Abstract
Sex-specific trait expression is frequently associated with highly variable, condition-dependent expression within sexes and rapid divergence among closely related species. Horned beetles are an excellent example for studying the molecular basis of these phenomena because horn morphology varies markedly among species, between sexes, and among alternative, nutritionally-cued morphs within sexes. In addition, horns lack obvious homology to other insect traits and provide a good opportunity to explore the molecular basis of the rapid diversification of a novel trait within and between species. Here we show that the sex-determination gene doublesex (dsx) underlies important aspects of horn development, including differences between sexes, morphs, and species. In male Onthophagus taurus, dsx transcripts were preferentially expressed in the horns of the large, horned morph, and RNAi-mediated knockdown of dsx dramatically altered male horn allometry by massively reducing horn development in large males, but not in smaller males. Conversely, dsx RNAi induced ectopic, nutrition-sensitive horn development in otherwise hornless females. Finally, in a closely related species (Onthophagus sagittarius) that has recently evolved a rare reversed sexual dimorphism, dsx RNAi revealed reversed as well as novel dsx functions despite an overall conservation of dsx expression. This suggests that rapid evolution of dsx functions has facilitated the transition from a regular sexual dimorphism to a reversed sexual dimorphism in this species. Our findings add beetle horns to existing examples of a close relationship between dsx and sexual trait development, and suggest that dsx function has been coopted to facilitate both the evolution of environmentally-cued intrasexual dimorphisms and rapid species divergences in a novel trait.
Keywords: cooption, polyphenism, evolutionary novelty, interchangeability, weak linkage
Exaggerated secondary sexual traits are common products of sexual selection, often diverge rapidly between closely related species, and frequently exhibit condition-dependent expression (1). In extreme cases, conditional expression of secondary sexual traits is discrete and results in the production of alternative, intrasexual morphs (2). For instance, the exaggerated horns of horned beetles are typically present only in males; have diversified dramatically in size, shape, and location across thousands of species; and—among conspecific males—frequently develop in a conditional, nutrition-dependent manner. In such cases, only males with access to optimal feeding conditions during the larval stage grow into large, major males with fully developed horns, which are then used as weapons in aggressive encounters with conspecific males. In contrast, poor larval feeding conditions result in males developing into smaller, nonaggressive sneaker or minor morphs with disproportionately smaller horns (3, 4).
In this study, we investigated whether morph-, sex-, and species-specific development of secondary sexual traits, as seen in horned beetles, may arise through the use of a shared developmental genetic machinery. In particular, we focused on the somatic sex-determination gene doublesex (dsx), for three reasons. First, recent array-based studies on horned beetles of the genus Onthophagus suggested that differential dsx expression is associated with morph-specific development of horns. Specifically, relative to the abdominal epidermis of the same individual, dsx expression was consistently elevated in the horn tissue, but not the legs, of horned males (5, 6).
Second, previous studies have shown that dsx has a highly conserved function as the terminal gene in the sex determination pathway that regulates the sex-limited expression of downstream genes, leading to sexually dimorphic development and behavior across diverse insects (7–9). The structure and function of dsx are best understood from studies in Drosophila. In Drosophila melanogaster, a hierarchy of sex-determination genes acts to regulate the expression of sex-specific isoforms of the Dsx transcription factor, which in turn regulates the sex-limited expression of downstream cytodifferentiation genes responsible for the elaboration of sexually dimorphic traits (10). Comparative analyses have revealed that even though the sex-determination pathway upstream of dsx is divergent across insect orders, dsx itself is highly conserved. In particular, in all insects examined thus far, the key features of dsx structure and function appear to be conserved—specifically, the expression of male- and female-specific Dsx transcription factors generated through alternative splicing of an exon included in dsx transcripts in males and excluded in females (8).
Third, recent studies have also revealed that dsx is a nexus for the evolution of sexually dimorphic traits; all cases understood at the molecular level involve either changes in cis-regulatory sequences of dsx target genes (11, 12) or changes in the expression of dsx itself (13, 14). In addition, the recent finding that developing Drosophila are mosaic for dsx expression—and thus are mosaic for the potential for sexual differentiation—has led to increased appreciation for the potential of evolution through tissue-specific changes in dsx activity (15). Indeed, it was recently found that the diversification of sex combs in Drosophila species is correlated with changes in dsx expression patterns (14).
Here we investigated the gene structure, expression, and function of dsx in two congeneric species of horned beetles, Onthophagus taurus and Onthophagus sagittarius, which exhibit highly divergent patterns of phenotype expression. O. taurus has a dramatic intrasexual and intersexual dimorphism in horn development typical for the genus and thought to reflect the ancestral character state of horn development in Onthophagus. O. sagittarius shared a common ancestor with O. taurus ∼5 Mya, but has since evolved a pattern of horn development radically divergent from the ancestral pattern still seen in O. taurus, including the rare expression of a reversed sexual dimorphism as well as horn development in a novel location not used by O. taurus or more basal branches in the Onthophagus phylogeny (16). We sought to investigate the degree to which dsx might mediate the differential development of horns across different developmental and evolutionary contexts. Our results suggest that dsx functions to regulate sex-specific development of horns, that this function has been coopted to enable the nutritionally-cued development of alternative male morphs, and that dsx function has further diversified to facilitate rapid and extreme species divergences in the expression of a novel trait.
