Abstract
Chediak-Higashi syndrome (CHS) is a lethal disease caused by mutations that inactivate the Lysosomal Trafficking Regulator protein (LYST). Patients suffer from diverse symptoms including oculocutaneous albinism, recurrent infections, neutropenia and progressive neurodegeneration. These defects have been traced back to over-sized lysosomes and lysosome-related organelles (LROs) in different cell types. Here, we explore mutants in the Drosophila mauve gene as a new model system for CHS. The mauve gene (CG42863) encodes a large BEACH domain protein of 3535 amino acids similar to LYST. This reflects a functional homology between these proteins as mauve mutants also display enlarged LROs, such as pigment granules. This Drosophila model also replicates the enhanced susceptibility to infections and we show a defect in the cellular immune response. Early stages of phagocytosis proceed normally in mauve mutant hemocytes but, unlike in wild type, late phagosomes fuse and generate large vacuoles containing many bacteria. Autophagy is similarly affected in mauve fat bodies as starvation-induced autophagosomes grow beyond their normal size. Together these data suggest a model in which Mauve functions to restrict homotypic fusion of different prelysosomal organelles and LROs.
Introduction
Chediak-Higashi syndrome (CHS) is a rare autosomal recessive disease (1, 2) caused by mutations in the CHS1 gene, which encodes the Lysosomal Trafficking Regulator protein, LYST (3–5). Patients suffer from a wide array of symptoms that include albinism, bleeding diathesis, progressive neurological disorders and immunodeficiency (6, 7). Common causes of death during childhood are infections and a so-called “accelerated phase” during which lymphocytes proliferate and invade multiple critical organs (8–11). Bone marrow transplantation can suppress the lethal “accelerated phase” and improve symptoms related to hematopoietic cells (12), but still leaves patients vulnerable to the neurological problems often accruing in early adulthood (8, 13, 14).
Many CHS symptoms have been traced back to changes in lysosome-related organelles (LROs) in different cell types. Albinism in CHS is the external reflection of over-sized melanosomes, which fail to be normally dispersed to keratinocytes and also tend to clump in hairshafts (6). Similarly, the bleeding tendency of CHS patients reflects defects in platelet dense bodies that normally function to accelerate blood clotting (11). Enlarged secretory granules in lymphocytes are a diagnostic hallmark of CHS and have been linked to immunodeficiency (6).
The biochemical mechanisms leading to these defects in organelle size and function are still poorly understood. The molecular identification of the CHS gene (3, 15) revealed that it encodes LYST, an unusually large protein of almost 4000 amino acids containing pleckstrin-homology (PH), BEACH and WD40 domains in an arrangement typical of BEACH domain proteins. BEACH domains were first defined as highly conserved domains shared between the Beige and CHS-1 proteins. Mice with mutations in the beige locus closely mimic many cell biological phenotypes observed in CHS patients (2). BEACH domains have also been observed in several more distantly related proteins: including the Alfy/Bluecheese proteins which regulate cargo recruitment to autophagosomes (16) and Neurobeachin proteins which are required for synaptic function (17). The BEACH domain of Neurobeachin, together with its associated PH domain, forms a novel structure with a prominent groove that suggests it may be binding to a specific ligand (18). However, the molecular mechanism by which LYST and its homologs function to control the size of LROs (19, 20) remains unknown.
Here, we describe a mutation in mauve (mv), the Drosophila ortholog of CHS1. as a novel model system for CHS. We find that mv mutants display the characteristic enlargement of LROs, evident in several vertebrate models of CHS (21). Furthermore, one of the most important clinical aspects of CHS, the defect in innate immunity (11, 21), is also present in mv flies. Using this model system to probe the function of LYST homologs in anti-bacterial host defense, we find a critical requirement of Mauve late in phagocytosis to prevent an increase in the size of late phagosomes. This results in delayed degradation and accumulation of bacteria in mv hemocytes. Similarly during starvation-induced autophagy, autophagosomes grow beyond their normal size.
Results
The eye color defect inmv mutants reflects oversized pigment granules
Many mutations that alter endocytic trafficking in Drosophila also affect pigmentation of the compound eye by altering trafficking to LROs (22). We previously described a FLP/FRT-based screen that capitalized on this connection to identify membrane trafficking mutants in Drosophila (23). Here, we describe one of these mutants, originally called 3L-85 and later renamed to mv2 (the name we use from here on), based on molecular and genetic data detailed below. To minimize the effect of background mutants we focused on the hemizygous combination mv2 / Df(3L)ED4287 for the following phenotypic experiments.
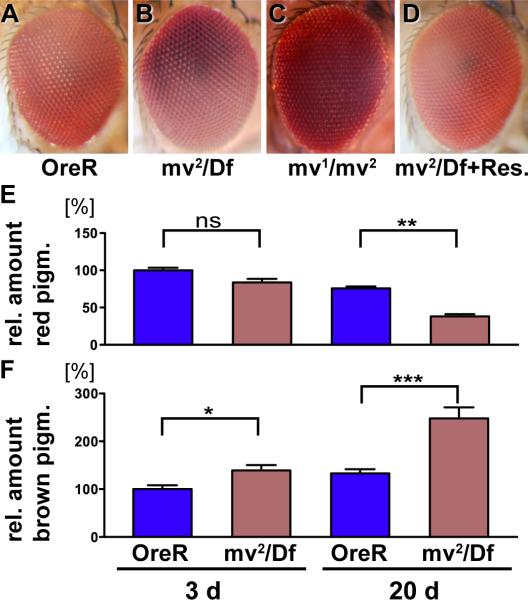
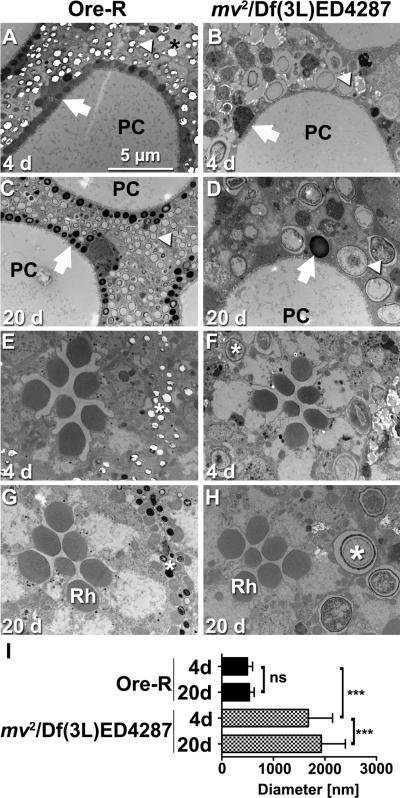
Compared to wild type, eyes homozygous for mv2 (data not shown) or hemizygous over deficiency Df(3L)ED4287 appeared brown (Fig. 1A,B), reflecting an increase in brown ommochromes and a decrease in red drosopterin pigments (Fig. 1E,F). These phenotypes were subtle in young flies and more pronounced in flies aged for 20 days or more. Ultrastructural analysis revealed another characteristic feature of this mutant. In wild-type eyes, primary pigments cells contain ommochrome-enriched, electron-dense pigment granules (24). These are reduced in number in mv2 /Df(3L)ED4287 eyes, but drastically increased in size (arrows Fig. 2A–D). This increase was even more pronounced for the electron-lucent granules that dominate the distal aspect of secondary pigment cells (arrowheads Fig. 2A–D). Their mean diameter was increased from 0.5±0.10 μm in wild type to 1.68±0.47 μm in mv2 / Df(3L)ED4287 eyes (Fig. 2 I, p<0.001), which corresponds to a more than 37-fold increase in their volume. In wild type, pigment granule size did not significantly increase in flies aged for 20 days (Figure 2C,I; 0.53±0.094 μm), b u t mv2 /Df(3L)ED4287 granules grew to a mean size of 1.93±0.47 μm by day 20 (Fig. 2D, p<0.001). This increase in the volume of mv granules corresponds to the addition of about one wild-type sized granule every day. Increased size of pigment granules could also be detected in the more proximal pigment granules situated between photoreceptor cells (marked by stars in Fig. 2 E–H). In contrast to the pigment granules, the structure and arrangement of the light-sensing organelles of photoreceptor cells, called rhabdomeres (Rh in Fig. 2G,H), appeared morphologically unaltered in young and old mv flies. This initial analysis indicated that mv mutants might have a defect in the control of LRO size, possibly due to the lack of a negative regulator for their fusion with other pigment granules or biosynthetic carrier vesicles. Alternatively, subsequent fissions that normally restore their size may require mv function.
