Abstract
New conformationally extended dipeptide surrogates based on an imidazo[1,2-a]pyridine scaffold are described. Efficient synthesis and incorporation into host peptides affords structures with native side-chain functionality and hydrogen bonding elements on one face of the backbone. Structural analysis by NMR suggests that model peptidomimetics adopt a β-strand-like conformation in solution.
β-Strands are key structural motifs in a number of protein-protein interfaces relevant to human disease.1 For example, the Akt-GSK3β,2 Ras-Raf1,3 and KRas-FTase4 interactions are all implicated in oncogenesis and involve the recognition of an extended or β-strand domain. Isolated β-strand peptides are also substrates for various proteolytic enzymes5 and major histocompatibility complex (MHC) proteins.6 Conformational mimicry of β-strands has thus gained increasing attention as a strategy toward new chemical probes and therapeutics.7
Synthetic templated β-strands/sheets often feature β-hairpin or macrocyclic motifs to impart conformational rigidity.7 In these cases, enhanced stability may require large scaffolding elements or complementary strands that are auxillary to a sequence of interest. The incorporation of constrained residues or backbone isosteres within extended host peptides represents a more ‘minimalist’ approach toward peptidomimetic drug candidates. The utility of hybrid peptides featuring β-strand prosthetics such as the Hao subunit,8 @-tide residues,9 and other dipeptide surrogates10 have been demonstrated in various applications. The development of artificial β-strands comprised entirely of non-peptide subunits has also emerged as an approach toward novel proteomimetic foldamers.11
Our interest in peptidomimetics targeting the Akt-GSK3β interaction12 led us to explore scaffolds that could be easily prepared and incorporated into host sequences, enforce an extended backbone conformation, and impart more ‘drug-like’ character onto short peptides. Here, we describe the design and synthesis of peptidomimetics incorporating a novel imidazo[1,2-a]pyridine scaffold. Structural analysis of model compounds by NMR suggests that hybrid imidazo[1,2-a]pyridine-based peptides adopt an extended conformation in solution. These studies lay the groundwork for the development of imidazo[1,2-a]pyridines as new core motifs for peptido- and proteomimetic drug design.
In our search for constrained templates with favorable pharmacokinetic properties, we were particularly attracted to the imidazo[1,2-a]pyridine core structure due to its presence in therapeutic agents such as the hypnotics zolpidem (Ambien®) and alpidem,13 and the vasodilator olprinone.14 We envisioned that imidazo[1,2-a]pyridines appropriately substituted with terminal amine and carboxy groups, and with side-chain diversity at the putative β carbon, could serve as ‘drug-like’ extended dipeptide surrogates. Figure 1 depicts the design of our scaffold as well as an overlay of a model tetrapeptide mimic with an idealized strand from an antiparallel β-sheet. Although the planarity of the aromatic core results in some deviation in main chain and side chain geometry, the imidazole nitrogen (H-bond acceptor) overlays reasonably well with the carbonyl oxygen in the native peptide strand. We thus pursued this scaffold as a potential backbone replacement for highly extended dipeptides. Despite a wealth of literature on the chemistry and pharmacology of imidazo[1,2-a]pyridines,15 to the best of our knowledge, these scaffolds have not previously been synthesized or studied in the context of peptide mimicry.
Figure 1.
Design of IP-based β-strand mimics.
