Abstract
Poor outcomes in diabetic patients are observed across a range of human tumors, suggesting that cancer cells develop unique characteristics under diabetic conditions. Cancer cells exposed to hyperglycemic insults acquire permanent aggressive traits of tumor growth, even after a return to euglycemic conditions. Comparative genome-wide mapping of hyperglycemia-specific open chromatin regions and concomitant mRNA expression profiling revealed that the neuregulin-1 gene, encoding an established endogenous ligand for the HER3 receptor, is activated through a putative distal enhancer. Our findings highlight the targeted inhibition of NRG1-HER3 pathways as a potential target for the treatment breast cancer patients with associated diabetes.
Keywords: PANIC-ATTAC, FAIRE-seq, metabolic imprinting
Diabetes (DM) is a risk factor for enhanced cancer progression (1, 2). DM is characterized by transient or persistent hyperglycemia due to inadequate insulin action and production (type 2 DM) or persistent insufficient insulin production from pancreatic β-cells (type 1 DM) (2). Because type 2 DM is more prevalent, most epidemiological studies were done with type 2 DM patients; in this context, it is challenging to distinguish between direct effects of hyperglycemia versus other confounding factors, such as hyperinsulinemia and chronic inflammation, frequently associated with these profound metabolic changes. Nevertheless, several groups of investigators have suggested that hyperglycemia per se is an independent risk factor for various types of human cancers, although the underlying mechanisms are still elusive (3, 4).
Results and Discussion
Cancer Cells with a History of Hyperglycemia Grow Faster in Euglycemic Hosts Compared with Naïve Cells.
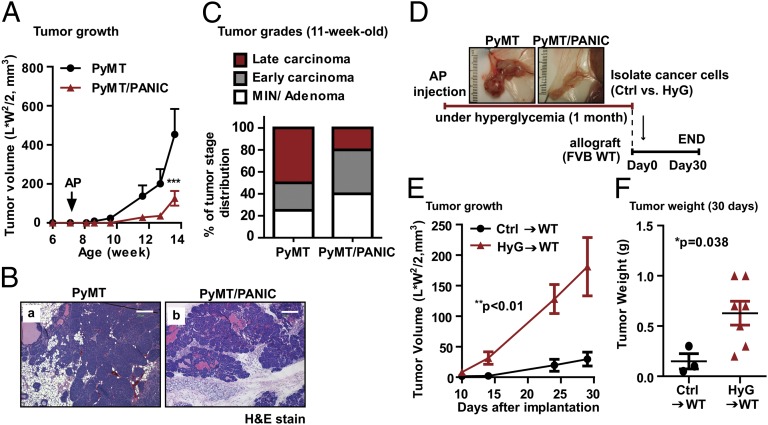
To study the specific contributions of hyperglycemia to breast cancer, we used a PyMT/PANIC [PANIC-ATTAC mice (Pancreatic Islet Cell Apoptosis Through Triggered Activation of Caspase-8) (5) in the background of MMTV-PyMT mice (6)] mouse model to induce hyperglycemia in mammary tumors. Hyperglycemia was brought about by inducible apoptosis of pancreatic β-cells after dimerizer injection (Fig. S1 A and B). The dimerizer was maintained throughout the initial tumor progression to avoid regeneration of β-cells. The resulting decrease of insulin levels (Fig. S1C) triggers hyperglycemia, effectively mimicking type 1 DM or late stages of type 2 DM. Because of the prevailing catabolic conditions in the absence of insulin, body weights of mice were significantly decreased with a reduction of body fat mass in PyMT/PANIC mice compared with PyMT mice (Fig. S1 D and E). Furthermore, mammary tumor growth was significantly attenuated in PyMT/PANIC mice compared with PyMT mice (Fig. 1A); thus, tumor progression from mammary intraepithelial neoplasias (“MINs”) to late carcinoma was significantly delayed as judged by whole mount staining (Fig. S1F) and histological analysis using H&E staining (Fig. 1 B and C), despite dramatically up-regulated availability of circulating glucose. Consistent with our observations, several type 1 DM rodent models have been reported to show attenuated tumor growth (7, 8). This reduced tumor growth is likely due to the lack of insulin, a factor with intrinsic mitogenic properties (9). Of interest, gene expression profiles in tumor tissues analyzed by cDNA microarray indicated that oncogenic pathways driven by components such as HER1 (also known as EGF-R), Ras, NF-κB, and antiapoptotic signals were highly up-regulated under these hyperglycemic conditions (Fig. S2 A and B) despite the fact that tumor growth was attenuated in PyMT/PANIC mice.
Fig. 1.
Hyperglycemic memory in mammary cancer cells result in the malignant progression. (A) Tumor growth for PyMT and PyMT/PANIC mice. Hyperglycemia was induced by dimerizer injection (AP20187; 0.5 μg/g per day for 5 d by i.p administration) in 7-wk-old PyMT/PANIC mice. Tumor growth was assessed by caliper measurements twice a week. n = 6 per group. ***P < 0.001 vs. PyMT by two-way ANOVA. (B) Representative images of H&E stain for PyMT and PyMT/PANIC. (Scale bars: 200 μm.) (C) Tumor grading was determined by using H&E-stained slides for 11-wk-old PyMT and PyMT/PANIC mice. n = 10 per group. (D) Schematic for the implantation of cancer cells taken from PyMT and PyMT/PANIC into isogenic wild-type hosts. (E and F) Tumor growth for hyperglycemia experienced cancer cells (HyG)-bearing mice compared with control cells (Ctrl)-bearing mice. Either HyG or Ctrl cancer cells (0.5 × 106 per mouse) were implanted into the mammary adipose tissues of wild-type mice and monitored tumor growth by caliper measurement (E). Data represent mean ± SEM (n = 5 per group). **P < 0.01 vs. Ctrl by two-way ANOVA. Tumor weights (F) were determined at 30 d after implantation. *P = 0.038 vs. Ctrl by unpaired Student’s t test.
