed match of hIL6 is functional, and is responsible for miR-608 repression of hIL6 expression (Figure 3D). To verify the specificity of each miRNA to its target, we generated a chimeric hIL6 by exchanging the corresponding region of hIL6 with the target region of miR-1293 in vIL6 (Figure 3E–F) and demonstrated that the miR-1293 target region of vIL6 could convert miR-1293-resistant hIL6 into miR-1293-sensitive hIL6 (Figure 3G), concluding that vIL6 and hIL6 are separately regulated by miR-1293 and miR-608 through their binding to specific seed matches in the corresponding ORF.
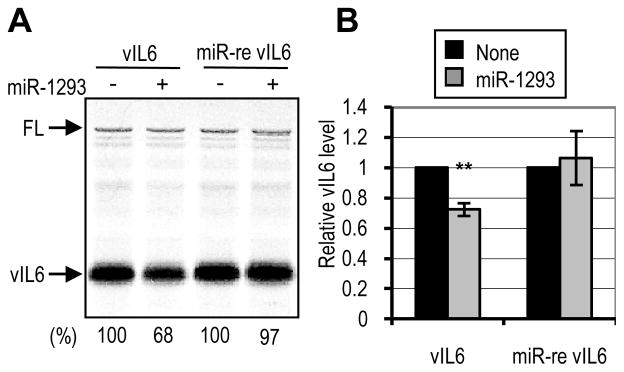
To discriminate the specificity of miR-1293 in repression of vIL6 protein expression from possible indirect effects such as off-targets or RNA instability, the inhibition of vIL6 translation by miR-1293 was performed by an in vitro cell-free translation system using RRL, modified from published protocols [45,46]. When miR-1293 was added to the translation assays, a specific translational repression (~25% – 30%) was observed for wt vIL6 RNA, but not for miR-re vIL6 RNA (Figure 4). The FL expression which was used as an internal control was not affected by addition of miR-1293 to the assays. These results indicate a direct role for miR-1293 in the translational repression of vIL6 expression.
Figure 4.
Translational repression of vIL6 by miR-1293 in vitro. (A) In vitro translation assay was performed by using rabbit reticular lysate and miR-1293 over an in vitro transcribed wt vIL6 or miR-re vIL6 transcript. The numbers below the representative gel indicate the translation efficiency of vIL6 mRNA relative to an internal control, firefly luciferase (FL) mRNA. (B) Relative levels of vIL6 protein normalized to FL are shown from 5 separate reactions. Bars represent mean ± SD (n=5). **, P<0.01 (t-test).
Anti-miR-1293 and anti-miR-608 interrupt miR-mediated repression of IL6 expression
The finding that miR-re vIL6 could be more efficiently expressed than wt vIL6 in transfected cells indicates that the endogenous miR-1293 in these cells could be a natural inhibitor for IL6 expression. We therefore assessed first by qRT-PCR the copy numbers of miR-1293 and miR-608 in KSHV-positive JSC-1 and BCBL-1 PEL cells, and in KSHV-negative HEK293 cells. We found that each PEL cell contains ~12 copies of miR-1293 and 40–50 copies of miR-608, while each HEK293 cell has ~45 copies of miR-1293 and 150 copies of miR-608 (Table 1). To assess the suppressive effects of endogenous miR-1293 and miR-608 on vIL6 or hIL6 expression, we then cotransfected HEK293 cells with a vIL6 ORF expression vector along with anti-miR-1293 or a hIL6 ORF expression vector along with anti-miR-608 and demonstrated that the cotransfection with a miRNA-specific inhibitor remarkably increased vIL6 or hIL6 expression (Figure 5A). Quantitative analyses of vIL6 and hIL6 RNAs showed that the cells cotransfected with a miRNA-specific inhibitor exhibited an increased level of the respective mRNA over the cells receiving a non-specific anti-miR control (Figure 5B). These data indicate that endogenous miR-1293 and miR-608 are highly potent, despite their relative low abundance in the cells. The increased vIL6 or hIL6 expression were specific only for anti-miR-1293 or anti-miR-608, but not for anti-miR-214 (Fig. 5C), suggesting that ectopic miR-214 affects the expression of vIL6 and hIL6 (Fig. 5D) presumably via an indirect mechanism.