Figure 1. The mv eye color mutant has opposite effects on ommochromes and drosopterins pigments.
Micrographs of adult eyes (A–D) show that compared to the Oregon R wild type control (A), the hemizygous mv2 /Df(3L)ED4287 (B) and the heterozygous mv1/mv2 flies (C) exhibit a darkened eye color. Normal eye color is restored by the genomic rescue transgene (BAC CH322-23O09) (D). Determination of the levels of red drosopterin (E) and brown ommochrome (F) reveals small changes in the levels of these eye color pigments in 3-day old flies that increase by 20 days of age. Unlike in most eye color mutants, the levels of brown pigments are increased in mv eyes. Values are normalized to the mean level of pigments in 3-day old wild type flies. Statistics: (*** indicates p<0.001; **: p<0.01, *: p<0.05, ns: p>0.05).
Figure 2. Oversized pigment granules characterize mv eyes.
Electron micrographs show sections of wild type (A,C,E,G) and mv2 /Df(3L)ED4287 (B, D, F, H) eyes at day 4 (A,B, E,F) or day 20 (C,D,G,H). Distal sections (A–D) show pigment granules in primary (arrows) and secondary (arrowheads) pigment cells surrounding the pseudocone (PC) just beneath the lens (66). Proximal sections (E–H) reveal the normal arrangement and structure of rhabdomeres (Rh) in the center of ommatidia. Pigment granules (stars) in secondary and tertiary pigment cells surrounding each ommatidium are drastically enlarged in mv2 /Df(3L)ED4287 eyes (F,H). A bar graph (I) shows quantification of the increased diameter of electron-lucent pigment granules that dominate the distal aspect of secondary pigment cells (***: p<0.001, ns: p>0.05).
The mv gene encodes the Drosophila ortholog of LYST
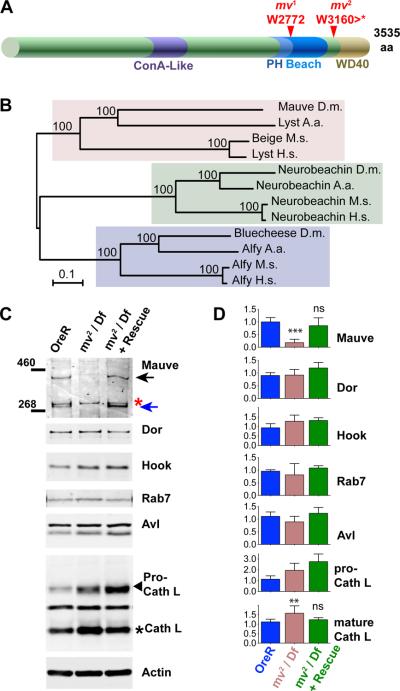
By non-complementation of deficiencies, the 3L-85 mutation was mapped to the chromosomal region 62B7–B12 and failed to complement the mv1 mutation (Fig. 1C) present on the inversion chromosome In(3LR)264 (25). Within the 62B7-B12 region, CG42863 encodes the Drosophila protein most similar to the mouse Beige and human LYST proteins. Their similarity is not restricted to the PH, BEACH and WD40 motifs which are shared by the BEACH domain protein family, but extends to the Concanavalin A-like lectin domain (26) which shares 16% identity and 32% similarity between Mauve and human LYST (Fig. 3A, B). Because of the characteristic CHS phenotype of oversized LROs, we tested CG42863 as a candidate gene for mv. Sequencing revealed a nonsense mutation at codon 3160 in the mv2 allele. Furthermore, a breakpoint in the inversion chromosome In(3LR)264, which harbors the mv1 allele (25), truncated CG42863 after 2771 amino acids (Fig. 3A). Similar truncations of human LYST cause severe CHS disease indistinguishable from larger truncations (27, 28).
Figure 3. The mv gene encodes the Drosophila ortholog of the BEACH domain protein LYST.
(A) Schematic indicates conserved structural features of the Mauve protein: the ConA-like domain including amino acids 1328 to 1626 and the arrangement of PH-BEACH and WD40 domains close to the C-terminus. The indicated mutations in the mv1 and mv2 alleles truncate these highly conserved domains. (B) A phylogenetic tree of BEACH domain proteins was calculated using the Neighbor-Joining method in the Mega5 software (64). Accession numbers for the orthologs from Homo sapiens (H.s.), Mus musculus (M.m.), Aedes aegypti (A.a.) and Drosophila melanogaster (D.m.), respectively, are for Alfy (H.s. AAN15137.1; M. m. NP_766470.2; A. a. EAT38266.1; D. m. Bluecheese: NP_608968.2), for Neurobeachin (H.s. CAH72182.1; M.m. CAC18811.1; A. a. XP_001649459.1; D. m.: NP_001138158.1), and for LYST (H.s. NP_000072.2; M. m. Beige NP_034878.2; A. a. XP_001650305; D. m. Mauve NP_647681.2). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. (C) Western blots of lysates from adult Oregon-R, mv2 /Df(3L)ED4287 and mv2 /Df(3L)ED4287 with a rescue transgene were probed with the indicated antibodies. Mauve-specific bands at (~400 and 300 kDa) are indicated by arrows. The red asterisk indicates a cross-reacting protein. (D) Quantification of western blots with indicated antibodies. At least four separate samples were used for each genotype (Oregon-R, mv2 /Df(3L)ED4287 and mv2 /Df(3L)ED4287 + rescue transgene). Note that the increased level of immature Pro-Cathepsin was not rescued by the mv transgene and therefore is unlikely to be due the mv mutation. Bars represent mean ± S.D.; ** P<0.001; ** P<0.01; and ns indicates P>0.05 compared to Oregon-R.
CG42863 is predicted to encode a protein of about 400 kDa. A protein of that size was recognized by antibodies raised against a C-terminal peptide of Mauve on western blots of lysates from wild type, but not mv2/Df(3L)ED4287 flies (black arrow in Fig. 3C). A second mauve-specific band of ~300 kDa (blue arrow) is likely to represent a degradation product. Mauve protein expression (Fig. 3C) and the mauve eye color phenotype (Fig. 1D) were rescued by a genomic transgene containing CG42863 (BAC CH322-23O09). These changes were specific for Mauve, as several other proteins required for normal lysosomal delivery were not significantly altered in mv mutants (Fig. 3C,D). Based on these findings, we re-named the 3L-85 allele mv2. Together these data indicate that mv mutants are due to loss-of-functions mutations in the Drosophila ortholog of the human LYST and mouse beige genes and, therefore, may constitute a useful model system for CHS disease.