We wanted to ask the question whether cancer cells retain a memory of these unusual metabolic conditions even after they are returned to the euglycemic state. These long-lasting effects imprinted upon transient exposure to excessively high carbohydrate levels are referred as “hyperglycemic memory.” Notably, previous work established that hyperglycemia under different conditions can actively trigger epigenetic modifications resulting in long-lasting changes in gene expression (10). To test this hypothesis in the context of our mammary tumor setting, cancer cells isolated from hyperglycemic conditions (“HyG”) in the PyMT/PANIC mice and control cancer cells (“Ctrl”) isolated from euglycemic PyMT mice were implanted into isogenic and euglycemic wild-type mice and tumor growth was monitored (Fig. 1D). Despite the fact that HyG cells grew more slowly in the original hyperglycemic conditions, they display a significantly more aggressive growth behavior in the new euglycemic setting relative to the Ctrl-cells (Fig. 1 E and F). This more aggressive growth pattern indicates that the hyperglycemic environment epigenetically imprinted oncogenic signals in cancer cells and resulted in a malignant progression even after prolonged presence in euglycemic wild-type hosts.
Identification of Neuregulin-1 as a Key Factor Mediating Hyperglycemic Memory Effects in Cancer Cells.
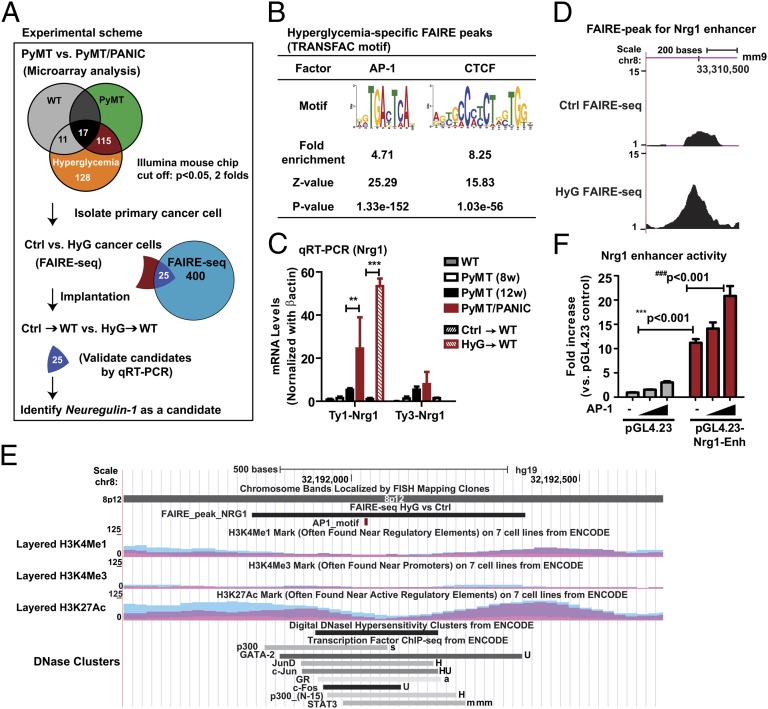
To identify epigenetic changes to the genome of breast cancer cells that are induced by hyperglycemia, we used an integrated genomic approach (Fig. 2A). We surmised that changes affecting gene regulation would result in conformation changes of chromatin in genomic regions harboring regulatory elements, e.g., promoters, enhancers, and insulators. Therefore, we used formaldehyde-assisted isolation of regulatory elements followed by deep sequencing (FAIRE-seq) (11) to map global changes in open chromatin conformation in HyG cells compared with Ctrl cells. We identified 400 FAIRE regions that have a more open chromatin conformation in HyG cells. De novo motif prediction and motif enrichment analysis (Fig. 2B) revealed that binding motifs for both AP-1 transcription factors and CCCTC-binding factor (CTCF; a key insulator protein essential for the maintenance of transcriptional units in metazoan genomes; ref. 12), are very significantly enriched in hyperglycemia-associated FAIRE peaks. This finding suggests that AP-1 and CTCF may be key players in maintaining a chromatin conformation of gene regulatory elements that confer the hyperglycemic memory of cancer cells (Fig. 2B). Genes associated with hyperglycemia-specific FAIRE peaks were cross-referenced with cDNA microarray data to identify epigenetically imprinted candidate genes (Fig. 2A). We filtered genes modulated by hyperglycemia per se in nontransformed cells identified by microarray analysis of mammary gland tissues from PANIC-ATTAC mice and wild-type mice (Fig. 2A). This integrated approach identified 25 candidate genes (Fig. S3A) that were validated by quantitative RT-PCR (qRT-PCR) of cDNA generated from tumors of PyMT/PANIC and PyMT mice (Fig. S3B), and tumors from HyG and Ctrl xenograft tumors (Fig. S3C).
Fig. 2.
