Abstract
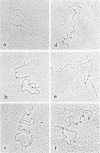
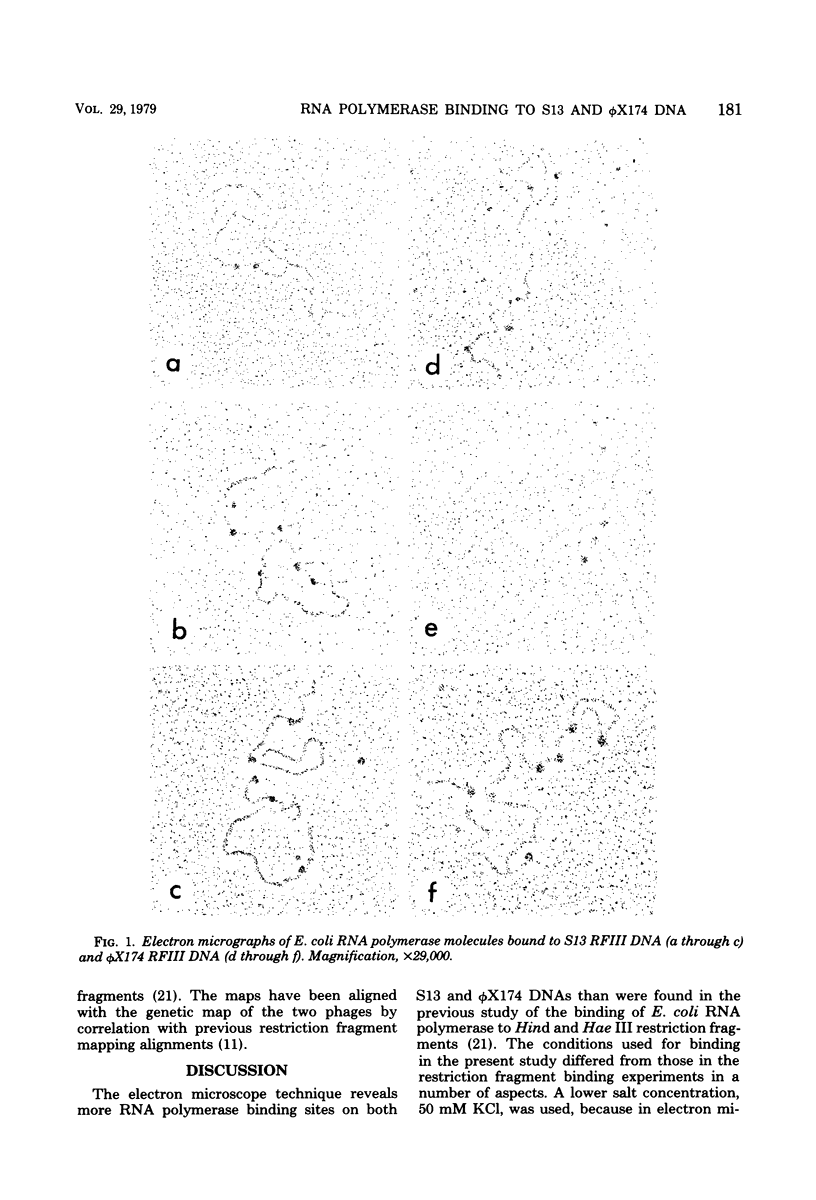
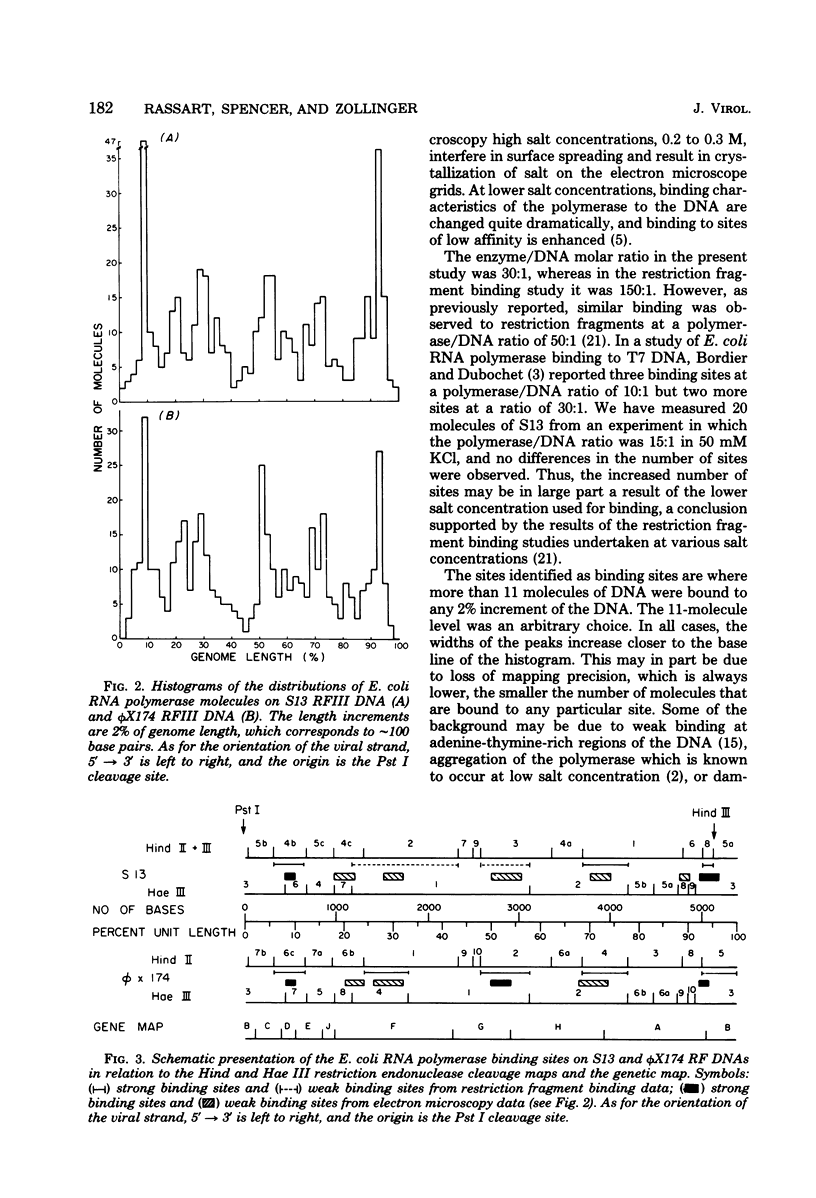
Complexes between Escherichia coli RNA polymerase and bacteriophage S13 and phage phiX174 replicative form III DNAs have been shown to form at specific locations on the phage genomes. The major locations on S13 have been mapped at 8 to 10 and 92 to 96% of the genome length, starting from the unique Pst I cleavage site. The locations correspond to the beginnings of genes D and B, respectively. Four minor locations map at 18 to 22, 28 to 32, 50 to 56, and 70 to 74% of the genome. The 70 to 74% site corresponds to the beginning of the A gene. The major locations on phiX174 are at 8 to 10, 50 to 54, and 92 to 94% of the genome. The 50 to 54% site is at the start of the H gene and has an equivalent minor site on S13, but it is not a promoter site. Three minor sites on phiX174, at 20 to 24, 26 to 32, and 68 to 74% of the genome, correspond to sites on S13. The data confirm the locations of sites identified by restriction fragment binding experiments (E. Rassart and J. H. Spencer, J. Virol. 27:677--687, 1978) and the assignment of putative promoters at the start of genes A, B and D.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axelrod N. Transcription of bacteriophage phiX174 in vitro: analysis with restriction enzymes. J Mol Biol. 1976 Dec 25;108(4):771–779. doi: 10.1016/s0022-2836(76)80116-7. [DOI] [PubMed] [Google Scholar]
- Berg D., Chamberlin M. Physical studies on ribonucleic acid polymerase from Escherichia coli B. Biochemistry. 1970 Dec 22;9(26):5055–5064. doi: 10.1021/bi00828a003. [DOI] [PubMed] [Google Scholar]
- Bordier C., Dubochet J. Electron microscopic localization of the binding sites of Escherichia coli RNA polymerase in the early promoter region of T7 DNA. Eur J Biochem. 1974 May 15;44(2):617–624. doi: 10.1111/j.1432-1033.1974.tb03519.x. [DOI] [PubMed] [Google Scholar]