Mauve affects a late step in bacterial phagocytosis
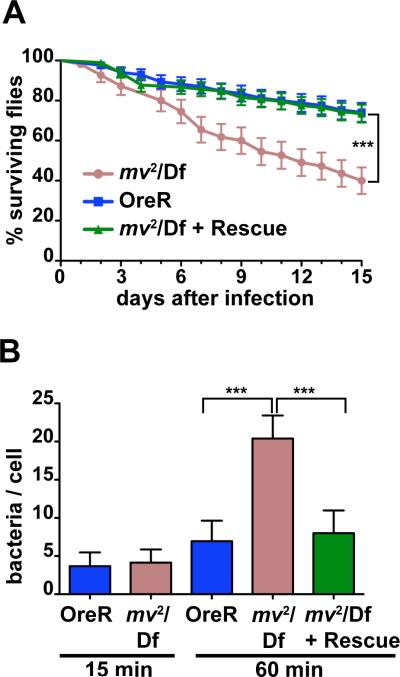
One of the most important clinical aspects of CHS is the vulnerability of patients to bacterial infections (7). To test whether this phenotype extended to the Drosophila model as well, we infected adult flies with low doses of E. coli bacteria that were well tolerated by wild-type flies (29). By contrast, mv2 /Df(3L)ED4287 flies exhibited significantly reduced survival (Fig. 4A, p<0.001 Log-Rank). Normal survival was restored by the genomic rescue transgene.
Figure 4. The mv mutant affects innate immunity.
(A) Survival of mv2 /Df(3L)ED4287 after injection of E. coli is significantly reduced compared to mv2 /Df(3L)ED4287 flies carrying a rescue transgene or OregonR flies. (B) Bar graph shows the number of live bacteria in larval hemocytes cultured from OreR, mv2 /Df(3L)ED4287, or mv2 /Df(3L)ED4287 larvae with a rescue transgene after 15 or 60 min of continued phagocytosis (*** indicates p<0.001).
This susceptibility to infections could be due to defects either in the cellular response to bacterial invaders or in the signaling pathways that induce release of antibacterial peptides (30). After infection with E. coli, expression levels of genes encoding antibacterial peptides increased in mv2 /Df(3L)ED4287 flies similarly in level to wild type flies, although with more variability (data not shown). This indicated that the defect in innate immunity was not primarily due to a failure in the signaling pathways required for the induction of antibacterial peptides.
To test for defects in the cellular response we investigated phagocytosis in primary hemocytes isolated from wild type or mv2/Df(3L)ED4287 larvae. Initial uptake of bacteria into macrophage-like hemocytes was not altered as measured by the number of live bacteria present in isolated hemocytes after 15 min of phagocytosis (Fig. 4B). After 60 min of phagocytosis, the number of bacteria in wild-type hemocytes had only slightly increased consistent with a balance between continued phagocytosis and degradation of bacteria. By contrast, 60 min of phagocytosis resulted in significantly increased numbers of live bacteria in mv2/Df(3L)ED4287 hemocytes (Fig. 4B; p<0.001), suggesting a defect in the degradation of phagocytosed bacteria. Importantly, this effect of mv on bacterial survival was reversed by a rescue transgene (Fig. 4B).
Direct imaging of bacteria after their uptake in hemocytes yielded similar results. GFP-labeled bacteria were incubated for 20 min with wild-type and mv hemocytes and phagocytosis was allowed to progress for various chase periods. After a 5-min chase, wild-type and mv hemocytes contained 5.0±3.0 bacteria (n= 49 cells for wt and 63 cells for mv). After a 10 min chase, we detected 5.0±3.5 bacteria in wild-type and 9.2±5.0 bacteria in mv hemocytes (n= 42 cells for wt and 41 cells for mv). The difference further increased after a 15 min chase with 5.0±3.0 bacteria in wild-type compared to 13.2±7.0 in mv hemocytes (p<0.001; n= 59 cells for wt and 61 cells for mv).
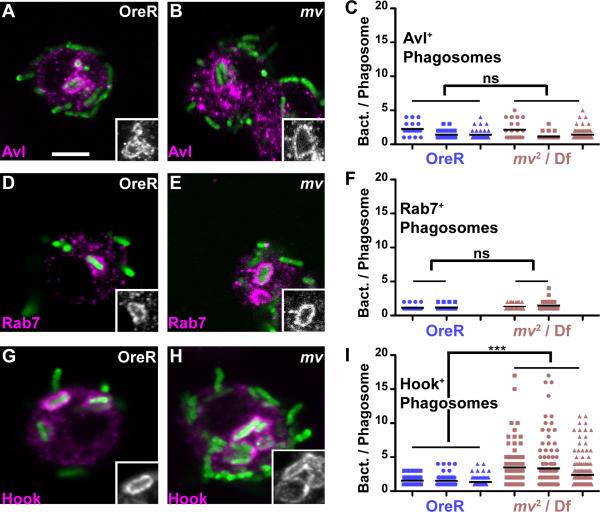
Interestingly, we noticed that at late time points bacteria in mv hemocytes often appeared in clumps causing us to wonder whether this reflected a specific stage of phagosome maturation. Avl labels early phagosomes that are enriched after a 5-min chase (29). These early Avl-positive phagosomes appeared unchanged in mv hemocytes (Fig. 5A–C). Similarly, Rab7-positive phagosomes (31) were unaffected in mv2 /Df(3L)ED4287 hemocytes (Fig. 5D–F) indicating a normal progression of phagosome maturation to this intermediate stage.
Figure 5. Mauve is required late in phagocytosis.
Micrograph show hemocytes isolated from wild-type (A,D,G) or mv2/Df(3L)ED4287 (B,E,H) larvae. Hemocytes were allowed to phagocytose heat-inactivated, GFP-labeled E. coli for 5 min (A–C), 10 min (D–F), or 15 min (G–I) and were fixed and immunostained for Avl (A,B) Rab7 (D,E) or Hook (G,H). Insets show the outline of phagosomes positive for each of the markers. The number of bacteria detected in individual phagosomes positive for the indicated markers are shown in graphs (C,F,I) were each dot represents one individual phagosome and each column is derived from an independent experiment. In each column, the line indicates the mean (***: P<0.001; ns: P>0.05).
Characteristic differences emerged, however, when bacteria were chased further into Hook-positive phagosomes. Maturation of phagosomes to this late stage requires longer chase times (15 min) and the activity of the fob gene (29). Strikingly, we observed a significant increase in the number of bacteria per phagosomes outlined by the late phagosome marker Hook (Fig. 5 G–I; p<0.001). While in wild-type hemocytes only 9.4% of Hook-positive phagosomes contained three or more bacteria, in mv2 /Df(3L)ED4287 hemocytes more than 36% of phagosomes fell into this category. 20% of the Hook-positive phagosomes in mv hemocytes contained five or more bacteria, which was never observed in wild type.
These oversized phagosomes appeared only late in phagocytic maturation, as earlier Avl or Rab7 positive phagosomes rarely contained more than three and never more than 5 bacteria (Fig. 5C,F). To exclude a contribution from bacterial division all experiments reported in Figures 5 and 6 were performed with heat-killed bacteria.
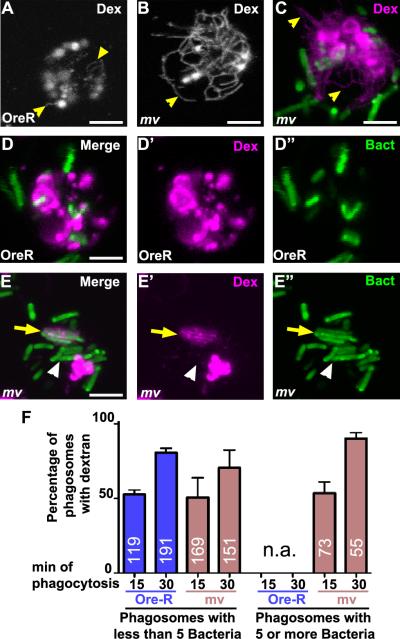
Figure 6. Phagosomes in mv hemocytes are enlarged before they fuse with lysosomes.