Identification of Nrg1 as a candidate gene engaged in the hyperglycemic memory effects in mammary cancer cells. (A) Integrated genomic approach that identified Nrg1. Microarray analysis was performed for RNA from tumors of PyMT/PANIC and PyMT mice as well as from mammary adipose tissues of PANIC-ATTAC and wild-type mice (Fig. S2). The subset of genes specifically modulated by hyperglycemia in tumors was retained for further analysis. Global analysis of open chromatin conformation was performed by FAIRE-seq with primary cancer cells isolated from tumor tissues of PyMT (Ctrl) and PyMT/PANIC (HyG). Twenty-five candidate genes were selected by cross-comparison of microarray data and FAIRE-seq data. qRT-PCR was used to validate the expression of candidate genes in tumors of PyMT/PANIC and PyMT and of HyG and Ctrl xenograft tumors (Fig. S3). (B) Enrichment analysis for AP-1 and CTCF motifs in hyperglycemia-specific FAIRE peaks. (C) qRT-PCR for Nrg1 mRNA levels. β-actin was used as a control. Data represent mean ± SEM **P < 0.01, ***P < 0.001 by two-way ANOVA. n = 6–8 per group. (D) The FAIRE peak in the HyG chromatin indicates a hyperglycemia-specific open chromatin region that may represent an active enhancer for Nrg1. The position of the FAIRE peak in the mouse genome (assembly mm9) in the University of California Santa Cruz (UCSC) Genome Browser is indicated. The y axis represents the number of the normalized FAIRE read frequencies. (E) The homologous region of the putative Nrg1 enhancer in the human genome. The position of the region in the human genome (assembly Hg19) in the UCSC Genome Browser is indicated. The conserved putative AP-1 motif is highlighted in red. Binding regions for multiple transcription factors, the levels of histone modifications and DNaseI hypersensitivity as obtained from ENCODE are shown. (F) Luciferase reporter assay in CHO cells. Enhancer element was inserted in the reporter vector containing a minimal promoter (pGL4.23), and assay was determined at 2 d after transfection in the presence or absence of the AP-1 transcription factor. Data represent mean ± SEM three independent experiments were performed. ***P < 0.001 and ###P < 0.001 by two-way ANOVA.
Based on literature analysis of the known functions of the 25 candidate genes, we selected Nrg1 as a putative key regulator of tumor cell growth for further analysis (Fig. 2A). Nrg1 encodes a ligand for HER3/ErbB3, which is a member of the epidermal growth factor family (EGFR) of receptor tyrosine kinases (13). Overexpression of HER3 has been reported for multiple common solid tumors, such as prostate, ovarian, and breast cancers (14, 15). Nrg1 mRNA levels were markedly increased (20-fold) in tumors that originated from hyperglycemic conditions (Fig. 2C). Nrg1 overexpression was maintained when these tumors were transplanted to euglycemic mice (Fig. 2C and Fig. S3 B and C). FAIRE-seq analysis identified a region 190 kb 5′ to the transcription start site of Nrg1 gene that had a more open chromatin conformation in the HyG cells (Fig. 2D). A putative AP-1 binding motif was identified in this FAIRE region (Fig. S4A), suggesting that this region may harbor a distal enhancer of Nrg1. Importantly, the FAIRE region is evolutionarily conserved based on a DNA sequence similarity of 82% between mouse and human, which is very high for an intergenic DNA region (Fig. S4A). The homologous fragment in the human genome is located 213 kb 5′ to the transcription start site of the NRG1 gene. Using ENCODE data, we found that this region is bound by AP-1 transcription factors such as Jun and Fos in multiple human cell lines, has high DNaseI hypersensitivity, and is flanked in multiple cell lines by nucleosomes with high levels of histone modifications (H3K4 monomethylation and H3K27 acetylation) that are characteristic for enhancers (Fig. 2E). Collectively, these findings suggest that the HyG FAIRE peak associated with Nrg1 identified an AP-1 binding enhancer with conserved function in human cells.
To test whether the identified Nrg1 FAIRE region has enhancer activity, we cloned the entire region into a luciferase reporter construct with a minimal-promoter (pGL4.23) and analyzed luciferase expression from this construct at different levels of ectopic AP-1 expression in CHO cells. The promoter activity of the luciferase construct was markedly increased by this enhancer element and was further activated by the presence of the AP-1 transcription factor (Fig. 2F).
Because the NRG1 gene generates various isoforms through alternative splicing and differential use of promoter sites (16), we explored which isoforms are primarily affected by hyperglycemic memory effects. NRG1 isoforms are subdivided into three groups, based on their N-terminal domain structure: heregulin (HRG, type I), glial growth factor (GGF, type II), and sensory and motor neuron derived factor (SMDF, type III) (16). The mRNA levels of type I Nrg1 in tumor tissues were slightly increased over the course of tumor progression but markedly augmented by hyperglycemic exposure in PyMT mice in comparison with type III Nrg1 (Fig. 2C). This increase was sustained in HyG tumors compared with Ctrl tumors implanted into wild-type mice (Fig. 2C). Furthermore, RNA-seq analysis with RNAs isolated from HyG tumors compared with Ctrl tumors confirmed that mRNA sequences encoding type I Nrg1 are specifically increased in HyG tumors (Fig. S5A). Homologous sequences found in human Nrg1 gene (Fig. S5B) encode several proneuregulin isoforms, which include HRG-β1b, HRG-β1c, HRG-β1d, HRG-β1, HRG-β2, and HRG-α (Fig. S5C). Therefore, we focused our analysis in the context of hyperglycemic imprinting events in breast cancer on type I Nrg1, also called heregulin (17), and will refer to it as Nrg1. NRG1 has been implicated in various types of human cancers because of its potent effects on cell proliferation, survival, invasion, and angiogenesis (18). In addition, it directly binds to HER3, which leads to the formation of heterodimers with other HER receptors (mostly HER2, also known as ErbB2 or Neu). These heterodimers further activate the downstream signaling pathways, such as PI3K, ERK, and NF-κB (19). As such, the functional involvement of NRG1 in breast cancer progression has been widely demonstrated, although its key regulatory aspects remain to be elucidated; when and how NRG1 activation takes place remains unclear. In this context, our findings strongly suggest that hyperglycemia may be a critical driving force to permanently lead to an activation of the NRG1-HER signaling axis in malignant tumor progression.
