Abstract
Although centromere function is highly conserved in eukaryotes, centromere sequences are highly variable. Only a few centromeres have been sequenced in higher eukaryotes because of their repetitive nature, thus hindering study of their structure and evolution. Conserved single-copy sequences in pericentromeres (CSCPs) of sorghum and maize were found to be diagnostic characteristics of adjacent centromeres. By analyzing comparative map data and CSCP sequences of sorghum, maize, and rice, the major evolutionary events related to centromere dynamics were discovered for the maize lineage after its divergence from a common ancestor with sorghum. (i) Remnants of ancient CSCP regions were found for the 10 lost ancestral centromeres, indicating that two ancient homeologous chromosome pairs did not contribute any centromeres to the current maize genome, whereas two other pairs contributed both of their centromeres. (ii) Five cases of long-distance, intrachromosome movement of CSCPs were detected in the retained centromeres, with inversion the major process involved. (iii) The 12 major chromosomal rearrangements that led to maize chromosome number reduction from 20 to 10 were uncovered. (iv) In addition to whole chromosome insertion near (but not always into) other centromeres, translocation and fusion were found to be important mechanisms underlying grass chromosome number reduction. (v) Comparison of chromosome structures confirms the polyploid event that led to the tetraploid ancestor of modern maize.
Keywords: centromere inactivation, chromosome number changes, grass genome evolution, paleopolyploidy
Evolutionary changes in plant genome structure occur at numerous levels, from nucleotide substitution to chromosomal rearrangement and polyploidy. Plant centromeres are large structural components that play an essential role in chromosome segregation. The gain, loss, and movement of centromeres are expected to have huge impacts on genome evolution and the independent segregation of genes in the genome. Previous studies have shown that centromeric regions, and their surrounding repeat-rich pericentromeres, are particularly dynamic portions of plant genomes. These centromeric/pericentromeric regions can experience whole-chromosome insertions that cause chromosome fusion (1, 2) and also contain fast-evolving genomic domains that exhibit high rates of unequal recombination (3–5). In flowering plants, different centromeres from an individual genome generally contain the same types of simple tandem repeat arrays (centromeric satellites) and centromeric retrotransposons (CRs) (6, 7). Although the sequences of centromeric satellites vary considerably between species, some CR families are quite conserved across the grass family. However, the locations and copy numbers of the repeats are hypervariable even between haplotypes of the same centromere (4), so it is challenging to investigate centromere evolution by direct comparison of sequences (5).
The nuclear genome of maize has the most complex structure of any yet studied in depth (8). Sharing a common ancestor with sorghum approximately 12 Mya (9), the progenitor of the modern maize genome is believed to have originated from a tetraploid ancestor (10, 11). The interactions of primary mechanisms of genome instability, namely DNA breakage, recombination, and transposition, have driven the rapid and dramatic changes in genome composition and structure in the diploidizing Zea lineage (8, 12). Recently, several studies have attempted to reconstruct the karyotypic history of the Zea lineage by comparing current genetic and/or physical maps (11, 13, 14). Wei et al. (11) and Salse et al. (14) identified the relationships between genomic blocks in maize and those of other grasses. These studies agreed that maize underwent a whole genome duplication resulting from hybridization of two ancestral genomes, each with 10 chromosomes per haploid gamete. However, detailed analysis of the sites and natures of chromosome rearrangements involved in this 20-to-10 process have not been determined. The sequenced Zea mays (15) and Sorghum bicolor (16) genomes now make it possible to uncover the precise chromosomal rearrangements that have led to the current maize genome structure.
In this study, correspondences are built between centromeres of sorghum and maize through conserved single-copy sequences in pericentromeres (CSCPs). These markers, in conjunction with evidence from comparative maps and centromeric repetitive DNA, were also used to identify inactive ancient centromeres in the maize genome, and all of the major rearrangements leading to the current maize chromosome complement were discovered.
Results and Discussion
Comparative Mapping and Its Limitations.
Sorghum-maize, rice-sorghum, and rice-maize comparative maps were constructed by using genomic sequences to investigate the general history of genomic rearrangement in these lineages (SI Appendix, Figs. S1 and S2). From a stable rice genome perspective, the maps indicated that the most pronounced rearrangements in the sorghum-rice comparison were two chromosome insertions (“fusions”) that reduced the chromosome number from 12 to 10: (i) ancestral chromosome 10 inserted into chromosome 3 to form sorghum chromosome 1 and (ii) ancestral chromosome 9 inserted into chromosome 7 to form sorghum chromosome 2 (SI Appendix, Fig. S1). These two insertions also can be seen in the rice-maize comparison. These events occurred either in the shared Andropogonae lineage before the divergence of maize and sorghum from a common ancestor or they were fissions that occurred in the Orzya lineage. Previous work based on genetic (17) and physical (11, 14) map comparisons identified these same relationships between rice and sorghum chromosomes. Consistent with the Devos and Gale (17) genetic maps with a low density of shared markers, our high resolution analysis based on genome sequences indicates that event (ii) is a chromosome insertion of ancestral chromosome 9 into 7, while disagreeing with two more recent studies (11, 14) that reconstructed sorghum chromosome 2 by end-to-end fusion of ancestral chromosomes 9 and 7 (see SI Appendix for details). Aside from these two events, our results indicate that most of the differences between maize and rice and between maize and sorghum are rearrangements that occurred specifically in the Zea lineage, a conclusion that is consistent with previous work based on genetic and physical maps (11, 14, 17).
