Abstract
Circadian rhythms are generated by well-conserved interlocked transcriptional feedback loops in animals. In Drosophila, the dimeric transcription factor CLOCK/CYCLE (CLK/CYC) promotes period (per), timeless (tim), vrille (vri), and PAR-domain protein 1 (Pdp1) transcription. PER and TIM negatively feed back on CLK/CYC transcriptional activity, whereas VRI and PDP1 negatively and positively regulate Clk transcription, respectively. Here, we show that the α isoform of the Drosophila FOS homolog KAYAK (KAY) is required for normal circadian behavior. KAY-α downregulation in circadian pacemaker neurons increases period length by 1.5 h. This behavioral phenotype is correlated with decreased expression of several circadian proteins. The strongest effects are on CLK and the neuropeptide PIGMENT DISPERSING FACTOR, which are both under VRI and PDP1 control. Consistently, KAY-α can bind to VRI and inhibit its interaction with the Clk promoter. Interestingly, KAY-α can also repress CLK activity. Hence, in flies with low KAY-α levels, CLK derepression would partially compensate for increased VRI repression, thus attenuating the consequences of KAY-α downregulation on CLK targets. We propose that the double role of KAY-α in the two transcriptional loops controlling Drosophila circadian behavior brings precision and stability to their oscillations.
Introduction
Circadian rhythms synchronize animal physiology and behavior with the day/night cycle. They are generated by a complex transcriptional network of interlocked feedback loops. The architecture of this network and many of its components are conserved between insects and mammals (Emery and Reppert, 2004).
In Drosophila, the dimeric transcription factor CLOCK/CYCLE (CLK/CYC) is at the center of this network (Hardin, 2005; Zhang and Emery, 2012). It promotes period (per) and timeless (tim) transcription. PER and TIM form dimers that are phosphorylated by several kinases: DOUBLETIME (DBT), CASEIN KINASE II, NEMO, and SHAGGY (Kloss et al., 1998; Price et al., 1998; Martinek et al., 2001; Lin et al., 2002; Akten et al., 2003; Ko et al., 2010). Once properly phosphorylated, PER and TIM move into the nucleus in which they interact with CLK/CYC. They initiate repression first on the chromatin and then displace CLK/CYC from their binding sites (E-boxes) (Menet et al., 2010). PER/TIM repression requires DBT, which stably binds PER (Kloss et al., 2001; Yu et al., 2009). This first transcriptional loop plays a particularly important role in the generation of 24 h period rhythms. Modulating this loop is the transcriptional repressor CLOCKWORK ORANGE (CWO), which recognizes the same E-boxes as the CLK/CYC dimer (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). cwo transcription is itself regulated by CLK/CYC.
The second feedback loop is somewhat simpler. CLK/CYC transactivate the vrille (vri) and PAR-domain protein 1 (pdp1) genes (Cyran et al., 2003; Glossop et al., 2003). PDP1 feeds back positively on the Clk promoter, whereas VRI antagonizes the activity of PDP1 by competing for the same binding sites. The phase of PDP1 and VRI protein rhythms differ by a few hours, hence permitting Clk transcription to oscillate. The importance of Clk transcription rhythms remains uncertain, because they are not necessary for 24 h period behavioral rhythms (Kim et al., 2002). However, PDP1 and VRI levels are crucial for proper CLK expression (Blau and Young, 1999; Zheng et al., 2009). For example, a mutation that specifically abolishes the PDP1ε isoform results in low CLK levels, loss of PER and TIM cycling, and arrhythmic behavior (Zheng et al., 2009). Interestingly, forced expression of CLK in pdp1ε mutants restores PER and TIM rhythms, without restoring behavioral rhythms. Thus, the CLK/VRI/PDP1 loop is particularly important for circadian output pathways.
This transcriptional network is present in all tissues with circadian rhythms, including the neurons driving circadian behavior. There are ∼150 circadian neurons in the fly brain (Nitabach and Taghert, 2008). For the control of rhythmic locomotor activity, the most important are the PIGMENT DISPERSING FACTOR (PDF)-positive small ventral lateral neurons (sLNvs). Indeed, these cells drive circadian locomotor behavior in constant darkness (DD) (Renn et al., 1999; Blanchardon et al., 2001; Stoleru et al., 2005). In their absence, or in the absence of the neuropeptide PDF, flies are usually arrhythmic or show weak short period rhythms. Here we show that KAYAK (KAY)—the Drosophila homolog of the transcription factor c-FOS—modulates the circadian transcriptional network in sLNvs.
Materials and Methods
Plasmid constructs.
The DNA sequence of the exon encoding the N-terminal region of KAY-α was amplified by PCR from Drosophila melanogaster genomic DNA with the following primers: kay-α forward, 5′-CGTAGCGAATTCATGATTGCACTAAAGGCCACC-3′; and kay-α–BstBI reverse, 5′-TCGAACTTCGAAGTTGCCGAGTTGTCTGCCCATTTGAAGGG-3′. The rest of the KAY-α coding sequence was amplified from BDGP cDNA clone SD04477 with the following primers: kay-α–BstB1 forward, 5′-GATCTGTTCGAAACCGGCCAGAGTGTTCTCAC-3′; and kay-α reverse, 5′-CATCTCCTCGAGTTATAAGCTGACCAGCTTGGA-3′. The two BstB1 primers introduce silent mutations that create a BstB1 restriction site that was used to clone in a three-fragment ligation the whole kay-α cDNA in the EcoRI and XhoI restriction sites of pAc5.1/V5–HisB (pAc for short). This construct (and all the other constructs generated by PCR) was confirmed by sequencing. We noticed two coding differences between our sequence and that of Flybase: six additional nucleotides are found in a stretch of glutamine-encoding codons in our sequence. This adds two glutamines to that glutamine repeat (Glu124–129 becomes Glu124–131). In addition, the Ala15 codon is changed to Pro15. These coding changes were found in independent clones and must thus represent polymorphisms. The kay-α cDNA was subsequently transferred to pUAST to make a pUAST–kay-α construct.
Genomic Drosophila pseudoobscura DNA was used to amplify the exon coding for KAY–trunc and KAY-α. The following primers were used: Psesro-5.1, 5′-GTCGAATTCATGATTGCCATAAAGTCCATC-3′; Psesro-3.1, 5′-GAGTTACTCGAGCTAGGGCATACTTACATGTCT-3′; and Psesro-3.2, TCGAACTTCGAAGTTGCCGAGATGTCTTTGTATCACTTGCCG. Note that we amplified an extra 24 nucleotides compared with the predicted Drosophila pseudoobscura KAY-α coding region, because those eight N-terminal codons are obviously conserved between Drosophila melanogaster and pseudoobscura. To generate the chimeric kay-α construct in pUAST, a three-fragment ligation was made with the PCR product obtained with Psesro-5.1 and Psesro-3.2. The pUAST–kay–trunc construct was generated by cloning the PCR product obtained with Psesro-5.1 and Psesro-3.1. These two constructs were injected in embryos to generate transgenic flies.
per–Ebox–luc, pAc–Clk, pAc–β-galactosidase, pAc–VP16–CWO, Clk–luc, CMV–vri–VP16, CMV–Pdp1ε, and CMV–Renilla were described previously (Darlington et al., 1998; Cyran et al., 2003; Lim et al., 2007). tim–luc and tim–mut–luc were generated by amplifying the proximal promoter region of the tim promoter with the following primers: 5′tim, 5′-CTAGCTGGTACCGAGTGCACAGAAACGTTCTG-3′; 3′tim, 5′-GCTACGACGCGTCTGAAAGTAGTTTTAAGAATATTTG-3′; ap1mut1, 5′-CTGCGACTCGAGGTGTAAGCACTCTCTTTAAGAT-3′; and ap1mut2, 5′-CGTCGACTCGAGCTGGTCTTTCTCTCAGTGTT-3′. The amplification products were cloned in the KpnI and MluI sites of pGL3. The kay-α–promoter–luc construct was made as follows. A 323 bp fragment ∼300–600 bp upstream of the transcription start site of kay-α was amplified from genomic DNA and ligated into pGL3 vector through KpnI and MluI restriction sites: primer forward, 5′-CGGGTACCATGTCTGGCTAGCGAAAAGC-3′; primer reverse, 5′-CCGACGCGTTTCACGCTGATGAGTCAACC-3′.