Neuregulin-1 Plays a Crucial Role as a Mediator of Hyperglycemic Memory Effects in Cancer Cells.
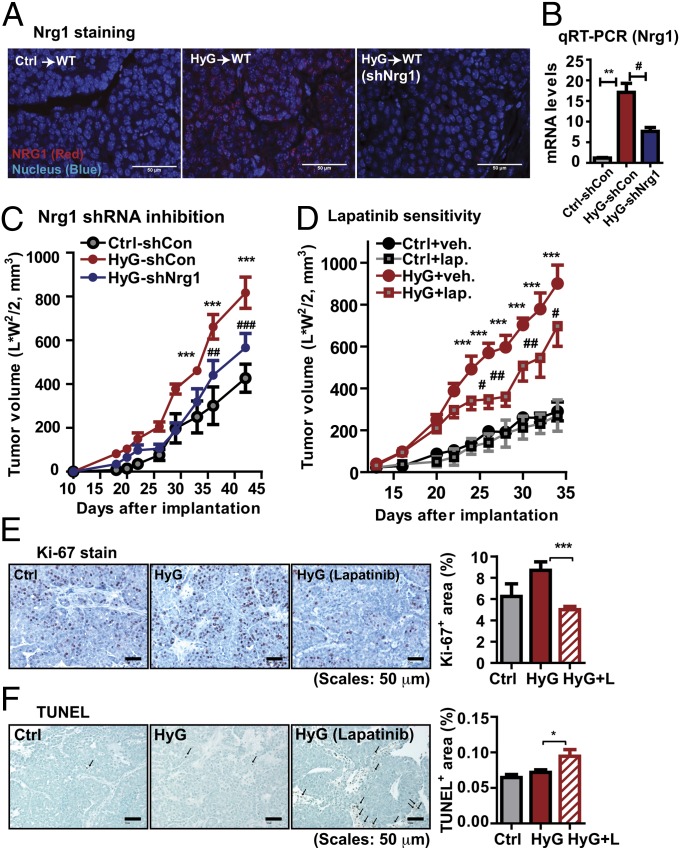
NRG1 can impact on cancer cells in various ways, depending on the HER receptor status. A NRG1-HER3 autocrine loop is essential to potentiate HER signaling and is seen in the context of drug resistance of HER-targeted therapies such as trastuzumab (a HER2-targeted monoclonal antibody) in HER2-amplified tumors (20–22). In addition, the NRG1-HER3 loop in HER2-negative tumors contributes to drug sensitivity in the context of HER2-targeted therapies (14, 15, 23). Furthermore, NRG1 per se is a mitogenic factor sufficient to promote tumor growth, regardless of HER receptor status (24). PyMT-derived tumors used in this study are HER2 amplified (25). Considering this HER2 amplification, we explored whether NRG1-HER signaling pathways are enhanced and persisted in the HyG tumors. Nrg1 immunostaining clearly showed that HyG tumors expressed significantly higher levels of Nrg1 than Ctrl tumors that showed no detectable Nrg1 (Fig. 3A). Consistently, Nrg1 levels in PyMT/PANIC were higher than those in PyMT mice (Fig. S6A) and HER receptors, including HER2 and HER3, which were further activated in tumors in PyMT/PANIC mice compared with PyMT as determined by immunoblotting (Fig. S6 B and C). This Nrg1 upregulation suggests that hyperglycemia-induced Nrg1 potentiates HER signaling via an autocrine pathway.
Fig. 3.
Nrg1 acquisition in HyG-tumors enhances tumor malignancy. (A) Nrg1 immunostaining, showing a strong Nrg1 signal in HyG-tumors, but not in Ctrl-tumors. shRNA-mediated Nrg1 knock down in HyG tumors decreases Nrg1 levels. (Scale bars: 50 μm.) (B) Total RNA was prepared from the tumor tissues of scrambled shRNA-infected Ctrl cells (Ctrl-shCon), scrambled shRNA-infected HyG cells (HyG-shCon), and Nrg1 shRNA-infected HyG cells (HyG-shNrg1) bearing mice. mRNA levels for Nrg1 were determined by qRT-PCR. mRNA levels were normalized with β-actin and represented as mean ± SEM (n = 8 per group). **P < 0.01 Ctrl-shCon vs. HyG-shCon; #P < 0.05 vs. HyG-shCon vs. HyG-shNrg1 by unpaired Student’s t test. (C) Tumor growth for the Nrg1 knock downed HyG-tumors, showing a reduced growth rate compared with HyG-tumors. ***P < 0.001 vs. Ctrl-shCon; ##P < 0.01, ###P < 0.001 vs. HyG-shCon by two-way ANOVA. n = 8 per group. (D) Lapatinib treatment (100 mg/kg per day by oral gavage) of HyG-tumor bearing mice, showing that a RTK inhibitor attenuated growth of HyG tumor (n = 10), but less affected of Ctrl tumors (n = 8). ***P < 0.001 vs. Ctrl; #P < 0.05 and ##P < 0.01 vs. HyG by two-way ANOVA. (E) Proliferation indices were determined by Ki-67 staining. Quantified results represent mean ± SEM (n = 5 per group). Three different fields per sample were analyzed. ***P < 0.01 vs. HyG by unpaired Student’s t test. (F) Apoptosis was determined by TUNEL assays. Quantified results represent mean ± SEM (n = 3 per group). Three different fields per sample were analyzed. *P < 0.05 vs. HyG by unpaired Student’s t test.