Results and Discussion
dsx Gene Structure and Expression Are Conserved in O. taurus Relative to Other Insects.
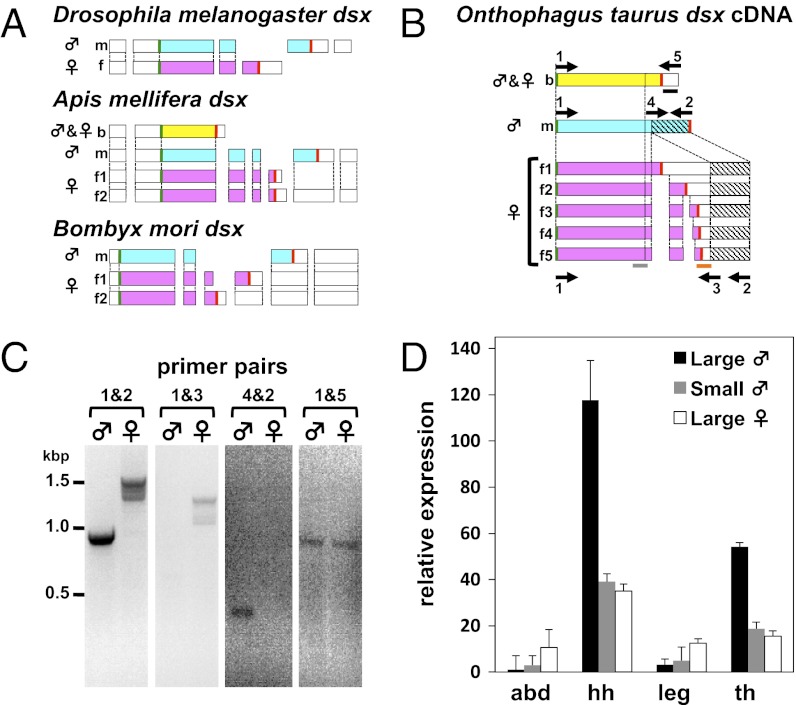
We first investigated the gene structure and function of dsx in O. taurus, a species with a typical intrasexual and intersexual dimorphism in horn development (Figs. 1 and 2). The O. taurus dsx gene architecture and inferred protein sequence are similar to those in other insects (Fig. 1 A and B). Specifically, O. taurus expresses sex-specific dsx mRNA isoforms (Fig. 1 B and C), with one isoform found only in males (Otdsxm), and at least five isoforms found only in females (Otdsxf1–5). In addition, we detected a single dsx isoform that was expressed in both sexes (Otdsxb; Fig. 1 B and C).
Fig. 1.
Structure and expression of O. taurus dsx. (A) dsx transcripts in D. melanogaster (Top), A. mellifera (Middle), and B. mori (Bottom). The gene structures of transcripts are indicated by rectangles, with ORFs shaded either blue (male), pink (female), or yellow (non–sex-specific isoform). Start and stop codons are indicated by green and red lines, respectively. (B) Cloned cDNA and inferred ORFs of O. taurus dsx, with the same color coding as in A. The 3′ region of Otdsxm is shared with five Otdsxf isoforms (shaded by hatched lines). Note that D. melanogaster dsx, A. mellifera dsx, and B. mori dsx are shown based on genomic structures, whereas Onthophagus dsx is reconstructed based on cDNA sequence matches across transcripts. Putative splice donor and acceptor sites in Otdsxf2–5 are shown in Fig. S1. The horizontal solid gray bar on O. taurus dsx indicates the region used to generate dsxC dsRNA and used for qRT-PCR. Orange and black solid horizontal bars indicate the regions used to generate dsxf dsRNA and dsxb dsRNA to knockdown female and sex-shared dsx splice variants, respectively. Arrows and numbers indicate primers used to assess gene structures. (C) RT-PCR results confirm sex-specific splicing pattern of O. taurus dsx. The RT-PCR primer pairs correspond to those shown in B. (D) Relative expression of dsx (qRT-PCR, estimates relative to actin; 1 indicates dsx expression in the abdominal epithelium of large males) in the indicated tissues [abd, abdominal epithelium; hh, head horn; leg, all six legs combined; th, thoracic (pronotal) horn] of O. taurus large males (black bars), small males (gray bars), and large females (white bars).
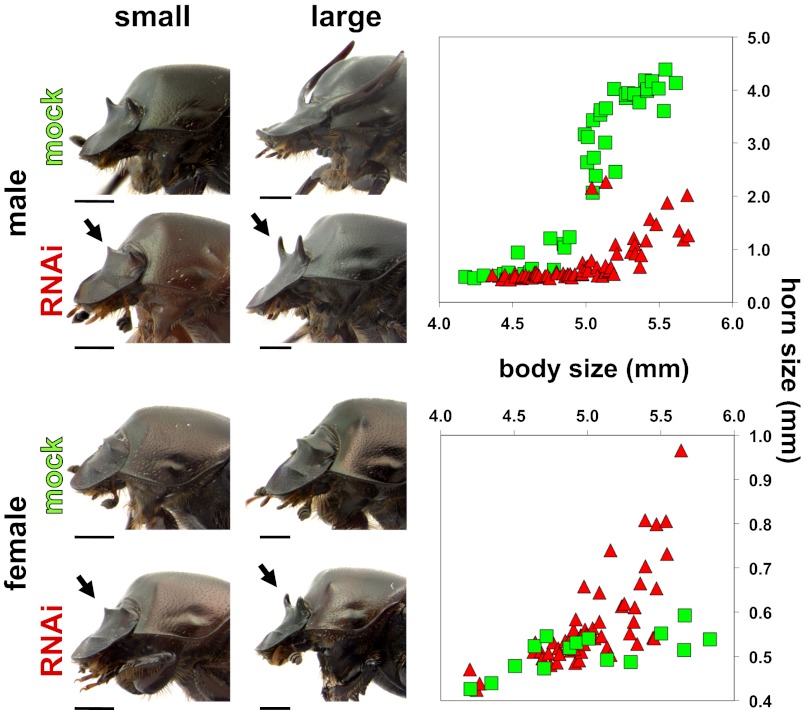
Fig. 2.
