Abstract
Identification of genomic regulatory elements is essential for understanding the dynamics of cellular processes. This task has been substantially facilitated by the availability of genome sequences for many species and high-throughput data of transcripts and transcription factor (TF) binding. However, rigorous computational methods are necessary to derive accurate genome-wide annotations of regulatory sites from such data. SwissRegulon (http://swissregulon.unibas.ch) is a database containing genome-wide annotations of regulatory motifs, promoters and TF binding sites (TFBSs) in promoter regions across model organisms. Its binding site predictions were obtained with rigorous Bayesian probabilistic methods that operate on orthologous regions from related genomes, and use explicit evolutionary models to assess the evidence of purifying selection on each site. New in the current version of SwissRegulon is a curated collection of 190 mammalian regulatory motifs associated with ∼340 TFs, and TFBS annotations across a curated set of ∼35 000 promoters in both human and mouse. Predictions of TFBSs for Saccharomyces cerevisiae have also been significantly extended and now cover 158 of yeast’s ∼180 TFs. All data are accessible through both an easily navigable genome browser with search functions, and as flat files that can be downloaded for further analysis.
INTRODUCTION
The study of gene regulatory networks is a central area of systems biology, and one of the crucial steps in the reconstruction of gene regulatory networks is the identification of functional regulatory sites in cis-regulatory regions genome wide. During the past decades, a combination of developments in experimental and computational methodologies has dramatically improved our ability to identify the binding sites of transcription factors (TFs) on a genome-wide scale. On the experimental side, the development technologies such as chromatin immuno-precipitation followed by micro-array hybridization or next-generation sequencing (ChIP-chip and ChIP-seq) [e.g. (1)] has made it possible to comprehensively identify short genomic regions at which particular TFs are bound in a given experimental condition. In parallel, protein array technology (2) can be used in high throughput to map the binding specificities of TFs in vitro. At least as important as these experimental developments have been the development of computational methodologies for the inference of TF binding specificities and the mapping of TF binding sites (TFBSs). The most advanced current methodologies typically make use of rigorous Bayesian probabilistic methods to analyze high-throughput biological data, and incorporate comparative genomic analysis to assess the functionality of putative sites, often involving explicit models of sequence evolution and the effects of natural selection, see (3) for a review.
Our group has been involved for more than a decade in the development of computational methodologies for the inference of TF binding specificities and annotation of functional regulatory sites genome wide (3–11). Using these methods, we have been curating a number of resources in our SwissRegulon online database (12), including collections of position-specific weight matrices (WMs), promoters and predictions of TFBSs across proximal promoter regions genome wide in a number of model organisms. At the time of our original report on the SwissRegulon database, the database contained TFBS predictions for yeast and 17 prokaryotic organisms. As we will detail below, in the intervening 5 years, the database has been significantly extended in several ways. Most importantly, SwissRegulon now contains annotations of functional TFBSs for 190 regulatory motifs, representing the binding specificities of ∼340 TFs, across proximal promoters genome wide in both human and mouse. In addition, the TFBS annotations for Saccharomyces cerevisiae have been significantly extended, and now include predictions for 158 of yeast’s ∼180 TFs (13), making it the most comprehensive annotation of genome-wide binding sites available for any organism. We have also completely overhauled the user interface, implementing a new version of the genome browser, and adding a number of search functionalities that significantly improve user friendliness. Besides allowing users to browse annotations for promoters, genes or regulators of interest, we also make all data available for download in flat file format, including promoter annotations, curated collections of WMs and all TFBS predictions. Finally, the SwissRegulon web site also provides access to various tools that allow users to analyze their own data, including source code of software used in the TFBS predictions (5,11), and online tools.
RESULTS
TFBS annotations in 17 prokaryotic genomes
The TFBS annotations for prokaryotic genomes have remained largely unchanged compared with the previously reported version of SwissRegulon. With the exception of Escherichia coli, the predictions in the 16 other prokaryotic genomes are based on an algorithm for identifying sequence segments in intergenic regions that are under purifying selection (IRUS) described previously (6). For E. coli, we curated a set of WMs using our algorithm for probabilistic clustering of sequencess (PROCSE) (4) as described previously (12), and used MotEvo to predict TFBSs in intergenic regions genome wide.
S. cerevisiae TFBS annotation
Using a combination of data from ChIP-chip and in vitro binding assays, we have curated a collection of WMs representing the binding specificities of 158 TFs from S. cerevisiae (14). We constructed multiple alignments of all intergenic regions in S. cerevisiae, i.e. all sequence segments between annotated coding regions, with the orthologous regions in Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii and Saccharomyces bayanus. Using our recently updated MotEvo algorithm (11), we then predicted functional regulatory sites for these 158 TFs, consisting of >400 000 sites. Notably, using this annotation we uncovered several striking features of the ‘grammar’ of yeasts regulatory code (10), and we have also used this annotation to investigate the effects of cis-regulatory polymorphisms on gene expression (14). Of particular interest, in that study, we also provided evidence that the predicted regulatory interactions between TFs and target promoters, based on our TFBS predictions, are up to an order of magnitude more accurate than those obtained directly from ChIP-chip experiments.