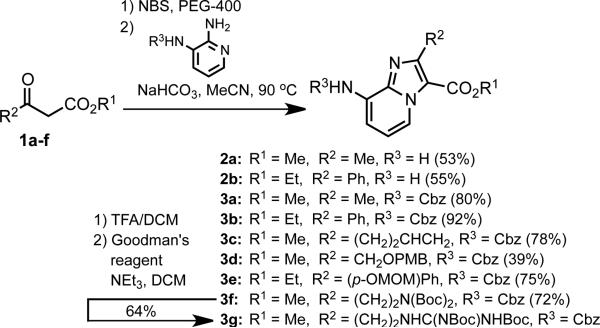
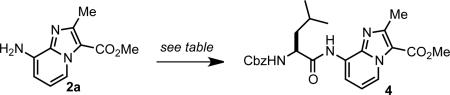
Scheme 1 depicts the synthesis of imidazo[1,2-a]pyridine (IP) dipeptide surrogates via monobromination of acetoacetate derivatives and condensation with 2,3-diaminopyridine. Although various methods exist for the α-bromination of β-ketoesters, we found reaction of 1 with 1.1 equiv of N-bromosuccinimide in PEG-400 to be the most convenient. Optimal yields of IP scaffolds 2 were obtained by directly heating the crude α-bromo-β-ketoesters with 1.0 equiv 2,3-diaminopyridine in the presence of NaHCO3. We found that the yields of imidazopyridine formation could be increased by using mono-Cbz-protected 2,3-diaminopyridine16 in the condensation reaction. To show that a wider array of substituents could be similarly incorporated, we also prepared orthogonally-protected dipeptide surrogates featuring butenylglycine (3c), homoserine (3d), tyrosine (3e), ornithine (3f), and arginine (3g) side chains.
Scheme 1.
Synthesis of orthogonally-protected IP-based extended dipeptide surrogates.
We next studied the incorporation of our IP building blocks into short peptides using conventional coupling reactions. Condensation of 2a with Cbz-Leu-OH proved challenging, presumably due to the low nucleophilicity of the IP aryl amine. As shown in Table 1, a variety of standard conditions afforded poor yields of the desired amide (entries 1-3). The use of EDC with catalytic DMAP (entries 4 and 5) led to an improvement in yield, but was attended by significant racemization of the Leu residue.17 Surprisingly, we found that omission of an auxilliary base resulted in higher conversions. Optimal results were obtained by using 2.0 eq. of both EDC and Cbz-Leu-OH without any added base to give 4 in 91% yield and >98:2 er (entry 7). Other coupling reagents such as PyBOP, COMU, DEPBT, and DCC consistently gave inferior results (entries 8-11).
Table 1.
N-terminal coupling of IP scaffold 2a.
| ||
|---|---|---|
| entry | conditionsa | yieldb |
| 1 | 1.2 equiv Z-Leu-OH, 1.2 equiv HBTU, HOBt, NEt3, MeCN | trace |
| 2 | 1.2 equiv Z-Leu-OH, 1.2 equiv PyBOP, NEt3, DMF | trace |
| 3 | 1.2 equiv Z-Leu-OH, 1.2 equiv EDC, HOBt, NEt3, DCM | trace |
| 4 | 1.2 equiv Z-Leu-OH, 1.2 equiv EDC, 0.2 eq. DMAP, DCM | 33c |
| 5 | 2.0 equiv Z-Leu-OH, 2.0 equiv EDC, 0.2 eq. DMAP, DCM | 58c |
| 6 | 1.2 equiv Z-Leu-OH, 1.2 equiv EDC, DCM | 59 |
| 7 | 2.0 equiv Z-Leu-OH, 2.0 equiv EDC, DCM | 91 |
| 8 | 2.0 equiv Z-Leu-OH, 2.0 equiv PyBOP, DCM | 33 |
| 9 | 2.0 equiv Z-Leu-OH, 2.0 equiv COMU, DCM | 44 |
| 10 | 2.0 equiv Z-Leu-OH, 2.0 equiv DEPBT, DCM | 26 |
| 11 | 2.0 equiv Z-Leu-OH, 2.0 equiv DCC, DCM | 77 |
All reactions carried out for 24 h at rt.
Isolated yields.
HPLC analysis of a phenylalanyl derivitive showed significant epimerization of the Leu chiral center.
