Abstract
Many clinical studies incorporate genomic experiments to investigate potential associations between high-dimensional molecular data and clinical outcome. A critical first step in the statistical analyses of these experiments is that the molecular data are pre-processed.
This article provides an overview of pre-processing methods, including summary algorithms and quality control metrics for microarrays. Some of the ramifications and impact pre-processing methods have on the statistical results are illustrated.
The discussions are centered around a microarray experiment based on lung cancer tumor samples with survival as the clinical outcome of interest. The procedures that are presented focus on the array platform utilized in this study. However, many of these issues are more general and are applicable to other instruments for genome-wide interrogation.
The discussions here will provide insight into the statistical challenges in pre-processing microarrays used in clinical studies of cancer. These challenges should not be viewed as inconsequential nuisances but rather as important issues that need to be addressed so that informed conclusions can be drawn.
1 Introduction
In recent years there has been a surge of genome-wide experiments using high throughput technologies included as companions to clinical studies in cancer. This situation reflects an increased understanding in the cancer research community of the valuable additional information that can be garnered from these experiments. From a practical standpoint, these technologies have been made practical through a reduction in overall cost and the availability of improved software and hardware computing resources. In addition, cancer researchers can conduct preliminary investigations on publicly available data, and use online facilities for querying annotation and biological pathway information for better understanding of the findings of genome-wide experiments.
High throughput technologies enable interrogation of characteristics of the genome such as SNP polymorphisms [1], DNA copy number changes [2] and mRNA expression levels [3]. Traditionally, the statistical objective for the experiments using these technologies is the investigation of potential associations between a large number of molecular markers with clinical endpoints such as tumor response or time to death. The corresponding statistical analyses generally fall into three categories: 1. Association studies whose aim is the construction of panels of interesting genes [4] or biological pathways [5]; or 2. Prognostic or prediction studies whose aim is the construction of models based on molecular markers to classify patients with respect to clinical endpoints. Throughout this article, we shall use the term "features" to refer to molecular markers on the array platforms. 3. Class discovery studies whose aim is to discover clusters based on molecular data.
On many array platforms, each feature is quantitatively represented by several measures of intensity. To carry out statistical analyses, these intensities need to be adequately pre-processed. This involves reducing a very large set of intensities to a matrix of summary measures so that each feature is quantified using a single representative measure (e.g., expression; ref. 6). The statistical methodology literature is heavily geared towards the development of new and assessment of existing statistical methods based on the summary measures. These are often referred to as high-level analyses. Likewise, in the cancer research literature the statistical method section is mainly devoted to descriptions of the high-level analyses with only a token description of the pre-processing methods. In both cases pre-processing, or low-level analyses, is relegated to the status of a nuisance factor thought to be of little importance. George [7] provides an overview of statistical issues arising in the application of genomics and biomarkers in clinical trials. In this article, we will illustrate some of the challenges and explore the implications of the pre-processing on the conclusions.
We being by considering an example using a data set originally analyzed and discussed by Beer et al. [8]. They conduct an extensive set of analyses for investigating the association between overall survival and features from Affymetrix hu6800 chip. This data set has been analyzed extensively in the literature including in a paper by Jung et al. [9] who conduct an analysis using a rank-covariance estimator, which can be thought of as a robust non-parametric counterpart of univariate Cox regression, to identify features associated with survival. The top ten features according to this analysis method, ranked according to the family-wise error rate adjusted P-values [10], are listed in Table 1 using summary measures obtained from three different pre-processing methods: robust multichip algorithm (RMA; refs. 11,12), MAS5 [13] and a method used by the Beer et al (described in supplementary document for ref. 8). At the 10% level, there is one significant feature (CD8B) based on the RMA method three significant features (RAFTLIN,TMSB4X,SLC2A1) based on the MAS5 method and two significant features (RAFTLIN,NP) based on the Beer et al method. Often, features are excluded based on non-phenotypic criteria during the pre-processing method. For this illustration, we employed the filter used by Beer et al. The results are also sensitive to the choice of the filter.
Table 1.
The top ten genes based on an analysis of the Beer et al. data using the method described in Jung et al [9] (P-values refer to the family-wise error adjusted rates).
| RMA | MAS5 | BEER | |||
|---|---|---|---|---|---|
| Symbol | P-value | Symbol | P-value | Symbol | P-value |
| CD8B | 0.0697 | RAFTLIN | 0.0245 | RAFTLIN | 0.0187 |
| SLC2A1 | 0.1270 | TMSB4X | 0.0465 | NP | 0.0993 |
| CCR2 | 0.2111 | SLC2A1 | 0.0559 | KLHDC3 | 0.2968 |
| PLD3 | 0.2224 | IHPK1 | 0.3312 | TMSB4X | 0.3808 |
| RAFTLIN | 0.2433 | MLL | 0.3414 | CXCL3 | 0.4084 |
| HNRPL | 0.2787 | NP | 0.3492 | SELP | 0.4441 |
| BCL2 | 0.3106 | PRKACB | 0.4494 | STX1A | 0.5026 |
| PFKP | 0.3223 | <NA> | 0.4787 | SEC31L1 | 0.5068 |
| STX1A | 0.3610 | E2F4 | 0.5528 | PRKACB | 0.5355 |
| INPP5D | 0.3690 | P2RX5 | 0.5846 | PBXIP1 | 0.5571 |
In the discussions to follow, we will focus our attention on illustrating some of the challenges regarding pre-processing within the framework of the Beer at al example data. Although the discussions will focus on Affymetrix RNA arrays, as a consequence of choosing this example, many of the concepts apply to other types of microarrays.