To determine whether the signature up-regulation of Nrg1 in HyG tumors is a critical feature setting HyG tumors apart from Ctrl tumors, we reduced Nrg1 levels by using a shRNAs approach. Lentiviral-mediated infection of HyG cancer cells with shRNAs targeting Nrg1 reduced both protein and mRNA levels by 50% over the tumor progression in vivo (Fig. 3 A and B). Accordingly, tumor growth rates were attenuated in HyG tumors subjected to Nrg1 reduction, effectively bringing it back to control levels (Fig. 3C). We subsequently examined drug sensitivity for HyG tumors with the HER1/HER2 dual-RTK inhibitor lapatinib, one of the most selective RTK inhibitors available and used to treat HER2-amplified metastatic breast cancers (26). Lapatinib also inhibits the activated NRG1-HER3 autocrine loop (14). HyG and Ctrl cells bearing mice were given daily either vehicle or lapatinib (OG, 100 mg/kg per day) starting 2 wk after implantation. HyG tumors treated with lapatinib grew at a slower rate than HyG tumors in the vehicle group, whereas the growth of Ctrl tumors was significantly less affected (Fig. 3D); there are however some remaining PI3K and ERK signals in this setting (Fig. S6 D and E). Ki67 staining showed that proliferation indices were approximately 40% decreased (Fig. 3E), whereas apoptosis was slightly increased in lapatinib-treated HyG tumors compared with vehicle-treated HyG tumors (Fig. 3F). These results suggest that lapatinib treatment attenuates tumor aggressiveness caused by Nrg1 induction in HyG-tumors.
Primary Cancer Cells Acquire Malignancy Through Prolonged Exposure of Hyperglycemia.
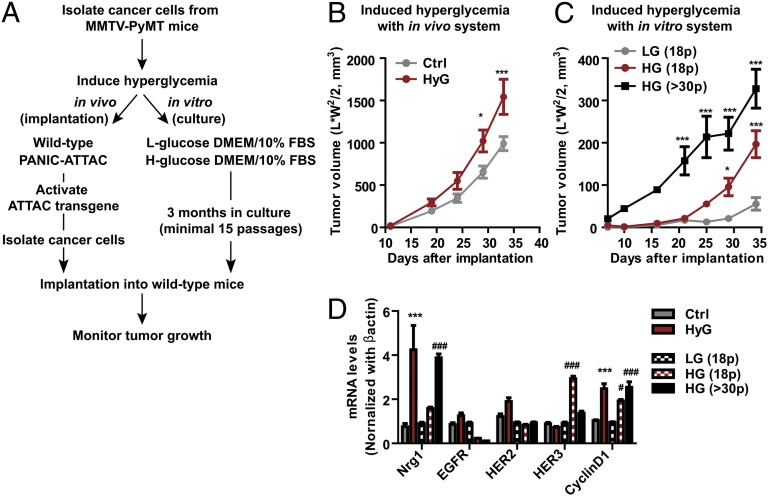
Most of established cancer cell lines are grown in media containing high levels of glucose. As such, it is challenging to test whether the NRG1-HER3 loop described in a subset of cancer cells (14) is intrinsic or acquired because of prolonged culture under hyperglycemic conditions. To address this issue, we used primary cancer cells freshly isolated from the tumors of PyMT mice under euglycemic conditions. These cells have basal-level Nrg1 expression. We then exposed these cells to hyperglycemia either in vivo or in vitro by either using hyperglycemic PANIC-ATTAC mice or euglycemic wild-type mice as hosts, or by culturing the cells in vitro with media containing low-glucose (1 g/L) or high-glucose (4.5 g/L) supplemented with 10% FBS, respectively (Fig. 4A). All cells grown under these different glycemic conditions were then harvested and implanted into euglycemic wild-type hosts, and tumor growth was monitored. Indeed, cancer cells exposed to hyperglycemia either in vivo or in vitro grew faster in the euglycemic hosts (Fig. 4 B and C). This enhanced growth clearly indicates that the hyperglycemic environment per se is actively involved in the programming of a more malignant growth behavior, and the phenotype is not due to another hormonal change secondary to hyperglycemia. We also assessed the mRNA levels for Nrg1 and HER receptors, including EGFR, HER2, and HER3, and cyclinD1 as proliferation indices. Nrg1 levels were consistently increased in cancer cells after hyperglycemic challenges both in vivo and in vitro; as a result, proliferation indices as judged by cyclinD1 levels were increased (Fig. 4D). The mRNA levels for HER receptors were less consistent, depending on the conditions and periods of hyperglycemic challenges. Nevertheless, it is clear that the cancer cells acquire malignant traits under hyperglycemic conditions. However, we cannot rule out the possibility that a subset of cancer cells, adapted to high glucose conditions, is clonally selected for further proliferation.