Table 1.
The copy numbers of miR-1293 and miR-608 in PEL and HEK293 cells.
| Cell line | miR -1293 | miR -608 |
|---|---|---|
| JSC -1 | 11.66±2.96 | 50.49±4.31 |
| BCBL-1 | 11.43±3.38 | 41.52±1.54 |
| HEK293 | 43.15±1.91 | 148.50±14.27 |
The copy numbers of miR-1293 and miR-608 in the indicated cell lines were determined by TaqMan real-time RT-PCR.
Figure 5.
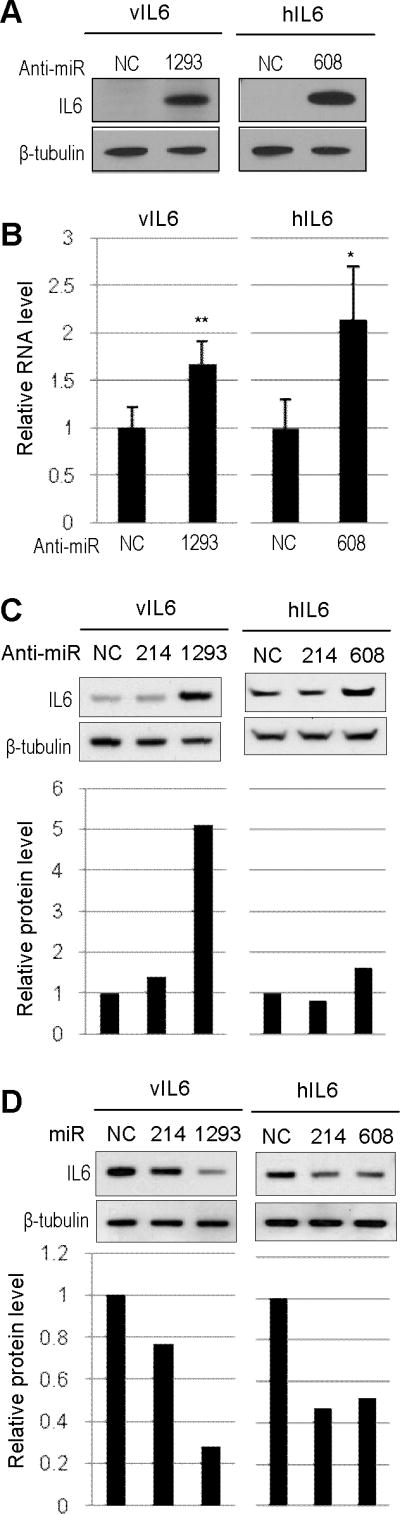
Enhancement of vIL6 or hIL6 expression by specific miRNA inhibitors. (A and B) Inhibition of endogenous miR-1293 or miR-608 in HEK293 cells to promote expression of vIL6 or hIL6. HEK293 cells were co-transfected with 200 nM of indicated anti-miRs along with 100 ng of FLAG-vIL6 (Left) or FLAG-hIL6 (Right). Protein samples and total RNA were prepared 24 h after transfection. Protein samples were analyzed by Western blot with anti-FLAG antibody for vIL6 or hIL6 expression or anti-β-tubulin antibody for sample loading (A). The amount of vIL6 or hIL6 RNA in the corresponding RNA samples was analyzed by qRT-PCR assays (B). Bars represent means ± SD (n=3). ** and * indicate P<0.01 and P<0.05 (t-test), respectively (B). (C and D) Inhibition of endogenous miR-214 did not affect vIL6 or hIL6 expression (C), but ectopic expression of miR-214 did do (D). See other details in (A).