2 Pre-processing: From Image to Measure
To carry out high level statistical analyses, raw imaging data are first quantified as intensities in the hybridization of sample to probe. The data then go through a series of pre-processing steps to generate a summary measure for each feature. These steps consist of some or all of the following.
Background Correction: For DNA-based arrays, it is important to apply adjustments for background noise that can result from non-specific hybridization, incomplete washing of the slide, or other technical artifacts in the generation of scanned images. These corrections are done at the probe level to remove spatial effects within each chip.
Normalization: As with many other lab measurements, the collection of intensities must be globally standardized such that features are comparable across all chips.
Summary Measure Calculation: When an array platform contains several probes for each feature, a summary measure must be obtained in order to quantify the amount of RNA expression, the change in DNA copy number or call the genotype.
Filtering: Some features should be excluded from the association studies. For example, features that are housekeeping or control genes are for quality control purposes and should be excluded from high level analyses. Filtering is also often used to reduce the number of features in the final analyses by removing features which for example have relatively low variability across the samples.
The Affymetrix oligonucleotide array used by Beer at al is a common platform for measuring mRNA expressions level. Affymetrix arrays are comprised of short sequences (25 base pairs in length) that are synthesized directly to glass slides using a photolithographic process. This technique can produce high-density chips with hundreds of thousands of unique oligomers. This allows multiple probes, collectively termed a "probeset", to represent a single feature on the array. A probeset typically consists of anywhere from five to twenty probe-pairs that correspond to distinct sequences within the transcript. Each probe-pair consists of a "perfect match" (PM) probe and a "mismatch" (MM) probe where the nucleotide in the 13th position is switched.
Pre-processing of Affymetrix arrays commonly involves generating a summary measure for each probeset. Affymetrix has released a series of algorithms [MAS4.0, MAS5.0 (ref. 13) and PLIER (ref. 14)), that quantify expression from increased binding to PM over MM probes. However, there is considerable debate as to whether MM probes detect only non-specific hybridization, and alternative algorithms have been proposed by academic investigators. For example, Model Based Expression Index (MBEI) proposed by Li and Wong [15,16], uses parametric multiplicative models for probe-specific rates of hybridization. This is defined for PM intensities only, the difference in PM and MM, or both. Robust Multichip Algorithm (RMA), proposed by Irizarry et al [11,12], employs parametric background correction followed by quantile normalization and robust fitting of a log-linear additive model based on PM only. GeneChip RMA (GCRMA) [17] extends the RMA algorithm to use probe sequence information in estimating non-specific hybridization during background correction. Numerous other algorithms have been employed in the literature including the method used in Beer et al [8].
With the increasing number of pre-processing methods, control experiments have been performed and made available as benchmarks for evaluating the relative performance. This includes dilution and mixture experiments [18], and spike-in experiments including the Affymetrix Latin Square data sets. In order to develop a standardized approach for comparing the various methodologies, the Affycomp project [19,20] has provided an application to apply each algorithm to the Affymetrix Latin Square data sets. The relative performance of these methods are assessed using a number of metrics for quantifying accuracy and precision, reflecting a bias-variance tradeoff among these methods.
3 Quality Control and Outlier Detection
Quality control at the array level is a critical step in detecting potential outliers and batch differences. Here we describe two different approaches. Plots are used to visualize aberrant hybridization patterns and to display poor correlation within the set(s) of arrays under investigation. Also, the quality of the arrays can be quantified using heuristic measures. In many cancer studies, replicate arrays cannot be run for defective arrays due either cost constraints or a lack of additional biospecimens. At the same time, poor quality arrays should not have undue influence on differential expression or classification algorithms. Therefore, one must be cautious in determining the level of stringency for excluding arrays from the analyses.
