Abstract
The evolutionary importance of meiosis may not solely be associated with allelic shuffling caused by crossing-over but also have to do with its more immediate effects such as gene conversion. Although estimates of the crossing-over rate are often well resolved, the gene conversion rate is much less clear. In Arabidopsis, for example, next-generation sequencing approaches suggest that the two rates are about the same, which contrasts with indirect measures, these suggesting an excess of gene conversion. Here, we provide analysis of this problem by sequencing 40 F2 Arabidopsis plants and their parents. Small gene conversion tracts, with biased gene conversion content, represent over 90% (probably nearer 99%) of all recombination events. The rate of alteration of protein sequence caused by gene conversion is over 600 times that caused by mutation. Finally, our analysis reveals recombination hot spots and unexpectedly high recombination rates near centromeres. This may be responsible for the previously unexplained pattern of high genetic diversity near Arabidopsis centromeres.
When considering the population genetic impact of recombination, classical theories predominantly concentrate on the impact of allelic shuffling, mediated by crossing-over, and the effect this has on linkage disequilibrium and, in turn, the effect the fate of one allele has on its genomic neighbors (1). However, when programmed double-strand breaks (DSBs) are introduced into chromosomes to initiate meiotic recombination, both crossover (CO) and noncrossover (non-CO) recombination events can occur. Non-CO mechanisms, such as synthesis-dependent strand annealing (SDSA), typically result in gene conversion (2). Gene conversion skews segregation rates of alleles and thus has immediate effects on allele frequencies. Although such direct consequences of recombination are generating more interest (3), relatively little is known about the rates of gene conversion, although its long-term impact on sequence evolution is thought to be profound and phylogenetically widespread (4, 5). Despite this, in the construction of CO maps from linkage disequilibrium data, gene conversion events are typically ignored, being treated as though they were genotyping errors.
Although many studies across diverse taxa have investigated the abundance and distribution of COs during meiosis, few studies have resolved gene conversion rates, largely because such analysis is challenging. Based on tiling microarray data, an average of 90.5 COs and 46.2 non-COs were observed per meiosis in yeast, matching an estimate of 140–170 DSBs per meiosis (6). This contrasts with what is seen in mammals, where gene conversion events considerably outnumber CO events (7).
Investigations in Arabidopsis have resulted in highly consistent estimates as regards CO events with under 10 (3.74–8.3) per meiosis (8–11). Similarly, the most recent report, using next-generation sequencing (NGS), revealed 9 COs per meiosis (9). According to the analyses in humans and yeast, meiotic gene conversion events typically have tract lengths less than 2 kb (12, 13), commonly smaller (9). The small size of gene conversions makes them all but impossible to be detected in nearly all of the prior recombinational analyses for our species, as markers were on average usually hundreds of kilobases or a few megabases apart. NGS approaches can potentially be influential in this arena allowing markers every few hundred base pairs to be used. The one recent NGS analysis suggested there to be as many CO events as gene conversions (9), making Arabidopsis more like yeast. This direct estimate, however, disagrees with indirect inferences. An immunolocalization study (14) suggests in excess of 200 recombination events per meiosis, whereas another (15) suggests a more modest 120–140. Assuming that these events mostly reflect non-CO recombination events, this suggests a considerable excess of gene conversion compared with CO.
Here, we use NGS to provide a robust direct estimate of the rate of gene conversion in Arabidopsis. The above discrepancy between direct and indirect estimates of gene conversion rates may reflect little more than the difficulty of detecting gene conversion events through NGS if sequence quality is poor. Both density and accuracy of sequence are critical to detect a full spectrum of recombination events. With this concern foremost, we sequenced 40 Arabidopsis F2 plants and their parents, Col and Ler, with unique sequencing strategies, incorporating high coverage, replicate independent sequencing, and long paired-end reads in long inserts. These strategies reveal abundant gene conversions in accord with, or possibly in excess of, the prior indirect estimates. We incidentally discover an unexpectedly rich world of recombination in and around centromeres. This may help resolve a prior paradox of Arabidopsis biology, namely why it is that its centromeres are unusual in having high levels of diversity (16, 17). Hot spots for recombination are also identified.