Considering that a low amount of vIL6 mRNA is constitutively expressed in a subset of PEL that do not express other KSHV lytic genes[20], KSHV-infected JSC-1 and Bac36 cells with no reactivation were analyzed for expression of vIL6 and hIL6 in response to anti-miR-1293 and anti-miR-608 transfection. Increased expression of vIL6 (Figure 6A) and hIL6 (Figure 6B) was obtained in the cells transfected with anti-miR-1293 or anti-miR-608 when compared to the cells transfected with a non-specific anti-miR control. This observation could be duplicated in BCBL-1 cells (data not shown). However, we noticed that hIL6 expression in JSC-1 cells was increased only by 10% in the presence of anti-miR-608 over the cells receiving an anti-miR control. Although the increase was significant, the narrow increase in JSC-1 cells generally with low transfection efficiency seen for other B cells was expected for hIL6 because the 3′ UTR of hIL6 is targeted additionally by miR-26 and let-7 [8,9]. Confocal microscopy of vIL6 expression in KSHV-positive Bac36 cells showed enhanced expression of vIL6 after transfection of anti-miR-1293, relative to the cells receiving an anti-miR control (Figure 6C). Together with the results from gain-of-function of miR-1293 and miR-608 (Figure 3), the loss-of-function of miR-1293 and miR-608 in the presence of their corresponding inhibitors provide further evidence that the PEL cells limit the expression of both vIL6 and hIL6, respectively, by less abundant miR-1293 and miR-608 which exhibited a minimal reduction during KHHV lytic induction (Fig. S2).
Figure 6.
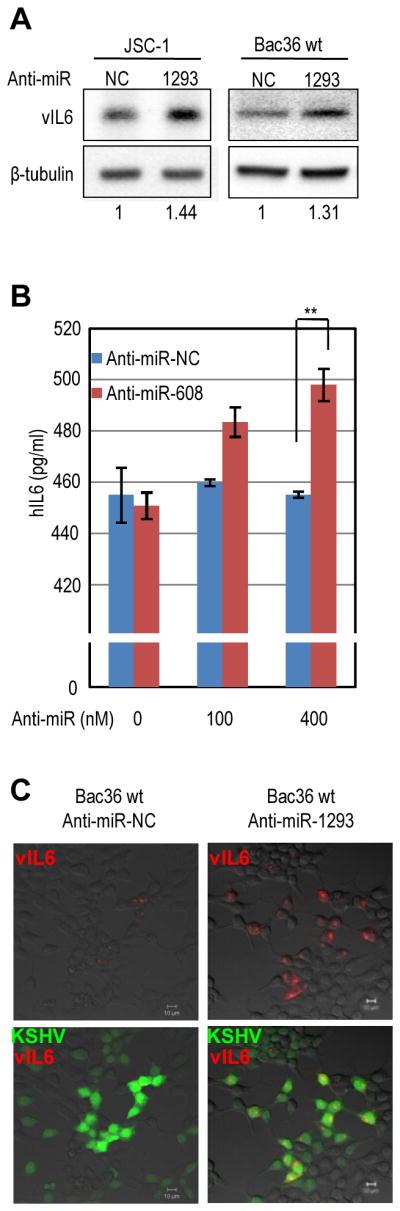
Blockade of miR-1293 or miR-608 function by a miRNA-specific inhibitor to increase vIL6 or hIL6 expression in KSHV-infected cells. (A) Increased expression of vIL6 in KSHV+ JSC-1 cells and Bac36 wt cells by a miR-1293 antimir. JSC-1 cells and Bac36 cells with a wild type KSHV genome (Bac36 wt) were transfected, respectively, with 400 nM and 200 nM of indicated anti-miRs. Protein samples prepared 3 days after transfection were immunoblotted with anti-vIL6 or anti-β-tubulin antibodies. The numbers below panels indicate a relative level of vIL6 expression after being normalized to β-tubulin. (B) Increased expression of hIL6 in KSHV+ JSC-1 cells by a miR-608 antimir. JSC-1 cells were transfected with indicated amount of anti-miRs. Culture media were collected 3 days after transfection. An ELISA assay was conducted to detect secreted hIL6. Bars represent means ± SD (n=3). ** indicates P<0.01 (t-test). (C) Confocal microscopy images of increased vIL6 expression in KSHV+ Bac36 wt cells transfected with an anti-miR-1293 inhibitor. Immunofluoresence assay using an anti-vIL6 antibody was carried out over the cells transfected with an indicated anti-miR. Bac36 cells with a wt KSHV genome expressing GFP were merged with the cells expressing vIL6 (red) shown in the bottom panels. Scale bar represents 10 μm.