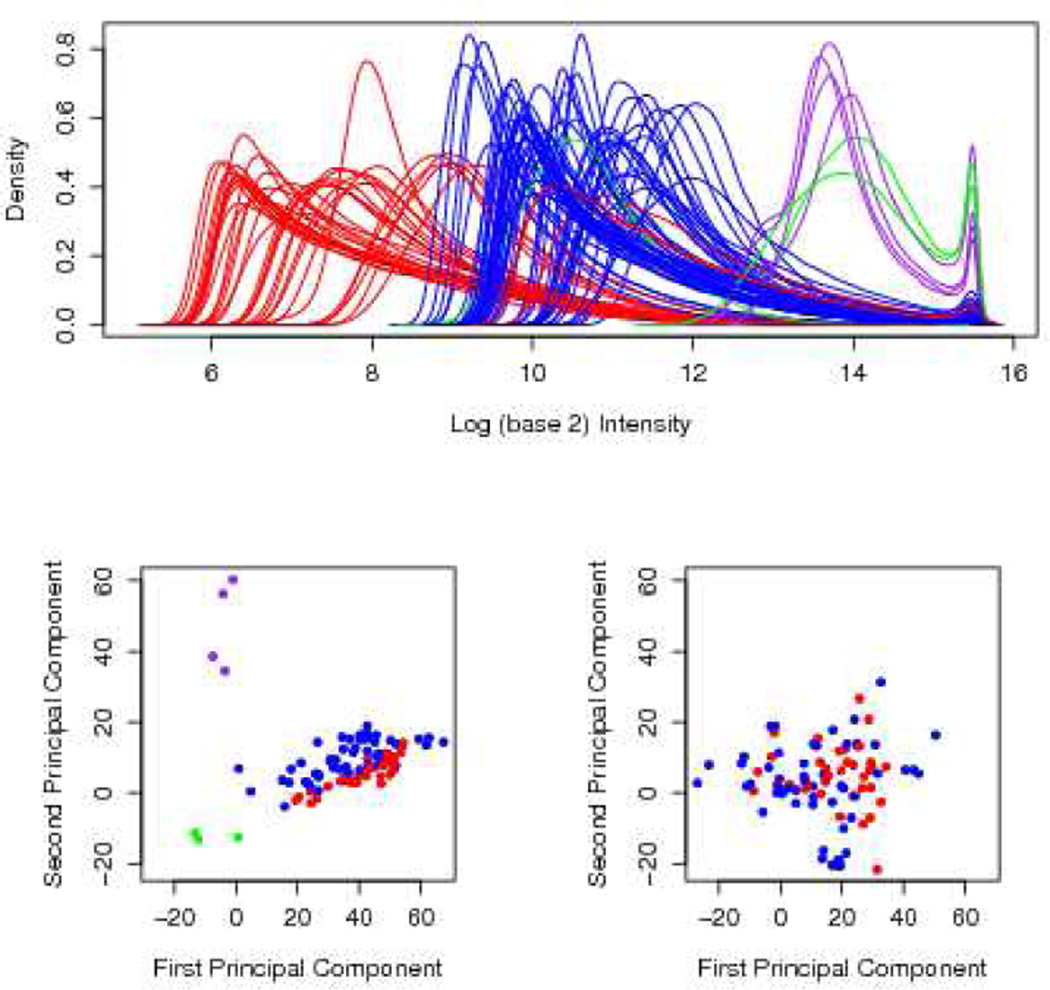
Plots of density estimates and principal components [21] are useful means of visualizing the data at both the probe intensity and transcript level. In an extensive quality assessment analysis of the Beer et al [8] microarray data set, we have employed these graphical devices to identify a batch effect and two sets of outliers in the data and their impact on several popular pre-processing algorithms. Some of the findings of this investigation are shown in Figure 1 using the colors red and blue, to represent the batch effect, and colors purple and green to label the two sets of outliers. In the top panel of this Figure, the density estimates of the PM intensities are drawn for each array. A distinct difference in the distribution of the intensities may be noted between the two batches of arrays (shown in red and blue). Likewise, the outlier arrays (purple and green) have intensity profiles that are much brighter than others (red or blue). In order to assess whether pre-processing would remedy these global effects, a plot of the first two principal components is produced. The results based on RMA pre-processing is shown in the bottom-left panel in Figure 1. The four clusters observed at the probe level are still present despite pre-processing. Next, the influence of the outlier arrays is examined by their removal from the RMA pre-processing procedure. The corresponding PC plot is shown in the bottom-right panel of Figure 1. The segregation of the two batches has vanished suggesting that the presence of these outliers prevents RMA from correcting the observed global difference.
Figure 1.
The panel in the top row illustrates the density estimates of the probe intensities for each of the 96 CEL files from Beer [8]. The PCA plots of expression values obtained when all arrays are RMA pre-processed (bottom-left panel) and when seven outlier arrays are removed (bottom-right panel)
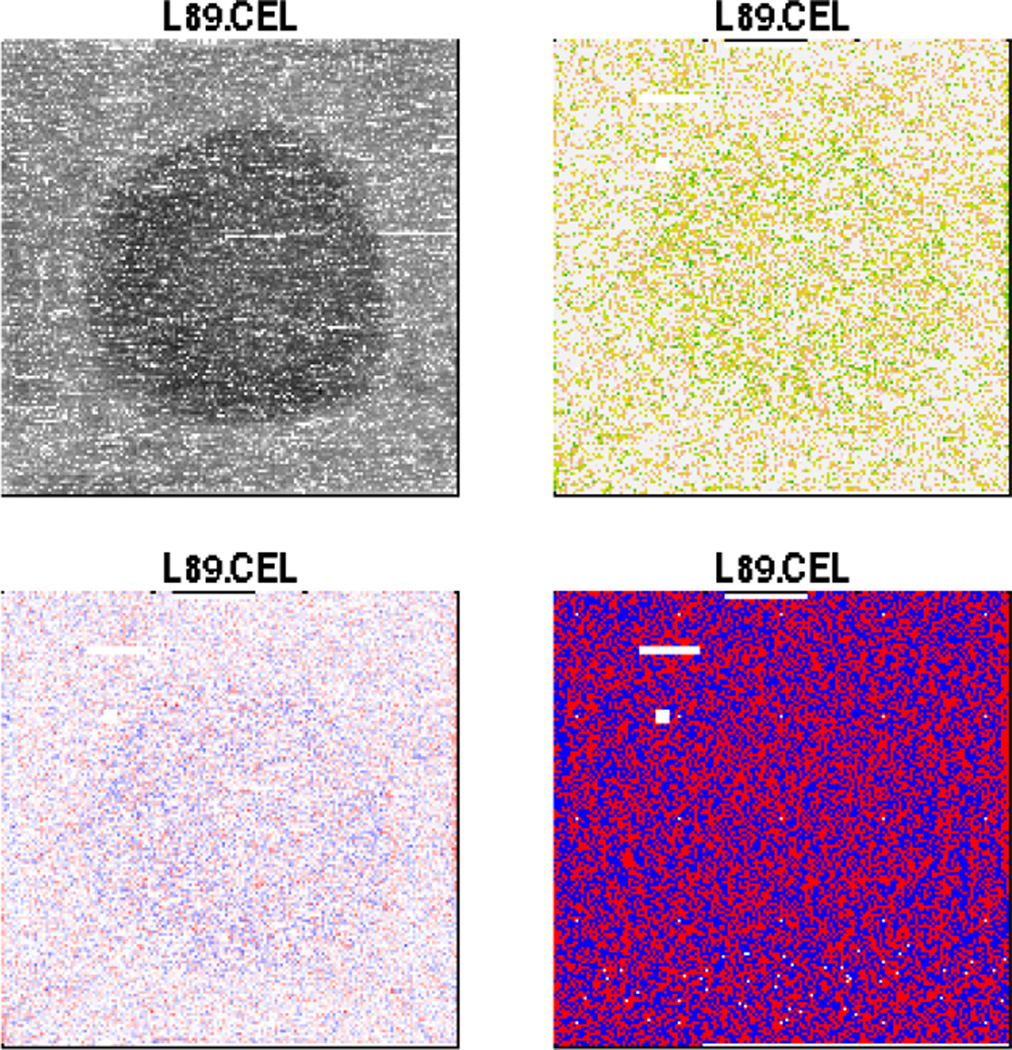