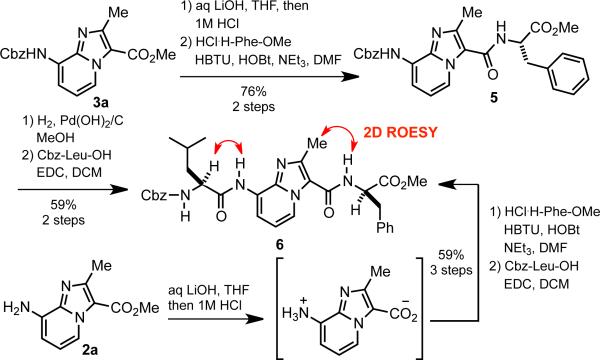
Hydrolysis of the ester in 4 required extended reaction times in the presence of hydroxide. Prolonged exposure to aqueous LiOH also resulted in an unacceptable degree of amide bond scission. We found that this issue could be circumvented by using Cbz-protected building block 3a for peptide assembly in the C−N direction (Scheme 2). The urethane bond in 3a proved stable to hydrolysis with LiOH. Condensation of the resulting crude acid with HPhe-OMe was followed by hydrogenolysis and coupling to Cbz-Leu-OH to give tetrapeptide mimic 6 in 45% yield over 4 steps.
Scheme 2.
Synthesis and selected ROESY correlations (in red) of tetrapeptide mimic 6.
The observed lack of aryl amine reactivity in the presence of various standard amidation reagents (Table 1, entries 1-3) also led us to explore the direct condensation of a fully unprotected IP derivative. We found that hydrolysis of 2a followed by neutralization afforded the crude amino acid zwitterion, which could be used directly in the subsequent coupling. Reaction with H-Phe-OMe in the presence of HBTU, HOBt, and NEt3 gave the desired amide exclusively, without any detectable homocoupling of the IP scaffold. The phenylalanyl intermediate was then reacted with Cbz-Leu-OH and EDC to provide 6 in an improved 59% overall yield from 2a.
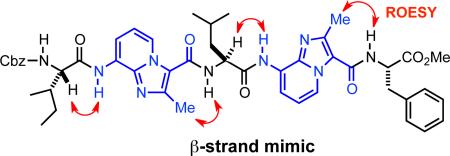
With a short model peptide in hand, we used NMR to determine whether 6 adopts an extended conformation in solution. The 1H NMR of 6 in CDCl3 was well resolved and showed no evidence of rotational isomers. The CHα proton resonances of both the Leu and Phe residues also appeared 0.3−0.5 ppm downfield of their expected values in an unstructured peptide.18 This is consistent with the chemical shift of β-strand/sheet CHα protons relative to those in random coil peptides. Most important, the 2D ROESY spectrum of 6 exhibited a correlation between the IPMe and PheNH protons, as well as between LeuCHα and IPNH (Scheme 2). These correlations strongly support trans amide bond configurations across tetrapeptide mimic 6.
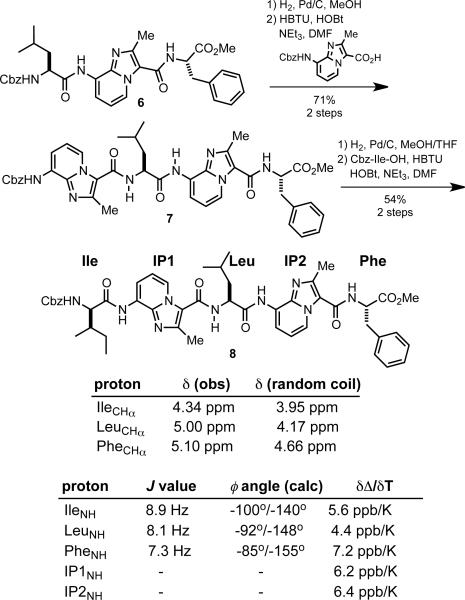
We then synthesized heptapeptide mimic 8 featuring two IP scaffolds embedded within a larger strand (Scheme 3). Compound 8 again exhibited a well-resolved 1H NMR spectrum that could be readily assigned using 2D COSY. The absence of NMR signals corresponding to minor rotational isomers was particularly notable due to the presence of 1 urethane and 4 amide bonds. Each of the amino acid CHα signals in 8 showed pronounced downfield shifts in CDCl3 relative to their corresponding random-coil values.18 In addition, the NH-CHα coupling constants obtained for the Ile and Leu residues were in the 8–9 Hz range, consistent with the values typical of a β-strand conformation. These coupling constants were used to calculate possible φ dihedral angles of the Ile, Leu, and Phe residues in 8 using the Pardi modification to the Karplus equation.19 Amino acid residues in idealized parallel and antiparallel β-sheets exhibit φ values of approximately –119° and –139°, respectively (see Scheme 3).7 The corresponding torsions in 8 are thus in good agreement with a β-strand-like conformation.