- CHANDLER B., HAYASHI M., HAYASHI M. N., SPIEGELMAN S. CIRCULARITY OF THE REPLICATING FORM OF A SINGLE-STRANDED DNA VIRUS. Science. 1964 Jan 3;143(3601):47–49. doi: 10.1126/science.143.3601.47. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Hutchison C. A., 3rd, Edgell M. H. Isolation and genetic localization of three phi-X174 promoter regions. Nat New Biol. 1973 Jun 20;243(129):233–236. doi: 10.1038/newbio243233a0. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Giacomoni P. U., Delain E., Le Pecq J. B. Electron microscopy analysis of the interaction between Escherichia coli DNA-dependent RNA polymerase and the replicative form of phage fd DNA. 1. Mapping of the binding sites. Eur J Biochem. 1977 Aug 15;78(1):205–213. doi: 10.1111/j.1432-1033.1977.tb11731.x. [DOI] [PubMed] [Google Scholar]
- Goodchild B., Spencer J. H. Mapping of the single cleavage site in S13 replicative form DNA of Hemophilus influenzae (Hind III) restriction enzyme. Virology. 1978 Feb;84(2):536–539. doi: 10.1016/0042-6822(78)90270-2. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Ojamaa K. M., Spencer J. H. Fragmentation of bacteriophage S13 replicative form DNA by restriction endonucleases from Hemophilus influenzae and Hemophilus aegyptius. Virology. 1976 May;71(1):312–324. doi: 10.1016/0042-6822(76)90115-x. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Fujimura F. K., Hayashi M. Mapping of in vivo messenger RNAs for bacteriophage phiX-174. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3519–3523. doi: 10.1073/pnas.73.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEINSCHMIDT A. K., BURTON A., SINSHEIMER R. L. ELECTRON MICROSCOPY OF THE REPLICATIVE FORM OF THE DNA OF THE BACTERIOPHAGE PHI-X174. Science. 1963 Nov 15;142(3594):961–961. doi: 10.1126/science.142.3594.961. [DOI] [PubMed] [Google Scholar]
- Koller T., Sogo J. M., Bujard H. An electron microscopic method for studying nucleic acid-protein complexes. Visualization of RNA polymerase bound to the DNA of bacteriophages T7 and T3. Biopolymers. 1974 May;13(5):995–1009. doi: 10.1002/bip.1974.360130514. [DOI] [PubMed] [Google Scholar]