A visual examination, provided by the Bioconductor package affyPLM, of one of the arrays drawn in purple is shown in Figure 2. In the top-left panel a raw image of the chip is shown. The remaining three panels plot various types of residuals obtained after subtracting off a probe-level linear model (additional details for this method are found in section 3.5 of Bolstad et al [22]). A distinct spatial artifact is visible in the middle of this chip and the other three purple arrays (images not shown).
Figure 2.
The top-left panel shows raw image of the (purple) outlier chips. The remaining three panels plot various types of residuals obtained after subtracting off a probe-level linear model.
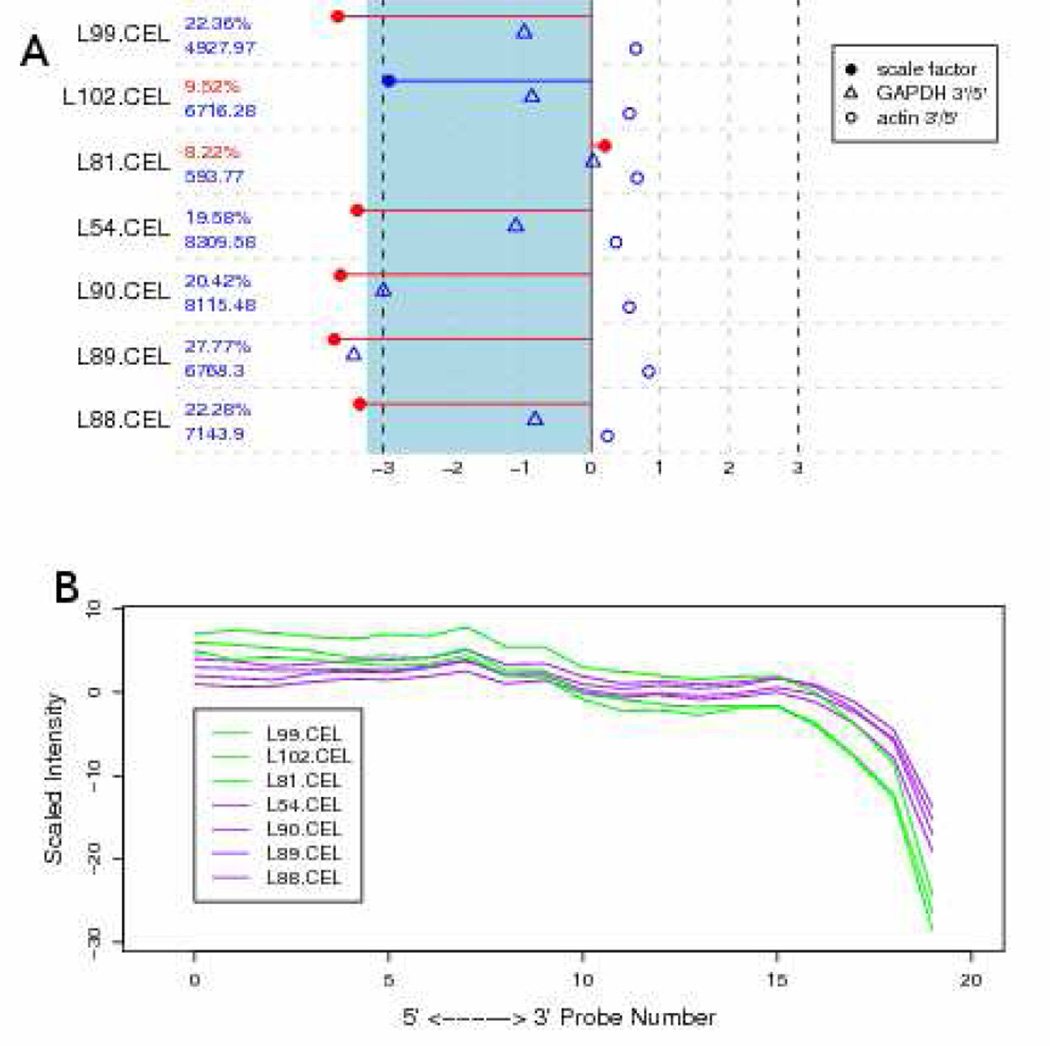
Summary measures of QC are provided in Figure 3 for the outlier arrays. The top panel provides several common measures in a graphical display supplied by the Bioconductor package simpleaffy. These include: a) the percentage of probesets with intensities above background, or "present" as defined by Affymetrix [23]; b) the average background intensity of the array; c) the scale factor and an acceptable range displayed as a shaded interval on the log2-scale; d) 3’/5’ ratio GAPDH; and e) 3’/5’ ratio of beta-actin. For each measure, values that exceed typical ranges of acceptability are highlighted in red. These ratios are commonly used measures of sample quality, where elevated levels indicate the integrity of starting RNA, efficiency of cDNA synthesis and/or transcription of cRNA in running arrays. Thus, global patterns of RNA degradation can also be plotted using functions supplied be the affy package from Bioconductor (Figure 3b), where the average probe inensity of all probesets are ordered from the 5’ to 3’ end. This plotting function is supplied by the affy package from Bioconductor as a means of evaluating global RNA degradation patterns in the samples. These results demonstrate that the outlier samples were not immediately detectable from summary measures of QC alone.