pAc–Jra was made by ligating the Jun-related antigen (Jra) cDNA (from BDGP clone LD25202) into pAc. The kay-γ cDNA was obtained from BDGP clone SD04477. pAc–kay-γ was made by ligating this cDNA into the EcoR1 and XhoI sites of pAc. pAc–kay-α was already described above. pAc–myc–kay-α was made by ligating a myc–kay-α PCR fragment into pAc through EcoRI and XhoI. Primers used for PCR were 5′-GACGAATTCATGGAGGAGCAGAAGCTGATCTCAGAGGAGGACCTGGCGGCCGCAATTGCACTAAAGGCCACCGA-3′ and kay-α reverse. An NotI site was introduced between the myc tag and kay-α. pAc–myc–kay-β was made by ligating a kay-β fragment to the NotI and BstB1 site of pAc–myc–kay-α. Primers to amplify the kay-β fragment were as follows: Not1–kay-β forward, 5′-CAACGCGGCCGCGAAAGTCAAAGTGGAGCGC-3′; and kay-β–BstBI reverse, 5′-TCGAACTTCGAAGTTGCCGAGGATAAGATTGCGCGTCGGTG-3′. To make pAc–kay–trunc–V5, kay–trunc with mutated stop codon was amplified from genomic DNA using the following primers: kay-α forward primer; and kay–trunc–V5 reverse, 5′-CGGCCTCGAGTATCGTACGCACTTAACTA-3′. This fragment was ligated into the EcoRI and XhoI restriction sites of pAc. The stop codon was mutated in a way such that the V5 in the pAc vector is in-phase with kay–trunc. In a similar way, pAc–kay-α–V5 was made using reverse primer 5′-CTAGACTCGAGTATAAGCTGACCAGCTTG-3′. CMV–myc–kay-α, CMV–myc–kay-β were made by ligating the corresponding kay sequence in pAc–myc–kay-α and pAc–myc–kay-β into pcDNA3.1 vector through EcoRI and XhoI restriction sites. kay-γ sequence from pAc–kay-γ was ligated into pcDNA3.1 through EcoRI and XhoI to make CMV–kay-γ.
pAc–VP16–kay-α was made by ligating kay-α sequence into the pAc–VP16 vector (generous gift from Dr. R. Allada, Northwestern University, Evanston, IL) using EcoRI and XbaI.
pAc–kay-α–basicΔ was made by ligating two PCR fragments amplified from pAc–kay-α into pAc vector through EcoRI, Acc65I, and XhoI restriction sites. This generates an ∼60 bp deletion of the basic region. Primers used were as follows: kay-α forward, 5′-TAGACTGGTACCCTGCTCCTCCTCCGGGGTCA-3′; and kay-α reverse, 5′-ATCGATGGTACCGTGGACCAGACCAACGAGCT-3′.
Drosophila stocks and transgenic flies.
Flies were reared on a standard cornmeal/agar medium at 25°C under a light/dark (LD) cycle. Pdf–GAL4 and Rh1–GAL4 flies were described previously (Renn et al., 1999; Mollereau et al., 2000). Pdf–GAL4 was meiotically recombined with UAS–dicer2 (Dietzl et al., 2007) to generate a Pdf–GAL4, UAS–dicer2 stock (PGD). Pdf–GAL4 or PGD was then crossed with NIG15507–R2 (kayR2) or NIG15507–R4 (kayR4) from the National Institute of Genetics (NIG) Stock Center (Mishima, Japan) to get Pdf–GAL4 /+; kayR2/+ or Pdf-GAL4 /+; kayR4/+ and PGD/+; kayR2/+ flies. A stable PGD/CyO; kayR2/TM6B line was made and crossed to different kay-α/trunc rescue lines for the cross-species rescue experiments. We also tested RNAi transgenes directed against all full-length KAY isoforms: NIG15509–R1 and R2 (NIG stock center) and VDRC6212 (Vienna Drosophila RNAi Center, Vienna, Austria). JraRNAi and luciferase RNAi (lucRNAi) lines are from the Transgenic RNAi Project at Harvard Medical School (Boston, MA). A stable PGD/CyO; lucRNAi/TM6B line was generated and crossed to per0;;UAS–per (Grima et al., 2004) to generate per0; PGD/+; UAS–per/lucRNAi flies. per0; PGD/+; UAS–per/kayR2 flies were generated similarly.
The pUAST–kay-α construct from D. melanogaster and the pUAST–kay-α and pUAST–kay–trunc from D. pseudoosbcura were introduced in flies by Genetic Services, using standard P-element-mediated germ-line transformation. A stable stock of Rh1–GAL4/CyO; UAS–kay-α/TM6B was generated and crossed with kayR2/TM6B to get Rh1–GAL4/+; kayR2/UAS–kay-α flies.
Behavioral assays.
Locomotor activities of adult males were monitored individually in Drosophila activity monitors (Trikinetics) in Percival I-36LL incubators (Percival Scientific). For LD cycle, light intensity was ∼500 lux in the light phase. Behavior data were collected using Drosophila Activity Monitor program (Trikinetics). Behavioral period was determined for each fly by analyzing the data from the first day to the fifth day in DD using FaasX (courtesy of F. Rouyer, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). Average actograms were generated by a signal processing toolbox for MATLAB using function “dam_panels” (MathWorks) (courtesy of J. Levine, University of Toronto, Mississauga, Ontario, Canada) (Levine et al., 2002). Only flies that survived the whole run were analyzed.
Adult brain immunocytochemistry and quantification of fluorescent signal.
Fly brains were immunostained and imaged essentially as described previously (Zhang et al., 2010). Primary antibodies included the following: mouse anti-PDF (1:400), rabbit anti-PER (1:1500; generous gift from Dr. M. Rosbash, Brandeis University, Waltham, MA), an affinity-purified guinea pig anti-TIM (1:100) (Rakshit et al., 2012), anti-rabbit PDP1 (1:400; generous gift from Dr. J. Blau, New York University, New York, NY), guinea pig anti-VRI (1:10,000), and guinea pig anti-CLK (1:2500) (generous gifts from Dr. P. Hardin, Texas A&M University, College Station, TX). Secondary antibodies included the following: anti-mouse Cy5, anti-rat Cy3, anti-rabbit FITC, and anti-guinea pig Cy3 (1:200 dilution; Jackson ImmunoResearch). Mounted brains were scanned using a Carl Zeiss LSM5 Pascal confocal microscope using 40× water lens. Images are single Z sections in Figures 2A,D and 5 and are digitally projected Z stacks in Figure 2C.
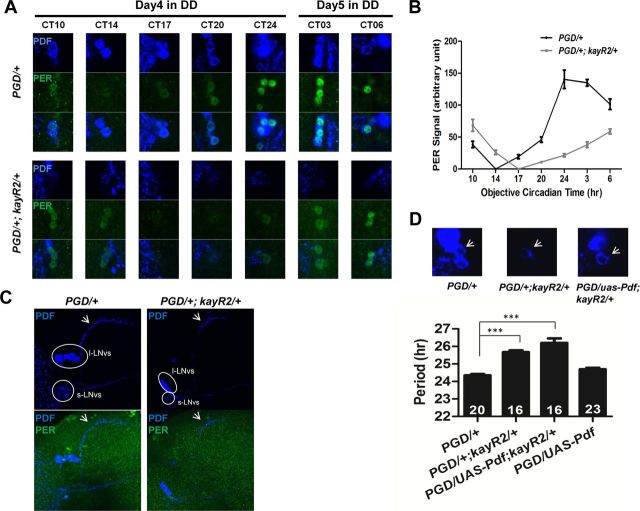
Figure 2.
Altered PER rhythms and reduced PDF levels in pacemaker neurons of kay-αRNAi flies. A, Confocal images of brains from control and kay-αRNAi (PGD/+; kayR2/+) flies immunostained with PER and PDF antibodies. Flies were entrained to a LD cycle for 3 d and then released in DD. Fly brains were dissected at indicated CTs during the fourth subjective night and fifth subjective day. Representative sLNvs are shown. B, Quantification of PER signals after subtraction of background signal. At CT14 for control flies and CT17 for kay-αRNAi flies, PER signals are indistinguishable from background; thus, they are set to “0” on the plot. Error bars correspond to SEM. C, The sLNvs develop normally in kay-αRNAi flies. Fly brains were dissected at CT24 on the first day in DD after 3 d of standard LD cycle and were immunostained for PDF and PER. Images are Z-stack projections of confocal images. Neuronal processes from the sLNvs to the dorsal brain (arrows) appear indistinguishable between PGD/+ and PGD/+; kayR2/+ flies in morphology. Circled are the regions containing cell bodies of large or small LNvs. PGD/+; kayR2/+ flies have only very weak PDF staining in the cell bodies of sLNvs. D, Restoring PDF levels does not rescue the long period phenotype. Overexpressing Pdf in kay-αRNAi flies restored PDF levels in sLNvs (the third panel, white arrow) but did not rescue the long period phenotype (bar 3), which is thus not attributable to low PDF levels but to a defective pacemaker. One-way ANOVA, p < 0.0001. Tukey's multiple comparison test. ***p < 0.001.
Figure 5.