- Le Talaer J. Y., Kermici M., Jeanteur P. Isolation of Escherichia coli RNA polymerase binding sites on T5 and T7 DNA: further evidence for sigma-dependent recognition of A-T-rich DNA sequences. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2911–2915. doi: 10.1073/pnas.70.10.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. DNA regions essential for the function of a bacteriophage fd promoter. Nucleic Acids Res. 1977 Jul;4(7):2213–2222. doi: 10.1093/nar/4.7.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. Studies on bacteriophage fd DNA. II. Localization of RNA initiation sites on the cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):33–44. doi: 10.1016/0022-2836(75)90333-2. [DOI] [PubMed] [Google Scholar]
- Pollock T. J., Tessman I. Units of functional stability in bacteriophage messenger RNA. J Mol Biol. 1976 Dec 25;108(4):651–664. doi: 10.1016/s0022-2836(76)80110-6. [DOI] [PubMed] [Google Scholar]
- Portmann R., Sogo J. M., Koller T., Zillig W. Binding sites of E. coli RNA polymerase on T7 DNA as determined by electron microscopy. FEBS Lett. 1974 Sep 1;45(1):64–67. doi: 10.1016/0014-5793(74)80811-2. [DOI] [PubMed] [Google Scholar]
- Rassart E., Spencer J. H. Localization of Escherichia coli RNA polymerase binding sites on bacteriophage S13 and phiX174 DNA: alignment with restriction enzyme maps. J Virol. 1978 Sep;27(3):677–687. doi: 10.1128/jvi.27.3.677-687.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. II. In vitro initiation sites of phiX174 transcription. J Mol Biol. 1976 Jun 5;103(4):699–710. doi: 10.1016/0022-2836(76)90204-7. [DOI] [PubMed] [Google Scholar]
- Spencer J. H., Cerný R., Cerná E., Delaney A. D. Characterization of bacteriophage S13 suN15 single-strand and replicative-form deoxyribonucleic acid. J Virol. 1972 Jul;10(1):134–141. doi: 10.1128/jvi.10.1.134-141.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanami M., Sugimoto K., Sugisaki H., Okamoto T. Sequence of promoter for coat protein gene of bacteriophage fd. Nature. 1976 Mar 25;260(5549):297–302. doi: 10.1038/260297a0. [DOI] [PubMed] [Google Scholar]
- Takeya T., Takanami M. Isolation of DNA segments containing promoters from bacteriophage T3 DNA. Biochemistry. 1974 Dec 17;13(26):5388–5394. doi: 10.1021/bi00723a022. [DOI] [PubMed] [Google Scholar]
- Zollinger M., Guertin M., Mamet-Bratley M. D. A new electron microscopic method for studying protein-nucleic acid interactions. Anal Biochem. 1977 Sep;82(1):196–203. doi: 10.1016/0003-2697(77)90148-8. [DOI] [PubMed] [Google Scholar]