Scheme 3.
Synthesis of heptapeptide mimic 8 and conformational analysis by 1H NMR.
The ROESY data obtained earlier for 6 suggested that the IPMe 1H NMR signal could serve as a useful reporter of local conformation. As with 6, we observed ROESY correlations between the IP1Me-LeuNH and IP2Me-PheNH protons in heptapeptide mimic 8, indicating the presence of trans amide bonds between these residues (Figure 2). Additional correlations between IleCHα–IP1NH and LeuCHα–IP2NH further supported an extended structure for 8 in solution. Dilution studies with 8 indicated a negligible chemical shift dependance on concentration, ruling out the presence of dimeric species that may give rise to intermolecular correlations (see Supporting Information).
Figure 2.
ROESY spectrum of 8 (500 MHz in CDCl3) highlighting key Overhauser correlations.
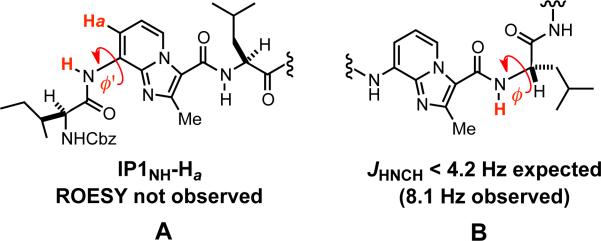
Conformers with IP scaffold φ’ torsions at or near 0°, exemplified by A in Figure 3, would be expected to exhibit a strong Overhauser correlation between IP1NH and Ha. The absence of (non-vicinal) ROESY cross-peaks involving either protons Ha/a’ or Hc/c’ suggests that stable turn conformations are not significantly populated on the NMR time-scale. While 2D NMR does not rule out the posibility of acute Ile, Leu, or Phe φ angles, the JHNCH coupling constants observed for 8 (see Scheme 3) are not consistent with structures such as B (Figure 3). Moreover, the position of the Ile, Leu, and Phe NH resonances in the 1H NMR spectrum of 8 (< 7.0 ppm) argue against their participation in intramolecular H-bonds resulting from tight turns.20 We also carried out Δδ/ΔT experiments in DMSO-d6 and found that each of the 5 NH protons in 8 exhibited a high (> 4.0 ppb/K) temperature dependence on chemical shift consistent with solvent accessibility (see Scheme 3 and Supporting Information).21 Taken together, these data provide compelling evidence that heptapeptide mimic 8 adopts an extended conformation in CDCl3.
Figure 3.
Structures of possible turn conformations of 8.
In summary, we have described the design and synthesis of novel extended dipeptide surrogates based on substituted imidazo[1,2-a]pyridine scaffolds. These building blocks can be readily prepared from β-ketoesters and incorporated into host peptides in either protected or unprotected forms. The efficient synthesis of model peptidomimetics harboring these scaffolds affords structures that adopt β-strand-like conformations in solution. We anticipate that the imidazo[1,2-a]pyridine subunit will find utility as a probe of peptide conformation, and as a dipeptide surrogate with enhanced drug-like character. Efforts toward IP-based β-strand mimics capable of disrupting protein-protein interactions are currently underway in our laboratory and will be reported in due course.