miR-1293 deficiency in the mantle zone of lymph nodes provides an environment for vIL6 expression
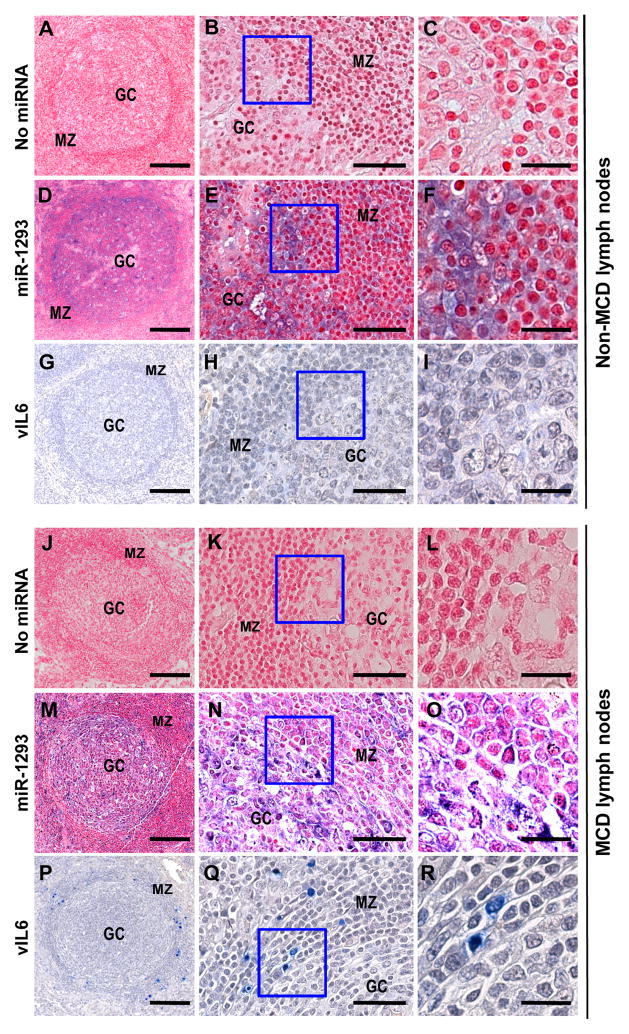
As assessed by immunohistochemical staining, vIL6 is mostly expressed in the mantle zones of lymph nodes in patients with MCD [28]. We hypothesized that this differential expression of vIL6 in lymph nodes might be associated with differential distribution of miR-1293. We next conducted ISH to determine miR-1293 distribution in lymph nodes in correlation with vIL6 expression. As shown in Figure 7, tissue sections of normal (A–I) and MCD lymph nodes (J–R) did not manifest any hybridization signal when no hybridization probe was used in the control sections (A–C; J–L). By contrast, the normal and MCD lymph node sections hybridized with a miR-1293 probe revealed that miR-1293 was primarily expressed in the germinal center (GC) (D–F; M–O), with the hybridization signals mainly in the cytoplasm (E–F; N–O), whereas cytokine-inducible miR-155 which targets activation-induced cytidine deaminase (AID) and transcription factor Pu.1 expressed in GC B cells for somatic hypermutation and class-switch recombination to diversify antibody repertoire [47–50] was mainly distributed, as suggested [39], in the mantle zone (MZ) region (Figure S3). Normal lymph nodes expressed no vIL6 (G–I), but MCD lymph nodes expressed vIL6 primarily in the enlarged MZ region (P–R). Thus, the relative deficiency of miR-1293 in MZ was consistent with a permissive environment for vIL6 expression in the specified region (Fig. 7, P–R) as reported [28]. Therefore, our results provide a possible biological mechanism for the selective expression of vIL6 in the MZ of lymph nodes from patients with KSHV-associated MCD (KSHV-MCD).
Figure 7.