Results
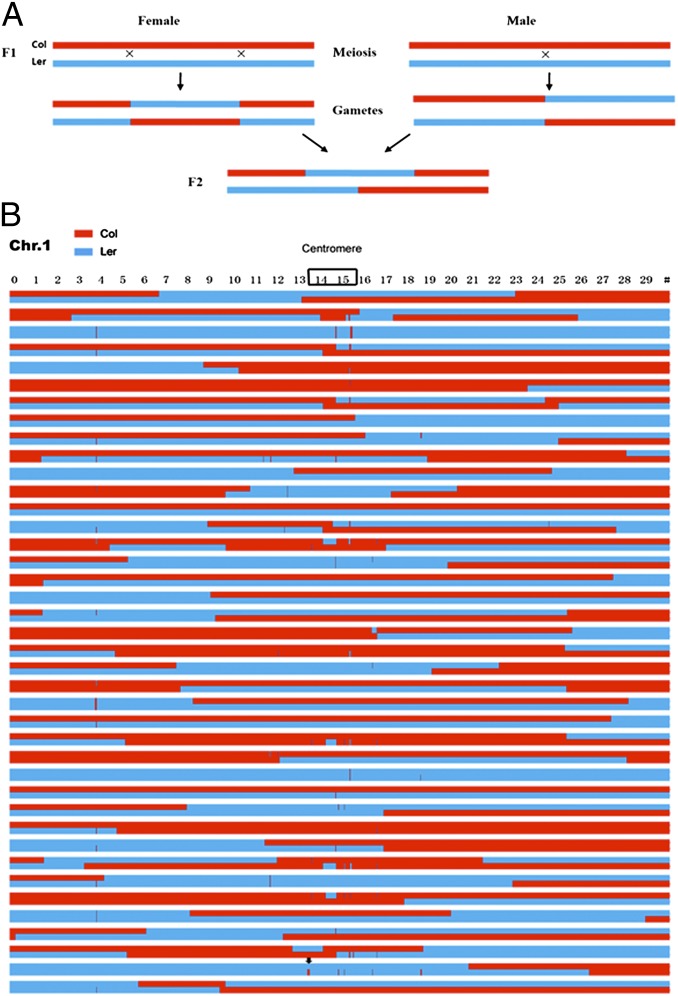
We crossed two inbred strains (Col and Ler) to generate F1 hybrids. These F1s were self-crossed to generate an F2 (Fig. 1A). We infer a recombination event (CO or gene conversion) in the F1 meiosis when in the F2 progeny a run of markers from one strain switches to those from the other (Methods). Were a recombination event to occur with matching breakpoints in both male and female meiosis, our approach could be misleading. However, with abundant (>300,000) and well-scattered markers and sparse recombination events (e.g., <1,000) in a diploid plant, assuming a random occurrence model, the probability of two events occurring between the same two markers is roughly equal to 1,000/300,0002 = 1.1 × 10−8. We can thus identify almost every recombinational event (examples in Fig. 1B, compendium in SI Appendix, Fig. S1).
Fig. 1.
Schemed patterns (A) and examples (B) of COs (>10 kb) in F2 plants. The red and blue bars represent the chromosomes of Col and Ler, respectively. One recombination of 40 F2 chromosome pairs (chromosome 1) is shown as an example from male and female meiosis. The CO highlighted with an arrow is shown in further detail in SI Appendix, Fig. S5.
Identification of Accurate Markers.
To guarantee accuracy of markers, multiple stringent strategies were used. First, 33 of 40 F2s, from Col/Ler F1 heterozygotes, were independently library-constructed and sequenced two to three times with high coverage (2 × 21.2× or 3 × 32.3×) and long paired-end reads (2 × 100 bp in 500-bp inserts; SI Appendix, Tables S1 and S2). The other seven F2s were sequenced only once (SI Appendix, Table S1). With high sequence quality and the addition of a second or third round, SNP calling and recombination block identification are expected to be almost 100% accurate (SI Appendix, Tables S3 and S4 and Dataset S1). Second, each of four Ler and three Col plants was sequenced two to three times with high coverage (up to 3 × 31.5× per plant). These sequences, combined with each parental genome being sequenced with 824× coverage in the 40 F2 plants (SI Appendix, Table S1), allow us to construct two accurate parental genomes, based on the well-sequenced Col. Third, three software packages (Novoalign, Shore, and Stampy) were used to independently call SNPs against the reference. These filters resulted in a total of 586,231 SNPs identified by at least two of the software packages and 41,743 1- to 3-bp Ler deletions, these being identified by Shore alone. As a negative control for recombination events, we sequenced a mixture of Col and Ler DNA (SI Appendix, Table S4).
To be yet more stringent, we further refined a gold standard set. Only markers (i) identified by all three softwares (deletions only by Shore), (ii) observed in >80% of 75 (=7 + 31 × 2 + 2 × 3) sequenced F2 genomes in corresponding heterozygous regions, and (iii) concordant with the 461,070 identified previously (18), were considered to be adequate. This set comprises 373,614 SNPs and 41,743 1- to 3-bp Ler deletions, a total of 415,357 markers, representing on average 1 every 289 bp. These gold standard markers were used to detect COs and gene conversions. Over 3,000 of these markers were sampled for PCR amplification followed by Sanger sequencing confirming >99.7% of them (Dataset S1). From the initial 586,231 SNPs, the 212,617 eliminated in the second round of processing to generate the 373,614 gold standard SNPs, the less reliable SNPs, were used to corroborate the identified gene conversions.
Estimates of CO Rates Accord with Lower Resolution Studies.
The blocks of runs of markers from a given genome are expected to come in a variety of lengths dependent on the manner of their creation. Spans >10 kb we assume to be the outcome of crossing-over. To enable comparison with prior studies (8–10, 12, 13), we group these events into long (>500 kb) and short spans (10–500 kb). The average number of long blocks are limited (8.4 or 3.6 cM/Mb per genome; Table 1) and consistent with prior reports for which a 500-kb interval was about the limit of resolution (8–11). The position of every long CO is unique (SI Appendix, Fig. S1 A–E). The number (28.8 per genome) of small blocks identified is about four times greater than the long ones.