In this report, we name the 20 sorghum-like maize progenitor chromosomes as 1A, 1B, 2A, and 2B, for example, following the numbering system initiated by Schnable and coworkers (18). This numbering system grouped the 20 progenitor chromosomes into two groups (maize1 and maize2) according to gene loss bias in the two sets of chromosomes: the 10 chromosomes with more retained genes were assigned to maize1, the other 10 to maize2 (18). In our numbering scheme, “A” corresponds to maize1 and “B” to maize2. The chromosome numbers in our studies are assigned according to the current sorghum chromosome number designations.
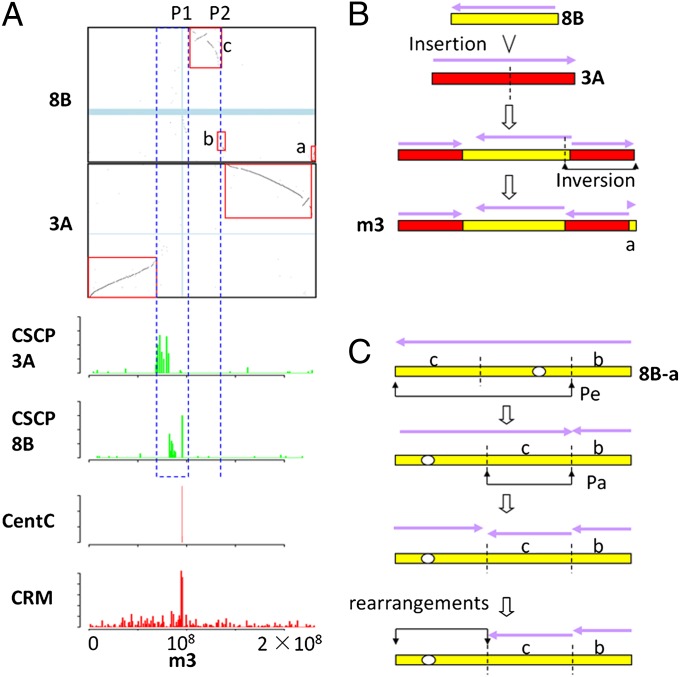
Highly detailed comparative maps, based on whole-gene sets of sorghum and maize (SI Appendix, Fig. S3), provide a rich source of information about the relationships of chromosomes in these two species. The lines within columns of the sorghum-maize comparative map indicate the conserved synteny in the genomes and permit identification of the basic nature of the rearrangements that created the current maize chromosomes from sorghum-like chromosomal ancestors. For example, panel m3-3A (see SI Appendix, Fig. S3 for details of the naming scheme) and m3-8B depict gene synteny blocks, but all other images in column 3 only contain scattered dots caused by paralogous homologies and/or segmental movement of one or a few genes during genome evolution (SI Appendix, Fig. S3). The counterparts of sorghum chromosome 3 were located at 0–68.6 Mb and 140–227.4 Mb on current maize chromosome 3, whereas sorghum chromosome 8 homology was found at 95.7–140 Mb and 227.7–230.6 Mb (Fig. 1A) on the current maize chromosome 3. Hence, maize chromosome 3 apparently originated by the insertion of ancestral chromosome 8B into ancestral chromosome 3A, followed by an inversion from 140.6 Mb to 227.4 Mb (Fig. 1B).
Fig. 1.
An example of the reconstruction of rearrangements that led to chromosome number reduction and centromere movement in the Zea lineage. (A) The x axis and y axis of each comparative map represents maize and sorghum chromosome nucleotide positions, respectively. Maize chromosomes are denoted by “m”. The sorghum chromosome name is replaced by our designation of an ancestral chromosome name. Graphs show dot plots of gene set (Ks ≤ 0.35) comparisons between the indicated chromosomes. Red boxes mark synteny blocks. Below the boxes, the distribution of CSCPs (green) and of centromeric satellite repeats (red) are aligned according to the shared maize chromosome (x axes). Vertical and horizontal light blue bars mark the locations of centromeres. The blue dashed box marks the synteny gap at 68.6–95.7 Mb of m3. (B) Reconstruction of the origin of m3 based on the synteny pattern in the comparative map. (C) Movement of cen8B can be determined from CSCP and CentC/CRM distribution and from the synteny pattern. “8B-a” means chromosome 8B excluding region “a.” “Pa” designates a paracentromeric inversion, and “Pe” indicates a pericentromeric inversion.