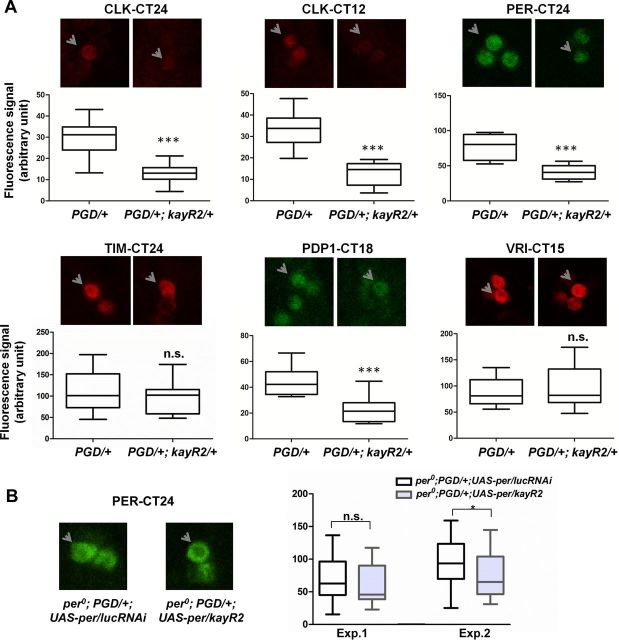
Altered circadian protein expression in kay-αRNAi flies. A, KAY-α downregulation alters expression of several circadian proteins. Flies were entrained to a standard LD cycle for 3 d and then released in DD. Fly brains were dissected on the first day of DD at the expected peak time point of the protein measured, followed by immunocytochemistry (kay-αRNAi flies were dissected ∼1–1.5 h after control flies to correct for differences in period length). Representative staining images and quantifications are shown. Arrows point to sLNvs in the focal plane. Quantification of protein levels are represented by boxes and whiskers in which whiskers show the minimum and maximum values, boxes show the middle 50% of the values, and horizontal lines in the boxes show the median. Two to four independent experiments were performed. CLK, PER, and PDP1 levels were markedly reduced in kay-αRNAi in all experiments. VRI levels were unaffected. TIM levels were reduced in kay-αRNAi flies in all three experiments, but only in one experiment was that decrease statistically significant. Student's t test. ***p < 0.001; n.s., not significant. B, KAY-α regulation of PER levels is strongly dependent on the per promoter. When PER was expressed independently of its promoter with Pdf–GAL4 in a per0 background, KAY-α downregulation had weak effects on PER levels. Quantifications of two independent experiments are shown. In the first one, the average 13% decrease in PER level in kay-αRNAi flies was not statistically significant, but the average 22% decrease in the second experiment was marginally significant (Student's t test, p = 0.04). Thus, posttranscriptional regulation of PER might have a weak contribution to its protein decrease in kay-αRNAi flies.
Fluorescent signals for circadian proteins were quantified using NIH ImageJ v1.42q (http://rsb.info.nih.gov/ij). We subtracted background signal from the signal intensity of each neuron. Background signal was determined by taking the mean signal of three surrounding fields of each neuron. Brains with similar background intensities were quantified. For each time point, at least eight neurons from three independent brain hemispheres were quantified.
Cell culture, cell transfection, and luciferase assay.
Drosophila S2R+ cells were maintained in SFX growth medium (HyClone) supplemented with 9% fetal bovine serum and 1% penicillin–streptomycin (Invitrogen). Cells were seeded in 12-well plates. When reaching 70–90% confluence, they were transiently transfected using Cellfectin II (Invitrogen) as detailed in the instructions of the manufacturer. Luciferase reporter constructs were transfected at 50 ng/well. β-Galactosidase (100 ng) was transfected in each well to normalize transfection efficiency. Empty pAc vector was added so that each well was transfected with same amount of total DNA. Two days after transfection, cells were washed and lysed. Luciferase activity and β-galactosidase activity were measured separately in a 96-well plate using a Microtiter Plate Luminometer. β-Galactosidase activity was measured with the Galacto-light Plus kit (Invitrogen). When involving RNAi in S2R+ cells, 7.5 μg/well double-stranded RNAs (dsRNAs) were added to cell culture medium 1 d before transfection. For each data point, at least three independent experiments were performed.
HEK293 cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum. Cells were seeded in 24-well plates. When reaching 70–90% confluence, they were transiently transfected using Lipofectamine 2000 reagent (Invitrogen) according to the instructions of the manufacturer. Clk–luc was transfected at 20 ng/well. CMV–Renilla (3 ng) was transfected in each well to normalize transfection efficiency. Empty pcDNA3.1 DNA was added so that each well was transfected with the same amount of total DNA. One day after transfection, dual luciferase assay were performed according to the instructions of the manufacturer (dual luciferase reporter assay system catalog #32788; Promega) in a 96-well plate using Veritas Microplate Luminometer. For each experiment, at least three independent experiments were performed.
S2R+ cell immunocytochemistry.
S2R+ cells were seeded on coverslips in six-well plates and transfected with pAc–Clk and/or pAc–kay-α. Two days after transfection, cells were washed twice in PBS and then fixed in 4% paraformaldehyde in PBS for 20 min. Cells were then washed in PBT (PBS plus 0.1% Triton X-100) and blocked in 10% normal donkey serum for 30 min at room temperature. After that, each coverslip was incubated with 150 μl of primary antibodies [guinea pig anti-CLK at 1:3000, rabbit anti-KAY-α/TRUNC at 1:50, and mouse anti-lamin at 1:50 (generous gift from V. Budnik, University of Massachusetts Medical School, Worcester, MA)] at 4°C overnight. Coverslips were then washed and incubated with secondary antibodies (anti-guinea pig Cy3, anti-rabbit FITC, and anti-mouse Cy5 at 1:200 dilution) for 4 h at room temperature. After washing in PBT, coverslips were mounted onto slides in Vectashield Mounting Media and visualized under confocal microscope using a 25× oil lens. Figure 3C shows digitally projected Z stacks.
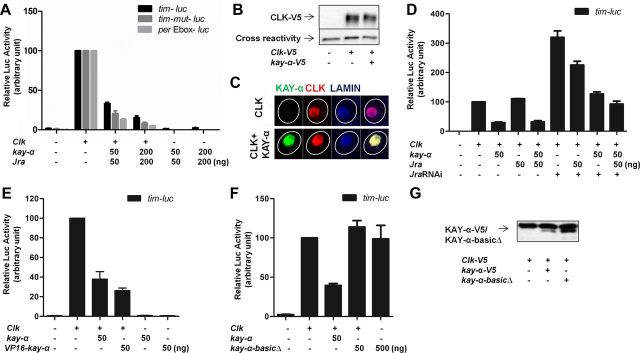
Figure 3.
KAY-α represses CLK/CYC transactivation. Drosophila S2R+ cells were transfected as indicated. Luciferase activity was measured 2 d after transfection. A β-galactosidase-expressing plasmid was cotransfected to normalize transfection efficiency. The relative luciferase activity with Clk was set to 100 on each graph. A, KAY-α represses CLK activation of the tim promoter and the per E-box. The proximal tim promoter with a wild-type or mutagenized AP-1 binding site and multimerized per E-boxes were cloned in the pGL3 luciferase reporter vector to make tim–luc, tim–mut–luc, and per–Ebox–luc. Jra was cotransfected with kay-α. B, KAY-α does not alter CLK expression level in S2R+ cells. Cell lysates were immunoblotted with anti-V5 antibody. CLK–V5 protein level was quantified. CLK–V5 levels did not change whether or not kay-α was transfected in three independent experiments. CLK-V5 levels in the absence of KAY-α were set at 100. In the presence of KAY-α, relative CLK amount was 108 ± 20, n = 3. C, KAY-α does not alter CLK subcellular localization in S2R+ cells. Representative immunostaining showing CLK localization in the nucleus in the presence or absence of KAY-α. More than 95% of cells expressing or not KAY-α show this primarily nuclear CLK localization. Very rarely, and in both cells with or without KAY-α, CLK showed both nuclear and cytoplasmic localization. Lamin stains inner nuclear membrane. Circles outline the cell bodies. D, The repression of CLK activation by KAY-α does not require JRA. S2R+ cells were treated with dsRNAs targeting Jra 1 d before transfection to knock down endogenous JRA. Even in the presence of these Jra dsRNAs, KAY can still repress CLK/CYC activation. E, VP16–KAY-α cannot activate the proximal tim promoter. The activation domain of VP16 was fused to the N terminal of KAY-α to generate VP16–KAY-α. The fusion protein could not activate tim–luc but still repressed the activity of CLK. F, KAY-α DNA binding domain is required for CLK repression. The basic region of KAY-α was deleted to generate a kay-α–basicΔ construct, which was not able to repress CLK activity. G, The removal of the DNA binding domain of KAY-α does not affect its stability. Expression of wild-type and kay-α–basicΔ was comparable on Western blots.
dsRNA synthesis.
We produced dsRNAs by in vitro transcription of a PCR-generated DNA template containing the T7 promoter sequence on both ends. A ∼500 bp Jra fragment was amplified from genomic DNA with the following primers: forward, 5′-TTAATACGACTCACTATAGGGAGAGGCTCGCTGGATCTGAAC-3′; and reverse, 5′-TTAATACGACTCACTATAGGGAGAGATGCAGCCACACGGTTA-3′.
We performed in vitro RNA transcription with Ambion MEGAscript T7 kit (catalog #AM1334) following the instructions of the manufacturer. After 2 h of 37°C incubation of the reaction, 1 μl of DNase was added to 20 μl of reaction and incubated for 15 min to degrade DNA template. Then, the reaction was stopped and single-stranded RNAs were precipitated, washed, and resuspended in water. Annealings of single-stranded RNAs were done by performing the following program: (1) 65°C, 30 min; (2) 65°C, 1 min, 1°C/cycle for 50 cycles. We confirmed the quality and size of dsRNAs on agarose gel.
Antibody production, coimmunoprecipitation, and Western blot.