Figure 3.
Graphical representations of summary measures for array quality. A) Output from the QC reports generated by Bioconductor/simpleaffy for the 6 outlying arrays identified in Figure 1. Percent present and average background are printed, and the scale factor, beta-actin 3’/5’ ratio, and GAPDH 3’/5’ ratio are plotted on the log2 scale. Values that cross typical thresholds are displayed in red. B) RNA degradation plots from Bioconductor/affy. For each transcript, probe pairs are ordered from 5’ to 3’, and the average position-specific PM value is plotted for each array to indicate any global patterns of sample degradation.
It is important to mention that the point of this discussion is not to suggest the removal of any array that appears to be an outlier from the statistical analyses. Rather, we set out to summarize the effect of a set of outliers on the performance of a popular pre-processing method for this specific data set. Removal of the outliers seemingly improves the performance of this pre-processing method. However, removal of an outlier array results in removing a patient from the statistical analyses may result in bias. More specifically, unless the array is deemed to be technically defective beyond a reasonable doubt, its removal from the analyses is not something that can be recommended. The emphasizes the importance of employing statistical methodology that is robust with respect to outliers.
4 Challenges in the Prospective Setting
As microarrays and other high-throughput biotechnologies are increasingly used in the study of cancer therapeutics, a particular interest has been the identification of genomic signatures that classify tumor subtypes according to clinical outcome. Retrospective analyses of tumor samples have generated genomic signatures for many types of cancer, including lymphoma [24], breast [25] and lung carcinomas [26]. Clinical trials are required to evaluate and properly validate the prognostic or predictive capability of signatures, and much effort has gone into developing design strategies for testing and validating genomic in a prospective manner [27–30]. However, there has been little discussion on the ramifications of pre-processing algorithms used in the development of each signature in the prospective setting.
Many of the common pre-processing and QC strategies presented above are designed to interrogate simultaneously the full set of samples in a study. This includes the model-based approaches to summarizing expression at the transcript level (e.g., RMA, MBEI and PLIER), QC plots for visualizing outliers and batch effects, and the QC summary measures for relative scale factor and average background. All pre-processing could be limited to single-array methods. However, the relative performance of these methods in Affycomp suggests that this would result in decreased precision and accuracy. Adaptations to the summarization approaches of MBEI and RMA have been proposed whereby probe-level parameters are first estimated by a training set, and then applied to the incoming sample as fixed effects [16,31]. As an alternative approach, one employs a set of âœstandardizationâ. samples that are selected prior to initiation of the trial; then, each microarray collected over the course of the trial is pre-processed in conjunction with the set. In this way, the incoming sample informs the probe-level parameter estimates, yet the principle of exchangeability is maintained. Whether a training or standardizing set is used in generating post-processed data, it is critically important that the samples are representative of the patient population. However, the adequacy of set must be determined by the investigators, and will depend on the experimental design such that universal standards have not been identified in the field. The following simulations represent one mechanism whereby quality of post-processed data can be assessed once a standardization set is in place.