Micrographs show hemocytes isolated from wild-type Oregon-R (A, D) or mv2/Df(3L)ED4287 (B, C, E) larvae. Lysosomes and their tubular extensions (yellow arrowheads) were labeled by a 20-min pulse and 3-hour chase of Alexa594-labeled dextran. (C to E) Alexa594-dextran loaded hemocytes were allowed to phagocytose GFP-labeled E. coli for 15 min and images were taken for an additional 15 min. In most cells, tubular lysosomes were visible only in optical sections close to the coverslip and much less so at more apical levels where phagosomes were enriched. Panel C shows a Z-projection to show both levels in a single image. In mv hemocytes (E) some phagosomes with more than 5 bacteria were positive for dextrans (yellow arrows), but not all (white arrowheads). Scale bars: 4 μm in A to C and 3 μm in D and E. (F) Quantification of percentages of phagosomes with less than five or five or more bacteria that were labeled with pre-loaded dextran. Phagocytosis was allowed to proceed for 15 or 30 min (as indicated) followed by 15 min of imaging. No phagosomes with 5 or more bacteria were detected in Ore-R hemocytes (n.a.). Bars represent means ± S.D. The number of phagosomes evaluated is indicated for each condition.
We considered two mechanisms that could contribute to the appearance of over-sized late phagosomes in mv mutant hemocytes. First, reduced degradative activity of lysosomes may result in the accumulation of multiple bacteria in lysosomes after their fusion with Hook-positive phagosomes. However, the level of mature Cathepsin L, which requires lysosomal activity for its maturation, is slightly increased in mv2 /Df(3L)ED4287 flies (Fig. 3C,D, p<0.01), and normal levels are restored by the rescue transgene (Fig. 3C,D, ns compared to wild type). This argues against any loss of lysosomal function in mv hemocytes and may simply reflect an increase in the size of lysosomes. Furthermore, other Drosophila mutants with lysosomal dysfunction show significant neurodegeneration in aged compound eyes (32–34). By contrast, 20 d-old adult eyes of mv2 /Df(3L)ED4287 flies did not exhibit any signs of neurodegeneration (Fig. 2). Similarly, degradation of the Boss or Delta ligands in developing eye imaginal discs requires normal lysosomal function (33, 34) and proceeds without change in mv2 /Df(3L)ED4287 eyes (data not shown). These data suggest that lysosomal dysfunction in mv mutant cells makes only a minor contribution, if any, to the defect in phagosome maturation.
A second model that could explain the oversized phagosomes is a function of Mauve in restricting the homotypic fusion of late phagosomes. To test this possibility directly, we labeled lysosomes by allowing cells to internalize Alexa594-labeled dextran followed by a 2-hour chase (Fig. 6). As previously observed in live hemocytes, but not fixed samples (35), the resulting dextran-labeled lysosomes often exhibit tubular extensions (Fig. 6 A). These lysosomal tubules appeared especially pronounced in mv hemocytes (Fig. 6B,C). A similar change in lysosomal morphology has recently been described for beige macrophages (19).
We wondered whether fusion with these extended lysosomes was the origin of the phagosomes containing more then four bacteria that were uniquely present in mv hemocytes. Therefore we examined co-localization of these dextran-labeled lysosomes with phagocytosed bacteria (Fig. 6D–F). About half of the phagosomes with less then five bacteria co-localized with dextran in wild type (53±3%) and mv hemocytes (51±13%) after 15 min of phagocytosis (Fig. 6F). After 30 min, the fraction of small phagosomes (less than five bacteria) that was labeled with dextran increased to 80 ± 3% in wild type and 71±12% in mv hemocytes. This increase is consistent with the time-dependent delivery of phagosomes to the pre-labeled lysosomes in wild-type and mv hemocytes.
Large phagosomes harboring five or more bacteria could be detected already after a 15-min chase in mv hemocytes, but not in wild type. At that time point, 54±8% of these large phagosomes (n=73) were labeled by dextran indicating they had already fused with lysosomes (yellow arrows Fig. 6E). Importantly, the remaining 46% of these large phagosomes lacked the dextran label (white arrowheads, Fig. 6E). Therefore, in mv hemocytes these late oversized phagosomes formed before reaching the pre-labeled lysosomes. This does not reflect an inability to fuse with lysosomes as after 30 min of phagocytosis more than 90% of these large phagosomes were labeled with dextran. Together, these findings are consistent with the notion that oversized phagosomes in mv hemocytes arise by homotypic fusion of late phagosomes before they fuse with lysosomes. We do not know whether this apparent increase in fusion reflects an actual increase in the fusion of late phagosomes. Alternatively, homotypic fusions of late phagosomes may be common in all hemocytes and balanced by fission events of wild-type but not mv phagosomes. Direct visualization of such fusion events awaits time-lapse microscopy using live-cell markers specific for such late phagosomes. The data shown here are consistent with either of these two models in which Mauve functions to restrain the size of late phagosomes.
Autophagy is impaired in mv flies
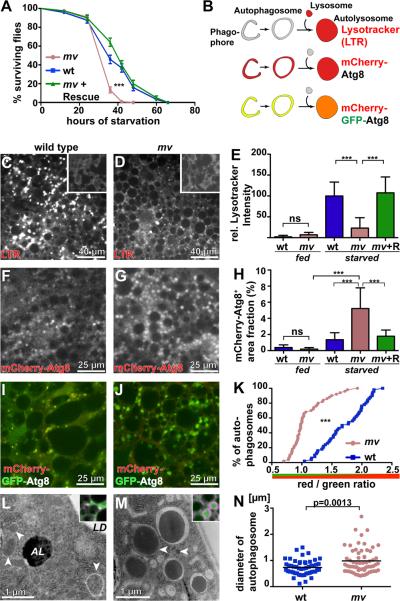
Given the changes in phagosome maturation, we wondered whether loss of mv function might also affect autophagosomes. Autophagy constitutes an alternative pathway to lysosomes in which autophagosomes deliver cytoplasmic components to the lysosome to be degraded. On an organismal level, autophagy has been shown to be critical for resistance to starvation stress (36, 37). When we tested the starvation resistance of mv2 /Df(3L)ED4287 flies, they died significantly faster than wild type after the withdrawal of food (Fig 7A, p<0.001 Log-Rank). Normal resistance to starvation stress was restored by a genomic mv transgene (p=0.16 Log-Rank compared to wild type).
Figure 7. Mauve is required late in autophagosome maturation.
(A) Survival of mv after onset of starvation is significantly reduced (p<0.001 Log-Rank) compared to wild type or mv flies carrying a rescue transgene. (B) Schematic of markers highlighting different stages of autophagosome maturation. Micrographs show larval fat bodies from wild type (C, F, I, L) or mv (D, G, J, M) larvae that at 96 h after egg laying were starved for 4 hours (62). (C, D) Fat bodies were stained with lysotracker (LTR) and the integrated lysotracker intensity for fed (insets, C,D) and starved larvae was measured and normalized to starved Oregon R intensity (E). (F, G) mCherry-Atg8 was expressed in fat bodies and the graph (H) shows the area fraction of mCherry-Atg8-positive autophagosomes. (I, J) mCherry-GFP-Atg8 was expressed in fat bodies and the graph (K) shows a cumulative plot of red to green ratio for individual phagosomes. (L, M) Electron micrographs of autophagosomes (arrowheads), autolysosomes (AL) and lipid droplets (LD) detected in larval fatbodies after a 4-h starvation. Insets show starved fat bodies expressing mCherry-Atg8, stained for Rab7. (N) The scatter blot depicts the diameters of individual autophagosomes. *** indicates p<0.001, ns is not significant.