Effects of dsx RNAi on horn development in adult O. taurus males and females. (Left) Representative animals obtained after mock (control) injections (Upper) and dsx dsRNA injections (Lower). Small individuals are shown on the left; large individuals, on the right. Anterior is to the left, and filled arrows indicate locations of head horn development of RNAi individuals. (Scale bars: 1 mm.) (Right) Bivariate plots of body size (x-axis) and head horn length (y-axis) for male (Upper) and female (Lower) O. taurus. Mock-injected and RNAi individuals are plotted as green squares and red triangles, respectively. The dsx RNAi substantially reduced horn expression in large males (T115 = 6.82, P < 0.001) and induced conspicuous ectopic head horns in females (ANOVA, F2,104 = 9.35, P < 0.001).
Each of the Otdsxf transcripts has female-specific sequences, including translation termination codons, that are absent from the Otdsxm and Otdsxb transcripts (Fig. S1 A and B). Thus, the organization of female-specific transcripts is most similar to that of the silkmoth Bombyx mori, which also expresses multiple dsxf transcripts that encode DsxF protein isoforms with different C-terminal sequences (8, 17) (Fig. S1C). Unlike in Drosophila, the male dsx isoform shares all of its putative coding exons with the female isoforms (Fig. 1B and Fig. S1A). We found that when translated, all sex-specific isoforms have all of the functional domains commonly present in other insects, including the oligomerization domains (ODs) 1 and 2 (Fig. S1 B–D). Specifically, overall identity of translated sequences is 46% for the male-specific isoform, 45–47% for DsxF1, and 46–47% for DsxF2 compared with B. mori. The predicted OD1 (also known as DM domain) and OD2 share 93% and 55% sequence identity, respectively, with the corresponding regions in D. melanogaster. In contrast, the dsx transcript shared by both sexes (Otdsxb) includes at least one exon that is absent from any of the other isoforms and lacks obvious similarities to published dsx sequences (Fig. S1B). Moreover, the Otdsxb transcript does not encode OD2 (Fig. S1 A and B). A sex-shared insect dsx isoform, also lacking OD2, has so far been reported only from the honeybee (18), and its functional significance, if any, remains unclear. Combined, these data suggest that male- and female-specific Otdsx isoforms may function as sex-specific transcription factors, but also raise the possibility that non–sex-specific functions may be performed by an additional, distinct dsx isoform whose expression is shared across sexes.
dsx Controls Sex-Specific Development of Horns in O. taurus.
To investigate possible functions of male, female, and nonspecific dsx isoforms, we used RNAi to knock down dsx expression. We then used morphometric measurements on the resulting pupae and adults to quantify the effects of dsx knockdown on male and female horn development compared with individuals receiving mock double-stranded RNA (dsRNA) injections and untreated WT individuals. We first injected dsRNA directed to the transcript region unique to Otdsxb, the isoform expressed in both sexes (indicated by the black bar in Fig. 1B and Fig. S1A). Compared with mock injection, Otdsxb dsRNA had no apparent effects on horn development (Fig. S2), genital development, or any other phenotypic trait in males or females. These findings suggest that this isoform may lack a function during sexual differentiation, at least to the limits of detection of our experiment.
We then investigated possible functions of Otdsxm and Otdsxf. Because these isoforms are spliced strictly in a sex-specific manner, we used dsRNA directed to the common region to knock down the expression of Otdsxm in males and Otdsxf in females. We refer to this construct as OtdsxC dsRNA (indicated by the gray bar in Fig. 1 and Fig. S1A). We replicated this approach using dsRNA directed to a gene region specific to female isoforms (Otdsxf dsRNA, indicated by the orange bar in Fig. 1B and Fig. S1A). However, because all coding regions of Otdsxm are also contained within the transcripts of female-specific isoforms, it was not possible to design a corresponding dsRNA construct that would affect only Otdsxm.
Injecting OtdsxC dsRNA into third instar larvae resulted in decreased expression of dsx in both sexes (Fig. S3A), defects in the development of male genitalia (Fig. S3 B and C), and obvious alterations to horn development in both sexes (detailed below), validating the effectiveness of our approach. Specifically, knocking down dsx dramatically affected the sexually dimorphic development of horns in O. taurus. Large adults normally express sexually dimorphic head horns, such that large males have prominent paired horns on the posterior head, whereas females feature a small continuous ridge in the corresponding location. Otdsx knockdown dramatically reduced the development of head horns in large males (Fig. 2; t test, T115 = 6.82, P < 0.001), whereas knocking down dsx in large females induced the development of paired ectopic head horns (Fig. 2; ANOVA, F2,104 = 9.35, P < 0.001). Injecting Otdsxf dsRNA, which specifically targets Otdsxf isoforms only, similarly induced paired ectopic head horns in females but left male horn development completely unaffected (Fig. S4). Collectively, these results indicate that Otdsxm promotes horn development in large males, whereas Otdsxf inhibits horn formation in females, and that reduced expression of either Otdsxm or Otdsxf causes males and females to default to a similar small-horned phenotype.