Supplementary Material
Acknowledgment
We gratefully acknowledge financial support from NIH (CA167215). We thank Dr. Edwin Rivera (USF) for assistance with 2D NMR acquisitions and Dr. Lukasz Wojtas (USF) for carrying out X-ray diffraction studies on 2a.
Footnotes
Supporting Information Available. Experimental procedures and structural characterization of compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a Dou Y, Baisnée P-F, Pollastri G, Pécout Y, Nowick J, Baldi P. Bioinformatics. 2004;20:2767. doi: 10.1093/bioinformatics/bth326. [DOI] [PubMed] [Google Scholar]; b Somers WS, Phillips SEV. Nature. 1992;359:387. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]; c Puglisi JD, Chen L, Blanchard S, Frankel AD. Science. 1995;270:1200. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]; d Derrick JP, Wigley DB. Nature. 1992;359:752. doi: 10.1038/359752a0. [DOI] [PubMed] [Google Scholar]; e Colon W, Kelly JW. Biochemistry. 1992;31:8654. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Nat. Struct. Mol. Biol. 2002;9:940. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 3.Nassar N, Horn G, Herrmann C, Block C, Janknecht R, Wittinghofer A. Nat. Struct. Biol. 1996;3:723. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- 4.a Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Biochemistry. 1998;37:16601. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]; b Long SB, Casey PJ, Beese LS. Structure. 2000;8:209. doi: 10.1016/s0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 5.a Tyndall JDA, Fairlie DP. J. Mol. Recog. 1999;12:363. doi: 10.1002/(SICI)1099-1352(199911/12)12:6<363::AID-JMR478>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]; b Fairlie DP, Tyndall JDA, Reid RC, Wong AK, Abbenante G, Scanlon MJ, March DR, Bergman DA, Chai CLL, Burkett BA. J. Med. Chem. 2000;43:1271. doi: 10.1021/jm990315t. [DOI] [PubMed] [Google Scholar]
- 6.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Nature. 1993;364:33. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 7.Loughlin WA, Tyndall JDA, Glenn MP, Hill TA, Fairlie DP. Chem. Rev. 2010;110:PR32. doi: 10.1021/cr900395y. [DOI] [PubMed] [Google Scholar]
- 8.a Nowick JS, Chung DM, Maitra K, Maitra S, Stigers KD, Sun Y. J. Am. Chem. Soc. 2000;122:7654. [Google Scholar]; b Nowick JS, Cary JM, Tsai JH. J. Am. Chem. Soc. 2001;123:5176. doi: 10.1021/ja010220s. [DOI] [PubMed] [Google Scholar]
- 9.Phillips ST, Rezac M, Abel U, Kossenjans M, Bartlett PA. J. Am. Chem. Soc. 2002;124:58. doi: 10.1021/ja0168460. [DOI] [PubMed] [Google Scholar]
- 10.a Blomberg D, Brickmann K, Kihlberg J. Tetrahedron. 2006;62:10937. [Google Scholar]; b Qian Y, Blaskovich MA, Saleem M, Seong CM, Wathen SP, Hamilton AD, Sebti SM. J. Biol. Chem. 1994;269:12410. [PubMed] [Google Scholar]; c Qian Y, Marugan JJ, Fossum RD, Vogt A, Sebti SM, Hamilton AD. Bioorg. Med. Chem. 1999;7:3011. doi: 10.1016/s0968-0896(99)00252-7. [DOI] [PubMed] [Google Scholar]; d Grandy D, Shan J, Zhang X, Rao S, Akunuru S, Li H, Zhang Y, Alpatov I, Zhang XA, Lang RA, Shi DL, Zheng JJ. J. Biol. Chem. 2009;284:16256. doi: 10.1074/jbc.M109.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Martin SF, Austin RE, Oalmann CJ, Baker WR, Condon SL, de Lara E, Rosenberg SH, Spina KP, Stein HH, Cohen J, et al. J. Med. Chem. 1992;35:1710. doi: 10.1021/jm00088a005. [DOI] [PubMed] [Google Scholar]; f Cluzeau J, Lubell WD. Pept. Sci. 2005;80:98. doi: 10.1002/bip.20213. [DOI] [PubMed] [Google Scholar]; g Hanessian S, McNaughton-Smith G, Lombart HG, Lubell WD. Tetrahedron. 1997;53:12789. [Google Scholar]