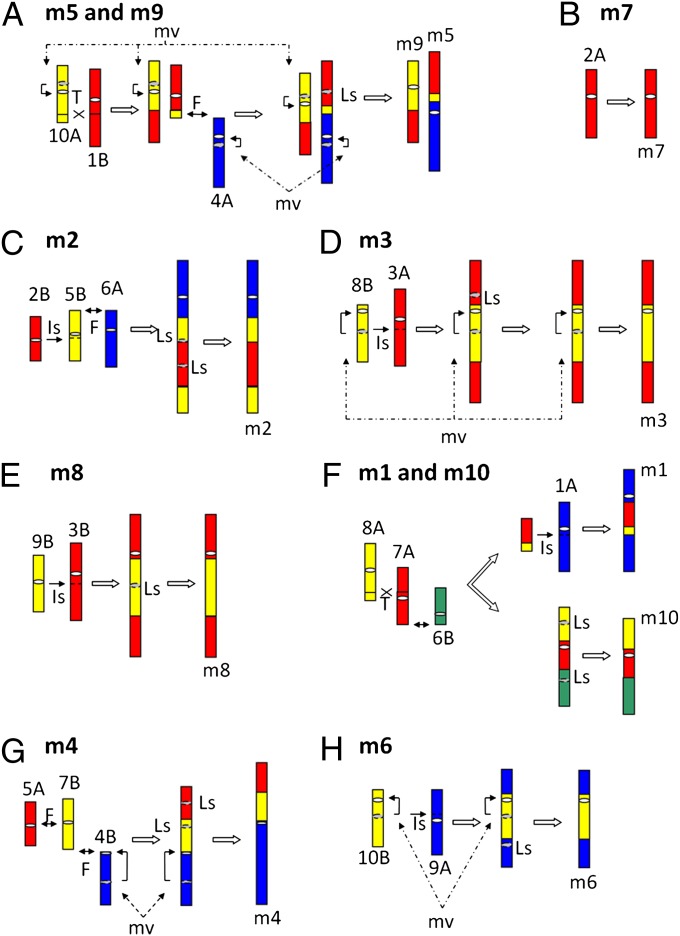
Besides whole-chromosome insertion, interchromosome reciprocal translocation and apparent end-to-end chromosome fusion were also demonstrated by the detailed comparative map. McClintock discovered long ago that regular chromosomes do not usually fuse, because of the properties of telomeres, but low frequencies of telomere fusion with broken chromosome ends have been observed (19). Alternatively, some of these apparent reciprocal translocations may have been caused by ectopic recombination, where the lost terminal (and thus acentric) portions were below the level of detection in this analysis. Devos and Gale (17) demonstrated that maize chromosome 5 contains synteny blocks that are homologous and largely colinear with rice chromosomes 2, 10, 3, and 6. Their analysis also indicated that maize chromosome 9 largely consists of one arm representative of rice chromosome 6 and another arm homologous to rice chromosome 3. As shown in SI Appendix, Figs. S1 and S3, these relationships are given the following designations with our naming system: Maize chromosome 5 consists of ancestral chromosomes 4A, 1B, and 10A, whereas maize chromosome 9 is formed by ancestral chromosomes 1B and 10A. Hence, these differences are rearrangements that occurred in the Zea lineage after its divergence from the Sorghum lineage, as confirmed by the current absence of these rearrangements (and, thus, excellent full-length synteny) between the rice and sorghum chromosomes. The synteny patterns in panels m5-1B, m9-1B, m5-10A, and m9-10A confirm these origins and also indicate the evolutionary events that led to these current maize chromosomes. This study indicates that an apparent reciprocal translocation event occurred between ancestral chromosomes 1B and 10A (SI Appendix, Fig. S4A). One of the products was the progenitor of current maize chromosome 9, whereas the other fused with ancestral chromosome 4A to become the progenitor of maize chromosome 5 (Fig. 2A). From this analysis, it is not possible to determine whether the fusion occurred before or after the translocation.
Fig. 2.
A comprehensive model of Zea chromosome evolution during the descent of maize from a tetraploid progenitor. The diagrams show the simplest model of how 20 sorghum-like ancestral chromosomes rearranged to form the current 10 maize chromosomes through T (translocation), Is (Insertion), F (chromosome fusion), Ls (centromere loss), and mv (centromere/pericentromere movement). Dashed lines on the chromosomes represent target sites of chromosome insertion. Arrows with dashed lines mark events that may have happened at several possible time points and, thus, do not necessary follow the order depicted. Gray dashed ellipses mark the locations of ancient centromeres. For simplicity, this figure is not drawn to scale. Scale at the conserved segments can be seen in SI Appendix, Fig. S3. The current chromosome drawings do have accurate centromere placement, so long and short arms can be discerned.
The above examples indicate three rearrangements that reduced four ancestral chromosomes to two current maize chromosomes. In the full analysis, 12 rearrangement events were identified that led to the 10 chromosomes that occupy a maize haploid nucleus from two sets of 10 sorghum-like chromosomes. These rearrangements consisted of two interchromosome reciprocal translocations, five whole-chromosome insertions, and five apparent end-to-end chromosome fusions (Fig. 2, Table 1, and SI Appendix, Fig. S5).
Table 1.