Fig. 4.
High levels of glucose confer malignancy to cancer cells. (A) Schematic diagram of hyperglycemic challenges to primary cancer cells in vivo and in vitro. (B) Upon secondary implantation into euglycemic hosts, tumor growth for hyperglycemia-exposed primary cancer cells originally grown in PANIC-ATTAC mice was faster in comparison with those isolated from wild-type mice. Cancer cells were exposed to hyperglycemia for 1 mo in vivo. Tumor volume represents mean ± SEM (n = 8 per group). *P < 0.05, ***P < 0.001 vs. Ctrl by two-way ANOVA. (C) Primary cancer cells isolated from PyMT mice were cultured with different glucose concentrations [low (LG, 1 g/L) vs. high glucose (HG, 4.5 g/L)] supplemented with 10% FBS for 3 mo (over 15 passages). These cells were then implanted into wild-type mice and tumor growth was monitored. Tumor volume represents mean ± SEM (n = 8–9 per group). *P < 0.05, ***P < 0.001 vs. LG (18 passages) by two-way ANOVA. (D) Total RNA was isolated from cancer cells from tumors grown in PANIC-ATTAC mice (HyG) or wild-type mice (Ctrl) as well as those cultured with either low glucose (18 passages) or high glucose (18 and more than 30 passages) media. mRNA levels for Nrg1, EGFR, HER2, HER3, and cyclinD1 were determined by qRT-PCR. Results represent mean ± SEM (n = 5 per group). ***P < 0.001 vs. Ctrl; #P < 0.05, ###P < 0.001 vs. LG (18p) by two-way ANOVA.
Breast Cancer Patients with a History of Hyperglycemia Express High Levels of Neuregulin-1 in Tumor Tissues.
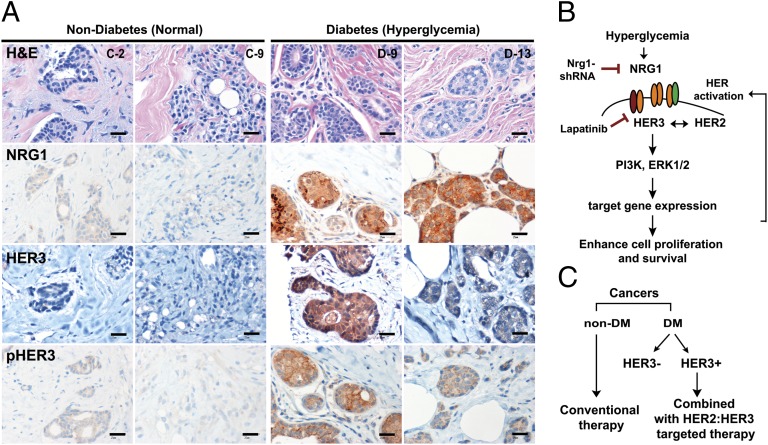
To validate whether these findings have any clinical relevance, we determined whether we could observe increased NRG1 levels in tumor sections from DM breast cancer patients compared with breast cancer patients with no indication of previous hyperglycemia. Twenty-five human patient samples (control n = 12 vs. DM n = 13) were included in this analysis. We did not control for age, body mass index, or antidiabetic treatment regimens (Table S1), but only selected on the basis of fasting glucose levels. NRG1 immunostaining showed that NRG1 levels were elevated in 11 of 13 DM-diagnosed breast cancer patients, whereas almost none of the euglycemic patients showed signals (NRG1+ patients; 84.6% in DM vs. 8.3% in non-DM) (Fig. 5A and Fig. S7A). In addition, approximately half of NRG1+ DM-patients expressed and activated HER3 (Fig. 5A and Table S1), suggesting that hyperglycemia is likely to activate the entire NRG1 mediated oncogenic pathway, such as HER signals. We appreciate that our study included only a relatively small number of subjects, and larger studies will be required to establish how universal our findings are for mammary and other tumors. However, given that fasting glucose levels were our only inclusion criterion and that we have obtained rather unambiguous “on/off” type responses suggests that these results are likely to hold up at larger scale as well. We do not yet know what the NRG1 status is for those individuals that are merely glucose intolerant but do not display frank diabetes.
Fig. 5.
DM breast cancer patients present a strong NRG1 signal and HER3 activation compared with non-DM patients regardless HER2 status. (A) Representative images for H&E, NRG1, HER3, and pHER3 staining in both groups (C-2 and C-9 for control vs. d-9 and d-13 for DM; Table S1). (Scale bars: 25 μm.) (B) Summary of this study. (C) Proposed therapeutic application for DM-breast cancer patients.
We have shown that hyperglycemia induces permanent changes in tumor cells, leading to a more aggressive growth behavior. This hyperglycemic effect is achieved by epigenetic up-regulation of the NRG1 gene, a ligand for the EGFR family. NRG1 activation results in more malignant tumor progression through the formation of a NRG1-HER3 autocrine loop to engage the HER signaling pathways. This effect was attenuated by reducing NRG1 levels, or with the RTK inhibitor lapatinib (Fig. 5B), suggesting that among several hyperglycemia-induced epigenetic modifications, the NRG1-mediated effects are almost single-handedly responsible for the observed changes. From a therapeutic perspective, our results suggest that DM patients with previous or current episodes of hyperglycemia that display HER3 overexpression may benefit from a combined treatment with HER2:HER3 targeted therapies (Fig. 5C); hence, a patient history of DM is a useful predictive biomarker to determine the therapeutic strategies beyond the HER2-amplified status.