A synthesized peptide corresponding to amino acids 186–200 of KAY-α was used for the immunization of rabbits at Cocalico. Affinity-purified antisera were used for KAY-α detection.
For coimmunoprecipitation experiments, HEK293 cells were seeded in 100 mm Petri dishes. When reaching 70–90% confluence, cells were transfected with 5 μg of CMV–vri–VP16, 6 μg of CMV–myc–kay-α, or both. Empty pcDNA3.1 DNA was added to equalize the total amount of DNA transfected. One day after transfection, cells were lysed in Passive Lysis Buffer (1 ml of buffer per Petri dish; Promega) for 20 min, and supernatant was collected after centrifugation. Protein G Sepharose beads (catalog #17-0618-01; GE Healthcare) were incubated with anti-MYC antibody (catalog #11667149001; Roche) for 1 h at room temperature (2.5 μl of antibody per 15 μl of beads). Then 15 μl of these beads were added to 1 ml of supernatant and incubated overnight at 4°C with gentle agitation. Beads were then washed and resuspended in 1× SDS-PAGE loading buffer and boiled, ready for Western blotting.
Western blot were performed essentially as described previously (Emery et al., 1998). Equal loading and quality of protein transfer were first verified by Ponceau Red staining and then by the intensity of cross-reacting bands. Primary antibodies included the following: mouse anti-MYC at 1:1000, rabbit anti-KAY-α/TRUNC at 1:1000, rabbit anti-KAY main body (Dfos-112AP; FabGennix) at 1:500, mouse anti-V5 (Invitrogen) at 1:5000, and guinea pig anti-VRI at 1:5000. Secondary antibodies conjugated with HRP from Jackson ImmunoResearch were used at 1:10,000 dilution, except for goat anti-guinea pig IgG–HRP (1:5000; Abcam).
In Figure 3B, signals on the film were digitalized using IR-LAS-1000 Lit V2.12 (Fujifilm) and quantified with Image Gauge V4.22. CLK–V5 signals were normalized to the cross-reacting signals, and the ratio without kay-α–V5 was set to 100.
Results
kay-α downregulation lengthens circadian behavioral rhythms
In a previous misexpression screen, we looked for flies that remained rhythmic under constant light (LL) with the idea of isolating novel genes that regulate the CRYPTOCHROME (CRY) input pathway (Dubruille et al., 2009). CRY is a key circadian photoreceptor that binds to TIM and triggers its proteasomal degradation after blue-light photon absorption, hence resetting the circadian molecular pacemaker (Emery et al., 1998; Stanewsky et al., 1998; Lin et al., 2001; Busza et al., 2004). Wild-type flies are arrhythmic in LL because CRY is constantly activated, whereas cry mutant flies remain rhythmic, as if they were in DD (Emery et al., 2000). However, it is known that overexpressing pacemaker genes can also make flies rhythmic in LL (Murad et al., 2007; Stoleru et al., 2007; Dubruille et al., 2009). Thus, our screen had the potential of identifying both pacemaker and light input genes. We previously identified the chromatin remodeling protein KISMET as a regulator of CRY photoresponses (Dubruille et al., 2009). The present study focuses on the role of kay—another candidate gene isolated with our LL screen—in the circadian pacemaker.
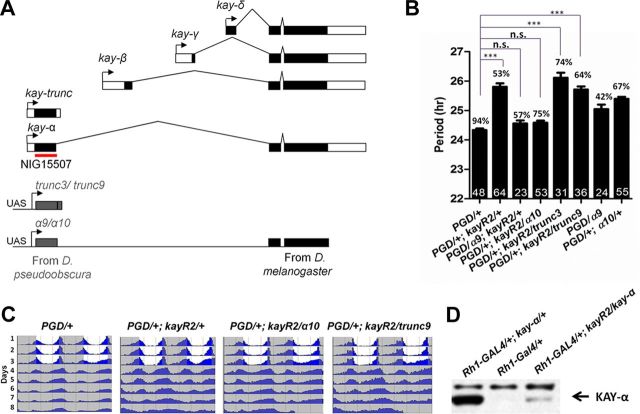
kay, the Drosophila homolog of c-fos, encodes a bZip transcription factor (Zeitlinger et al., 1997). To understand the role kay might play in the regulation of Drosophila circadian rhythms, we decided to test kay loss-of-function flies. Because severe kay mutants are embryonic lethal (Zeitlinger et al., 1997), we used an RNAi approach to knockdown kay. There are five KAY isoforms in D. melanogaster (Fig. 1A) (Hudson and Goldstein, 2008). Full-length isoforms α, β, γ, and δ are generated through alternative promoters. The four alternative first exons encode specific N-terminal domains. The other exons are common to all four full-length isoforms, which thus share the same DNA binding domain (bZIP domain). The fifth isoform is encoded only by the exon specific to the α isoform and thus does not contain the bZIP domain. This truncated isoform was previously called KAY–SRO, because the shroud (sro) mutation was initially mapped to its encoding exon (Hudson and Goldstein, 2008). Because it is now known that the sro mutation affects another gene (Niwa et al., 2010), we renamed this isoform KAY–TRUNC. The P-element [EP(3)3084] we isolated in our LL screen is inserted about 1 kb upstream of the exon specific to the KAY-α/TRUNC isoforms. We therefore expressed a dsRNA targeting specifically these isoforms, encoded by the NIG15507 transgene. Expression of this dsRNA was lethal with widely expressed drivers, such as tim–GAL4 (Kaneko et al., 2000). We therefore used the Pdf–GAL4 driver, which is expressed only in the circadian neurons that drive circadian behavior in DD, the sLNvs (Renn et al., 1999). We observed an ∼1-h-long period phenotype in DD with two independent NIG15507 insertions (kayR2 and kayR4) (Table 1). Period was slightly longer with the kayR2 insertion. For the rest of this study, we used this insertion. Coexpressing DICER2 (DCR2) with dsRNAs is known to enhance RNAi effects (Dietzl et al., 2007). Consistently, the period phenotype was slightly enhanced when DCR2 was coexpressed with the dsRNAs. These RNAi flies showed a period lengthening of ∼1.5 h (Fig. 1B, bars 1 and 2, C). The proportion of arrhythmic flies was also increased in the presence of DCR2.
Figure 1.
Downregulating kay-α lengthens the period of free-running circadian behavior in DD. A, Organization of the kay locus in D. melanogaster. kay is predicted to produce five isoforms. The dark boxes indicate coding sequences, and open boxes indicate noncoding sequences. Transgene NIG15507 generates dsRNAs targeting both the α and trunc isoforms. The red line indicates the region targeted by dsRNA. Constructs for cross-species rescue experiments are shown on the bottom of the panel. α9, α10, trunc3, and trunc9 are insertions of UAS-controlled transgenes that generate kay mRNAs resistant to the NIG15507 dsRNAs. The region targeted by NIG15507 dsRNAs was replaced with homologous D. pseudoobscura sequences. B, Downregulation of kay-α lengthens circadian behavior period. Bars 1 and 2, Flies expressing dsRNAs targeting kay-α and kay–trunc under the Pdf–GAL4 driver have ∼26-h-long period rhythms (control, 24.4 h). PGD: Pdf-GAL4, UAS-dcr2. Bars 3–6, The kay RNAi phenotype can be rescued with the kay-α construct resistant to the dsRNAs but not with the kay–trunc construct. Bars 7 and 8, The rescue is not explained by a period shortening caused by expression of the chimeric kay-α, because its expression in wild-type flies does not shorten circadian behavioral rhythms (it actually slightly lengthens them). Error bars correspond to SEM. Digits in the bar are the numbers of tested flies. Percentage of rhythmicity is indicated above the bars. One-way ANOVA, p < 0.0001. Tukey's multiple comparison test. ***p < 0.001; n.s., not significant at level of 0.05. C, Double-plotted actograms showing the average activity for each genotype. Flies were entrained in standard LD cycle for 3 d and then released in DD. D, The NIG15507-R2 transgene can inhibit KAY-α expression in vivo. Fly head extracts were immunoblotted with an anti-KAY-α/TRUNC antibody. Strong immunoreactivity was observed when KAY-α was misexpressed in the eyes with the Rh1–GAL4 driver (lane 1), whereas endogenous KAY-α was undetectable (lane 2). dsRNAs generated by NIG15507-R2 transgene were able to dramatically knock down the overexpression of KAY-α (lane 3).
Table 1.