Chromosome structural rearrangements in the Zea lineage
| Maize chromosome | Ancestral chromosome | Fates of ancient centromeres | Events changing chromosome no. |
| 1 | 1A | Cen1 | Translocation between 7A and 8A; insertion of one recombinant into 1A; fusion of another recombinant with 6B |
| 7A | No cen* | ||
| 8A | No cen* | ||
| 10 | 6B | Lost | |
| 7A | Lost | ||
| 8A | Cen10 | ||
| 2 | 2B | Lost | Insertion of 2B into 5B; fusion with 6A |
| 5B | Lost | ||
| 6A | Cen2 | ||
| 3 | 3A | Lost | Insertion of 8B into 3A |
| 8B | Cen3 | ||
| 4 | 4B | Cen4 | Two fusions among 4B, 5A, and 7B |
| 5A | Lost | ||
| 7B | Lost | ||
| 7 | 2A | Cen7 | |
| 8 | 3B | Cen8 | Insertion of 9B into 3B |
| 9B | Lost | ||
| 5 | 1B | Lost | Translocation between 1B and 10A; fusion of one recombinant with 4A |
| 4A | Cen5 | ||
| 10A | No cen* | ||
| 9 | 1B | No cen* | |
| 10A | Cen9 | ||
| 6 | 9A | Lost | Insertion of 10B into 9A |
| 10B | Cen6 |
*The designation “No cen” indicates that this ancestral fragment contributing to the current maize chromosome did not have any centromere-related sequences or (it is assumed) centromere function in the ancestral chromosome.
These comparative maps provide a framework to infer the events responsible for chromosome number evolution. However, they gave little information about precise rearrangements sites, because these mostly appear to have occurred in gene-poor regions, specifically centromeric/pericentromeric DNA. As can be seen from SI Appendix, Fig. S3, for instance, synteny gaps of 10–30 Mb were found in comparisons involving all maize centromeres/pericentromeres and in some other regions. In some cases, it was possible to identify the ancestral source of a current centromere according to the centromere location in a current sorghum chromosome, but not in all cases. Taking maize chromosome 3 as an example, the synteny gap at 68.6–95.7 Mb (region P1 in Fig. 1A) covered current maize centromere 3. This centromere-containing gap might come from a centromere/pericentromere of ancestral chromosome 3A or ancestral chromosome 8B or both. Even for those regions where comparative maps provided a strong indication of centromere origin, more direct evidence is still desirable to help substantiate any proposed model.
CSCPs.
One of the most interesting characteristics of plant pericentromeric regions is the presence of a few low-copy-number sequences, including functional genes (20, 21). Many of these genes are conserved across species, in both sequence and colinear order. These centromeric/pericentromeric conserved sequences were termed CSCP pairs when found in the centromeric/pericentromeric regions of both maize and sorghum. Because they are found flanking a given centromere/pericentromere, they provide a relatively precise centromere location. A total of 3,144 pairs of CSCPs were identified, covering approximately 832 kb and 820 kb in the sorghum and maize genomes, respectively (SI Appendix, Table S1). CSCP pairs were found to be dispersed across a broad range of centromeric/pericentromeric domains. The lengths of these sequences in the two genomes exhibited highly similar unimodal distributions, with a mean of approximately 250 bp (SI Appendix, Fig. S6). By comparing the coordinates of CSCPs with available annotation data, it was found that CSCPs are mainly genic sequences and unannotated sequences (Table 2). Hence, this analysis suggests that these unannotated sequences, given their conservation, are functional (perhaps as RNA genes or genes that encode very small peptides). This observation is consistent with previous results showing that coding regions of grass genomes exhibit strong conservation for tens of millions of years, whereas intergenic regions rapidly decay and are deleted within just a few million years (22). Because each centromere/pericentromere has numerous distinctive CSCPs, these sequences allow the relatively precise characterization of the relationships between the centromeres/pericentromeres in the maize and sorghum genomes.
Table 2.
The annotation of CSCPs in sorghum and maize
| Genome | Category | Percentage |
| Sorghum | Overlapping gene | 62.1 (69.3) |
| Near gene* | 7.2 | |
| Transposable element | 4.7 | |
| Unannotated | 26 | |
| Maize | Overlapping gene | 69.8 (73.8) |
| Near gene* | 4 | |
| Transposable element | 0 | |
| Unannotated | 26.2 |
Numbers in parentheses are the combined percentage of the overlapping and near gene categories.
*<500 bp up- or downstream of annotated gene models.
CSCPs Reveal Loss and Retention of Ancient Centromeres.