Materials and Methods
Mice.
All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center. Genotyping for MMTV-PyMT and PANIC-ATTAC mice was performed by using PCR as in previous studies (5, 6). All animals used in this study were in a pure FVB background. AP20187 for in vivo dimerization of the FKBP-caspase-8 fusion was purchased from Clontech.
Implantation of Cancer Cells.
To isolate primary tumor cells, tumor tissues were excised from female mice and minced by using a razor blade in PBS. Tumors were incubated with collagenase type III (1.25 mg/mL) and hyaluronidase (1 mg/mL) for 2 h at 37 °C. Cell suspension was centrifuged at 1,000 × g for 10 min, and pellets were incubated with 0.25% trypsin for 10 min at 37 °C. The cell suspension was centrifuged at 1,000 × g for 10 min, and pellets were incubated with DNaseI (20 U/mL) for 10 min at 37 °C, cell pellets were washed with PBS containing 5% (vol/vol) serum. Cells were seeded into culture dishes in growth medium, DMEM (Mediatech) containing 10% (vol/vol) FBS (Gemini Bio Products) after passing through 40-µm nylon filter (Fishe). Freshly isolated cells were used in this study. Recipient animals were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg), and cancer cells were implanted into mammary adipose tissues by intraductal injection. Tumor growth was monitored once a week starting 2 wk after implantation.
Microarray.
Total RNA was extracted from tumor tissue from 12-wk-old PyMT and PyMT/PANIC (n = 12 per group). Microarray experiments were performed by the University of Texas Southwestern microarray core facility. The Mouse Illumina Bead Array platform (47K array) (Illumina) was used in this study. Gene lists and pathway analyses of the data sets were performed by using Ingenuity Pathway Software (Ingenuity Systems). Gene profiling data are available from GEO (www.ncbi.nlm.nih.gov/geo) under accession no. GSE40361.
FAIRE-seq.
FAIRE was performed as reported (27). Barcoded libraries of FAIRE DNA from HyG and Ctrl cells were generated with the SOLiD Fragment Library Barcoding Kit (Applied Biosystems), and 35-nt single-end reads were generated with the SOLiD4 system (Applied Biosystems). The primary sequencing data were directly translated from color space to mapped DNA sequence reads in the human reference genome (NCBI37/mm9 assembly) by using LifeScope (v.2.10). During the alignment, three filter steps were applied to remove low quality, ambiguous, and redundant reads. Four hundred HyG-specific FAIRE regions were identified as genomic regions with a significant read enrichment and binding peak profile in the HyG FAIRE reads over the Ctrl reads by using the Model-based Analysis of ChIP-Seq (MACS) software tool (28) with 10% FDR.
De novo motif discovery analysis for HyG FAIRE regions was performed with the Multiple EM for Motif Elicitation (MEME) software tool (29). High quality motifs were aligned against transcription factor motifs retrieved from JASPAR (30) and TRANSFAC (31) by using the TOMTOM software tool (32) to identify known transcription factor motifs that match the MEME predicted motifs. We analyzed the enrichment of the identified AP-1 and CTCF motifs. For this analysis, we determine the frequency of the motifs FAIRE regions and for 75,000 random sets of the same sample size by using Motif Scanner. The motif enrichment score is calculated as the ratio of the motif frequency in the FAIRE region set and the mean motif frequency in 75,000 random sets. The Z value and the statistical significance (P value) of the enrichment score is calculated based on the variance and the mean obtained from the 75,000 random simulations.
Potential protein-coding target genes associated with the identified FAIRE regions were identified based on the distance of their transcription start sites (TSSs) according to their RefSeq annotation in the mm9 assembly to HyG FAIRE peaks. Genes whose TSSs were within 50 kb or nearest to a FAIRE peak were called as target genes. FAIRE-seq data are available from GEO (www.ncbi.nlm.nih.gov/geo) under accession no. GSE40949.
Luciferase Assay.
The NRG1 enhancer element was isolated by PCR from mouse genomic DNA and subsequently subcloned into pGL4.23 reporter vector (Promega). Either control or NRG1 enhancer containing reporter plasmids were transiently transfected into CHO cells with or without AP-1 (c-jun and c-fos) transcription factor. Cell lysates were harvested and analyzed for luciferase reporter assays by following the manufacturer’s protocol (Applied Biosystems; Dual-Light luminescent reporter gene assay).
qRT-PCR.
Total RNA was isolated by using the RNeasy kit (Qiagen) following tissue homogenization in TRIzol (Invitrogen). Total RNA (1 μg) was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed in the Roche Lightcycler 480. The primers used in this study are listed in Table S2.
shRNA Lentivirus.
shRNA constructs for control and NRG1 were purchased from Open Biosystems. Lentivirus was produced with HEK293T cells and concentrated with Lenti-Concentrator (Clontech Laboratories) by following the manufacturer’s protocol.
Histology of Human and Mouse Tissues.