Distribution of endogenous miR-1293 and vIL6 in lymph nodes. In situ hybridization was performed on lymph node sections from a patient with HIV-associated folicular hyperplasia (non-MCD, A–I) or with MCD (J–R) by using no specific probe (A–C and J–L) or a miR-1293 detection probe (D–F and M–O). The purple color indicates miR-1293. After NBT/BCIP reaction, tissue sections were counterstained with the FastRed nuclear staining reagent. The same tissue sections on separate slides were immunostained by an anti-vIL6 mouse monoclonal antibody for vIL6 (blue color) protein expression (G–I and P–R). MZ, mantle zone; GC, germinal center. (B, E, H, K, N and Q), a representative region showing both MZ and GC from (A, D, G, J, M and P), respectively, was imaged at higher magnification. (C, F, I, L, O and R), the regions in the box of (B, E, H, K, N and Q) were further enlarged, respectively. The images in (N )and (O) were adjusted for better contrast of the fastRed in MZ. Scale bars represent 200, 50 and 20 μm from the left to the right in each panel sets of three.
Discussion
In this report, we have demonstrated that miRNAs play an important role in the expression of both vIL6 and hIL6. We found that both vIL6 and hIL6 contain a miRNA binding site within their ORF, that these sites are bound by cellular miR-1293 and miR-608 respectively, and that these bound miRNAs serve to suppress the translation of the respective proteins. Consistent with this observation, the scope of functional miRNA-mRNA interactions have been recently expanded from RNA 3′ UTRs to include the coding regions of the targeted RNAs [51,52].
During virus infection, one conceivable mechanism for host cells to defend against virus invasion is to inhibit virus pathogenicity by host cell miRNAs [52–55]. It is possible that miR-1293, which suppresses KSHV vIL6, has developed in part to serve this purpose. KSHV evades the immune system and coexists in a host for many years through a latent gene repertoire and remaining quiescent. Regulation of vIL6 by miR-1293 may aid in this process.
The genes encoding miR-1293 (GeneID: 100302220) and miR-608 (GeneID: 693193) have been mapped to chromosomes 12q13.12 and 10q24.31, respectively. To date, little is known about their expression and function. We found that miR-1293 and miR-608 are expressed at low copy number in HEK293, JSC-1 and BCBL-1 cells and exhibit only slight reduction in PELs during lytic KSHV infection. The ability of endogenous miR-1293 and miR-608 to suppress vIL6 and hIL6 in these cells was verified with individual miRNA inhibitors, anti-miR-1293 and anti-miR-608, suggesting that both miR-1293 and miR-608 are highly potent and do not need to be present in a high copy number to regulate gene expression. The levels of miR-1293 can vary substantially among B cells located in anatomically different parts of a lymph node. Although miR-1293 function in B cell regulation remains unknown, it may play a role in the differential regulation of vIL6 in KSHV-MCD.
KSHV-MCD is a rare lymphoproliferative disorder characterized by polyclonal proliferations of KSHV-infected mature B cells with plasmablastic features that involve in lymph node tissues at multiple sites. Patients with MCD overexpressing vIL6 manifest high fevers, weight loss, anemia, and low white blood cell counts[56,57]. In affected lymph nodes of patients with MCD, a number of B cells are infected with KSHV, and a sizable percentage of these express viral lytic genes[28,57] and another sizable subset of these express vIL6 but not other lytic KSHV genes. Histologic evaluation of lymph nodes in KSHV-MCD demonstrates that vIL6 expression is specifically limited to MZ, home of non-responding B cells with a naive phenotype [28,58]. Until now, the reason for this differential expression remains unclear[21,28]. In this report, we provide evidence that a selective lack of miR-1293 in the MZ of lymph nodes may provide an environment permissive for vIL6 expression and a possible explanation for this unique feature of KSHV-MCD.