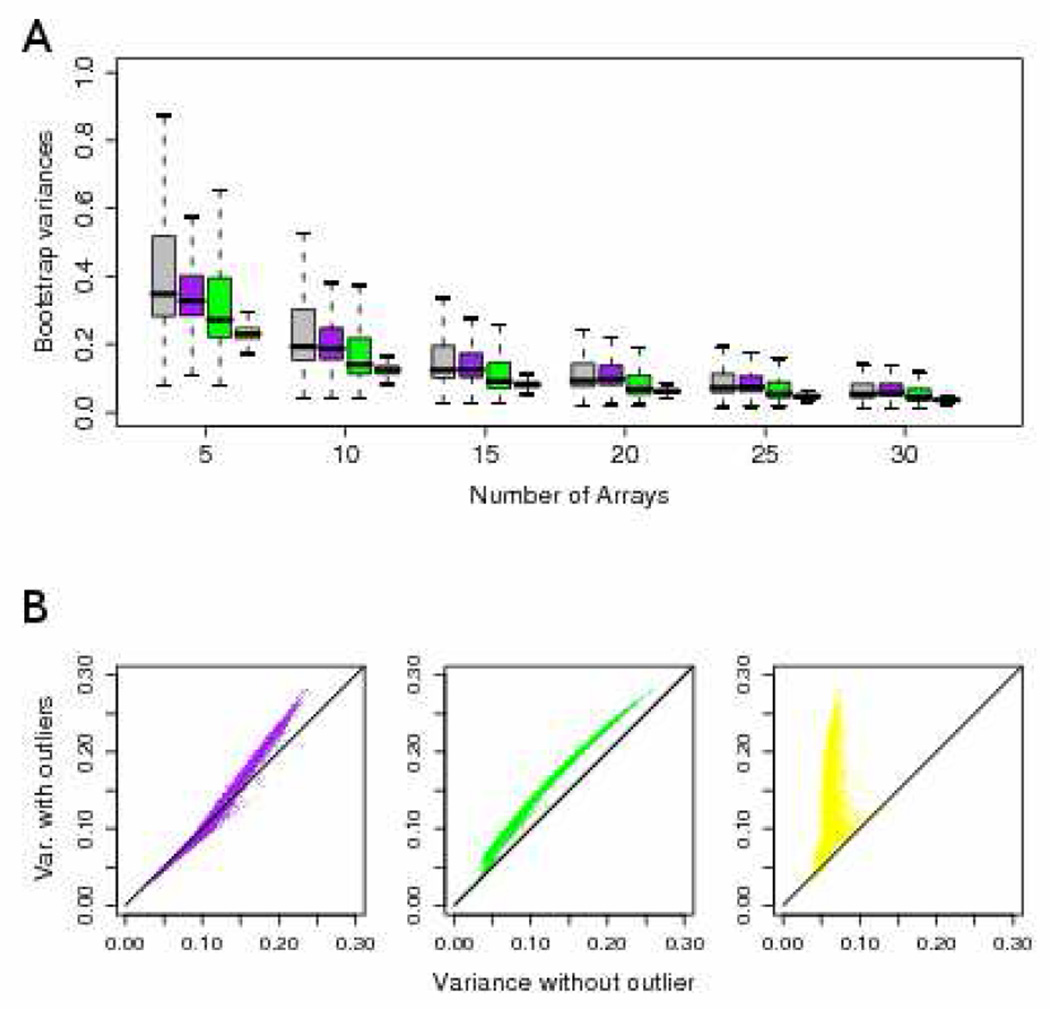
One important determination in selecting a standardizing set, is the minimum necessary number of arrays. We conducted a comprehensize bootstrap analysis of the Beer et al data set, to evaluate the sensitivity of post-processed data to set size. For each array, standardizing sets of N = 5, 10, 15, 20, 25 or 30 were randomly selected, and the RMA pre-processing algorithm is applied. Probeset-specific variances in expression are computed from 200 bootstrap replicates, and then averaged across all arrays. Boxplots in Figure 4. demonstrate that variability is substantially reduced when the standardizing set consists of at least twenty arrays. Furthermore, variances are attenuated when one or both sets of outlier samples are removed, and variance stabilization appears to be achieved when all seven outlier samples are removed (lower-right panel). These results illustrate the sensitivity of RMA to standardizing set size and outlier arrays. Further analyses are required a using data sets with technical replicates and spike-in genes to evaluate precision and accuracy.
Figure 4.
Bootstrap variance estimates for expression values generated from RMA. A) Boxplots of the average variance in expression across all arrays when pre-processed with 200 random standardizing sets of size N = 5, 10, 15, 20, 25, 30. Arrays were either selected from the full set of samples from Beer et al. [8], after removing outlier set 1 (purple), outliers set 2 (green), or both (yellow). B) Scatterplots of bootstrap variance estimates from standardization sets of size N = 20 with or without removing outlier samples.
5 Computational Tools
Pre-processing of microarray data is a computationally intensive task requiring access to appropriate computing hardware and software. There are a number of commercial and open-source products that can be used to carry out the pre-processing steps presented in this paper. The analyses presented using the example data set were performed using the open-source R [32] statistical environment along with packages from Bioconductor [33]. The affy package provides functions for MAS5 and RMA pre-processing and gcrma and plier packages provide functions for GCRMA and PLIER pre-processing. The expresso function in affy allows the user to mix and match from a set of pre-defined background correction and normalization methods. The reduced models in MBEI have been implemented in the freely available software dCHIP.
6 Discussion
In this article, we have reviewed several pre-processing and quality-control methods and applied them to an example data set microarrays in lung cancer. For a detailed and accessible discussion on various aspects of pre-processing, the reader may refer to the monograph by Simon et al [34] and the articles by Quakenbush [35], Hoffmann et al [36], McClintick et al [37], McClintick and Edenberg [38], Jones et al [39] and Seo and Hoffman [40]. The results in our paper indicate the presence of outlier arrays and batch effect in the data. A number of post-processing methods have been proposed to address these issues. Suárez-Fariñas et al [41] propose a corrective method for removing spatial artifacts. For batch effect correction, Johnson et al [42] propose an empirical Bayes method and Benito et al [43] propose a method using support vector machines.