Clinical histology samples were obtained after approval by the University of Texas Southwestern Medical Center Institutional Review Board and in accordance with an assurance filed with and approved by the Department of Health and Human Services. Informed consent was documented in writing for all subjects. Formalin-fixed paraffin-embedded mouse tissue sections were stained by using anti-mouse Ki-67 antibody (Dako Cytomation), and TUNEL assays were performed according to the manufacturer’s protocol (Trevigen). H&E staining were performed by John Shelton at the University of Texas Southwestern Medical Center. Human patient samples were obtained from the University of Texas Southwestern Tissue Resource. Evaluation of NRG1, HER3, pHER3, HER2, ER, PR, p53, and Ki67 expression in breast tumors was performed by standardized automated immunohistochemistry methods using 5-μm sections of formalin fixed paraffin embedded tumor tissue and analyzed by computer-assisted image analysis in the Department of Pathology at the University of Texas Southwestern Medical Center. NRG1 (H210; Santa Cruz Biotechnology), HER3 (C17; Santa Cruz Biotechnology), pHER3 (21D3; Cell Signaling), HER2 (4B5; Ventata Medical System), ER (SP1; Ventata Medical System), PR (1E2; Ventata Medical System), Ki-67 (3O-9; Ventata Medical System), and p53 (DO-7; Ventata Medical System) were used for human samples.
Statistical Analyses.
Data are presented as mean ± SEM. Data were analyzed by ANOVA followed by Newman–Keuls multiple comparison test or by Student’s t test, as appropriate with GraphPad Prism v.5 software.
Supplementary Material
Acknowledgments
We thank Jie Song and Steven Connell for technical assistance, Rahul Kollipara for computational analyses, and members of the P.E.S., Unger, Clegg and Gupta laboratories for helpful discussions. We were supported in part by National Institutes of Health (NIH) Grants R01-DK55758, R01-CA112023, and P01DK088761 (to P.E.S.); and NIH Grant DK081182 (to Jay Horton) and Cancer Prevention and Research Institute of Texas (CPRIT) Grant R1002 (to R.K.). J.P. was supported in part by Department of Defense Fellowship USAMRMC BC085909. R.K. is a CPRIT Scholar in Cancer Research of the Cancer Prevention and Research Institute of Texas and a John L. Roach Scholar in Biomedical Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE40361 (cDNA microarray data) and GSE40949 (FAIRE-seq microarray data)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214400109/-/DCSupplemental.
References
- 1.Barone BB, et al. Long-term all-cause mortality in cancer patients with preexisting diabetes mellitus: A systematic review and meta-analysis. JAMA. 2008;300(23):2754–2764. doi: 10.1001/jama.2008.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigneri P, Frasca F, Sciacca L, Pandini G, Vigneri R. Diabetes and cancer. Endocr Relat Cancer. 2009;16(4):1103–1123. doi: 10.1677/ERC-09-0087. [DOI] [PubMed] [Google Scholar]
- 3.Stattin P, et al. Prospective study of hyperglycemia and cancer risk. Diabetes Care. 2007;30(3):561–567. doi: 10.2337/dc06-0922. [DOI] [PubMed] [Google Scholar]
- 4.Jee SH, et al. Fasting serum glucose level and cancer risk in Korean men and women. JAMA. 2005;293(2):194–202. doi: 10.1001/jama.293.2.194. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZV, et al. PANIC-ATTAC: A mouse model for inducible and reversible beta-cell ablation. Diabetes. 2008;57(8):2137–2148. doi: 10.2337/db07-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuson JC, Legros N. Influence of insulin deprivation on growth of the 7,12-dimethylbenz(a)anthracene-induced mammary carcinoma in rats subjected to alloxan diabetes and food restriction. Cancer Res. 1972;32(2):226–232. [PubMed] [Google Scholar]
- 8.Cocca C, et al. Suppression of mammary gland tumorigenesis in diabetic rats. Cancer Detect Prev. 2003;27(1):37–46. doi: 10.1016/s0361-090x(02)00130-7. [DOI] [PubMed] [Google Scholar]
- 9.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8(12):915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 10.Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6(12):665–675. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- 11.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 14.Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell. 2011;20(2):158–172. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Q, et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell. 2010;17(3):298–310. doi: 10.1016/j.ccr.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falls DL. Neuregulins: Functions, forms, and signaling strategies. Exp Cell Res. 2003;284(1):14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 17.Holmes WE, et al. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256(5060):1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- 18.Stove C, Bracke M. Roles for neuregulins in human cancer. Clin Exp Metastasis. 2004;21(8):665–684. doi: 10.1007/s10585-004-6917-6. [DOI] [PubMed] [Google Scholar]
- 19.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: Receptor heterodimerization in development and cancer. EMBO J. 2000;19(13):3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee-Hoeflich ST, et al. A central role for HER3 in HER2-amplified breast cancer: Implications for targeted therapy. Cancer Res. 2008;68(14):5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 21.Holbro T, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100(15):8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritter CA, et al. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin Cancer Res. 2007;13(16):4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 23.de Alava E, et al. Neuregulin expression modulates clinical response to trastuzumab in patients with metastatic breast cancer. J Clin Oncol. 2007;25(19):2656–2663. doi: 10.1200/JCO.2006.08.6850. [DOI] [PubMed] [Google Scholar]
- 24.Atlas E, et al. Heregulin is sufficient for the promotion of tumorigenicity and metastasis of breast cancer cells in vivo. Mol Cancer Res. 2003;1(3):165–175. [PubMed] [Google Scholar]
- 25.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina PJ, Goodin S. Lapatinib: A dual inhibitor of human epidermal growth factor receptor tyrosine kinases. Clin Ther. 2008;30(8):1426–1447. doi: 10.1016/j.clinthera.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(Web Server issue):W369–W373. [DOI] [PMC free article] [PubMed]
- 30.Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wingender E, Dietze P, Karas H, Knüppel R. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.