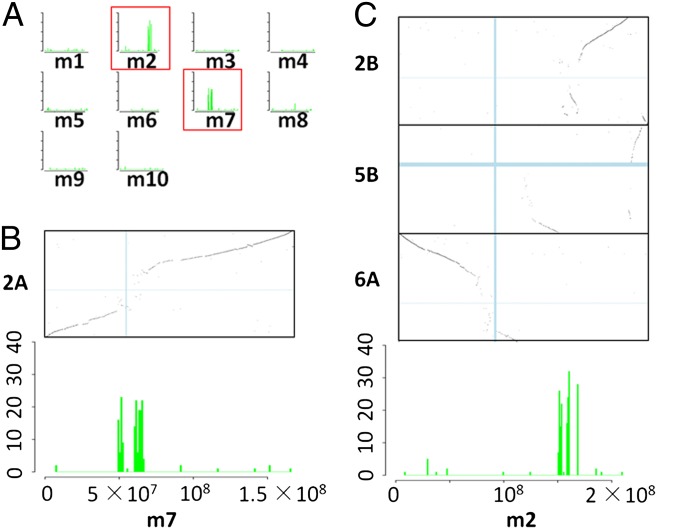
The distribution of CSCPs was investigated relative to maize and sorghum centromeric/pericentromeric regions (SI Appendix, Fig. S7). These analyses indicated the exact correspondence between centromeres/pericentromeres (SI Appendix, Fig. S8), thereby indicating 10 ancestral centromeres that were lost during the descent of maize from its tetraploid ancestor. Fig. 3 depicts an example of CSCP analysis. Using CSCPs in the centromere/pericentromere of sorghum chromosome 2 as query, most related CSCPs in maize were found on chromosomes 2 and 7 (Fig. 3A). According to the comparative maps (SI Appendix, Fig. S3), maize chromosome 7 evolved directly from ancestral chromosome 2A. The CSCP distribution peak (Fig. 3B) in current maize chromosome 7 was located at 49.7–66 Mb, thereby completely overlapping the centromere/pericentromere. This pattern provides direct evidence that the centromere/pericentromere of maize chromosome 7 and that of sorghum chromosome 2 are orthologous.
Fig. 3.
Examples of the detection of current and ancient centromeric/pericentromeric regions in maize. CSCP distribution reveals the locations of current and ancient centromeres. (A) The distribution of maize CSCPs (green) related to sorghum chromosome 2 CSCPs. (B) CSCP distribution in m7 and comparative map m7-2A are aligned according to m7 (x axes). CSCP peaks represent ancestral centromere/pericentromere 2A, which was observed to overlap with current maize cen7 (vertical light blue bar). This pattern indicates that current maize cen7 is the descendant of ancestral centromere cen2A. (C) Distribution of CSCPs in m2 and comparative map m2-2B, m2-5B, and m2-6A are aligned according to m2 (x axes). CSCP peaks represent remnants of centromere/pericentromere 2B. The synteny pattern suggests numerous rearrangements. Current maize cen2 overlaps with centromere/pericentromere 6A (SI Appendix, Fig. S8B). These results indicate the loss of cen2B.
Maize chromosome 2 originated by the insertion of ancestral chromosome 2B into ancestral chromosome 5B and the fusion of ancestral chromosome 6A with this recombinant (Fig. 3C). The synteny gap covering sorghum cen2 suggests that the centromere/pericentromere of ancestral chromosome 2B was located here, whereas the gene synteny pattern at 150–170 Mb indicated extensive intrachromosome rearrangement. The CSCPs distributed in this chromosome as three peaks spanned the region 151–169 Mb, and the peaks just inside the synteny gaps result from additional rearrangements (Fig. 3C). In short, the evidence from the comparative map and from CSCPs were consistent, and both supported the model that the ancient cen2B was lost in association with numerous intrachromosomal rearrangements.
Checking the positions of all CSCP peaks, it was found that (i) the location of all current and ancient centromeres/pericentromeres could be traced by CSCP analysis and (ii) most CSCP peaks corresponded to current and/or ancient centromeres/pericentromeres (SI Appendix, Figs. S7 and S8). Two peaks that were not located in ancient or current centromeres/pericentromeres and some dispersed CSCPs might be artifacts caused by genome sequence assembly errors or may be remnants of more ancient and/or smaller rearrangements that were not analyzed in this study (SI Appendix, Fig. S8). Overall, the corresponding relationships of centromeres/pericentromeres in the maize and sorghum genomes revealed that six of the lost centromeres came from ancestral chromosomes 1B, 2B, 3A, 6B, 7B, and 8A, whereas the other four were lost from ancestral chromosomes 5A, 5B, 9A, and 9B (Table 1 and Fig. 2). These data indicate that both centromeres from the ancestral chromosome pairs 4 and 10 are retained, on the current chromosomes 5 (4A), 4 (4B), 9 (10A), and 6 (10B) of maize. Hence, although maize1 and maize2 homeologues exhibited very different biases in gene loss (“fractionation”) leading to a genome that is now much closer to a diploid in gene content (18), there is no obvious relationship between the chromatids that experienced preferential gene loss and the chromatids that lost or retained their centromeres as functional in the current maize genome.
CSCPs and Centromeric Repetitive DNA Reveal Centromere Movement.
The first molecular-level study of centromere movement in plants was conducted on the sequenced rice cen8 (4). The comparison of centromeric/pericentromeric domains on the orthologous chromosome 8s of two Oryza species that last shared a common ancestor approximately 8 Mya revealed that cen8 of Oryza sativa was physically shifted by a recent inversion within the centromere/pericentromere. Our study of maize chromosomes has found that intrachromosomal inversions caused long-distance movement of centromeres in at least five chromosomes, with “long-distance” defined as rearrangements that extend outside the centromere/pericentromere to include adjacent synteny blocks.