- 11.a Smith AB, Guzman MC, Sprengeler PA, Keenan TP, Holcomb RC, Wood JL, Carroll PJ, Hirschmann R. J. Am. Chem. Soc. 1994;116:9947. [Google Scholar]; b Angelo NG, Arora PS. J. Am. Chem. Soc. 2005;127:17134. doi: 10.1021/ja056406z. [DOI] [PubMed] [Google Scholar]; c Wyrembak PN, Hamilton AD. J. Am. Chem. Soc. 2009;131:4566. doi: 10.1021/ja809245t. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Jamieson AG, Russell D, Hamilton AD. Chem. Commun. 2012;48:3709. doi: 10.1039/c2cc30295k. [DOI] [PubMed] [Google Scholar]; e Raghuraman A, Ko E, Perez LM, Ioerger TR, Burgess K. J. Am. Chem. Soc. 2011;133:12350. doi: 10.1021/ja2033734. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Ko E, Liu J, Perez LM, Lu G, Schaefer A, Burgess K. J. Am. Chem. Soc. 2010;133:462. doi: 10.1021/ja1071916. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Chandrasekhar S, Babu BN, Prabhakar A, Sudhakar A, Reddy MS, Kiran MU, Jagadeesh B. Chem. Commun. 2006:1548. doi: 10.1039/b518420g. [DOI] [PubMed] [Google Scholar]
- 12.a Ranatunga S, Del Valle JR. Bioorg. Med. Chem. Lett. 2011;21:7166. doi: 10.1016/j.bmcl.2011.09.079. [DOI] [PubMed] [Google Scholar]; b Ranatunga S, Liyanage W, Del Valle JR. J. Org. Chem. 2010;75:5113. doi: 10.1021/jo1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langtry HD, Benfield P. Drugs. 1990;40:291. doi: 10.2165/00003495-199040020-00008. [DOI] [PubMed] [Google Scholar]
- 14.Mizushige K, Ueda T, Yukiiri K, Suzuki H. Cardio. Drug Rev. 2002;20:163. doi: 10.1111/j.1527-3466.2002.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 15.Enguehard-Gueiffier C, Gueiffier A. Mini Rev. Med. Chem. 2007;7:888. doi: 10.2174/138955707781662645. See also: Shao N, Pang G-X, Yan C-X, Shi G-F, Cheng Y. J. Org. Chem. 2011;76:7458. doi: 10.1021/jo201273p., and references therein.
- 16.Choi JY, Plummer MS, Starr J, Desbonnet CR, Soutter H, Chang J, Miller JR, Dillman K, Miller AA, Roush WR. J. Med. Chem. 2012;55:852. doi: 10.1021/jm201349f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hydrogenolysis of 4 (from entries 4 and 5) and HBTU/HOBt-mediated coupling to Cbz-Phe-OH afforded a ~83:17 mixture of diastereomers by HPLC.
- 18.Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 19.Two possible torsions for each J value result from the inherent ambiguity of the Karplus relationship. Values were calculated according to the parametrization described in: Pardi A, Billeter M, Wuthrich K. J. Mol. Biol. 1984;180:741. doi: 10.1016/0022-2836(84)90035-4.
- 20.Nowick JS, Smith EM, Pairish M. Chem. Soc. Rev. 1996;25:401. [Google Scholar]
- 21.a Kessler H. Angew. Chem. Int. Ed. 1982;21:512. [Google Scholar]; b Llinas M, Klein MP. J. Am. Chem. Soc. 1975;97:4731. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.