Table 1.
Numbers and types of recombination events per meiosis
| Track length | ≥500 kb | ≥100 kb | ≥10 kb | 2–10 kb | 20 bp to 2 kb | 2–19 bp |
| Chr 1 | 2.3 | 3.2 | 7.1 | 6.2 | 62.1 | 84.4 |
| Chr 2 | 1.6 | 3.3 | 8.8 | 6.9 | 48.7 | 33.2 |
| Chr 3 | 1.7 | 2.6 | 9.4 | 6.7 | 52.9 | 69.1 |
| Chr 4 | 1.1 | 1.6 | 5.6 | 4.2 | 39.1 | 54.1 |
| Chr 5 | 1.7 | 2 | 6.3 | 6.2 | 62.5 | 80.1 |
| Total | 8.4 | 12.7 | 37.2 | 30.2 | 265.3 | 320.9 |
Forty, 31, and 33 F2 plants were used for the analyses of ≥10-kb, 20-bp to 10-kb, and 2- to 19-bp tracks, respectively.
With 20 or more markers (77.4, on average), every CO can be clearly identified. However, a gene conversion event may involve relatively few markers. False-positive gene conversion events are thus a possibility and the estimate of the number of gene conversion events will be sensitive to the stringency of analysis. We start by attempting to define lower bounds for the total number of gene conversion events (Table 2 and SI Appendix, Tables S5 and S6).
Table 2.
Numbers and types of gene conversions in 31 F2 plants
| Sample | Direction of the gene conversion |
Total | ||
| Col (%)→Het | Het→C or L | Ler (%)→Het | ||
| 5 | 106 (15.1) | 150 | 54 (14.0) | 310 |
| 6 | 27 (15.3) | 60 | 149 (22.9) | 236 |
| 7 | 62 (22.6) | 115 | 76 (18.7) | 253 |
| 14 | 61 (17.1) | 248 | 94 (18.5) | 403 |
| c42 | 47 (39.8) | 27 | 46 (7.6) | 120 |
| c45 | 77 (24.9) | 49 | 63 (11.5) | 189 |
| c48 | 69 (23.7) | 40 | 92 (15.4) | 201 |
| c51 | 70 (18.7) | 56 | 173 (23.6) | 299 |
| c52 | 91 (24.4) | 55 | 82 (14.2) | 228 |
| c57 | 6 (7.3) | 53 | 81 (12.0) | 140 |
| c61 | 80 (56.3) | 50 | 16 (3.8) | 146 |
| c62 | 134 (39.7) | 36 | 259 (13.7) | 429 |
| c63 | 80 (21.7) | 54 | 405 (44.1) | 539 |
| c64 | 61 (44.1) | 40 | 38 (7.9) | 139 |
| c65 | 58 (19.2) | 67 | 82 (19.5) | 207 |
| c66 | 45 (12.1) | 134 | 34 (5.3) | 213 |
| c73 | 75 (21.8) | 154 | 15 (4.0) | 244 |
| c81 | 31 (19.8) | 30 | 175 (42.3) | 236 |
| c82 | 106 (22.5) | 45 | 236 (45.2) | 387 |
| c83 | 81 (17.1) | 69 | 49 (12.7) | 199 |
| c84 | 9 (7.4) | 59 | 159 (27.0) | 227 |
| c85 | 141 (68.8) | 22 | 44 (5.8) | 207 |
| c87 | 93 (27.6) | 44 | 222 (29.9) | 359 |
| c88 | 59 (14.4) | 33 | 86 (20.8) | 178 |
| c89 | 76 (19.6) | 122 | 146 (24.7) | 344 |
| c90 | 82 (31.1) | 47 | 133 (16.8) | 262 |
| c91 | 70 (21.1) | 29 | 470 (49.9) | 569 |
| c92 | 55 (15.4) | 51 | 170 (30.6) | 276 |
| c93 | 157 (47.1) | 39 | 42 (9.5) | 238 |
| c94 | 34 (15.2) | 20 | 185 (54.2) | 239 |
| c95 | 97 (30.2) | 35 | 75 (24.7) | 207 |
| Average | 72.3 | 65.6 | 127.5 | 265.3 |
| ABP | 0.252 | 0.540 | 0.208 | 1 |
| S/C | 63/63 | 42/20 | 21/20 | 126/104 |
| Estimated | 286.9 | 146.3 | 583.8 | — |
The gene conversion track length, the farthest distance between two or more markers, is 20–2,000 bp. Het, Col (C), and Ler (L) represent heterogeneous, Col- and Ler-homologous backgrounds, and their average background proportions (ABPs) are 0.540, 0.252, and 0.208, respectively. The individual proportions of Col and Ler background are shown in parentheses. The first number in S/C means the numbers of sampled gene conversions for PCR amplifications, and the second denotes the true gene conversions verified by Sanger sequencing. The estimated gene conversions are equal to the following: Average gene conversions × Confirmed/Sampled gene conversions/Proportion of corresponding background in each column.
Estimating Lower Bounds for the Number of Gene Conversion Events.