In recent work (E. A. Ozonov and E. van Nimwegen, submitted for publication), we have been using rigorous biophysical models to analyze the competition between TFs and nucleosomes for binding to DNA, and have investigated to what extent observed nucleosome positioning in yeast can be explained by the competitive binding of TFs. The results of these investigations include predicted nucleosome occupancies, as well as occupancies of individual TFs in YPD genome wide, i.e. including across coding regions. These genome-wide TF occupancy profiles, as well as experimentally measured nucleosome occupancies (15), are also made available through SwissRegulon.
Human and mouse data
Promoterome
Our genome-wide predictions of TFBSs in human and mouse originated in our analysis work in the context of the FANTOM4 project (7,16,17). As part of this project, deepCAGE sequencing of transcription start sites (TSS) across different tissues in human and mouse was obtained in high throughput. We developed several novel procedures to analyze these TSS data and obtained hierarchical mammalian ‘promoteromes’ consisting of individual TSSs and transcription start clusters (TSCs) of nearby TSSs that are co-expressed across different conditions (8). We have further extended this set of human and mouse promoters to include promoters of transcripts that are not expressed in the cell types sampled by the deepCAGE data. In particular, we included 5′ ends of messenger RNAs from the University of California at Santa Cruz (UCSC) database (18), which were mapped to the human and mouse genomes using the BLAST-like alignment tool (BLAT) (18). To avoid transcripts whose 5′ ends are misaligned, we filtered out those for which >25 bp at the 5′ end of the transcript were unaligned. Subsequently, we then integrated the TSCs based on the deepCAGE data with these 5′ ends using the following iterative clustering procedure: at each step the nearest pair of clusters is fused, with the constraint that there can be at most one TSC per cluster (because different TSCs by construction are not co-expressed), and that the distance between merged clusters cannot be >150 bp (i.e. we use a distance cut-off roughly corresponding to the amount of DNA wrapped around a single nucleosome). The resulting reference set of promoters contains ∼36 000 promoters in human and ∼34 000 promoters in mouse. The promoteromes are available both in flat file format and through the genome browser.
Regulatory motifs
Using a combination of data from the JASPAR (19) and TRANSFAC (20) databases, other motifs from the literature, and motifs obtained using our own analysis of ChIP-chip and ChIP-seq data, we curated a data set of 190 position-specific WMs that represent the binding specificities of ∼340 TFs in both human and mouse. The curation included reducing redundancy by fusing WMs that are similar, have dominantly overlapping binding sites or are associated with TFs sharing highly similar DNA-binding domains. WMs were also refined by iteratively performing TFBS predictions in all proximal promoters genome wide (as described later). A detailed description of the curation procedure was provided in the supplementary materials of (7), and an updated version is provided in the documentation section of the web site. Both the WM collections and the associated mapping to human and mouse TFs are available in flat file format.
TFBS predictions in proximal promoters
For every promoter in the collection, we extracted 1 kb of DNA sequence from the genome, centered on the most highly expressed TSS of the promoter, and extracted orthologous sequences from human, mouse, rhesus macaque, dog, cow, horse and opossum, using pairwise genome mappings from the UCSC database (18). The sets of orthologous sequences were then multiply aligned with T-Coffee (21), and TFBSs were predicted on these multiple alignments.
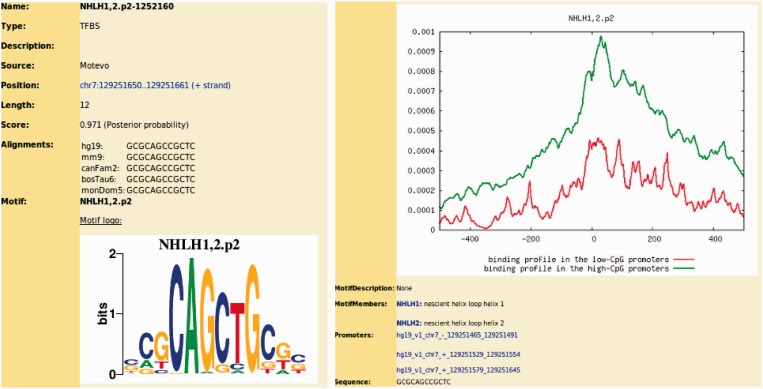
To predict TFBSs for each of the 190 mammalian regulatory motifs, we used our MotEvo algorithm (11). As described in (8), promoters in human and mouse naturally fall into two categories according to their sequence composition: promoters associated with CpG islands, and those not associated with CpG islands. We separated the multiple alignments associated with each class of promoters and performed TFBS predictions separately for each class. We have observed that different TFs have clearly distinct preferences in the positioning of their sites with respect to the TSS, and our TFBS predictions take these preferences explicitly into account. For each WM and each promoter class, we initialize a position-dependent prior distribution with a uniform distribution and perform an initial round of TFBS predictions using MotEvo. Using expectation maximization, we then iteratively update the position-dependent prior distribution of site frequencies and the TFBS predictions, until the position-dependent prior converges. Figure 1 (right panel) illustrates the inferred position-dependent profile for the motif NHLH1,2. Note that sites for this motif are more abundant in high-CpG promoters than in low-CpG promoters, and that, especially in high-CpG promoters, the sites preferentially occur immediately downstream of the TSS.