KAY-α downregulation
| Genotype | n | % of rhythmic flies | Period average ± SEM | Power average ± SEM |
|---|---|---|---|---|
| y w | 122 | 74.59 | 23.81 ± 0.03 | 76.08 ± 3.76 |
| w1118 | 23 | 91.30 | 23.96 ± 0.05 | 99.51 ± 6.78 |
| pdf-GAL4/+ | 103 | 89.32 | 24.45 ± 0.03 | 70.59 ± 2.86 |
| kayR2/+ | 103 | 72.82 | 24.29 ± 0.04 | 69.62 ± 4.60 |
| Pdf–GAL4 /+; kayR2/+ | 73 | 91.78 | 25.62 ± 0.06*** | 76.78 ± 3.91 |
| kayR4/Y | 63 | 85.71 | 24.01 ± 0.05 | 80.48 ± 3.91 |
| kayR4 /Y; Pdf–GAL4/+ | 32 | 71.88 | 25.20 ± 0.08*** | 37.70 ± 2.92 |
Behavior of control flies and flies expressing dsRNAs targeting kay-α/trunc in DD at 25°C. One-way ANOVA, p < 0.0001. Tukey's multiple comparison test,
***p < 0.001 when compared with Pdf–GAL4/+.
We generated an antibody directed against the KAY-α Ν-terminal region. Unfortunately, this antibody was not sensitive enough to detect endogenous KAY-α expression by Western blotting or immunohistochemistry. Thus, to verify that the NIG15507 dsRNAs are indeed able to downregulate KAY-α expression, we overexpressed KAY-α specifically in the eyes with the Rhodopsin1–GAL4 (Rh1-GAL4) driver (Mollereau et al., 2000) in the presence or absence of the kay-α dsRNAs. KAY-α overexpression was dramatically reduced in the presence of the RNAi (Fig. 1D). Thus, the NIG15507 dsRNAs efficiently inhibit KAY-α expression.
Although the NIG15507 dsRNAs lengthen circadian period by ∼1.5 h, dsRNAs targeting all full-length KAY isoforms resulted only in modest period lengthening (up to 0.5 h; Table 2). Thus, either these dsRNAs are inefficient at repressing KAY expression or repression of the truncated KAY isoform plays a major role in the long period phenotype. To determine whether the long period phenotype is attributable to KAY-α or to KAY–TRUNC knockdown, we performed a cross-species rescue experiment (Fig. 1A). We generated UAS–kay-α and UAS–kay–trunc transgenes expressing kay mRNAs resistant to the NIG15507 dsRNAs by replacing the region targeted by these dsRNAs with homologous but sufficiently divergent D. pseudoobscura sequences (Langer et al., 2010). We expressed these constructs in flies expressing NIG15507 dsRNAs. We found that the chimeric pseudoobscura/melanogaster KAY-α rescued the long period phenotype (6 of 10 lines tested fully rescued, one rescued partially), although the pseudoobscura KAY–TRUNC did not (none of the nine lines tested could rescue) (Fig. 1B,C; Table 3). Thus, we conclude that the long period phenotype observed in flies expressing the NIG15507 dsRNAs in PDF-positive LNvs is caused by KAY-α downregulation. Therefore, this KAY isoform regulates circadian behavior. The increase in arrhythmicity was not rescued, however, and might thus be attributable to off-target effects. For the rest of the manuscript, flies expressing NIG15507 dsRNAs (and DCR2) in PDF neurons will be simply referred to as kay-αRNAi flies.
Table 2.
Downregulation of all full-length KAY isoform
| Genotype | n | % of rhythmic flies | Period average ± SEM | Power average ± SEM |
|---|---|---|---|---|
| PGD/+ | 31 | 83.87 | 24.95 ± 0.07 | 67.66 ± 4.32 |
| PGD/+; NIG15509–R1/+ | 46 | 97.83 | 25.46 ± 0.06*** | 34.20 ± 5.10 |
| NIG15509–R1/+ | 30 | 96.67 | 24.23 ± 0.10 | 77.57 ± 4.49 |
| PGD/+; NIG15509–R2/+ | 31 | 83.87 | 25.32 ± 0.08* | 77.75 ± 5.27 |
| NIG15509–R2/+ | 15 | 60.00 | 24.23 ± 0.18 | 69.88 ± 8.53 |
| PGD/+; VDRC6212/+ | 8 | 62.50 | 25.34 ± 0.06 | 38.62 ± 7.28 |
| VDRC6212/+ | 19 | 78.95 | 24.27 ± 0.09 | 81.63 ± 7.36 |
Behavior of control flies and flies expressing dsRNAs targeting all full-length kay isoforms in DD at 25°C. One-way ANOVA, p < 0.0001. Tukey's multiple comparison test,
***p < 0.001,
*p < 0.05 when compared with PGD/+.
Table 3.
Cross-species rescue of KAY-α downregulation
| Genotype | n | % of rhythmic flies | Period average ± SEM | Power average ± SEM |
|---|---|---|---|---|
| y w | 76 | 84.21 | 23.74 ± 0.05 | 69.47 ± 3.01 |
| PGD/+ | 80 | 87.50 | 24.39 ± 0.04 | 55.07 ± 2.69 |
| PGD/+; kayR2/+ | 86 | 51.16 | 25.75 ± 0.10 | 50.96 ± 3.98 |
| PGD/α1; kayR2/+ | 22 | 59.09 | 25.56 ± 0.21 | 45.49 ± 5.33 |
| PGD/+; kayR2/α2 | 20 | 60.00 | 25.42 ± 0.14 | 64.13 ± 8.43 |
| PGD/α3; kayR2/+ | 31 | 51.61 | 24.72 ± 0.10+ | 53.29 ± 4.35 |
| PGD/+; kayR2/α4 | 22 | 77.27 | 24.68 ± 0.06+ | 70.38 ± 6.05 |
| PGD/+; kayR2/α5 | 34 | 85.29 | 25.17 ± 0.07++ | 55.41 ± 3.51 |
| PGD/α6; kayR2/+ | 40 | 52.50 | 24.22 ± 0.08+ | 44.76 ± 4.75 |
| PGD/+; kayR2/α7 | 16 | 62.50 | 25.18 ± 0.10 | 55.12 ± 5.95 |
| PGD/α9; kayR2/+ | 23 | 56.52 | 24.56 ± 0.10+ | 49.52 ± 5.60 |
| PGD/+; kayR2/α10 | 53 | 75.47 | 24.59 ± 0.07+ | 47.06 ± 2.14 |
| PGD/α11; kayR2/+ | 18 | 33.33 | 24.62 ± 0.23+ | 48.68 ± 5.95 |
| PGD/trunc1; kayR2/+ | 23 | 52.17 | 25.75 ± 0.18 | 52.35 ± 6.53 |
| PGD/+; kayR2/trunc2 | 35 | 37.14 | 25.40 ± 0.21 | 46.11 ± 2.98 |
| PGD/+; kayR2/trunc3 | 31 | 74.19 | 26.11 ± 0.17 | 39.49 ± 3.11 |
| PGD/+; kayR2/trunc4 | 18 | 44.44 | 26.04 ± 0.46 | 54.59 ± 7.03 |
| PGD/trunc5; kayR2/+ | 14 | 50.00 | 26.03 ± 0.24 | 52.19 ± 10.52 |
| PGD/trunc6; kayR2/+ | 13 | 23.08 | 25.57 ± 0.17 | 37.00 ± 3.38 |
| PGD/+; kayR2/trunc7 | 22 | 18.18 | 26.53 ± 0.29 | 34.18 ± 5.76 |
| PGD/trunc8; kayR2/+ | 35 | 37.14 | 26.77 ± 0.19 | 39.82 ± 4.80 |
| PGD/+; kayR2/trunc9 | 36 | 63.89 | 25.73 ± 0.11 | 45.67 ± 3.31 |
Behavior of kay-αRNAi flies expressing chimeric pseudoobscura/melanogaster kay-α or kay–trunc in DD at 25°C. In bold are the lines that show statistically significant rescue of the period lengthening caused by kay-α/trunc downregulation (7 of 10 α lines, 0 of 9 trunc lines). One-way ANOVA, p < 0.0001. Bonferroni's multiple comparison test was done on the following pairs: all rescue lines versus PGD/+ and all rescue lines versus PGD/+; kayR2/+. α = 0.05. Lines with period means that are not significantly different from that of PGD/+ are considered as fully rescued (+). Lines with period means that are significantly different from both PGD/+ and PGD/+; kayR2/+ and are less than that of PGD/+; kayR2/+ are considered as partially rescued (++).
Reduced PER and PDF levels in the sLNvs of kay-αRNAi flies
To begin to understand how KAY-α might control circadian behavior, we measured PER levels in PDF-positive sLNvs of kay-αRNAi flies. These flies and control flies were first entrained to a standard LD cycle for 3 d and then released in DD. Fly brains were dissected at different circadian times (CT) during the fourth subjective night and fifth subjective day and immunostained with PER and PDF antibodies. In control flies, PER peaked near CT24 as expected (Fig. 2A,B). However, in kay-αRNAi flies, the peak was between CT6 and CT10. This delay on day 4–5 of DD fits well with the ∼1.5 h period lengthening observed behaviorally with these flies. We therefore conclude that the long period phenotype of kay-αRNAi flies is attributable to a slow-running pacemaker in the sLNvs. We also noted that overall PER levels were reduced by approximately half at peak level in kay-αRNAi flies. KAY-α thus positively affects PER levels.
Interestingly, we also observed that PDF levels were severely reduced at all time points in the sLNvs of kay-αRNAi flies (Fig. 2A). Nevertheless, PDF was detectable in the dorsal projections of sLNvs, and these projections appeared anatomically normal, indicating that the sLNvs develop normally when KAY-α is downregulated (Fig. 2C). As a circadian neuropeptide, PDF is required for normal rhythm amplitude and circadian period length in DD (Renn et al., 1999). Absence of PDF leads to arrhythmicity or short behavioral period rhythms. It thus seemed unlikely that the long period phenotype we observed would be caused by low PDF levels. To confirm this, we restored PDF levels in the sLNvs of kay-αRNAi flies with a UAS–Pdf transgene (Renn et al., 1999). Circadian behavior kept its long period (Fig. 2D). This demonstrates that low PDF levels are not the cause of the long period phenotype seen in kay-αRNAi flies.