To define a stringent set we require that each gene conversion tract must be between 20 bp and 2 kb and contain two or more of the gold standard markers, each of which must, in addition, be identified in all independently sequenced genomes for the same F2 individual (the seven F2 genomes sequenced only once were excluded from this analysis). In addition, we require that these gene conversion tracts must be consistent with the slightly less reliable SNPs (Methods). Even with these severe filters, we identified 265.3 gene conversion tracts per meiosis (Table 2). The analysis of the two negative controls showed that the error rates for gene conversion identification to be 0–5% (SI Appendix, Table S4), consistent with PCR results in Table 2. Our analysis hence largely confirms the prior higher estimates based on immunolocalization data (14) and suggests that at a minimum 90% of recombination events are resolved as gene conversions.
Confirmation of Lower Bounds.
To confirm these estimates we sequenced 126 regions containing gene conversion events from two randomly sampled F2 plants (c52 and c66). This confirmed 100% of them in Col-homozygous regions, an average of 72.3 per genome in these regions. As the individual proportions of Col and Ler background (Table 2) are significantly correlated with their gene conversion numbers (r = 0.645 and 0.797; P < 0.01, respectively), we can extrapolate the numbers seen for the Col homozygous background based on a random occurrence model. Given that the Col-homozygous regions account for 25.2% of the sequence, 286.9 gene conversions (=72.3/0.252) are predicted genome-wide (Table 2). Notably, the two samples (c94 and c95 in Table 2 and SI Appendix, Table S1), sequenced thrice with ∼100× coverage per plant, produce similar gene conversion numbers when the same extrapolation form Col homozygous regions is used (288.5 = 65.5/0.227).
Given difficulties in unambiguously assigning sequence to repeat rich areas of the genome, we checked that our estimates are not repeat-associated mapping artifacts. We determined for all gene conversions identified in Table 2 whether they are in repeat or nonrepeat regions. The majority are fully or partially in nonrepeat regions, suggesting that they are not products of repeat-associated mapping errors (SI Appendix, Tables S6 and S7). Even if we assume that only gene conversion events in nonrepeated regions can be identified unambiguously, we find that there are ∼161 gene conversion events. As nonrepeat regions account for 77.56% of the genome, this suggests there to be 207 gene conversion events per genome (assuming a random-occurrence model).
Inclusion of Very Short and Long Tracts Modestly Increases Gene Conversion Number Estimates.
The above analyses ignore possible very short (<20 bp) and long gene conversion (>2 kb) tracts. Applying the strict requirements above, but requiring the tracts to be 20 bp to 10 kb long (rather than 20 bp to 2 kb), we detect a further 30 tracts that likely reflect gene conversion events (Table 1). If we add in very small (2-19 bp) but well-described gene conversion tracts (SI Appendix, Table S5), an additional 73 gene conversion events are estimated. For this analysis, we again require two reliable markers in the span and consistency on adding in intervening but less reliable SNPs. We detected 18.5 small gene conversion events in Col homozygous background, 100% of which were confirmed by Sanger sequencing. Based on the equation in Table 2, 73 gene conversions per genome are expected. Including these longer and shorter events thus increases the estimate 35% to 390 gene conversions per meiosis (∼287 + 30 + 73).
Estimating Upper Bounds for the Number of Gene Conversion Events.
In the above analyses, we ignore the possible gene conversion tracts supported by few markers. These are harder to confidently estimate. A total of 2,377 possible incidences of gene conversion per meiosis was identified in a set where either one in the first round of sequencing or one or more markers in second round are required to define a gene conversion event (SI Appendix, Table S8). This provides one upper estimate.
An alternative estimate can be deduced via extrapolation. When focusing on the number of markers involved in each CO and gene conversion, there is a smooth distribution relating the number of occurrences to the number of markers involved (SI Appendix, Fig. S2). Assuming that the gene conversion tracts with four or more markers are real, we can fit a frequency curve that, by extrapolation, can give a prediction for the gene conversion tracts with one to three markers as ∼2,800 per plant. Together with the 207 tracts with more than three markers (used to define the frequency distribution), there may thus be >3,000 gene conversion events, i.e., 80 times more gene conversion events than COs. When gene conversions in repeat regions are discarded from the set from which extrapolation is based, there are still >2,000 gene conversion events genome-wide, meaning 50 times more gene conversion events than COs.
Assuming an upper bound of around 3,000 gene conversion events, we conclude that between 90 and 99% of recombination events are gene conversion events. The higher estimate is somewhat in excess of the most extreme prior indirect estimate. For estimates of the mean tract length and proportion of the genome that are part of such tracts (19), see SI Appendix, Table S9. In terms of the total span of DNA recombined, the impact of COs is greater than gene conversion, even using our upper estimate, as the span of each CO event is so long.
Evidence of Abundant Pericentromeric Recombination.