Cytokine-induced miR-155 in B and T cells has a role in GC development and function [47] by targeting AID [48] and Pu.1 [49] expressed in GC B cells to diversify antibody repertoire [47–50,59]. KSHV at latent infection expresses miR-K11 bearing the same seed sequence as miR-155 [60,61]. We found that MCD lymph nodes with KSHV infection of naive B cells [62] express miR-155 primarily in MZ, but less in GC. Whether there is a defect in BIC expression to encode miR-155 [63] in the diseased GC B cells remains unknown. An increased miR-155 expression in MCD MZ cells might simply reflect KSHV infection and viral cytokine activation.
Supplementary Material
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research. We thank Natalia Pripuzova for construction of the vIL6 expression vector pNP4, Giovanna Tosato and John Nicholas for providing anti-vIL6 antibodies, Bert Vogelstein for providing RKO Dicer knockout cells, and other members of the Zheng laboratory for their assistance and critical comments.
Footnotes
Author contribution statement
JGK performed all experiments and drafted the paper; VM carried out the in situ miR-1293 hybridization of MCD lymph nodes; XW analyzed miR-214 function; MK conducted confocal microscropy; TSU and RY collected lymph nodes and tissue sections; ZMZ designed the research and drafted the paper. JGK, VM, XW, TSU, MK, RY, and ZMZ analyzed all data and finalized the paper.
Conflict-of-interest disclosure: Dr. Giovanna Tosato, the spouse of one of the authors, R.Y. is a co-inventor on a patent describing the measurement of KSHV vIL6. This invention was made when G.T. was an employee of the US Government under 45 Code of Federal Regulations Part 7. All rights, title, and interest to this patent have been assigned to the U.S. Department of Health and Human Services. The government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P.L. 99-502).
References
- 1.Kishimoto T. Interleukin-6: From basic science to medicine - 40 years in immunology. Ann Rev Immunol. 2005;23:1–21. doi: 10.1146/annurev.immunol.23.021704.115806. [DOI] [PubMed] [Google Scholar]
- 2.Rutsch S, Neppalli VT, Shin DM, et al. IL-6 and MYC collaborate in plasma cell tumor formation in mice. Blood. 2010;115:1746–1754. doi: 10.1182/blood-2009-08-237941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trikha M, Corringham R, Klein B, Rossi JF. Targeted anti-interleukin-6 monoclonal antibody therapy for cancer: A review of the rationale and clinical evidence. Clin Cancer Res. 2003;9:4653–4665. [PMC free article] [PubMed] [Google Scholar]
- 4.Paschoud S, Dogar AM, Kuntz C, et al. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer I, Schaljo B, Vogl C, et al. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita K, Takeuchi O, Standley DM, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliopoulos D, Hirsch HA, Struhl K. An Epigenetic Switch Involving NF-kappa B, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones MR, Quinton LJ, Blahna MT, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing Q, Huang S, Guth S, et al. Involvement of MicroRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 11.Vasudevan S, Tong YC, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 14.Cesarman E, Chang Y, Moore PS, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 15.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 16.Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 17.Aoki Y, Yarchoan R, Wyvill K, et al. Detection of viral interleukin-6 in Kaposi sarcoma-associated herpesvirus-linked disorders. Blood. 2001;97:2173–2176. doi: 10.1182/blood.v97.7.2173. [DOI] [PubMed] [Google Scholar]
- 18.Jones KD, Aoki Y, Chang Y, et al. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi’s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood. 1999;94:2871–2879. [PubMed] [Google Scholar]
- 19.Nador RG, Cesarman E, Chadburn A, et al. Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood. 1996;88:645–656. [PubMed] [Google Scholar]
- 20.Nicholas J, Ruvolo VR, Burns WH, et al. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 21.Parravicini C, Corbellino M, Paulli M, et al. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV-seronegative Castleman’s disease. Am J Pathol. 1997;151:1517–1522. [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee M, Osborne J, Bestetti G, et al. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science. 2002;298:1432–1435. doi: 10.1126/science.1074883. [DOI] [PubMed] [Google Scholar]
- 23.Uldrick TS, Wang V, O’Mahony D, et al. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without Multicentric Castleman disease. Clin Infect Dis. 2010;51:350–358. doi: 10.1086/654798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng HY, Song MJ, Chu JT, Sun R. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) J Virol. 2002;76:8252–8264. doi: 10.1128/JVI.76.16.8252-8264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng HY, Chu JT, Rettig MB, et al. Rta of the human herpesvirus 8/Kaposi sarcoma-associated herpesvirus up-regulates human interleukin-6 gene expression. Blood. 2002;100:1919–1921. doi: 10.1182/blood-2002-01-0015. [DOI] [PubMed] [Google Scholar]