Maize cen3 was found inside the synteny gap expending from 68.6 Mb to 95.7 Mb and another large sorghum-specific gap covers the centromere/pericentromere of chromosome 8 (P1 in Fig. 1A). The simplest explanation based on the comparative map is that maize cen3 originated from the ancestral cen3A and the centromeric/pericentromeric region of ancestral chromosome 8B was deleted. However, sequences in position P1 originated from two centromeres/pericentromeres: those at 68.6–81.3 Mb are from the centromeric/pericentromeric regions of ancient chromosome 3A and those at 82.2 Mb to 96 Mb are from the centromere/pericentromere of ancient chromosome 8B, where current maize cen3 resides. These patterns suggest that two inversions, one paracentric and one pericentric, within chromosome 8B moved the ancestral chromosome 8B centromere/pericentromere (Fig. 1C) from position P2 to position P1 in Fig. 1A. Hence, this result indicates that ancestral cen3A was functionally lost on the current maize chromosome 3, although the CSCPs flanking this now-inactive centromere are still present (Fig. 1A).
As described above, maize chromosome 9 originated from a reciprocal translocation of ancestral chromosomes 1B and 10A, and the other translocation product became current maize chromosome 5 (SI Appendix, Fig. S4A). One inversion encompasses the two synteny gaps indicated by position P1 and P2 in SI Appendix, Fig. S4B, and maize cen9 is found within P2. Extrapolating from the observed gene synteny indicated that gap P1 corresponds to the centromere/pericentromere of ancestral chromosome 10A, so an inversion is predicted to be responsible for the movement of the centromere/pericentromere from P1 to P2. Furthermore, CSCPs related to the centromere/pericentromere of sorghum chromosome 10 were found to exhibit two peaks in the two gaps. These observations indicate that the centromere/pericentromere of ancestral chromosome 10A was part of an inversion (SI Appendix, Fig. S4B) and now provides the centromere for modern maize chromosome 9. Repetitive DNA distribution further supported that this ancestral centromere/pericentromere region was involved in the inversion: Consistent with CSCPs, two peaks of CRMs with similar patterns of CRM insertion dates and family abundance were found at P1 and P2 (SI Appendix, Fig. S4B).
Although repetitive DNA analysis was occasionally useful to provide additional evidence that confirmed the positions of ancient and currently inactive centromere/pericentromeres, given the hyperinstability associated with unequal recombination inside centromeric core regions that has been demonstrated in rice (5), it is not surprising that the nonrepetitive and possibly functionally conserved CSCPs provide a much better indication of the locations of ancient centromeres. As shown in SI Appendix, Fig. S4C, CSCP distribution and the gene synteny pattern related to gaps P1, P2, and P3 indicate that maize cen5 moved from P3 to P1 (reconstruction of this movement is shown in SI Appendix, Fig. S9C), but CentC/CRMs do not show peaks at regions other than the current cen5. In the centromeres/pericentromeres related to the 10 lost centromeres/pericentromeres, only 1 (cen8A corresponding to 11–13Mb of chromosome 10) was found to contain relatively abundant CentC and/or CRMs (SI Appendix, Fig. S8J). The sorghum centromeric satellite pCEN38 had no match in the maize genome by using a BLASTN search at E value of 10−5, consistent with previous results that this satellite is missing from maize (23). Compared with satellites, CR elements are more conserved in grasses (6, 24). However, similar to CRMs, the sorghum CR elements showed no peaks in the majority of ancient centromeres/pericentromeres in maize. Given the removal of unused maize DNA at a half-life of less than 1 My (25, 26), then it seems likely that this centromere loss occurred several million years ago. Similarly, the residual CRMs at 51–54 Mb on chromosome 9 (P1 in SI Appendix, Fig. S4B) indicate that the movement of 10A also happened relatively recently.
The above suggestions that the loss of cen8A and the movement of 10A are recent events are based on the concurrence of centromeric repeats with CSCPs. We note that this estimation depends on the quality of assembly. The occurrence of these centromeric repeats at loci other than current functional centromeres can also be explained by assembly errors. Some of such peaks (54–58 Mb on chromosome 4, 96 Mb and 166 Mb on chromosome 8, and 34–36 Mb on chromosome 10 on chromosome 4) were obvious artificial peaks because these peaks disappeared when mapping CentC/CRMs to the new maize assembly, release 5b.60 (SI Appendix, Fig. S10). Regarding the peaks at 22–26 Mb on chromosome 7 and 78–80 Mb on chromosome 6, although both are at the same location in the original and subsequent assemblies, previous genetic mapping suggests that their locations are artifactual outcomes of genome misassembly (ref. 27 and personal communication with Gernot Presting).
Along with the centromere rearrangements on chromosomes 3, 5 and 9, long-distance movements of current centromeres on chromosomes 4 and 6 were also detected (SI Appendix, Fig. S9). All of the centromere/pericentromere movements could be simply explained by intrachromosomal pericentric or paracentric inversions.
Maize Chromosome Structure Evolution.