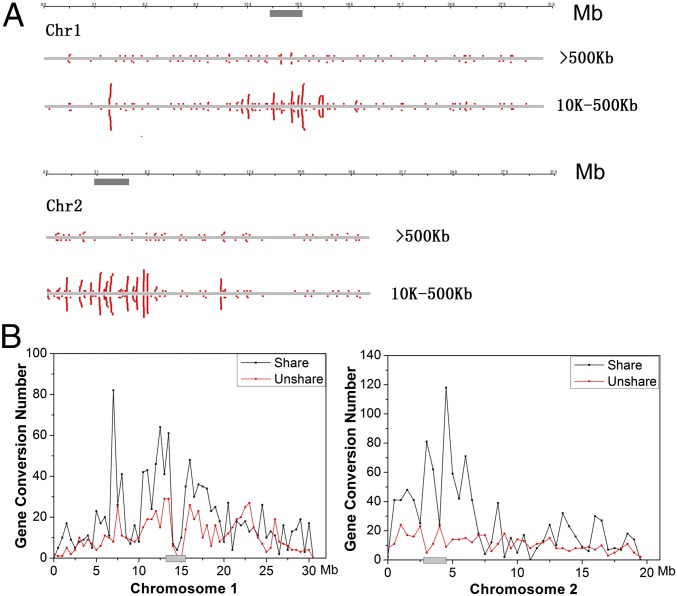
The large (>500 kb) and small (10-500 kb) CO blocks have quite different patterns of distribution on chromosomes. The long COs distribute almost randomly along chromosomes (Fig. 2A and SI Appendix, Figs. S1 A–E and S3A) their density, hence correlating with chromosomal length (P = 0.038; SI Appendix, Fig. S4A). This is as classically reported for coarsely resolved CO maps. Unexpectedly, of the small CO spans, 72.6% occur in pericentromeric regions (within 2 Mb; Figs. 1B and 2A and SI Appendix, Fig. S3B), classically considered to be recombinational deserts (20, 21).
Fig. 2.
Distribution of (A) COs and (B) gene conversions on chromosomes 1 and 2. The long (>500 kb) and small COs (10-500 kb) are showed separately. Shared and nonshared gene conversions (20 bp to 10 kb) are demonstrated by different lines. The centromere regions were represented as gray bars. Only those gene conversions in Table 2 were used.
Can we be confident that this unexpected result is not a build or sequencing artifact? In negative controls (SI Appendix, Table S4), no block with 10–500 kb was identified. These pericentromeric blocks thus cannot be explained by sequencing errors. Moreover, all or some of the markers in 71% of 10- to 500-kb blocks are contained in nonrepeat regions, which can be mapped without ambiguity. Furthermore, many of 10- to 500-kb blocks, including those in pericentromeric regions, are located within a pure Col or Ler background as shown in Fig. 1B, indicating that they are unlikely to be from mapping errors. In fact, the high mapping quality can be clearly displayed by the long paired-ends reads in 500-bp-long inserts at the border of CO transitions (SI Appendix, Fig. S5), which further indicates that they are neither artifacts nor rearrangements in Ler compared with Col. In addition, we examined by PCR followed by Sanger sequencing the markers for nine small COs, seven of which are pericentromeric. We confirm all of the markers for all of the nine blocks.
Using the most robustly defined gene conversion events, we observe a similar excess of gene conversion events near centromeres. To verify this, we used a single-stranded cloning strategy. This can resolve the sequence for each sister chromosome at the same region. Ten candidate gene conversions putatively near pericentromeric regions were analyzed by this strategy, with all 10 being confirmed as residing in proximity to centromeres (SI Appendix, Table S10).
Recombination Events Are in Domains of High Diversity and Low Gene Density.
Breakpoints of both COs and gene conversion events are often located in regions with high diversity (SI Appendix, Fig. S6). As regards gene conversion events, this is in part a definitional necessity (DSB followed by SDSA may occur in homozygous stretches but in the absence of variable markers involves no allelic gene conversion). However, through simulation we observe that there is more diversity than expected by chance, allowing for the definitional bias (SI Appendix, Fig. S6). Given the long span of CO events allied with our very high marker density, we can be confident at having identified all COs and thus have an unbiased assay of the location of breakpoints. The excess diversity in the vicinity of COs is thus neither easily accountable in terms of ascertainment bias nor definitional necessity.
Gene conversion and CO events both tend to be more prevalent in gene-poor regions and, as commonly reported (8, 10), tend to be intergenic (SI Appendix, Fig. S7). However, on average, 16.1 gene conversions, containing 32.3 nonsynonymous SNPs and 17.2 Ler deletions, are detected per meiosis. The rate of nonsynonymous conversion is ∼675 times higher than the nonsynonymous mutation rate reported in laboratory conditions [a total of 11 detected in five individuals for 30 generations (22)]. With 991 markers in intergenic DNA converted per meiosis, the effects on sequence affecting gene expression may be more profound. These numbers, however, apply to our F1 plants. For highly selfing wild populations of Arabidopsis thaliana (A. thaliana), the number of heterozygous sites in any given meiosis is likely to be low and hence the actual conversion rate also much lower.
Shared Recombination Events.