Similar sex-specific effects on horn growth were also observed in thoracic (pronotal) horns in pupae. Pupal thoracic horns are ubiquitous in Onthophagus and function as molting organs, designed to remove the rigid larval head capsule during the larval-to-pupal molt (19). Secondary cooption of pupal thoracic horns into the adult stage is believed to have enabled the evolution of adult thoracic horns seen now in a subset of extant species (19, 20). In O. taurus, pupae develop conspicuous thoracic horns that are completely resorbed before the pupal-adult molt through programmed cell death in both sexes (19, 21). Moreover, in this species, pupal thoracic horns are sexually dimorphic, with males having larger horns than females (21). dsx RNAi reduced pupal thoracic horn length in males (ANOVA, F2,156 = 12.75, P < 0.001), but increased it in females (ANOVA, F2,118 = 13.02, P < 0.001), again causing both sexes to converge toward an intermediate phenotype when dsx function was reduced (Fig. S5). In contrast, mock injections using nonsense dsRNA did not affect horn location and allometries in male and female O. taurus pupae or adults compared with untreated WT individuals (Fig. S6).
Nutrition Influences dsx Function in Regulating Sex-Specific Horn Development.
Normally, the development of head horns in males differs distinctly among morphs. Large, well-nourished males have prominent horns nearly 10-fold longer than those of smaller males that received suboptimal nutrition (Fig. 2). If dsx regulates horn development independent of nutritional conditions, we would predict dsx RNAi to result in the same relative reduction in horn length regardless of male body size. Instead, sensitivity to dsx knockdown increased dramatically with adult body size, ranging from no change to only modest reductions in horn length in small and medium-sized males to as much as an 80% reduction in horn length in the largest males (Fig. S7). A nutrition-dependent dsx RNAi response was also observed, albeit to a lesser extent, in females. Normally, the posterior ridge on the female head is proportional to body size (Fig. 2). Knocking down Otdsxf in large females resulted in the development of conspicuous ectopic horns similar in size to those small WT males and large Otdsxm RNAi males. However, the effect in small females was more subtle and involved the induction of distinct lateral points but without altering the overall height of the ridge. Similar results were obtained when female larvae were injected with female isoform-specific Otdsxf dsRNA (Fig. S3). These data support the hypothesis that the function of dsx in regulating sex-specific horn development is conditional on nutritional conditions.
Tissue-Specific and Nutrition-Dependent Expression of Otdsxm May Mediate dsx-Dependent Horn Development in Alternative Male Morphs.
How does nutrition-dependent regulation of horn development intersect with the dsx dependent regulation of horn development? Nutrition-dependent regulation could act independent, or downstream, of dsx for example, influencing the sensitivity to dsx regulation. Alternatively, nutrition-dependent regulation could act through dsx; for example, in well-nourished males, dsx expression could be up-regulated in body regions fated to undergo exaggerated growth. To test the latter hypothesis, we examined dsx expression in the developing horn (head, thorax) and nonhorn (leg, abdomen) tissues of large and small males as well as females. We found that Otdsxm expression is by far highest in the head horn primodia of large males (which exhibit the most dramatic growth response to increased nutrition), followed by the thoracic horn primordia of large males (which exhibit a less extreme response), but low in the head and thoracic horn primordia of small males and in the legs and abdomen regardless of male size. In contrast, in females Otdsxf levels remained more uniform across all tissue types (Fig. 1C). These findings are consistent with the model that the development of exaggerated horns in large males is associated with tissue-specific, nutrition-dependent expression of Otdsxm in the horn primordia of large males.
Reversed and Novel dsx Functions Underlie Recently Evolved Reversed Sexual Dimorphism in the Congener O. sagittarius Despite Conservation of dsx Expression.
We next investigated whether lineage-specific changes in dsx function may have mediated the evolution of radically different patterns of sex-specific trait expression, such as reversed sexual dimorphisms. Here we used O. sagittarius, a species that has evolved a radically divergent pattern of horn development not seen elsewhere in the genus. Recall that O. sagittarius shared a common ancestor with O. taurus ∼5 Mya, but has since evolved a pattern of horn development radically divergent from the ancestral pattern still seen in O. taurus. Most obvious is the expression of a reversed sexual dimorphism, with female O. sagittarius expressing single long medial thoracic and head horns that are lacking in males (Fig. 3). Careful analysis of the location of head horn development suggests that the location of female head horn development in O. sagittarius is homologous to that of the paired posterior head horns in male O. taurus (22). If and how female O. sagittarius use their horns in reproduction is presently unknown. In addition, adult male O. sagittarius express a pair of small head horns in a novel location near the front of the head. Anterior head horns present in O. sagittarius are not present in O. taurus or more basal branches in the phylogeny and thus are believed to have arisen since the divergence of O. sagittarius and O. taurus (16). Finally, and again in contrast to O. taurus, no dimorphism exists within male or female O. sagittarius. Instead, in both sexes, large individuals essentially represent proportionately enlarged versions of small individuals (21, 23), which reduces the need for allometric analyses in this species.