KAY-α represses CLK transactivation of the tim promoter and the per E-box
KAY is a bZip transcription factor. This class of transcription factors uses their leucine Zipper domains to form homodimers or heterodimers. The best-known partner of KAY is JRA (Bohmann et al., 1994), with which it forms the AP-1 complex that recognizes the consensus sequence TGAGTCA. In most cases, AP-1 functions as a transcriptional activator. Intriguingly, we found a perfect AP-1 binding site in the tim promoter. If KAY-α were to activate transcription through this site, it could explain the phenotypes seen in kay-αRNAi flies. Indeed, low TIM levels result in long period phenotypes and low PER levels (McDonald et al., 2001).
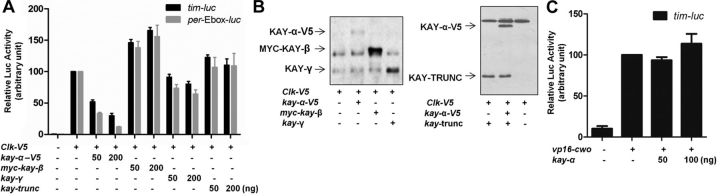
We tested this idea using a luciferase reporter assay. The proximal tim promoter was fused to a luciferase reporter gene and transfected into Drosophila S2R+ cells. Unexpectedly, coexpression of KAY-α and JRA did not activate the tim promoter at all (Fig. 3A). We reasoned that CLK might be needed as well, because it forms a crucial tim transactivator with CYC, which is endogenously expressed in S2R+ cells (Darlington et al., 1998). As expected, we saw a robust transactivation of the tim promoter when CLK was expressed. However, we did not see additional activation when coexpressing KAY-α and JRA with CLK. On the contrary, we actually observed a decrease in the activation of the tim promoter of CLK (Fig. 3A). Also unexpectedly, the AP-1 binding site in the tim promoter was dispensable for KAY-α repression of tim (Fig. 3A). Because per is another target gene of CLK/CYC, we tested a construct that only contains multimerized per E-boxes in front of the luciferase reporter gene. We found that KAY-α could also repress the activation of these E-boxes by CLK (Fig. 3A). This clearly shows that KAY-α represses CLK transactivation independently of AP-1 binding sites. Importantly, KAY-α-mediated repression was not the result of reduced CLK levels, nor was it attributable to retention of CLK in the cytoplasm (Fig. 3B,C). In conclusion, KAY-α represses the transactivation potential of CLK and might thus regulate a large subset of CLK target genes.
Given that AP-1 sites are not required for KAY-α repression, we wondered whether JRA is actually required. We observed that it made no difference to cotransfect or not a plasmid encoding JRA (Fig. 3D, bars 3 and 5). This suggests that JRA is not required for repression. To exclude the possibility that KAY-α uses endogenously expressed JRA to form the AP-1 complex, we treated the S2R+ cells with dsRNAs targeting Jra. This led to increased CLK transactivation, a phenomenon that we did not study further. Importantly, however, KAY-α could still repress efficiently the activation of the tim promoter by CLK (Fig. 3D, bars 6 and 8). Consistently, Jra downregulation by RNAi in flies did not lengthen circadian period (Table 4). Together, our data show that KAY-α represses CLK activation independently of JRA and AP-1 binding sites.
Table 4.
Knockdown of Jra in pacemaker neurons did not result in long period
| Genotype | n | % of rhythmic flies | Period average ± SEM | Power average ± SEM |
|---|---|---|---|---|
| yw | 16 | 62.50 | 23.84 ± 0.16 | 46.93 ± 6.53 |
| PGD/+ | 30 | 96.67 | 24.92 ± 0.04 | 56.96 ± 4.90 |
| PGD/+; JraRNAiJF01184/+ | 32 | 93.75 | 24.47 ± 0.05 | 57.71 ± 2.98 |
| PGD/+; JraRNAiJF01379/+ | 16 | 93.75 | 24.73 ± 0.11 | 65.37 ± 7.27 |
Behavior of control flies and flies expressing dsRNAs targeting Jra in DD at 25°C.
To determine whether KAY-α might directly bind to the E-box and compete for CLK binding, as does CWO (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008), we added a VP16 activation domain to KAY-α. In the case of CWO, this repressor protein is transformed into an activator by the addition of VP16 (Kadener et al., 2007; Lim et al., 2007). This was not the case with KAY-α. The VP16–KAY-α protein was unable to transactivate tim–luc and still functioned as a repressor in the presence of CLK (Fig. 3E). Thus, KAY-α probably does not bind directly the tim or per E-boxes. Nevertheless, its DNA binding domain is required for repression (Fig. 3F,G), because deletion of the basic region of the bZIP domain—responsible for interaction with DNA—completely eliminated repression (see Discussion).
We then asked whether the repression was specific to the KAY-α isoform. We therefore tested whether KAY-β, KAY-γ, and KAY–TRUNC could also repress CLK. Interestingly, none of these isoforms were able to repress CLK, although they were expressed at similar or higher levels than KAY-α (Fig. 4A,B). Thus, CLK repression is specific to the α isoform. This specificity strongly strengthens the notion that the repression of CLK by KAY-α is important to determine circadian period length, because specifically downregulating this KAY isoform is sufficient in vivo to lengthen circadian behavioral rhythms.
Figure 4.
CLK repression is specific to KAY-α. Drosophila S2R+ cells were transfected as indicated. A β-galactosidase-expressing plasmid was cotransfected to normalize transfection efficiency. The relative luciferase activity with Clk or VP16–cwo was set to 100 on the graph. A, CLK activation in the presence of different KAY isoforms. Neither KAY-β, KAY-γ, nor KAY–TRUNC can repress CLK activation of the tim promoter or the per E-box. B, KAY-β, KAY-γ, KAY–TRUNC, and KAY-α–BASICΔ are well expressed in S2R+ cells. Cell lysates were immunoblotted with anti-KAY-main body antibody (left) or anti-KAY-α/TRUNC antibody (right). C, KAY-α does not repress the activation of the tim promoter by VP16–CWO.
We next determined whether KAY-α repression was specific to CLK or could happen with any activator recognizing E-boxes. As mentioned previously, CWO is a repressive helix–loop–helix protein that competes with CLK for binding to E-boxes (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007; Richier et al., 2008). When CWO is fused to the strong VP16 transactivation domain, CWO becomes an activator. KAY-α could not repress the activation of the tim promoter by VP16–CWO (Fig. 4C). This indicates that KAY-α repression is specific to CLK. In summary, our transcription assays show that KAY-α specifically represses CLK-mediated transcription.
Both circadian transcriptional feedback loops are altered in kay-αRNAi flies
Our results in S2R+ cells appear to be paradoxical. If KAY-α is a CLK repressor, then its downregulation should increase PER levels rather than decrease them as observed in sLNvs (Fig. 2A,B). Thus, the role played by KAY-α might not be limited to repressing CLK activity. To understand better the impact of KAY-α on the circadian molecular pacemaker, we measured PER, TIM, PDP1, and VRI levels at expected peak times in kay-αRNAi flies during the first day of DD. We also measured CLK levels at two time points: CT12 and CT24. Overall CLK levels do not oscillate, but its phosphorylation does. CT12 and CT24 correspond to the peak for the hypo-phosphorylated and hyper-phosphorylated CLK isoforms, respectively (Yu et al., 2006). As expected, PER levels were reduced by ∼50% in kay-αRNAi flies, as observed on day 4–5 of DD (Fig. 2). When PER was expressed independently of its own promoter with Pdf–GAL4 in a per0 background, KAY-α downregulation had only weak effects on PER levels, further supporting the notion that KAY-α regulates PER transcriptionally (Fig. 5B). The small reduction in PER levels, significant only in one experiment, could indicate that KAY-α also weakly affect PER posttranscriptional regulation. This could be because two subunits of protein phosphatase 2A, which regulates PER phosphorylation and thus its stability, are under circadian control (Sathyanarayanan et al., 2004) and might therefore be misregulated in kay-αRNAi flies. PDP1 levels were also clearly reduced (∼40% on average), but VRI was not affected (Fig. 5A). TIM levels were lower in all three experiments that we performed (Fig. 5A), but this was statistically significant in only one of them. Overall, the strongest effects of KAY-α downregulation were on CLK levels, which were reduced by ∼60% in kay-αRNAi flies (Fig. 5A). The Clk promoter might thus be an important target of KAY-α.
KAY-α modulates VRI activity on the Clk promoter
PDP1 and VRI are critical activator and repressor of the Clk promoter, respectively. Interestingly, they have similar positive and negative effects on PDF expression, through as yet unknown mechanisms. In Pdp1ε mutants, both CLK and PDF levels are severely reduced in sLNvs (Zheng et al., 2009). PDF expression is also decreased when VRI is overexpressed, although Pdf mRNA appears unaffected. This indicates that VRI regulates the expression of a gene controlling PDF posttranscriptionally (Blau and Young, 1999). Because the strongest effects of kay-α downregulation are on CLK and PDF, we wondered whether KAY-α might interact with VRI and/or PDP1 and modulate their activity.