About 67% of gene conversions and 89.4% of the small COs are shared in two or more individuals and their track/spans lengths are on average 402 bp and 36 kb (98.5% of them <100 kb), respectively. This rate of sharing is significantly greater than the expected value (1.1 × 10−8) in a random-occurrence model. A repeatable CO span (26 kb long) is shown at 3- to 4-Mb position with a frequency of 18 of 80 chromosomes in Fig. 1B. Each of the shared gene conversion or small length CO loci is seen in 6.3 and 8.6% of individuals, respectively. In total, 59.3% of shared gene conversions/COs were located or partly located within nonrepeat regions, suggesting that the majority of them could not be repeat-associated mapping artifacts. Based on the trees for shared gene conversions or COs (SI Appendix, Fig. S8), every individual has a different set, suggesting independent occurrence. As expected given the location of small-sized CO events, the shared events are common around centromeres (Fig. 2 and SI Appendix, Fig. S9) and roughly coincident with the recombination hot spots reported recently (15). Sanger sequencing of PCR products, from unique pairs of primers in the Arabidopsis genome, confirmed that 17 putatively shared loci sampled (eight gene conversions or nine small COs) were indeed shared among different plants. SI Appendix, Fig. S10, shows two confirmed examples where the PCR and sequencing results from multiple pairs of primers, including ones crossing the border of the breakpoint, were consistently positive among many plants.
MEME analysis reveals that conserved motifs with several copies per sequence are often located in 300-bp regions surrounding a gene conversion (or CO) or within the converted sequences (SI Appendix, Table S11), which could be associated with the frequent occurrence of gene conversions or small COs at specific positions among individuals (23).
Distorted Segregation Ratios and Gene Conversion-Content–Biased Gene Conversions.
As with prior reports from interline crosses (8, 10), we find strong evidence for distorted segregation ratios (SI Appendix, Fig. S11), with three of the five chromosomes significantly different from Mendelian expectations (SI Appendix, Fig. S11). They are either Col-dominant (chromosomes 2 and 5) or Ler-dominant (chromosome 4). The underlying cause is unclear but may reflect meiotic drive, genes under selection for early viability (10) or genetic incompatibility (8).
Meiotic gene conversion is thought to be biased toward nucleotides G and C in the great majority of eukaryotes (4). If u is the number of AT→gene conversion SNPs per A or T and v the number of gene conversion→AT SNPs per G or C, then we can consider the ratio u/v (SI Appendix, Fig. S12). For the gene conversion tracts (20 bp to 2 kb), this is 1.22, which is significantly greater than the null (unity) (from randomization: P < 0.0001), consistent with biased gene conversion increasing the frequency of AT→gene conversion SNPs as seen in other taxa (4). By contrast, the 10- to 500-kb spans (u/v = 0.96) are no different (P = 0.18) from unity, suggesting that these spans might be COs rather than gene conversion events.
Discussion
Our analysis supports early indirect approximations to the number of gene conversions events, strongly rejecting the one prior NGS-based estimate, which suggested equal numbers of CO and gene conversion events (9). This rejection is rendered yet more robust by our conservative assumption that 10- to 500-kb events are COs, not gene conversions. The cause of these midsized blocks is, however, yet to be fully resolved. A few similarly sized recombination events were observed previously (9) and assumed to reflect an interference-free mode of crossing-over. Consistent with this, we find no evidence for interference for small COs (SI Appendix, Fig. S13). The same is true for the shared COs, which are almost only present in <100-kb spans. Similarly, we find no evidence for distorted G and C content, consistent with an absence of gene conversion. In principle, however, tracts a little over 10 kb may reflect gene conversions created by helicase-mediated resolution of double Holliday junctions (14, 24) (rather than through the SDSA mechanism), tract lengths for which are unknown or could be mitotic conversion events (13, 25). However, mitotic conversion rates are typically 104–105 lower than meiotic conversion (25), making the latter unlikely. Unfortunately, with segregation distortion common across the chromosomes, we cannot perform a segregation analysis and so cannot definitively conclude that these are CO events.
The commonality of gene conversion events has implications for population genetic inferences. Regular gene conversion events are likely to reduce the structure of linkage disequilibrium (26) and will have a strong effect on the distribution of nucleotide polymorphisms. Adding gene conversion to genetic models will make them more appropriate for the inference of population history from linkage disequilibrium (26). CO rate inference from linkage disequilibrium data are robust to moderate gene conversion rates (treating it as genotyping errors) and would have little or no problem were the recent (9) lower end estimate correct. With our new more extreme estimates, caution is advisable in application of such methods.
Why So Much Gene Conversion?
The abundant gene conversions in Arabidopsis suggest that plants are more like mammals (7) than yeast, the latter having relatively common crossing-over compared with gene conversion (6). This difference between taxa we suggest may reflect differences in repeat content, as repetitive sequences are a source of genomic instability during meiosis (27), owing to nonallelic homologous recombination (28). Compared with COs, non-CO [e.g., via SDSA (2)], which yields the most gene conversions, poses the least genomic threat among mechanisms that repair DSBs (29). Analysis of repeat poor genomes of multicellular species (e.g., Oikopleura) will be informative.
Mechanistically, the above suggestion may require that organisms scan the local sequence environment to determine how to resolve a DSB. In a related vein, Dooner (30) has suggested that the local diversity levels may mediate the choice between gene conversion and crossing-over following a DSB. However, we find no significant differences in the distribution of SNPs and indels around gene conversions and COs (t test, P = 0.42). Although this fails to support Dooner’s conjecture, our evidence is not decisive as we consider only small indels (≤3 bp), whereas Dooner’s hypothesis concerns larger indels in addition.