Genotypes: (A) wt: Oregon R, mv : mv2 /Df(3L)ED4287, mv + Rescue : P[acman] CH322-23O09 / +; mv2 /Df(3L)ED4287 (C–E) wild type: Oregon R, mv: mv2 /Df(3L)ED4287, mv + R: P[acman] CH322-23O09 / +; mv2 /Df(3L)ED4287 (F–H) wild type: w1118; spr-Gal4 / uas-mCherry-Atg8, mv: w1118; spr-Gal4 / uas-mCherry-Atg8; mv2 /Df(3L)ED4287, mv + R: w1118; spr-Gal4 uas-mCherry-Atg8 / P[acman] CH322-23O09; mv2 / Df(3L)ED4287 (I–K) wild type: w1118; spr-Gal4 / uas-mCherry-GFP-Atg8 mv: w1118; spr-Gal4 / uas-mCherry-GFP-Atg8; mv2 /Df(3L)ED4287 (L–M) wild type: Oregon R, (Inset: w1118; spr-Gal4 / uas-mCherry-Atg8), mv: mv2 /Df(3L)ED4287, (inset: w1118; spr-Gal4 / uas-mCherry-Atg8; mv2 /Df(3L)ED4287).
To further evaluate mv function during autophagy, we used probes (Fig. 7B) that specifically label early or late stages of autophagy in larval fat bodies at 96 hours after egg laying, a stage at which autophagy is reliably induced upon starvation (38, 39). Lysotracker (LTR) was used to label acidic amphisomes and autolysosomes that accumulate in larval fat bodies after 4 hours of starvation. Compared to wild type or mv2/Df(3L)ED4287 with a rescue transgene, mv2/Df(3L)ED4287 fat bodies exhibited reduced levels of starvation-induced lysotracker staining (Fig. 7C–E; p<0.001). This finding suggested that Mauve may be necessary for induction of autophagy or, alternatively, for the maturation of autophagosomes to autolysosomes. To distinguish between these possibilities we examined larvae that express mCherry-Atg8, which labels early and late stages of autophagy (40) (Fig. 7B). Starvation significantly increased mCherry-Atg8 positive structures in mv2/Df(3L)ED4287 fat bodies (Fig. 7F–H; p<0.001). mCherry-Atg8 positive autophagosomes were even increased compared to starved wild-type cells (Fig. 7F). These data suggest that mv is not required for the starvation-induced formation of autophagosomes, but for their normal maturation.
We further explored autophagosome maturation using the mCherry-GFP-Atg8 fusion protein. Because fluorescence from GFP, but not mCherry, is quenched by the low pH of autolysosomes, an increased ratio of red to green fluorescence from this fusion protein is a measure of autophagosome acidification after fusion with lysosomes (40). Determination of these ratios for mCherry-GFP-Atg8-positive structures in starved fat bodies revealed a significant green-shift in mv fat bodies compared to wild type (Fig. 7I–K; p<0.001). This indicates that either Mauve is required for early stages of autophagosome maturation such as the “closure” of the phagophore to an autophagosome or, alternatively, during the subsequent fusion of autophagosomes with lysosomes. Inspection of starved fat bodies by electron microscopy did not reveal an accumulation of early phagophores, but instead showed that autophagosomes were significantly increased in size (Fig. 7L–N; p=0.0013). These large Atg8-labelled autophagosomes were positive for Rab7 (insets in Fig. 7L,M), which is required for the final steps in autophagosome maturation (41). Together, these findings indicate that Mauve is required for a late step in autophagosome maturation. In its absence autophagosomes grow beyond their normal size and appear delayed in their maturation to fully acidified autolysosomes.
Discussion
Mutations that interfere with the function of human LYST are the molecular cause underlying CHS (1–5). Here, we show that phenotypes similar to those in CHS patients result from loss-of-functions mutations in mv, which encodes the closest homolog to LYST in the Drosophila genome. The original identification of the mv gene was based on its effect on eye color, which is changed in mv mutants due to their oversized pigment granules. This mirrors the oculocutaneous albinism of CHS patients caused by oversized and clumped melanosomes (28). Similarly, an important clinical symptom of CHS is the susceptibility to bacterial infections (7), which is also shared by mv mutants. Furthermore, in mv hemocytes we observed an increased tubular morphology of lysosomes, similar to the changes observed in beige macrophages (19). Together these morphological and phenotypic similarities support the notion that aspects of CHS can be modeled in mv flies. The unique set of molecular and genetic tools available in Drosophila suggests that this fly model will be useful for the analysis of the molecular mechanisms by which LYST homologs regulate membrane trafficking.
Recurring bacterial infections are among the most frequent clinical complications observed in CHS patients, but is not well understood how the cell biological defects cause the enhanced susceptibility of patients for infections (7). Defects in phagocytosis have been considered as a possible cause. For example, changes in the phagocytosis of Staphylococcus aureus by leukocytes have been detected in some CHS patients (42), but several other studies found no loss of phagocytic activity in leukocytes from CHS patients (28, 43, 44). To gain further insight into the role of LYST homologs in phagocytosis we used primary hemocytes cultured from Drosophila larvae. These cells have proven to be a useful, genetically tractable model system (45) with markers available for different stages of phagocytosis (29).
Our data indicate that Mauve is not required for the initial phagocytic uptake of bacteria into hemocytes, in agreement with previous work in human or mouse cells lacking LYST function (28, 43, 44). However, we observed that phagocytosed bacteria were amassing in oversized late phagosomes of mv hemocytes. Accumulation of bacteria in phagosomes of CHS leukocytes has previously been observed (42) and primarily attributed to intravacuolar bacterial proliferation. We show that even when heat-killed bacteria were used in phagocytosis assays, many more bacteria populated late phagosomes of mv compared to wild-type hemocytes. Furthermore, this difference was not reflective of an altered mode of initial uptake of the bacteria, as phagosomes positive for the early Avl and the intermediate Rab7 markers (29) exhibit the normal distribution of bacterial content. Interestingly, enhanced homotypic fusion and the resulting formation of “megasomes” is a hallmark of monocyte phagosomes containing Helicobacter pylori. This strategy is thought to contribute to the ability of these bacteria to evade the immune system and persist lifelong in human hosts (46). It is not known by which molecular mechanism H. pylori induces phagosome fusion and thus it is intriguing to speculate that these bacteria may inactivate LYST or an associated factor to promote the formation of megasomes.
Similar to our observations with mv, the LYST homolog LvsB has been proposed to function by preventing homotypic fusions of contractile vacuoles in Dictyostelium (47). Although cells mutant for lvsb displayed significantly enlarged contractile vacuoles, delivery of phagocytosed cargo to these vacuoles was not altered (47). Our observations on phagosome maturation also parallel those on the maturation of secretory lysosomes in CHS cytotoxic T lymphocytes described by Stinchcombe et al. (48), who showed that early steps in the biogenesis of these organelles proceeded indistinguishably from wild-type. Only late in their maturation did secretory lysosomes fuse to form the giant LROs characteristic of CHS.
Therefore, a straightforward explanation for our data is a function of Mauve, and other LYST homologs, late during the maturation of different LROs to either directly or indirectly suppress their homotypic fusion.
An equivalent function of Mauve may also explain the phenotypes we observe during starvation-induced autophagy in mv larval fat bodies. Two key observations are the reduced intensity of lysotracker staining and the increase in the area of mCherry-Atg8 positive structures. The latter is unlikely to reflect simply an increase in expression of mCherry-Atg8, which is driven by Gal4 under control of a heterologous promoter. Furthermore, an increase in size was visible for mCherry-Atg8 and Rab7-positive structures by immunofluorescence and for autophagosomes morphologically identified by electron microcopy (Fig. 7). Instead, these observations are straightforward to reconcile with the notion of Mauve functioning to restrain homotypic fusions of autophagosomes and the resulting increase in autophagosome size and ease of their detection in mv fat body cells.