Integrating information from comparative maps, CSCPs, and centromeric repetitive DNA, the major events in maize chromosome evolution over the last 10 My were delineated parsimoniously and are described in Table 1 and Fig. 2. All 10 sorghum chromosomes were found to have two counterparts in the maize genome, including four ancestral sorghum-like chromosomes that differed from those in maize by two reciprocal translocations (Fig. 2 A and F). The comparative maps strongly confirm that the modern maize genome has descended from a recent tetraploid ancestor.
Both comparative genetic mapping (17) and segmental chromosome sequence comparison (9) have indicated that the current maize genome arose from a tetraploid whose two ancestral genomes diverged from each other approximately 12 Mya. Physical map studies assumed that the two progenitors had the same number of chromosomes (2n = 20) (11, 14), without strong evidence to support this assumption. At the whole-genome sequence level, our analysis indicates that the reduction of chromosome number from 12 to 10, when comparing rice to sorghum, occurred before the divergence of maize and sorghum lineages, suggesting that an ancestral chromosome number of n = 10 for each Zea genome parent is a reasonable assumption. Furthermore, by showing how the two sets of chromosomes evolved through rearrangements to form the nuclear genome of modern maize, our data strongly support this 10-chromosome-progenitor model.
The known mechanisms underlying chromosome structural evolution consist of translocation, inversion, segmental duplication/insertion/excision, and fusion/fission. Given that intact chromosome ends are usually recalcitrant to any sort of merging with other DNA sequences, it is possible that most or all fusions are actually reciprocal translocations and that nonreciprocal translocations rarely if ever occur. To date, almost all descriptions of whole-chromosome insertions in the grass family are proposed to have occurred into centromeric/pericentromeric domains (1, 2, 13). Luo et al. (1) suggested that such insertions are the dominant mechanism of chromosome number reduction in grasses, providing an obvious mechanism for the loss of one centromere function due to its interruption by a whole chromosome insertion. The results in this study indicate at least one whole chromosome insertion that is near but not inside a centromere/pericentromere (SI Appendix, Fig. S11). These results, like those for apparent terminal chromosome fusions, require an explanation for loss of centromere function that does not involve insertional inactivation. In this same vein, Lysak et al. (28) suggested that multiple mechanisms may play roles in chromosome number evolution of Brassicaceae species, based on their comparative chromosome painting data.
Fusion, translocation, and insertion of ancestral chromosomes will often lead to abnormal chromosomes having two or more centromeres. However, more than one centromere in one chromosome usually causes chromosome breakage because different centromeres may be pulled toward different poles in telophase of mitosis and meiosis (29). However, such chromosomes should be stable if only one centromere remains active (30), or if the centromeres in the dicentric chromosome are physically close and coordinate their movement to one or the other pole (31). Inactivation of redundant centromeres have been found in maize and other grasses (30, 32, 33), and although the details underlying the inactivation are not clear yet, several recent studies have suggested that plant centromere function is an epigenetic process (30, 32–34). The molecular details of such an epigenetic loss, and whether it involves dominance of one centromere over another (as seen for instance with nucleoli in some nascent polyploids; ref. 35), remain unknown. Future experiments should allow characterization of the nature, rate, and direction of local genome change in the current residual centromeres that we have detected in the maize genome and those found by similar studies in other eukaryotic species.
Materials and Methods
Genome Sequences.
The genomic sequence and annotation data for Z. mays (release 4a.53) and S. bicolar (version 1.4) were downloaded from MaizeSequence (www.maizesequence.org/index.html) and DOE-JGI (www.jgi.doe.gov), respectively. The estimated positions of maize centromeres were directly extracted from Wolfgruber et al. (27). Based on the length information of centromere gaps provided by table S4 of Paterson et al. (16), the position of sorghum centromere gaps were detected in the corresponding pseudochromosomes, i.e., cen1: 33.8Mb-38.6Mb; cen2: 34.5Mb-34.6Mb; cen3: 35.3Mb-35.6Mb; cen4: 27.9Mb-32.2Mb; cen5: 38.7Mb-40.9Mb; cen6: 22.1Mb-22.2Mb; cen7: 31.6Mb-35.2Mb; cen8: 19.3Mb-21.8Mb; cen9: 27.5Mb-30.7Mb; and cen10: 29.0Mb-32.1Mb.
Sorghum-Maize Comparative Map.
The sorghum-maize comparative map was constructed from the ordered set of coding gene annotations. Using maize genes as query, their homologous genes in sorghum were defined by running BLASTP at an E value of 10−5. The top 2 matches with Ks values ≤0.35 were plotted because (i) the Ks values of all orthologous gene pairs (mutual best BLASTP hits) between the two genomes exhibited a unimodal distribution and Ks values ≤0.35 were included in the peak (SI Appendix, Fig. S12) and (ii) the large-scale synteny relationships between the two genomes were stable regardless of gene variation in Ks values. Matches with Ks value >0.35 only added scattered points and secondary synteny blocks (synteny blocks caused by paralogous gene pairs), but never changed the positions of primary synteny blocks (those caused by orthologous gene pairs).
Identification of Orthologous Synteny Blocks and Ks Analysis.