Fig. 3.
Effects of dsx dsRNA injection on horn development in O. sagittarius males (Upper) and females (Lower). Shown are lateral (Upper row) and frontal (Lower row) views of representative animals, with mock-injected individuals on the left and dsRNA-injected individuals on the right. Horn development did not differ between mock-injected individuals and WT individuals. Open triangles indicate anterior head horns in mock-injected males and the corresponding region in females. Orange triangles indicate posterior head horns in mock-injected females and the corresponding region in males. Filled triangles indicate thoracic horns. (Scale bars: 1 mm.)
We investigated structures and functions of male and female dsx isoforms in O. sagittarius. Sequencing cDNA clones and RT-PCR experiments revealed that O. sagittarius also expresses male- and female-specific dsx transcripts (Fig. S8). The inferred splicing patterns and translated protein sequences of male and female dsx isoforms are highly similar to those in O. taurus (Fig. S1 E and F). Thus, the reversed sexual dimorphism in horn growth in O. sagittarius is not related to reversal of the sex-specific splicing of dsx isoforms in developing horns. In contrast to our results for O. taurus, we failed to clone a dsxb isoform in O. sagittarius. However, this does not rule out the possibility that O. sagittarius also expresses this isoform.
We investigated possible functions of Osdsxm and Osdsxf using dsRNA injections targeting the same transcript region shared across isoforms as used in O. taurus. As with O. taurus, dsxC RNAi resulted in malformation of male genitalia (Fig. S3B), but resulted in a more complex suite of changes in horn development. Specifically, dsx knockdown in O. sagittarius revealed three major effects. First, dsx RNAi substantially reduced thoracic horn size in females and induced a small but conspicuous protrusion in the male prothorax (Fig. 3, filled triangles). This indicates that Osdsxm functions to inhibit growth of the thoracic horn, whereas Osdsxf functions to promote growth. Thus, the sex-specific functions of dsx in regulating thoracic horn growth are reversed in O. sagittarius compared with O. taurus.
Second, dsx RNAi had no obvious effect on the development of anterior head horns in males, but caused females to express a pair of ectopic, male-like anterior head horns (Fig. 3, open triangles). This indicates that Osdsxf functions to inhibit the growth of anterior head horns in females. Although the direction of the effect was the same as that seen in O. taurus females, the specific head region affected differed distinctly.
Third, dsx RNAi resulted in the development of surprisingly large, branched ectopic horns in the posterior head in males, and the transformation of the large single horn normally seen in females to a branched horn (Fig. 3, orange triangles). In male O. sagittarius, dsx thus functions to inhibit the growth of branched posterior head horns. This suggests that similar to the situation for the thoracic horn, the male-specific function of dsx in regulating posterior head horn development is reversed in O. sagittarius compared with O. taurus; dsxm promotes the development of paired posterior horns in male O. taurus, but inhibits it in O. sagittarius. In contrast, dsx RNAi in female O. sagittarius did not result simply in a reduction of the single medial head horn, as would be predicted if Osdsxf functioned exactly opposite to Otdsxf, that is, as a promotor of horn growth. Instead, Osdsxf promotes transformation of a branched into an unbranched posterior head horn. All of the phenotypes described above were observed in 100% of dsx RNAi individuals (n = 15 per sex), and in no control-injected individuals (n = 98).
Taken together, these findings suggest that dsx RNAi phenotypes observed in O. sagittarius reflect a mosaic of (i) reversed dsx functions relative to O. taurus and the presumed common ancestor, as seen in the regulation of thoracic horns (both sexes) and posterior head horns (males), and (ii) novel dsx functions, as seen in the regulation of anterior head horn size and posterior head horn shape in females.
dsx and the (Co)evolution and Diversification of Sexual and Male Dimorphism.
Our results significantly expand our knowledge of the range of evolutionary changes mediated by dsx. First, our findings add beetle horns to the list of secondary sexual traits whose development is regulated by sex-specific dsx splice forms. Moreover, our results suggest that the location, direction, and nature of dsx-mediated regulation of horn development have diversified rapidly among closely related species, enabling in at least one instance the evolution of a dramatic reversal of sexually dimorphic trait expression. For instance, whereas in O. taurus, Otdsxm functions to promote posterior head horns in males and Otdsxf inhibits them in females, their orthologs in O. sagittarius function to inhibit posterior head horns in males but promote their fusion into a single horn in females.
Second, our results suggest that, at least in O. taurus, Otdsx expression may be influenced by nutritional conditions, and that differential expression of Otdsxm may underlie the development of alternative, nutritionally-cued male morphs. These observations parallel in part earlier findings in Nasonia wasps, in which differential dsx expression in males has been proposed to underlie species-specific differences in male wing size (13). However, dsx expression in male Nasonia differs constitutively between species, whereas in male O. taurus it appears to be altered facultatively in response to nutrition. If correct, the use of dsx as a regulator of both sexual and male dimorphism also may explain the tight coevolution of both patterns of phenotype expression in Onthophagus reported by earlier phylogenetic studies (24), which concluded that 19 of 20 instances of change in sexual dimorphism (gain or loss) were paralleled by a corresponding gain or loss of male dimorphism. Whereas these earlier studies suggested shared endocrine mechanisms as the underlying cause of this striking covariation, our findings offer an alternative, or additional, explanation: in O. taurus, a species typical for the genus, Otdsxm appears to execute a dual function, promoting sex-specific horn development in males and facilitating the dimorphic development of horns in males of different body sizes. Thus, concerted gain and loss of sexual and male dimorphism may be a consequence of lineage-specific gain and loss of the role of dsx in the promotion of male horns.