Pericentromeric Recombination May Explain Prior Unusual Observations.
The apparently high-frequency pericentromeric recombination events may explain some prior data. First, our data could explain why the CO frequencies were seen, in lower resolution maps, to increase adjacent to the centromeres (8). When examining the distribution of all COs (Fig. 1B), more frequent recombination can be identified between two arms of chromosome 1, due to the denser distribution of small COs around the centromere. Given that many of these COs are double COs, they are unable to cause a recombination between the two arms (or part of the arms). When excluding those COs, however, the potential frequency can still be as high as about 1/4 on this chromosome, suggesting a partly free exchange between two arms.
Second, the finding of abundant recombination, including crossing-over, near centromeres helps resolve a prior paradoxical result. In many taxa, there is a positive correlation between intrapopulation diversity and genomically local recombination rates (3). In A. thaliana (16, 17) and the outbred A. lyrata (31), there is, unusually, high sequence diversity near centromeres. This has been considered contrary to classical theory as centromeres were assumed to have low CO rates, and thus prone to weak Hill–Robertson interference reducing diversity (31). Reduction of such interference under high CO rates may not, however, be the full explanation. A. thaliana is a near obligate selfer, and as such CO should have relatively little effect on Hill–Robertson-mediated diversity. Moreover, that we observe that the breakpoints of both COs and gene conversion events are often located in regions with high diversity (SI Appendix, Fig. S6) suggests instead that either (i) there is a preference for DSBs to occur in domains of high polymorphism or (ii) DSBs promote polymorphism. The latter may be mediated by a coupling between DSB repair and the mutation process (32) or reflect the activity of biased gene conversion, which can increase load at gene conversion hot spots even if inbreeding levels are very high (33). Biased gene conversion is supported by SNP analysis (see above) and from the finding that in the 100-bp sequences around the tracts of gene conversion, the gene conversion content (0.368) in shared loci, with two or more gene conversions among different individuals, is higher than that at unshared (0.345) or randomly sampled loci (0.348; P = 14 × 10−10). Recent evidence (22) supports a higher mutation rate in proximity to centromeres.
Materials and Methods
Plant Material.
The F1 seeds were obtained from female Col individuals crossed with Ler male plants, both of which were either from a single Col or Ler seed. Thousands of F2 plants were grown from seeds obtained from selfed F1 plants. Finally, 40 F2, 4 Ler, and 3 Col plants were used to extract DNA by the cetyltrimethyl ammonium bromide (CTAB) method for genome sequencing.
Resequencing.
Paired-end sequencing libraries with insert size of 500 bp were constructed for each plant according to the manufacturer’s instructions. Then 2 × 100-bp paired-end reads were generated on Illumina HiSEq 2000. Finally, 47 plants (SI Appendix, Table S1) were resequenced with >21.2× coverage and high quality for each by BGI-Shenzhen. To increase the accuracy, 2 parental plants, 33 F2 plants (7 of the 40 were without enough DNA) were independently constructed for libraries and sequenced two to three times with the same coverage (3 × 29.8× for each of parent, 3 × 32.3× per plant for 2 F2, and 2 × 21.2× for the remaining 31 F2; SI Appendix, Table S1). For the other five parental plants, an equal amount of DNA from any two of the five parental plants was mixed to perform resequencing, e.g., Sample_C1C2, C2L1, L1L2, L2L3, and L3C1 (C stands for Col and L for Ler). The two mixed Col-Ler samples (i.e., C2L1 and L3C1) were used as negative controls. The other parental samples were used as references for SNP calling.
Marker Identification.
The Col genome (TAIR9) was downloaded from TAIR Web site (ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes). The assembly Ler scaffolds, SNPs and indels were downloaded from 1001 Genomes (http://1001genomes.org/projects/assemblies.html). To identify reliable markers, SHORE (22), Novoalign (www.novocraft.com), and Stampy (34) were used to independently call SNPs against the reference Col genome for our sequenced parental plants.
Identification of CO Events.
Based on 415,357 markers, regions along chromosome pairs were converted into blocks of genotype H (heterozygosity), C (Col homozygosity), and L (Ler homozygosity) by searching for the genotype switching points, e.g., H→C, H→L, L→H, or L→C. The <500-kb blocks were first ignored to construct the genotype background (for details, see SI Appendix, Fig. S14).
Detection of Gene Conversions.
More strict criteria were applied to detect gene conversions. A quality evaluation for two or three rounds of independent sequencings was carried out for each chromosome pair. We split the genome into 300-kb nonoverlapping windows and for each window calculated the proportion of markers that were in disagreement between the independent sequencings. Only those windows with less than 5% disagreement were used. Thirty-one of 33 F2 plants have almost no regions with >5% difference. For the remaining two plants, the first round of sequencing had variable quality, whereas the second had much higher quality. We retained only domains with high quality in both (<5% discordance).