Other observations do not easily fit the notion of increased fusion events in starved mv fat bodies, however. First, we observed reduced lysotracker staining that typically labels acidic amphisomes and autolysosomes. One explanation for such an observation would be a reduced rate of the fusion of autophagosomes with late endosomes and lysosomes that yield the acidified amphisomes and autolysosomes. Defects in these fusion events have been observed for subunits of the HOPS complex, which is necessary for the fusion of lysosomes with different organelles (e.g. 33, 49). However, none of the experimental systems in which its function has been probed has indicated a requirement of LYST for fusion with lysosomes (see Fig. 6 and refs. 19, 47). Alternatively, after their fusion with lysosomes, the significantly increased size of autophagosomes may result in a dampened or delayed acidification of the resulting autolysosomes thus resulting in a reduced trapping of lysotracker dye in those hybrid organelles.
Such a change in acidification, whether due to the increased size of autophagosomes or due to another effect of Mauve function, could also explain our observation of a green-shift of mCherry-GFP-Atg8 labeled autophagosomes. This chimeric protein has been developed to measure cargo flux by following quenching of the pH-dependent GFP fluorescence as this indicator moves from autophagosomes, which have a pH similar to the cytosol, to acidified lysosomes (40). If mv autolysosomes fail to acidify as efficiently as in wild type, the mCherry-GFP-Atg8 indicator is predicted to exhibit the green-shift we observed. Thus, both the reduced lysotracker staining and green-shift of mCherry-GFP-Atg8 labeled autolysosomes are consistent with an acidification defect that may be a secondary effect of the exceptional size of mv autolysosomes.
A direct inhibitory effect of LYST homologs on limiting membrane fusion is a compelling model to explain the recurring theme of oversized organelles that are observed in the diverse CHS models ranging from CHS patient cells and beige mice (7, 11) to lvsB mutant Dictyostelium (50, 51) and now the Drosophila mv mutant. However, the biochemical mechanism by which LYST homologs execute this function is not clear. One set of possible mechanisms has been suggested based on results of two-hybrid screens that detected interactions between LYST and several proteins involved in regulating membrane fusion events, including subunits of SNARE complexes (52). Whether LYST homologs engage these or other elements of fusion machineries in vivo to suppress inappropriate fusion of LROs remains to be discovered.
The size of organelles is not only determined by the rate of membrane addition by fusions events, but also by the rate at which membranes are removed by fission events. The notion that LYST may contribute to membrane fission from lysosomes was first supported by the observation that LYST overexpression causes a reduction in the size of lysosomes (15). Furthermore, in lvsB mutant Dictyostelium cells, a defect in fission may also contribute to the failure of lysosomes to mature to post-lysosomes that fuse with the plasma membrane and recycle internalized cell surface proteins (50). Defects in fission also emerged as the major difference between beige and wild-type mouse cells when Durchfort and colleagues probed the kinetics with which lysosomes restored their steady-state size after acute disturbances (19).
Our data do not help to distinguish between these two models for the molecular function of LYST homologs. The dynamics of the appearance of oversized late phagosomes strongly points to a role of Mauve in suppressing homotypic fusion or promoting fission late during phagosome maturation. Similarly, oversized autophagosomes and pigment granules may reflect a direct role of Mauve in suppressing inappropriate homotypic fusion during maturation of these organelles in wild type cells. Alternatively, such phenotypes may be an indirect consequence of altered lysosome and LRO physiology. Distinguishing between these possibilities will require a better understanding of the molecular mechanism by which Mauve affects LRO size. The availability of the fly model may open new genetic and molecular approaches towards this goal.
Materials and Methods
Fly work
The screen for identification of eye color mutants has been described (23). A mauve rescue transgene was generated by inserting the P[acman] BAC CH322-23O09 (53) using PhiC31 integrase-mediated insertion at the landing site at 22A (Rainbow transgenics). Other fly strains used in this study were In(3LR)264, which harbors the mv1 allele (25), UAS-mCherry-Atg8, and UAS-mCherry-GFP-Atg8 (40), both of which were expressed in fat bodies using the srp-Gal4 driver (54). Df(3L)ED4287 and the 3L deficiency set for mapping of mauve where obtained from the Bloomington stock center. Adult flies were infected with E. coli as described (29). Lethality of the mv2 chromosome is due to a background mutant, as mv2 / Df(3L)ED4287 hemizygotes are viable, but female sterile as previously reported for mv1 (25).
Molecular biology
Molecular defects in mv1 and mv2 where determined after inverse or regular PCR amplification of the gene from each allele over Df(3L)ED4287. The mv1 allele constitutes one breakpoint of the inversion In(3LR)264 which creates a fusion with transforming acidic coiled-coil protein (TACC, CG9765), connecting codons 1-2772 of Mauve with nucleotide 853 of isoform 4 of TACC, thus deleting the BEACH and WD40 domains of Mauve.
Hemocyte culture
For phagocytosis and immunofluorescence analysis, hemocytes were harvested and cultured as described (29). For functional labeling of lysosomes, 1 mg/ml Alexa-594 Dextran (10kDa, Invitrogen) was added for 20 min and then chased for 2 hours (29). To measure phagocytosis of E. coli, hemocytes were harvested from third instar larvae and adjusted to 105 hemocytes/ml of Schneider's medium supplemented with 10% fetal bovine serum. A suspension of E. coli grown to mid log phase was added to the culture (at ~30 bacteria/hemocyte), followed by 15 min or 60 min incubation at 25°C. Cells were washed at 25°C in 10% fetal bovine serum containing Schneider's Drosophila media supplement with 20 μg/ml gentamicin to remove non-phagoctyosed bacteria. Cells were pelleted and then lysed with PBS containing 0.1% Triton and the number of released bacteria was determined by spreading the cell lysate on LB agar plates, and counting the number of bacterial colonies after incubation overnight at 37°C.
Histology
Adult eyes were imaged using a SteREO DiscoveryV12 microscope. Composite pictures were generated from projections of optical sections at multiple Z positions using CZFocus software. Transmission electron microscopy was performed on an FEI Tecnai G2 Spirit Biotwin (55).
For immunofluorescence detection of bacterial phagocytosis E. coli expressing GFP were grown to mid-log phase, diluted to OD600 = 0.5 and heat killed at 65°C for 20 minutes, pelleted and re-suspended at OD600 = 0.5 in Schneider's S2 cell medium.
Antibodies used for immunofluorescence were anti-Avl at 1:1000 (56), anti-Boss1 at 1:1000 (57), anti-Dl at 1:400 (58), anti-Hook at 1:1000 (59), and anti-Rab7 at 1:250 (60). We followed published protocols for the staining of eye discs (61), phagosomes in isolated hemocytes (29), and for the detection of organelles marked with lysotracker (38), mCherry-Atg8 (39, 62) or mCherry-GFP-Atg8 (40) in fed and starved fat bodies.
Fluorescence images were captured with a 63×, NA 1.4 lens on an inverted confocal microscope (LSM510 Meta; Carl Zeiss, Inc.) at room temperature (21°C). All digital images were imported into Photoshop (Adobe) and adjusted for gain, contrast, and gamma settings.
For the quantification of lysotracker, images were thresholded in ImageJ to detect autolysosomes and their staining intensity was integrated and expressed relative to the average integrated intensity of starved wild-type fat bodies. For the quantification of ATG8-positive structures, images were thresholded in ImageJ to detect autophagosomes and their area, as fraction of the total area, was determined in ImageJ.