More generally, our findings illustrate a phenomenon considered of critical importance in the evolution and diversification of novel traits: the lability of regulatory inputs, such as sexual identity or nutritional conditions, and target traits, such as genitalia or different horn types, linked through an otherwise highly conserved switch mechanism, dsx. This phenomenon, referred to as “interchangeability” (25) or “weak linkage” (26), is thought to facilitate the origin and diversification of form and function because it enables novel combinations of inputs and outputs without requiring their de novo evolution, while simultaneously producing functional and integrated phenotypes through the use of a conserved regulatory and developmental machinery. In the context of horned beetles, such interchangeability not only might have facilitated the evolution of sexual and male dimorphisms, but also might have occurred repeatedly and convergently for different horn types, further facilitating their subsequent diversification. For instance, both functional and phylogenetic studies suggest that head and thoracic horns developed, and have evolved, largely independent of one another (16, 24).
Despite this independent history, based on the present study, sex- and morph-specific development of both horn types appears to be regulated by dsx in extant onthophagine species, consistent with convergent cooption of dsx in the developmental regulation of different horn types. Furthermore, comparative morphological studies have shown that head and thoracic horns in the same species can differ widely in the degree to which their development is affected by sex and nutrition, suggesting that sex- and nutrition-dependent horn development can diversify independently for different body regions (21, 23, 24, 27). Taken together, our results paint an extraordinarily dynamic picture of dsx evolution and its role in the origin and diversification of sexual and male dimorphism in horned beetles.
Materials and Methods
Beetle Care.
Beetles used in this study were collected and reared essentially as described previously (28). In brief, O. taurus was collected near Bloomington, IN and Chapel Hill, NC, and O. sagittarius was collected near Kaneohe, Hawaii. Adults were kept at 25 °C (O. taurus) and 28 °C (O. sagittarius) in a sand/soil mixture at a 16:8 h light:dark cycle and fed homogenized cow manure twice a week. To obtain larvae, five females and three males were selected and kept in a small plastic container with packed, moist sand/soil mixture with cow manure and allowed to produce brood balls. Brood balls were collected after 8 d, and larvae were transferred to 12-well plates as described previously (29).
Cloning and Sequencing of Onthophagus dsx.
The dsx sequence information was first obtained from a previously published EST sequence (5). RT-PCR was used to amplify dsx cDNA fragments from both male and female RNA samples; primer sequences are listed in Table S1. Fragments were subsequently cloned into pSC-A vector using a StrataClone PCR Cloning Kit (Agilent). Sequencing reactions from cloned fragments were carried out using BigDye cycle sequencing chemistry (Life Technologies). All 12 cDNA fragments isolated in this work were sequenced and submitted to GenBank (accession nos. JN165757–JN165768).
dsRNA Generation and Injection.
dsRNA to knock down dsx splice variants was generated as described previously (30). In brief, the region of interest (Fig. 1) was cloned using a StrataClone PCR Cloning Kit, followed by BigDye sequencing. The vector containing the fragment was purified using a QIAprep Spin Miniprep Kit (Qiagen). The vector was then subjected to PCR using M13 forward and reverse primers, and the PCR product was used as a template for in vitro transcription. Forward and reverse RNA strands were produced using MEGAscript T7 and T3 Kits (Life Technologies), treated by DNase I to remove DNA template, precipitated by ethanol, denatured at 75 °C for 10 min, chilled on ice, and mixed at a 1:1 ratio by weight. The resulting mixture was incubated in a water bath at 80 °C until room temperature was reached. The concentration of the annealed RNA was measured, confirmed by gel electrophoresis, and stored at −80 °C until injection. Injections into Onthophagus larvae were carried out as described previously (30), with 0.5 µg of dsRNA injected into larvae during the first 5 d of the final, third instar. Approximately 200 O. taurus larvae and 150 O. sagittarius larvae were injected with dsxRNA constructs over the course of the experiment. The same procedure was performed to knock down female-specific variants, as well as the splice variant present in both sexes.
Rapid Amplification of cDNA Ends for dsx.
Rapid amplification of cDNA ends (RACE) was performed to obtain possible additional dsx splice variants in Onthophagus, using the FirstChoice RLM-RACE Kit (Life Technologies) in accordance with the manufacturer’s instructions. Specifically, the aim was to determine the existence of possible variants whose expression is shared between both sexes. Primers were designed for the same region used for dsxC dsRNA generation; primer sequences are listed in Table S1; 3′- and 5′-RACE were performed, and the products from males and females were cloned and sequenced as described above. Then primers were designed to amplify fragments containing the sequences obtained from both 3′- and 5′-RACE, to confirm that both RACE products were derived from the same splice variant.
Mock Injections.