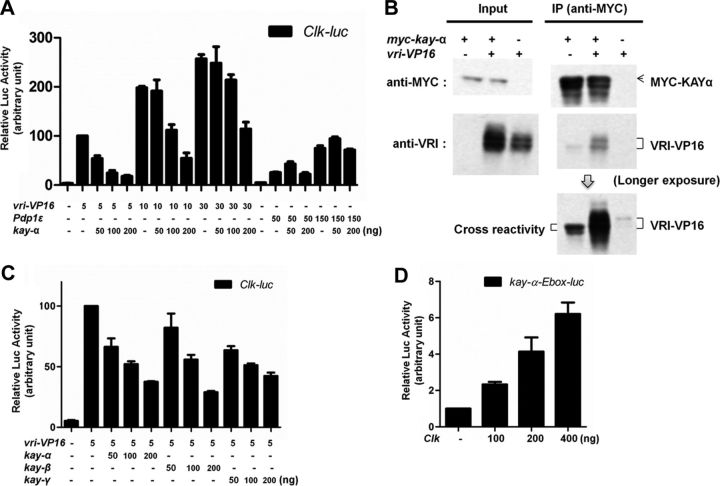
We turned to a well-established luciferase reporter assay in HEK293 cells, in which PDP1 and a VRI–VP16 fusion protein can both activate the Clk promoter. We cotransfected KAY-α with either PDP1 or VRI–VP16 (Cyran et al., 2003). Strikingly, we found that KAY-α could repress the activation of the Clk promoter by VRI–VP16 in a dose-dependent manner. Conversely, it was unable to block PDP1 activity (Fig. 6A). These results fit remarkably well with those of the dimerization prediction study of Fassler et al. (2002), in which they anticipated that KAY-α would interact with VRI, but not with PDP1, based on the amino acid content of their leucine Zipper.
Figure 6.
KAY-α interacts with and inhibits VRI. A, KAY-α blocks specifically VRI–VP16 activation of Clk promoter. HEK293 cells were transfected as indicated. Renilla luciferase was transfected to normalize transfection efficiency. Luciferase activity was measured 1 d after transfection. Relative luciferase activity with vri–VP16 was set to 100. VRI–VP16 activates the Clk promoter, as described previously (Cyran et al., 2003). The activation of the Clk promoter by VRI–VP16 was inhibited in a dose-dependent manner by KAY-α, but the activation of the Clk promoter by PDP1ε was unaffected. Error bars are SEM. B, KAY-α interacts with VRI–VP16 in HEK293 cells. HEK293 cells were transfected as indicated. Cell lysates were immunoprecipitated with anti-MYC antibody. Bound proteins were probed with anti-MYC and anti-VRI antibodies. VRI–VP16 was coimmunoprecipitated with MYC–KAY-α. C, KAY-β and KAY-γ can also repress the activation of the Clk promoter by VRI–VP16. HEK293 cells were transfected as indicated. A Renilla-expressing vector was cotransfected to normalize transfection efficiency. The normalized luciferase activity with vri–VP16 was set to 100 on the graph. Error bars are SEM. D, CLK can activate the kay-α promoter. A ∼300 bp kay-α promoter fragment containing an E-box was cloned in the pGL3 vector to generate kay–α-Ebox–luc. It can be activated by CLK. The normalized luciferase activity without Clk was set to 1 on the graph. Error bars are SEM.
To confirm that KAY-α indeed interacts with VRI, we performed a coimmunoprecipitation experiment. MYC–KAYα and VRI–VP16 were coexpressed in HEK293 cells. KAY-α was immunoprecipitated with an anti-MYC antibody. As expected, we found VRI–VP16 to coimmunoprecipitate with KAY-α (Fig. 6B). These results lead us to propose that KAY-α negatively regulates the repressive activity of VRI by forming a complex that is unable to recognize its targets, such as the Clk promoter.
Discussion
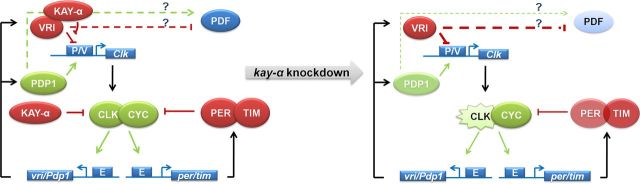
Transcriptional feedback loops play a critical role in the generation of circadian rhythms in most organisms. In Drosophila, as in mammals, two interlocked feedback loops produce antiphasic circadian transcriptional oscillations (Zhang and Emery, 2012). In this study, we have identified a novel transcriptional regulator of circadian rhythms in Drosophila: KAY-α. Our results indicate that it modulates both circadian feedback loops: it would be both a repressor of the CLK/CYC transactivation complex and an inhibitor of VRI-mediated repression (Fig. 7).
Figure 7.
A model for the role of KAY-α in the control of circadian behavior. Our results indicate that KAY-α affects both circadian transcriptional feedback loops. It inhibits VRI through direct physical contact and also represses CLK transactivation (left). When KAY-α is absent (right), VRI repression is enhanced (thicker red lines), which results in decreased CLK and PDF levels (lighter filling color). However, CLK activity also increases (zigzags). This mitigates the effects of increased VRI repression, and CLK/CYC targets are either weakly (TIM) or moderately (PER, PDP1) affected or not at all (VRI).
Interestingly, our results indicate that KAY-α directly binds to VRI. KAY and VRI are two of the 27 bZip proteins encoded by the D. melanogaster genome (Fassler et al., 2002). The leucine zipper domain is typically composed of four to five heptad repeats of amino acids, with the seven unique positions in the heptad labeled “a,” “b,” “c,” “d,” “e,” “f,” and “g.” These positions are critical for dimerization stability and specificity. By evaluating (1) the presence of attractive or repulsive interhelical “g–e” electrostatic interface and (2) the presence of polar or charged amino acid in the “a” and “d” positions of the hydrophobic interface, Fassler et al. (2002) predicted that KAY can dimerize with VRI but not PDP1. Our results confirm functionally this prediction. KAY-α blocks the ability of VRI to transactivate the Clk promoter when VRI is made an activator by the addition of a VP16 domain, but it does not interfere with PDP1 transactivation. Moreover, it forms a complex with VRI. By taking together our data and Fassler's prediction, we propose that KAY and VRI form a dimer through leucine Zipper interactions and that this dimer is not able to bind to the Clk promoter and other VRI/PDP1 target sequences. This model would explain why CLK and PDF levels are low in the sLNvs of kay-αRNAi flies. There would be an increase in VRI repressive activity in these flies (Fig. 7). It is interesting to note that PDF levels seem to be particularly low in the small LNvs but much less dramatically affected in the large LNvs (Fig. 2C). Thus, it appears that PDF levels are regulated through different mechanisms in small and large LNvs. This could be a reflection of the different role played by these cells in the control of circadian behavior and arousal (Shang et al., 2008; Sheeba et al., 2008).
The mechanism by which KAY-α regulates CLK/CYC transactivation is not yet clear. We could not detect a direct interaction between KAY-α and CLK/CYC by coimmunoprecipitation when these proteins were expressed in cell culture. Of course, this does not exclude the possibility that they interact in vivo or that they do so transiently. However, because these proteins belong to two different families of transcription factors (bZip and bHLH–PAS), direct interactions would not be predicted. It also seems unlikely that KAY-α competes with CLK/CYC for binding to the E-box. Indeed, a VP16–KAY-α fusion protein was not able to transactivate E-box containing promoters, whereas a CWO–VP16 fusion protein could (CWO does compete with CLK/CYC for E-box binding) (Kadener et al., 2007; Lim et al., 2007). In fact, KAY-α–VP16 still functioned as a CLK/CYC repressor. Nevertheless, the basic region necessary for DNA binding is needed for KAY-α repression. This suggests that KAY-α controls the expression of a gene important for CLK/CYC activity. Most likely, it promotes the expression of a CLK/CYC repressor. In any case, this repressive role for KAY-α probably explains why the expression of CLK/CYC targets is mildly affected by KAY-α downregulation and the resulting increased VRI repression. The decrease in repressive activity on the CLK/CYC transcription factor would compensate for the stronger VRI repression of the Clk promoter (Fig. 7). CLK/CYC targets include PDP1, which positively activates Clk. Particularly high affinity of PDP1 for the Clk promoter could explain why CLK levels are less affected than PDF by KAY-α downregulation. Also supporting an important role of CLK/CYC repression by KAY-α is the fact that this repression is specific to the α isoform. This result implies that the N-terminal domain of KAY proteins plays an important role in defining their respective function and the genes and proteins they are regulating. However, we note that repression of VRI–VP16 transactivation occurred with all full-length KAY isoforms tested (Fig. 6C). This is not entirely surprising, because this repression is most likely mediated through the leucine Zipper domain common to all isoforms, as discussed above.