For the quantification of bacteria in different stages of phagosomes, hemocytes were harvested and allowed to phagocytose GFP-tagged E. coli as described above. After the indicated times, cells were fixed and stained for Avl, Rab7 or Hook. Images were collected as Z-stacks, to ensure that only bacteria were counted that were contained within phagosomes outlined by the relevant marker.
Biochemistry
For western blots, two adult flies were crushed in 800 μl of lysis buffer (5% SDS, 50 mM Tris/HCl pH 6.8), incubated for two minutes at 95°C, and spun for 10 minutes at 20,000 g to remove cuticle and other debris. 10 μl of cleared lysate were run in each lane. Antibodies were raised in rabbits against the C-terminal 18 amino acids of Mauve and affinity purified with the same peptide (Open Biosystems). Other antibodies for western blots were against actin (JLA20) at 1:2,000 (Developmental Studies Hybridoma Bank), Rab7 at 1:500 (60), Hook at 1:5,000 (59), Dor at 1:2,000 and Avl at 1:10,000 (56) and Cathepsin L at 1:5,000, (R&D Systems). Bound antibodies were detected with far-red labeled secondary antibodies (Li-Cor) and quantified using an Odyssey imaging system (Li-Cor). For quantification, band intensities were normalized to the level of actin in each sample. The pre-stained HiMark molecular weight marker was from Invitrogen.
Levels of red (drosopterin) and brown (ommochromes) eye color pigments were measured as described (63).
Statistical Methods
Statistical significance was determined in Prism (Graph Pad Software Inc) using One-way ANOVA, followed by Tukey's test. Error bars indicate standard deviation (S.D.) in all figures unless indicated otherwise. P-values above 0.05 are considered not significant (ns).
Protein comparisons
Phylogeny was inferred using the Neighbor-Joining method using the Mega5 software (64). For each of the twelve BEACH domain proteins compared we used the C-terminal 2000 amino acids that contained the BEACH domains and other well-conserved features. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (65). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions with less than 95% site coverage were eliminated.
Acknowledgements
We thank Drs. Patrick Dolph, Hugo Bellen, Robin Hiesinger, Thomas Neufeld, and Harald Stenmark for fly lines and antibodies. We thank members of the Krämer lab for helpful discussions and comments to the manuscript. We are grateful to the Bloomington Stock Center for fly lines, and the Developmental Studies Hybridoma Bank for antibodies. We are grateful to Yoshie Sugiura and the Molecular and Cellular Imaging Facility at UT Southwestern for technical assistance. The authors declare no conflict of interest. This work was supported by grants from the National Institutes of Health (EY10199) and the NHARP (010019-0009-2009).
References
- 1.Huizing M, Anikster Y, Gahl WA. Hermansky-Pudlak syndrome and Chediak-Higashi syndrome: disorders of vesicle formation and trafficking. Thromb Haemost. 2001;86:233–245. [PubMed] [Google Scholar]
- 2.Spritz RA. Genetic defects in Chediak-Higashi syndrome and the beige mouse. J Clin Immunol. 1998;18:97–105. doi: 10.1023/a:1023247215374. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA, Detter JC, Blaydes SM, Brandt SJ, Chotai D, Hodgman C, Solari RC, Lovett M, Kingsmore SF. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature. 1996;382:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ, Jr., Perou CM, Boissy RE, Duyk GM, Spritz RA, Moore KJ. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat Genet. 1996;14:307–311. doi: 10.1038/ng1196-307. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Moore KJ, Nagle DL, Misumi DJ, Woolf EA, McGrail SH, Holmgren L, Brody TH, Dussault BJ, Jr., Monroe CA, Duyk GM, Pryor RJ, Li L, Justice MJ, Kaplan J. Identification of the murine beige gene by YAC complementation and positional cloning. Nat Genet. 1996;13:303–308. doi: 10.1038/ng0796-303. [DOI] [PubMed] [Google Scholar]
- 6.Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan J, De Domenico I, Ward DM. Chediak-Higashi syndrome. Curr Opin Hematol. 2008;15:22–29. doi: 10.1097/MOH.0b013e3282f2bcce. [DOI] [PubMed] [Google Scholar]
- 8.Blume RS, Wolff SM. The Chediak-Higashi syndrome: studies in four patients and a review of the literature. Medicine (Baltimore) 1972;51:247–280. [PubMed] [Google Scholar]
- 9.Conley ME, Henle W. Acyclovir in accelerated phase of Chediak-Higashi syndrome. Lancet. 1987;1:212–213. doi: 10.1016/s0140-6736(87)90020-1. [DOI] [PubMed] [Google Scholar]
- 10.Padgett GA, Reiquam CW, Gorham JR, Henson JB, O'Mary CC. Comparative studies of the Chediak-Higashi syndrome. Am J Pathol. 1967;51:553–571. [PMC free article] [PubMed] [Google Scholar]
- 11.Introne W, Boissy RE, Gahl WA. Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome. Mol Genet Metab. 1999;68:283–303. doi: 10.1006/mgme.1999.2927. [DOI] [PubMed] [Google Scholar]
- 12.Virelizier JL, Lagrue A, Durandy A, Arenzana F, Oury C, Griscelli C, Reinert P. Reversal of natural killer defect in a patient with Chediak-Higashi syndrome after bone-marrow transplantation. N Engl J Med. 1982;306:1055–1056. doi: 10.1056/nejm198204293061718. [DOI] [PubMed] [Google Scholar]
- 13.Sheramata W, Kott HS, Cyr DP. The Chediak-Higashi-Steinbrinck syndrome. Presentation of three cases with features resembling spinocerebellar degeneration. Arch Neurol. 1971;25:289–294. doi: 10.1001/archneur.1971.00490040015001. [DOI] [PubMed] [Google Scholar]
- 14.Sung JH, Stadlan EM. Neuropathological changes in Chediak-Higashi disease. J Neuropathol Exp Neurol. 1968;27:156–157. [PubMed] [Google Scholar]
- 15.Perou CM, Leslie JD, Green W, Li L, Ward DM, Kaplan J. The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J Biol Chem. 1997;272:29790–29794. doi: 10.1074/jbc.272.47.29790. [DOI] [PubMed] [Google Scholar]
- 16.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 17.Niesmann K, Breuer D, Brockhaus J, Born G, Wolff I, Reissner C, Kilimann MW, Rohlmann A, Missler M. Dendritic spine formation and synaptic function require neurobeachin. Nat Commun. 2011;2:557. doi: 10.1038/ncomms1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jogl G, Shen Y, Gebauer D, Li J, Wiegmann K, Kashkar H, Kronke M, Tong L. Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain. Embo J. 2002;21:4785–4795. doi: 10.1093/emboj/cdf502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durchfort N, Verhoef S, Vaughn MB, Shrestha R, Adam D, Kaplan J, Ward DM. The Enlarged Lysosomes in beige(j) Cells Result From Decreased Lysosome Fission and Not Increased Lysosome Fusion. Traffic. 2012;13:108–119. doi: 10.1111/j.1600-0854.2011.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kypri E, Schmauch C, Maniak M, De Lozanne A. The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic. 2007;8:774–783. doi: 10.1111/j.1600-0854.2007.00567.x. [DOI] [PubMed] [Google Scholar]
- 21.Ward DM, Shiflett SL, Kaplan J. Chediak-Higashi syndrome: a clinical and molecular view of a rare lysosomal storage disorder. Curr Mol Med. 2002;2:469–477. doi: 10.2174/1566524023362339. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd V, Ramaswami M, Kramer H. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 1998;8:257–259. doi: 10.1016/s0962-8924(98)01270-7. [DOI] [PubMed] [Google Scholar]