Mock (control) injections were carried out as described previously (30). In brief, animals were reared under the same conditions as RNAi-injected animals, but were injected with dsRNA from a 167-bp PCR product derived from a pBluescript SK vector. Transcription reactions, DNaseI treatment, and transcript annealing were performed as descrived above. A total of 1 µg of dsRNA was injected into larvae during the first 5 d of the final, third instar. Approximately 70 O. taurus and 80 O. sagittarius larvae received mock injections.
Allometric Measurements.
RNAi-treated and control pupae and adults were measured using a 2D image analysis setup consisting of a dissecting microscope (Leica) mounted with a digital camera (Scion) and ImageJ software (31). Thorax width served as a measure of pupal and adult body size. Head length and thoracic horn length were measured as described previously (21). Measurements were recorded to the nearest 0.01 mm.
Imaging.
Beetle images were collected using the same setup as used for allometric measurements. In most cases, 5–10 images with different focal planes were aligned and merged using Adobe Photoshop Creative Suite 4.
Statistical Analysis.
Allometric scaling relationships of head and pronotal horns were analyzed similarly to previous studies (21, 30, 32, 33). In brief, because female head horns and pupal thoracic (pronotal) horns of both sexes exhibit linear scaling relationships, effects of treatment (dsx dsRNA injections, mock injections, and WT) were compared using ANOVA, with horn length as the response variable and body size and treatment as model effects. Analyses were repeated for each sex, followed by pairwise comparisons using Tukey's HSD test. In males, in contrast, head horns exhibit a strongly sigmoidal allometry, and thus horn length residuals were compared across treatment groups. Residuals were calculated as the difference between horn lengths observed in a given individual and those expected for the individual's body size given the WT scaling relationship. Two-tailed t tests were used for pairwise comparisons.
Quantitative RT-PCR.
Quantification of dsx in WT animals.
Quantitative RT-PCR (qRT-PCR) was performed to detect dsx expression levels in O. taurus in different body regions. Head horns, pronotal horns, legs, and abdominal epithelium of ∼24-h-old pupae were dissected, and total RNA was purified using a Qiagen RNeasy Kit, followed by on-column genomic DNA digestion (Qiagen). Primers used to detect dsx were dsxqPCRF01 and dsxqPCRR01 (Integrated DNA Technology); sequences are listed in Table S1. dsx expression level was normalized using actin as an internal control. The same primer set was used for actin as in a previous study (34). Two-step qRT-PCR was performed using a QuantiTect Reverse-Transcription Kit and QuantiTect SYBR Green PCR Kit (Qiagen) on a Stratagene MX3000P system (Agilent). Three independent sets of tissue samples from large males, small males, and females (i.e., 4 tissues × 3 morph/sex × 3 biological replicates) were harvested and subjected to qRT-PCR, with two technical replicates for each sample. Equal volumes of fractions of all RNA samples were pooled and vacuum-concentrated, then treated along with all remaining samples. The mixed sample was serially diluted to obtain a dilution series ranging from 0.01 ng to 10 ng of template RNA, which was subsequently subjected to PCR to confirm that both genes amplified with similar PCR efficiency (28, 29). A 40-ng RNA sample was used in each PCR, with a final concentration of 0.3 µM for primer sets. Reaction conditions were 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 58 °C for 30 s, and 72 °C for 30 s. After PCR, the temperature was raised to 95 °C to observe dissociation curves. The ΔΔCt method was used to obtain relative expression levels of dsx across tissues, morphs, and sexes (35, 36). Relative dsx expression values were obtained in reference to the abdomen of large males. The same procedure was repeated three times with different sets of RNA samples, to confirm reliability of the experiments.
Quantification of dsx in control-injected and dsx dsRNA-injected animals.
Approximately 24-h-old pupae were used for validation of dsx knockdown by qRT-PCR. Total RNA was extracted from whole bodies of six large dsx dsRNA-injected males and females, as well as three large WT males and females, using TRI Reagent (Life Technologies), following the manufacturer’s standard protocol. qRT-PCR was performed separately on each independent set of samples. Each reaction was duplicated during the PCR step. A total of 100 ng of total RNA was treated and analyzed as described above. Knockdown efficiency was assessed relative to dsx expression levels detected in WT individuals. The experiment was repeated three times with different RNA samples.