Ks values were calculated for each pair of homologous gene pairs to help discriminate orthologous and paralogous synteny blocks. When a genomic block of maize matched more than one region in sorghum, the sorghum region that had the smallest average Ks value to the block was taken as its presumed orthologous region. Ks values of homologous genes were calculated by the yn00 program of the PAML package (36) according to the Nei-Gojobori method (37).
Detection of CSCP Positions in Sorghum and Maize.
We used the 10 centromeric/pericentromeric regions of sorghum as query to BLASTN against the maize genome at an E value of 10−5 and high-scoring segment pairs (HSPs) were extracted. Each HSP is composed of two members–one from sorghum; the other from maize. We then obtained CSCP pairs by selecting only one-to-one matches. A CSCP pair was only designated as such when the member from sorghum came from a centromeric/pericentromeric region and the member from maize was present only as a single hit.
Mining Centromere-Specific Repetitive DNAs.
Centromeric repetitive DNAs were found by two methods. First, all of the CentC units in the assembled maize genome were found by BLASTN, using the CentC set provided in ref. 15 as query. To find all CRMs, LTR_FINDER (38) and LTR_STRUC (39) were implemented to obtain all intact LTR elements in maize centromeric/pericentromeric regions, then these elements were compared with known CRM records provided in ref. 24 to find all intact CRMs. Subsequently, representative sequences of CentCs and CRMs were constructed according to ref. 40. Second, all centromeric repetitive DNAs were found by RepeatMasker (www.repeatmasker.org) using previously identified CentC (15) and CRM (24) sequences. These two approaches yielded identical results.
Supplementary Material
Acknowledgments
We thank Dr. Xiyin Wang of the Plant Genome Mapping Laboratory at the University of Georgia for help in drawing comparative maps. This research was supported by National Science Foundation Grant DBI-0607123, the Georgia Research Alliance and by the Giles Professorship at the University of Georgia (to J.L.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218668109/-/DCSupplemental.
References
- 1.Luo MC, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106(37):15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Initiative TIB. International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7282):763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 3.Lee HR, et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA. 2005;102(33):11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma J, Wing RA, Bennetzen JL, Jackson SA. Evolutionary history and positional shift of a rice centromere. Genetics. 2007;177(2):1217–1220. doi: 10.1534/genetics.107.078709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci USA. 2006;103(2):383–388. doi: 10.1073/pnas.0509810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, Birchler JA, Parrott WA, Dawe RK. A molecular view of plant centromeres. Trends Plant Sci. 2003;8(12):570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Wing RA, Bennetzen JL, Jackson SA. Plant centromere organization: A dynamic structure with conserved functions. Trends Genet. 2007;23(3):134–139. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Bennetzen JL. 2009. Maize Genome Structure and Evolution. Maize Handbook: Genetics and Genomics, eds Bennetzen JL, Hake S (Springer, New York), Vol 2, pp 179–200.
- 9.Swigonová Z, et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14(10A):1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitkus R, Doebley J, Lee M. Comparative genome mapping of sorghum and maize. Genetics. 1992;132(4):1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3(7):e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennetzen JL. Patterns in grass genome evolution. Curr Opin Plant Biol. 2007;10(2):176–181. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Wilson WA, et al. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics. 1999;153(1):453–473. doi: 10.1093/genetics/153.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salse J, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20(1):11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 16.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 17.Devos KM, Gale MD. Comparative genetics in the grasses. Plant Mol Biol. 1997;35(1-2):3–15. [PubMed] [Google Scholar]
- 18.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 2011;108(10):4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26(2):234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copenhaver GP, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286(5449):2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 21.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36(2):138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 22.Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W. Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol. 2004;7(6):732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Zwick MS, et al. Distribution and sequence analysis of the centromere-associated repetitive element CEN38 of Sorghum bicolor (Poaceae) Am J Bot. 2000;87(12):1757–1764. [PubMed] [Google Scholar]
- 24.Sharma A, Presting GG. Centromeric retrotransposon lineages predate the maize/rice divergence and differ in abundance and activity. Mol Genet Genomics. 2008;279(2):133–147. doi: 10.1007/s00438-007-0302-5. [DOI] [PubMed] [Google Scholar]
- 25.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci USA. 2002;99(14):9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14(5):860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfgruber TK, et al. Maize centromere structure and evolution: Sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 2009;5(11):e1000743. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lysak MA, et al. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA. 2006;103(13):5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewin B. Genes VII. New York: Oxford Univ Press; 2000. p. 560. [Google Scholar]
- 30.Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA. 2006;103(9):3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20(3):227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 32.Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102(28):9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb JC, Meyer JM, Birchler JA. A hemicentric inversion in the maize line knobless Tama flint created two sites of centromeric elements and moved the kinetochore-forming region. Chromosoma. 2007;116(3):237–247. doi: 10.1007/s00412-007-0096-6. [DOI] [PubMed] [Google Scholar]
- 34.Han F, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21(7):1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pikaard CS. The epigenetics of nucleolar dominance. Trends Genet. 2000;16(11):495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 37.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 38.Xu Z, Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(Web Server issue):W265-8. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy EM, McDonald JF. LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19(3):362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 40.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.