- 23.Haberman AS, Akbar MA, Ray S, Kramer H. Drosophila acinus encodes a novel regulator of endocytic and autophagic trafficking. Development. 2010;137:2157–2166. doi: 10.1242/dev.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark WS, Sapp R. Eye color pigment granules in wild-type and mutant Drosophila melanogaster. Canadian Journal of Zoology. 1988;66:1301–1308. [Google Scholar]
- 25.Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. Academic Press; San Diego, CA: 1992. [Google Scholar]
- 26.Burgess A, Mornon JP, de Saint-Basile G, Callebaut I. A concanavalin A-like lectin domain in the CHS1/LYST protein, shared by members of the BEACH family. Bioinformatics. 2009;25:1219–1222. doi: 10.1093/bioinformatics/btp151. [DOI] [PubMed] [Google Scholar]
- 27.Karim MA, Suzuki K, Fukai K, Oh J, Nagle DL, Moore KJ, Barbosa E, Falik-Borenstein T, Filipovich A, Ishida Y, Kivrikko S, Klein C, Kreuz F, Levin A, Miyajima H, et al. Apparent genotype-phenotype correlation in childhood, adolescent, and adult Chediak-Higashi syndrome. Am J Med Genet. 2002;108:16–22. [PubMed] [Google Scholar]
- 28.Zarzour W, Kleta R, Frangoul H, Suwannarat P, Jeong A, Kim SY, Wayne AS, Gunay-Aygun M, White J, Filipovich AH, Gahl WA. Two novel CHS1 (LYST) mutations: clinical correlations in an infant with Chediak-Higashi syndrome. Molecular genetics and metabolism. 2005;85:125–132. doi: 10.1016/j.ymgme.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Akbar MA, Tracy C, Kahr WH, Kramer H. The full-of-bacteria gene is required for phagosome maturation during immune defense in Drosophila. The Journal of Cell Biology. 2011;192:383–390. doi: 10.1083/jcb.201008119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 31.Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23:2501–2514. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dermaut B, Norga KK, Kania A, Verstreken P, Pan H, Zhou Y, Callaerts P, Bellen HJ. Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. The Journal of cell biology. 2005;170:127–139. doi: 10.1083/jcb.200412001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akbar MA, Ray S, Kramer H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol Biol Cell. 2009;20:1705–1714. doi: 10.1091/mbc.E08-03-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 35.Sriram V, Krishnan KS, Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. The Journal of cell biology. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes & development. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cumming RC, Simonsen A, Finley KD. Quantitative analysis of autophagic activity in Drosophila neural tissues by measuring the turnover rates of pathway substrates. Methods Enzymol. 2008;451:639–651. doi: 10.1016/S0076-6879(08)03235-7. [DOI] [PubMed] [Google Scholar]
- 38.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;90:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 42.Bellinati-Pires R, Salgado MM, Joazeiro PP, Carneiro-Sampaio MM. Delayed phagocytosis and bacterial killing in Chediak-Higashi syndrome neutrophils detected by a fluorochrome assay. Ultrastructural aspects. Mem Inst Oswaldo Cruz. 1992;87:575–581. doi: 10.1590/s0074-02761992000400018. [DOI] [PubMed] [Google Scholar]
- 43.Stossel TP, Root RK, Vaughan M. Phagocytosis in chronic granulomatous disease and the Chediak-Higashi syndrome. N Engl J Med. 1972;286:120–123. doi: 10.1056/NEJM197201202860302. [DOI] [PubMed] [Google Scholar]
- 44.Root RK, Rosenthal AS, Balestra DJ. Abnormal bactericidal, metabolic, and lysosomal functions of Chediak-Higashi Syndrome leukocytes. J Clin Invest. 1972;51:649–665. doi: 10.1172/JCI106854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 46.Borlace GN, Keep SJ, Prodoehl MJ, Jones HF, Butler RN, Brooks DA. A role for altered phagosome maturation in the long-term persistence of Helicobacter pylori infection. American journal of physiology Gastrointestinal and liver physiology. 2012;303:G169–179. doi: 10.1152/ajpgi.00320.2011. [DOI] [PubMed] [Google Scholar]
- 47.Harris E, Wang N, Wu Wl WL, Weatherford A, De Lozanne A, Cardelli J. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Molecular biology of the cell. 2002;13:656–669. doi: 10.1091/mbc.01-09-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinchcombe JC, Page LJ, Griffiths GM. Secretory lysosome biogenesis in cytotoxic T lymphocytes from normal and Chediak Higashi syndrome patients. Traffic. 2000;1:435–444. doi: 10.1034/j.1600-0854.2000.010508.x. [DOI] [PubMed] [Google Scholar]
- 49.Lindmo K, Simonsen A, Brech A, Finley K, Rusten TE, Stenmark H. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res. 2006;312:2018–2027. doi: 10.1016/j.yexcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Charette SJ, Cosson P. A LYST/beige homolog is involved in biogenesis of Dictyostelium secretory lysosomes. J Cell Sci. 2007;120:2338–2343. doi: 10.1242/jcs.009001. [DOI] [PubMed] [Google Scholar]
- 51.Cornillon S, Dubois A, Bruckert F, Lefkir Y, Marchetti A, Benghezal M, De Lozanne A, Letourneur F, Cosson P. Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J Cell Sci. 2002;115:737–744. doi: 10.1242/jcs.115.4.737. [DOI] [PubMed] [Google Scholar]
- 52.Tchernev VT, Mansfield TA, Giot L, Kumar AM, Nandabalan K, Li Y, Mishra VS, Detter JC, Rothberg JM, Wallace MR, Southwick FS, Kingsmore SF. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med. 2002;8:56–64. [PMC free article] [PubMed] [Google Scholar]
- 53.Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bataille L, Auge B, Ferjoux G, Haenlin M, Waltzer L. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development. 2005;132:4635–4644. doi: 10.1242/dev.02034. [DOI] [PubMed] [Google Scholar]
- 55.Sunio A, Metcalf AB, Kramer H. Genetic dissection of endocytic trafficking in Drosophila using a horseradish peroxidase-bride of sevenless chimera: hook is required for normal maturation of multivesicular endosomes. Mol Biol Cell. 1999;10:847–859. doi: 10.1091/mbc.10.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- 57.Krämer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- 58.Klueg KM, Parody TR, Muskavitch MA. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol Biol Cell. 1998;9:1709–1723. doi: 10.1091/mbc.9.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kramer H, Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol. 1996;133:1205–1215. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chinchore Y, Mitra A, Dolph PJ. Accumulation of rhodopsin in late endosomes triggers photoreceptor cell degeneration. PLoS Genet. 2009;5:e1000377. doi: 10.1371/journal.pgen.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sevrioukov EA, Walenta JH, Sunio A, Phistry M, Kramer H. Oligomerization of the extracellular domain of Boss enhances its binding to the Sevenless receptor and its antagonistic effect on R7 induction. J Cell Sci. 1998;111:737–747. doi: 10.1242/jcs.111.6.737. [DOI] [PubMed] [Google Scholar]
- 62.Juhasz G, Neufeld TP. Experimental control and characterization of autophagy in Drosophila. Methods Mol Biol. 2008;445:125–133. doi: 10.1007/978-1-59745-157-4_8. [DOI] [PubMed] [Google Scholar]
- 63.Ooi CE, Moreira JE, Dell'Angelica EC, Poy G, Wassarman DA, Bonifacino JS. Altered expression of a novel adaptin leads to defective pigment granule biogenesis in the Drosophila eye color mutant garnet. Embo J. 1997;16:4508–4518. doi: 10.1093/emboj/16.15.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 66.Wolff T, Ready DF. Pattern Formation in the Drosophila retina. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1993. pp. 1277–1325. [Google Scholar]