Only the 415,357 gold standard markers were used for gene conversion detection. After identification of candidate gene conversions, however, the other SNPs were used to distinguish whether the form of a gene conversion tract remains the same or changes to a different one when adding more SNPs between the gold standard markers. For example, a block was identified as pure Col (C-C-C) by three markers and two more non-gold standard SNPs were inserted between these markers. Imagine that the pattern was changed into C-H-C-H-C or C-H-H-C-C. In either case, the original candidate gene conversion was discarded. In the latter instance, a new gene conversion (C-C) was, however, accepted. A large number of gene conversion candidates were randomly sampled for checking via PCR and Sanger sequencing. Each pair of primers in all PCRs was unique in the well-sequenced Arabidopsis genome.
Estimation of Sequence Quality.
To ensure the quality of our sequences and genome assembly, we used numerous methods. First, two mixtures of Col and Ler DNAs were separately sequenced as negative controls (SI Appendix, Table S4) to identify false-positive recombination events (SI Appendix, Tables S2–S4). Second, there are multiple rounds of independent sequencing for 33 F2 plants, which can be used to estimate the numbers and types of gene conversions independently between the two sets of sequences for the same plant. The differences between multiple independent sequencings provide an estimate of the range of errors (SI Appendix, Table S3-2).
We also compared our results with those from Lu et al. (9). The genome sequences of four progeny from one meiosis provided by them were used to (i) compare the sequencing strategy and quality with this study (SI Appendix, Table S2) and (ii) identify the possible gene conversion events in the samples of Lu et al. (9) by the criteria used in this study for different sets of gene conversions (SI Appendix, Table S3-1). Note, although there were eight progeny in total, only four could be used.
Controlling for Repeats.
The repeat and nonrepeat sequences were grouped by both annotated transposable elements and RepeatMasker regions for Arabidopsis (www.repeatmasker.org) and calculated separately for recombination events to avoid the possible assembly problems in repeat regions.
Checking Borders.
We analyzed in detail the transition borders of 10–500k COs in sample c94 and c95 (each with 97× coverage). To directly see whether a CO might be incorrectly mapped or built (SI Appendix, Fig. S5), we considered instances where there are two or more markers within 400 bp of each other but with >100–200 bp between them. These were analyzed in detail for all of the paired reads (2 × 100 bp) in long inserts (500 bp). In total, there are only 12 COs with enough markers, enough space, and an unambiguous border to do such analysis [based on the software inGAP-sv (http://ingap.sourceforge.net)]. By this analysis, we confirmed all 12 COs. For a full list of recombination events, see Dataset S2.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) Grants 30930049, 30930008, and 31071062, and NSFC of Jiangsu Province Grant BK2011015 (to D.T. or J.-Q.C.). L.D.H. is a Royal Society Wolfson Research Merit Award holder.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/sra (accession no. SRA060909).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211827110/-/DCSupplemental.
References
- 1.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8(3):269–294. [PubMed] [Google Scholar]
- 2.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106(1):47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 3.Webster MT, Hurst LD. Direct and indirect consequences of meiotic recombination: Implications for genome evolution. Trends Genet. 2012;28(3):101–109. doi: 10.1016/j.tig.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Pessia E, et al. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol Evol. 2012;4(7):675–682. doi: 10.1093/gbe/evs052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innan H. A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics. 2002;161(2):865–872. doi: 10.1093/genetics/161.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454(7203):479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paigen K, Petkov P. Mammalian recombination hot spots: Properties, control and evolution. Nat Rev Genet. 2010;11(3):221–233. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomé PA, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 2012;108(4):447–455. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu P, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 2012;22(3):508–518. doi: 10.1101/gr.127522.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7(11):e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyota M, Matsuda K, Kakutani T, Terao Morita M, Tasaka M. Developmental changes in crossover frequency in Arabidopsis. Plant J. 2011;65(4):589–599. doi: 10.1111/j.1365-313X.2010.04440.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. Gene conversion: Mechanisms, evolution and human disease. Nat Rev Genet. 2007;8(10):762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 13.Judd SR, Petes TD. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics. 1988;118(3):401–410. doi: 10.1093/genetics/118.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chelysheva L, et al. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3(5):e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21(17):2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borevitz JO, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(29):12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317(5836):338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 18.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci USA. 2011;108(25):10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansai SPKT, Innan H. The rate and tract length of gene conversion between duplicated genes. Genes. 2011;2:313–331. doi: 10.3390/genes2020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talbert PB, Henikoff S. Centromeres convert but don’t cross. PLoS Biol. 2010;8(3):e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326(5956):1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 22.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18(12):2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44(2):212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3α and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008;4(12):e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee PS, et al. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5(3):e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall JD. Close look at gene conversion hot spots. Nat Genet. 2004;36(2):114–115. doi: 10.1038/ng0204-114. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11(3):182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vader G, et al. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477(7362):115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329(5987):82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dooner HK. Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell. 2002;14(5):1173–1183. doi: 10.1105/tpc.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawabe A, Forrest A, Wright SI, Charlesworth D. High DNA sequence diversity in pericentromeric genes of the plant Arabidopsis lyrata. Genetics. 2008;179(2):985–995. doi: 10.1534/genetics.107.085282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MA. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci USA. 2008;105(29):10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glémin S. Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression. Genetics. 2010;185(3):939–959. doi: 10.1534/genetics.110.116368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21(6):936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.