Supplementary Material
Acknowledgments
We thank E. Yoder and W. Anderson for helping with beetle care; W. Haines and R. Hoan for collecting beetles in the field; E. Snell-Rood, Z. Lai, J. Lopez, A. Cash, N. Rodibaugh, and X. Yu for assisting with qRT-PCR experiments; and the Indiana University Center for Genomics and Bioinformatics for allowing the use of their qPCR facilities. Funding for this study was provided by National Science Foundation Grant IOS 0820411 (to J.A. and A.P.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN165757–JN165768).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118589109/-/DCSupplemental
References
- 1.Emlen DJ. The evolution of animal weapons. Annu Rev Ecol Evol Syst. 2008;39:387–413. [Google Scholar]
- 2.Andersson MB. Sexual Selection. Princeton: Princeton Univ Press; 1994. pp. 1–599. [Google Scholar]
- 3.Moczek AP, Emlen DJ. Proximate determination of male horn dimorphism in the beetle Onthophagus taurus (Coleoptera: Scarabaeidae) J Evol Biol. 1999;12(1):27–37. [Google Scholar]
- 4.Moczek AP, Emlen DJ. Male horn dimorphism in the scarab beetle, Onthophagus taurus: Do alternative reproductive tactics favour alternative phenotypes? Anim Behav. 2000;59(2):459–466. doi: 10.1006/anbe.1999.1342. [DOI] [PubMed] [Google Scholar]
- 5.Kijimoto T, Costello J, Tang Z, Moczek AP, Andrews J. EST and microarray analysis of horn development in Onthophagus beetles. BMC Genomics. 2009;10:504. doi: 10.1186/1471-2164-10-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snell-Rood EC, et al. Developmental decoupling of alternative phenotypes: Insights from the transcriptomes of horn-polyphenic beetles. Evolution. 2011;65(1):231–245. doi: 10.1111/j.1558-5646.2010.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52(7):837–856. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 8.Shukla JN, Nagaraju J. Doublesex: A conserved downstream gene controlled by diverse upstream regulators. J Genet. 2010;89(3):341–356. doi: 10.1007/s12041-010-0046-6. [DOI] [PubMed] [Google Scholar]
- 9.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10(11):797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 10.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 11.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7(8):e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134(4):610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loehlin DW, et al. Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet. 2010;6(1):e1000821. doi: 10.1371/journal.pgen.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 2011;9(8):e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell, II: There is a time and place for sex. PLoS Biol. 2010;8(5):e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emlen DJ, Marangelo J, Ball B, Cunningham CW. Diversity in the weapons of sexual selection: Horn evolution in the beetle genus Onthophagus (Coleoptera: Scarabaeidae) Evolution. 2005;59(5):1060–1084. [PubMed] [Google Scholar]
- 17.Ohbayashi F, Suzuki MG, Mita K, Okano K, Shimada T. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 2001;128(1):145–158. doi: 10.1016/s1096-4959(00)00304-3. [DOI] [PubMed] [Google Scholar]
- 18.Cho S, Huang ZY, Zhang J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177(3):1733–1741. doi: 10.1534/genetics.107.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kijimoto T, Andrews J, Moczek AP. Programed cell death shapes the expression of horns within and between species of horned beetles. Evol Dev. 2010;12(5):449–458. doi: 10.1111/j.1525-142X.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 20.Moczek AP. The origin and diversification of complex traits through micro- and macroevolution of development: Insights from horned beetles. In: Jeffrey WR, editor. Current Topics in Developmental Biology. Vol 86. New York: Academic; 2009. pp. 135–162. [DOI] [PubMed] [Google Scholar]
- 21.Moczek AP. Pupal remodeling and the development and evolution of sexual dimorphism in horned beetles. Am Nat. 2006;168(6):711–729. doi: 10.1086/509051. [DOI] [PubMed] [Google Scholar]
- 22.Balthasar V. Monographie der Scarabaeidae und Aphodiidae der palaearktischen und orientalischen Region; Coleoptera: Lamellicornia. Prague: Tschechoslowakische Akademie der Wissenschaften; 1963. [Google Scholar]
- 23.Shelby JA, Madewell R, Moczek AP. Juvenile hormone mediates sexual dimorphism in horned beetles. J Exp Zoolog B Mol Dev Evol. 2007;308(4):417–427. doi: 10.1002/jez.b.21165. [DOI] [PubMed] [Google Scholar]
- 24.Emlen DJ, Hunt J, Simmons LW. Evolution of sexual dimorphism and male dimorphism in the expression of beetle horns: Phylogenetic evidence for modularity, evolutionary lability, and constraint. Am Nat. 2005;166(Suppl 4):S42–S68. doi: 10.1086/444599. [DOI] [PubMed] [Google Scholar]
- 25.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford Univ Press; 2003. [Google Scholar]
- 26.Gerhart J, Kirschner M. The theory of facilitated variation. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8582–8589. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moczek AP. Pupal remodeling and the evolution and development of alternative male morphologies in horned beetles. BMC Evol Biol. 2007;7:151. doi: 10.1186/1471-2148-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moczek AP, Nagy LM. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evol Dev. 2005;7(3):175–185. doi: 10.1111/j.1525-142X.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 29.Shafiei M, Moczek AP, Nijhout HF. Food availability controls the onset of metamorphosis in the dung beetle Onthophagus taurus (Coleoptera: Scarabaeidae) Physiol Entomol. 2001;26(2):173–180. [Google Scholar]
- 30.Moczek AP, Rose DJ. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc Natl Acad Sci USA. 2009;106(22):8992–8997. doi: 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11(7):36–42. [Google Scholar]
- 32.Emlen DJ. Artificial selection on horn length body size allometry in the horned beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae) Evolution. 1996;50(3):1219–1230. doi: 10.1111/j.1558-5646.1996.tb02362.x. [DOI] [PubMed] [Google Scholar]
- 33.Emlen DJ. Costs and the diversification of exaggerated animal structures. Science. 2001;291(5508):1534–1536. doi: 10.1126/science.1056607. [DOI] [PubMed] [Google Scholar]
- 34.Snell-Rood EC, Moczek AP. Insulin signaling as a mechanism underlying developmental plasticity: The role of FOXO in a nutritional polyphenism. PLoS ONE. 2012;7(4):e34857. doi: 10.1371/journal.pone.0034857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270(1):41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.