If a study in development plans to use information from a cancer gene list of signature constructed based on an older chip, then one has to decide how to map the features from the old chip to the newer chip. The mapping may be done by matching on gene symbols or homologs queried from public databases. However, one should be concerned that the marginal or joint distributions of the intensities may differ between the platforms and consequently the summary measures may not be comparable. More importantly, besides these potential statistical caveats, the matching may not be biologically relevant if the probe sequences for the features on the two chips do not match.
SNP arrays are used to interrogate DNA polymorphisms. The fact that the final outcome is not a continuous expression measure but rather numbers of copies of an allele for each feature, may give the erroneous impression that pre-processing of SNP arrays is more straightforward than that of say RNA microarrays. The genotypes are not determined but rather called based on intensities. The issues raised related to pre-processing for the RNA microarrays applies to these instruments as well.
In some cancer studies, multiple chips are produced for some of the patients. These could be replicates generated for quality control and reproducibility. Unless there is overwhelming and definitive evidence that a replicate chip is defective, it is likely to be inappropriate to exclude it from the analysis. As such, in the case of replicate arrays, it may be necessary to aggregate the arrays. One simple approach is to average the arrays for each patient across the features. The results discussed regarding the outliers in the Beer et al data set are part of a more extensive study we have carried out. One of the conclusions from this study is that the most influential aspect of outlier effects is the method used for background correction. This conclusion agrees with that of the Affycomp report [20].
As microarray experiments are increasingly used in cancer trials, the MicroArray Quality Control (MAQC) project was formed between the FDA and academic institutions to provide quality control tools [44]. In the first phase of MAQC, the relative performance of different array platforms was assessed using several large sets of technical replicates that were run across multiple sites and determined to be comparable [45]. Future efforts of the MAQC are to examine diagnostics of array reliability and to explore the utility of microarray technology in the development and validation of predictive models.
In summary, we outline a list of general recommendations.
The examples discussed illustrate that it is difficult to assess the quality of the data solely based on summary measures. For any study the investigators should be provided the files containing the raw data (e.g., Affymetrix *.CEL, Illumina *.idat or aCGH *.sproc files) rather than a spreadsheet with expressions.
Standardized quantitative quality control measures, such as those provided by the chip manufacturer are useful and should be considered as part of the pre-processing package. These are however not substitutes for graphical tools such those considered in this paper.
Often the physical file names of the arrays reveals experimental factors such as treatment assignment or cell line type. The lab generating the arrays should be blinded to the experimental factors to avoid unintentional induction of batch effects.
The lab should be asked to provide information (e.g., date or time) that can be used in the identification of potential batch effects.
In the case of post-treatment arrays, investigators should avoid confounding batch and experimental factors by not sending the specimens from each group of the factor in batches to the lab.
As Chau et al. [6] point out that sample processing can affect the quality of the biospecimens. Consequently batch effects may be introduced at the institution obtaining the biospecimens as well as at the repository responsible for receiving, storing and processing the biospecimens for shipment to the microarray lab. As such, it is important for the investigators to understand the flow of the biospecimens.
The pre-processing steps should be reproducible. For R and Bioconductor users, the Sweave [46] tool provides the facilities to simultaneously carry out and document the entire pre-processing procedure by intertwining R with the type-setting system LATEX [47]. The resulting document, which can be submitted as supplementary material, will also facilitate the manuscript review process. More importantly, this document will provide an important quality control component of the study.
Arrays could be generated based on various types of biospecimens such as frozen tumor tissue, paraffin embedded tumor tissue or cancer cell lines. Care should be taken before jointly pre-processing arrays based on different types of biospecimens. Many pre-processing algorithms require the provision of input parameters or thresholds. The defaults may not be appropriate for all tissues.
We have provided a glimpse of some of the basic challenges investigators face during the pre-processing phase of high dimensional molecular data in cancer studies. It is inappropriate, to designate these challenges as unimportant or inconsequential nuisances, especially considering the potential ethical ramifications of using models based these data to assign treatment in a prospective manner. These challenges should be welcome as an opportunity for additional research on the development of improved methodology and to pave the way for better understanding.
Summary Box.
To carry out statistical analyses, including inference and construction of prognostic or predictive models, using data from genome-wide microarray experiments from cancer studies, the molecular data first needs to be pre-processed.
Pre-processing consists of several steps including background correction, normalization and summarization.
The results from any given statistical analysis may not only differ with respect to the statistical testing and learning methods, but also on the pre-processing method employed.
Pre-processing may not necessarily alleviate artifacts or batch effects in the data both of which may have effects on the final results.
Although some comparative studies of pre-processing methods have been conducted, there is no consensus in the research community on which method to choose.
Further research is needed to develop improved methodology and to pave the way for better understanding of the potential ramification of pre-processing on the final results.
Acknowledgment
The authors thank two reviewers for providing insightful and helpful comments leading to substantial improvements of the manuscript.