- 26.Paulose-Murphy M, Ha NK, Xiang CS, et al. Transcription program of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) J Virol. 2001;75:4843–4853. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandriani S, Ganem D. Array-Based Transcript Profiling and Limiting-Dilution Reverse Transcription-PCR Analysis Identify Additional Latent Genes in Kaposi’s Sarcoma-Associated Herpesvirus. J Virol. 2010;84:5565–5573. doi: 10.1128/JVI.02723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parravicini C, Chandran B, Corbellino M, et al. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases - Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummins JM, He YP, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannon JS, Ciufo D, Hawkins AL, et al. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi’s sarcoma herpesvirus-containing supernatant. J Virol. 2000;74:10187–10193. doi: 10.1128/jvi.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renne R, Zhong WD, Herndier B, et al. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura H, Lu M, Gwack Y, et al. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majerciak V, Pripuzova N, Mccoy JP, et al. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8 alpha, and K8. 1 and the production of infectious virus. J Virol. 2007;81:1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Aoki Y, Jaffe ES, Chang Y, et al. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- 36.Aoki Y, Narazaki M, Kishimoto T, Tosato G. Receptor engagement by viral interleukin-6 encoded by Kaposi sarcoma-associated herpesvirus. Blood. 2001;98:3042–3049. doi: 10.1182/blood.v98.10.3042. [DOI] [PubMed] [Google Scholar]
- 37.Kang JG, Pripuzova N, Majerciak V, et al. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Promotes Escape of Viral and Human Interleukin-6 from MicroRNA-Mediated Suppression. J Virol. 2011;85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Meyers C, Guo M, Zheng ZM. Up-regulation of p18Ink4c expression by oncogenic HPV E6 via p53-miR-34a pathway. Int J Cancer. 2011;129:1362–1372. doi: 10.1002/ijc.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuovo GJ, Elton TS, Nana-Sinkam P, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protocols. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majerciak V, Yamanegi K, Allemand E, et al. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol. 2008;82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 44.Tang F, Hajkova P, O’Carroll D, et al. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem Biophys Res Commun. 2008;372:24–29. doi: 10.1016/j.bbrc.2008.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang BB, Love TM, Call ME, et al. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 46.Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex elF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 47.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 48.Teng G, Hakimpour P, Landgraf P, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muramatsu M, Kinoshita K, Fagarasan S, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 51.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 53.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng ZM, Tang S, Tao M. Development of resistance to RNAi in mammalian cells. Ann N Y Acad Sci. 2005;1058:105–118. doi: 10.1196/annals.1359.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aoki Y, Yarchoan R, Braun J, et al. Viral and cellular cytokines in AIDS-related malignant lymphomatous effusions. Blood. 2000;96:1599–1601. [PubMed] [Google Scholar]
- 57.Staskus KA, Sun R, Miller G, et al. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J Virol. 1999;73:4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katano H, Sato Y, Kurata T, et al. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi’s sarcoma, and multicentric Castleman’s disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottwein E, Mukherjee N, Sachse C, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skalsky RL, Samols MA, Plaisance KB, et al. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du MQ, Liu H, Diss TC, et al. Kaposi sarcoma-associated herpesvirus infects monotypic (IgM lambda) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood. 2001;97:2130–2136. doi: 10.1182/blood.v97.7.2130. [DOI] [PubMed] [Google Scholar]
- 63.Eis PS, Tam W, Sun L, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meads MB, Medveczky PG. Kaposi’s sarcoma-associated herpesvirus-encoded viral interleukin-6 is secreted and modified differently than human interleukin-6 - Evidence for a unique autocrine signaling mechanism. J Biol Chem. 2004;279:51793–51803. doi: 10.1074/jbc.M407382200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.