A question that remains to be answered is whether KAY-α levels are regulated by the circadian clock. Our antibodies were not sensitive enough to detect endogenous KAY-α protein levels, and, because of low mRNA levels in head extracts, we could not reliably measure kay-α mRNA levels by quantitative real-time PCR. Interestingly, the kay-α promoter actually contains a perfect E-box, and this promoter can be transactivated by CLK/CYC in cell culture (Fig. 6D). However, Abruzzi et al. (2011) did not detect CLK/CYC binding to the kay-α promoter in whole-head chromatin immunoprecipitation experiments, although CLK/CYC binds rhythmically to the kay-β promoter. CLK/CYC could thus influence at distance the kay-α promoter in vivo. It is also possible that CLK/CYC only bind to the kay-α promoter in specific tissues, such as the PDF-positive sLNvs.
This brings us to the question of the site of action of KAY-α. Our study focused on the sLNvs, because we could not broadly drive dsRNAs against KAY-α. KAY plays an important role during development and in many signal transduction cascades. Not surprisingly, lethality was observed with tim–GAL4, which is expressed in all peripheral circadian tissues. It is thus possible that KAY-α plays a role not just in pacemaker neurons but also in peripheral tissues. However, recent whole-genome expression studies have revealed a striking enrichment of transcription factors in PDF-positive circadian neurons (Nagoshi et al., 2010). This suggests that transcriptional regulation is particularly important for the function of these neurons. KAY-α might thus be specifically recruited in this complex transcriptional network to provide the sLNvs with their striking characteristic of being self-sustained pacemaker neurons that drive rhythms of other clock neurons in DD (Stoleru et al., 2005). By affecting both transcriptional loops, KAY-α might bring stability to the circadian pacemaker, particularly if it proved to be itself clock-controlled.
The transcriptional network controlling circadian rhythms is well conserved between mammals and Drosophila. Could FOS family members also control circadian period in mammals? There are four mammalian members of the FOS family: c-FOS, FOSB, FRA-1, and FRA-2. Light exposure during the subjective night strongly increases expression of c-FOS, FOSB, and FRA-2 in the suprachiasmatic nucleus (SCN), the brain structure that controls circadian rhythms in mammals (Aronin et al., 1990; Kornhauser et al., 1990; Schwartz et al., 2000). This induction might be important to phase shift circadian rhythms. Indeed, c-fos knock-out mice reset their circadian behavioral rhythms to a phase-shifted LD cycle more slowly (Honrado et al., 1996), and antisense oligonucleotides targeting both jun-b and c-fos block circadian responses to short light pulses in rats (Wollnik et al., 1995). Interestingly also, c-FOS and FRA-2 are rhythmically expressed in DD in the dorsomedial region of the SCN, which is particularly important for circadian rhythm generation (Schwartz et al., 2000). To our knowledge, only c-fos knock-out mice have been tested in DD, and the period length of their circadian behavior is normal (Honrado et al., 1996). This could be explained by redundancy with FRA-2, which can compete with c-FOS for the same DNA binding sites (Takeuchi et al., 1993). If FOS family members were important for generating circadian rhythms, it would be unlikely that they function identically to KAY-α in Drosophila. Indeed, although constructed with the same logic in flies and mammals, the feedback loop interlocked with the PER feedback loop is based on different transcription factors: bZIPs (VRI and PDP1) in flies (Blau and Young, 1999; Cyran et al., 2003) and nuclear receptors (RORα and REVERBα) in mammals (Preitner et al., 2002; Sato et al., 2004). It is thus unlikely that FOS family members would be able to directly bind to REVERBα as they do with VRI in Drosophila. However, it would be interesting to determine whether mammalian FOS proteins can modulate CLK/BMAL1 transactivation.
In summary, we have identified KAY-α as a novel transcriptional regulator of the circadian pacemaker that modulates both circadian transcriptional feedback loops. KAY-α contributes to the precision of the circadian timekeeping mechanisms and possibly also to its stability.
Footnotes
This study was supported by National Institute of General Medical Sciences R01 Grant GM 066777 (P.E.). We thank Tom Vu, D. Wentworth, and D. Szydlik for technical support. We are grateful to Dave Weaver for critical reading of our manuscript. We thank J. Blau, S. Reppert, and the Drosophila Genomics Resource Center for donating plasmids. We also thank the National Institutes of Genetics (Shizuoka, Japan), the Vienna Drosophila RNAi Center (Vienna, Austria), and Transgenic RNAi Project at Harvard Medical School (Boston, MA) for RNAi transgenic flies. We are grateful to J. Blau, P. Hardin, M. Rosbash, V. Budnik, and D. Bohmann for donating antibodies.
The authors declare no competing financial interests.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Aronin N, Sagar SM, Sharp FR, Schwartz WJ. Light regulates expression of a Fos-related protein in rat suprachiasmatic nuclei. Science. 1990;87:5959–5962. doi: 10.1073/pnas.87.15.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chélot E, Hardin PE, Préat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Bohmann D, Ellis MC, Staszewski LM, Mlodzik M. Drosophila Jun mediates Ras-dependent photoreceptor determination. Cell. 1994;78:973–986. doi: 10.1016/0092-8674(94)90273-9. [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors PER and TIM. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dubruille R, Murad A, Rosbash M, Emery P. A constant light-genetic screen identifies KISMET as a regulator of circadian photoresponses. PLoS Genet. 2009;5:e1000787. doi: 10.1371/journal.pgen.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Reppert SM. A rhythmic Ror. Neuron. 2004;43:443–446. doi: 10.1016/j.neuron.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- Fassler J, Landsman D, Acharya A, Moll JR, Bonovich M, Vinson C. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res. 2002;12:1190–1200. doi: 10.1101/gr.67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Honrado GI, Johnson RS, Golombek DA, Spiegelman BM, Papaioannou VE, Ralph MR. The circadian system of c-fos deficient mice. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1996;178:563–570. doi: 10.1007/BF00190186. [DOI] [PubMed] [Google Scholar]
- Hudson SG, Goldstein ES. The gene structure of the Drosophila melanogaster proto-oncogene, kayak, and its nested gene, fos-intronic gene. 2008;420:76–81. doi: 10.1016/j.gene.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 2000;43:207–233. doi: 10.1002/(sici)1097-4695(20000605)43:3<207::aid-neu1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci. 2010;30:12664–12675. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-w. [DOI] [PubMed] [Google Scholar]
- Langer CC, Ejsmont RK, Schönbauer C, Schnorrer F, Tomancak P. In vivo RNAi rescue in Drosophila melanogaster with genomic transgenes from Drosophila pseudoobscura. PLoS One. 2010;5:e8928. doi: 10.1371/journal.pone.0008928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, Hardin PE, Tanimura T, Ueda HR. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Wernet MF, Beaufils P, Killian D, Pichaud F, Kühnlein R, Desplan C. A green fluorescent protein enhancer trap screen in Drosophila photoreceptor cells. Mech Dev. 2000;93:151–160. doi: 10.1016/s0925-4773(00)00287-2. [DOI] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Niwa R, Namiki T, Ito K, Shimada-Niwa Y, Kiuchi M, Kawaoka S, Kayukawa T, Banno Y, Fujimoto Y, Shigenobu S, Kobayashi S, Shimada T, Katsuma S, Shinoda T. Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the “Black Box” of the ecdysteroid biosynthesis pathway. Development. 2010;137:1991–1999. doi: 10.1242/dev.045641. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Rakshit K, Krishnan N, Guzik EM, Pyza E, Giebultowicz JM. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol Int. 2012;29:5–14. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Richier B, Michard-Vanhée C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms. 2008;23:103–116. doi: 10.1177/0748730407313817. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Carpino A, Jr, de la Iglesia HO, Baler R, Klein DC, Nakabeppu Y, Aronin N. Differential regulation of fos family genes in the ventrolateral and dorsomedial subdivisions of the rat suprachiasmatic nucleus. Neuroscience. 2000;98:535–547. doi: 10.1016/s0306-4522(00)00140-8. [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernández MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Takeuchi J, Shannon W, Aronin N, Schwartz WJ. Compositional changes of AP-1 DNA-binding proteins are regulated by light in a mammalian circadian clock. Neuron. 1993;11:825–836. doi: 10.1016/0896-6273(93)90112-5. [DOI] [PubMed] [Google Scholar]
- Wollnik F, Brysch W, Uhlmann E, Gillardon F, Bravo R, Zimmermann M, Schlingensiepen KH, Herdegen T. Block of c-Fos and JunB expression by antisense oligonucleotides inhibits light-induced phase shifts of the mammalian circadian clock. Eur J Neurosci. 1995;7:388–393. doi: 10.1111/j.1460-9568.1995.tb00334.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Kockel L, Peverali FA, Jackson DB, Mlodzik M, Bohmann D. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 1997;16:7393–7401. doi: 10.1093/emboj/16.24.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Emery P. Molecular and neural control of insects circadian rhythms. In: Gilbert LI, editor. Insect molecular biology and biochemistry. New York: Academic; 2012. pp. 513–551. Chap 15. [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A. An isoform-specific mutant reveals a role of PDP1 epsilon in the circadian oscillator. J Neurosci. 2009;29:10920–10927. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]