References
- 1.Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhart DJ. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Research. 2000;10(8):1126–1137. doi: 10.1101/gr.10.8.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genetics. 1999;23(1):41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 3.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 4.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21(9):1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 6.Chau CH, McLeod H, Figg WD. Validation of analytical methods for biomarkers employed in drug development. Clin Cancer Res. 2008;18 doi: 10.1158/1078-0432.CCR-07-4535. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephen SL. Statistical issues arising in the application of genomics and biomarkers in clinical trials. Clin Cancer Res. 2008;18 in press. [Google Scholar]
- 8.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature Medicine. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 9.Jung SH, Owzar K, George SL. A multiple testing procedure to associate gene expression levels with survival. Statistics in Medicine. 2005;24(20):3077–3088. doi: 10.1002/sim.2179. [DOI] [PubMed] [Google Scholar]
- 10.Westfall PH, Young SS. Wiley Series in Probability & Mathematical Statistics: Applied Probability & Statistics. John Wiley & Sons; 1992. Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. ISBN 0471557617. [Google Scholar]
- 11.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 2003 Feb;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 13.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 14.Hubbell E. PLIER: An M-Estimator for Expression Array. Santa Clara, CA: Affymetrix Inc.; 2005. [Google Scholar]
- 15.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2(8):1–11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12(6):882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 18.Lemon WJ, Palatini JJT, Krahe R, Wright FA. Theoretical and experimental comparisons of gene expression indexes for oligonucleotide arrays. Bioinformatics. 2002;18(11):1470–1476. doi: 10.1093/bioinformatics/18.11.1470. [DOI] [PubMed] [Google Scholar]
- 19.Cope LM, Irizarry RA, Jaffee HA, Wu Z, Speed TP. A benchmark for affymetrix genechip expression measures. Bioinformatics. 2004 Feb;20(3):323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Wu Z, Jaffee HA. Comparison of affymetrix genechip expression measures. Bioinformatics. 2006 Apr;22(7):789–794. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- 21.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press; 1979. [Google Scholar]
- 22.Bolstad BM, Irizarry R, Gautier L, Wu Z. Preprocessing high-density oligonucleotide arrays. In: Gentleman RC, Carey VJ, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Statistics for Biology and Health) Springer-Verlag; 2005. [Google Scholar]
- 23.Affymetrix. Statistical Algorithms Description Document (whitepaper) Santa Clara, CA: Affymetrix Inc.; 2002. [Google Scholar]
- 24.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RCT, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nature Medicine. 2002;8(1):68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 25.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. National Acad. Sciences United States Am. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J. Clinical Oncology. 2006;24(31):5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 27.Simon R. Using genomics in clinical trial design. Clin Cancer Res. 2008;18 doi: 10.1158/1078-0432.CCR-07-4531. in press. [DOI] [PubMed] [Google Scholar]
- 28.Taylor JMG, Ankerst DP, Andridge RR. Validation of bio marker-based risk prediction models. Clin Cancer Res. 2008;18 doi: 10.1158/1078-0432.CCR-07-4534. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freidlin B, Simon R. Adaptive signature design: An adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clinical Cancer Research. 2005;11(21):7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 30.Simon R, Wang SJ. Use of genomic signatures in therapeutics development in oncology and other diseases. Pharmacogenomics J. 2006;6(3):166–173. doi: 10.1038/sj.tpj.6500349. [DOI] [PubMed] [Google Scholar]
- 31.Katz S, Irizarry RA, Lin X, Tripputi M, Porter MW. A summarization approach for Affymetrix GeneChip data using a reference training set from a large, biologically diverse database. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. R Development Core Team: R. ISBN 3-900051-07-0. [Google Scholar]
- 33.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini A, Sawitzki G, Smith C, Smyth G, Tierney L, Yang J, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. ISSN 1465-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and analysis of DNA microarray investigations. Springer-Verlag; 2004. [Google Scholar]
- 35.Quackenbush J. Microarray data normalization and transformation. Nature Genetics. 2002 Dec;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R, Seidl T, Dugas M. Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis. Genome Biology. 2002;3(7):research0033.1–research0033.11. doi: 10.1186/gb-2002-3-7-research0033. ISSN 1465-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4(1):4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClintick JN, Edenberg HJ. Effects of filtering by present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones L, Goldstein DR, Hughes G, Strand AD, Collin F, Dunnett SB, Kooperberg C, Aragaki A, Olson JM, Augood SJ, Faull RLM, Luthi-Carter R, Moskvina V, Hodges AK. Assessment of the relationship between pre-chip and post-chip quality measures for affymetrix genechip expression data. BMC Bioinformatics. 2006;7:211. doi: 10.1186/1471-2105-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo J, Hoffman EP. Probe set algorithms: is there a rational best bet? BMC Bioinformatics. 2006;7:395. doi: 10.1186/1471-2105-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suárez-Fariñas M, Pellegrino M, Wittkowski K, Magnasco M. Harshlight: a corrective make-up program for microarray chips. BMC Bioinformatics. 2005;6(1):294. doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 43.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, Marron JS. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20(1):105–114. doi: 10.1093/bioinformatics/btg385. ISSN 1367-4803. http://dx.doi.org/10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 44.Shi LM, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nature Biotechnology. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao WJ, Fang H, Kawasaki ES, Hager J, Tikhonova IR, Walker SJ, Zhang LA, Hurban P, de Longueville F, Fuscoe JC, Tong WD, Shi LM, Wolfinger RD. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nature Biotechnology. 2006;24(9):1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 46.Leisch F. Sweave: Dynamic generation of statistical reports using literate data analysis. In: Härdle W, Rönz B, editors. Compstat 2002 — Proceedings in Computational Statistics. Heidelberg: Physica Verlag; 2002. pp. 575–580. ISBN 3-7908-1517-9. [Google Scholar]
- 47.Lamport L. LaTeX: A Document Preparation System. second edition. Addison-Wesley; 1994. [Google Scholar]