Figure 1.
Information provided for each predicted site. Left panel: listed are the name of the regulatory motif (NHLH1,2), the identification number of the site (NHLH1,2-1252160), the source of the prediction (MotEvo), the length of the site (12 bp), its posterior probability (0.971), the sequence of the site and the orthologous sites from other organisms aligned to it. In this case, orthologous sites of this site occur in mouse (mm9), dog (canFam2), cow (bosTau6) and opossum (monDom5). A sequence logo of the WM is also shown. Right panel: position-dependent TFBS density for this motif. The figure shows the probability to find a TFBS for the TF NHLH motif as a function of position relative to TSS in both high-CpG (green) and low-CpG (red) promoters. Listed are also the TFs associated with the motif (NHLH1 and NHLH2), the promoters that are putatively driven by this site and finally the sequence of the site.
In the final predictions, each TFBS in each promoter is characterized by a posterior probability that rigorously incorporates the quality of its match to the WM, the evidence for purifying selection on this binding site from the multiple alignments and its position relative to TSS. For human (UCSC assembly hg19), MotEvo reported >1 320 000 sites in ∼36 000 promoters. For mouse (UCSC assembly mm9), MotEvo reported >1 180 000 sites in ∼34 000 promoters. We also provide regulatory site annotations for an older human assembly (UCSC assembly hg18), which, currently, is still used by a significant number of researchers. Clicking on a predicted site in the genome browser leads to a separate page with detailed information on the site, as shown in Figure 1. This allows users to, among other things, investigate the precise conservation of the site across mammals.
MotEvo prediction algorithm
The binding site predictions in SwissRegulon are made using our MotEvo software, which is an integrated suite of Bayesian probabilistic methods for the prediction of TFBSs and inference of regulatory motifs from multiple alignments of phylogenetically related DNA sequences (11). Similar to a few other methods for TFBS prediction (22,23), MotEvo uses an explicit evolutionary model for the evolution of sequence segments that are under purifying selection to maintain their affinity for a given TF. However, in contrast to these methods, MotEvo robustly deals with binding sites that are only conserved in a subset of the species, or that appear missing because of local errors in the multiple alignments, which strongly improves performance (11). In addition, MotEvo takes into account that there may be many segments in the multiple alignments that are under strong purifying selection, but not for any of the known regulatory motifs, and models this by rigorously integrating over the space of possible WMs. This ‘unknown functional element’ model is also used to predict putative regulatory sites for genomes for which no regulatory motifs are available. Finally, MotEvo can also be used to perform enhancer finding by searching for clusters of binding sites for a given subset of WMs, similar to the functionality provided by the Stubb algorithm (24), but generalized to arbitrary multiple alignments. The MotEvo software package is available for download from the SwissRegulon web site.
Quick search functionality
We provide a convenient way of searching through our collection of mammalian promoters and motifs from the main page of the SwissRegulon using case insensitive search by keyword. The keyword may be a gene name, a transcript identifier, an entrez gene identifier, a motif identifier or a general keyword that occurs in the description of the feature of interest, e.g. ‘transferase’ or ‘regulator of chromatin’. The user may also use partial keywords like ‘PAX’ to find matches for all PAX TFs.
The search results are presented in a hierarchical form, initially listing all organisms for which matches were found, and the number of matches is shown in each of the following categories: matches to motifs, matches to genes or transcripts associated with a promoter or matches within the description of genes associated with a promoter. Clicking on one of the categories of matches expands the list showing summary information for all matches. This summary information is in turn clickable and takes the user to pages with detailed information on the match. Matches to promoters are linked to the corresponding page in the genome browser, showing all predicted regulatory sites in the neighborhood of that promoter. Matches to gene names, transcript accessions and gene description are also linked to the corresponding NCBI pages.
Motifs are linked to pages with extensive information on the motif including a sequence logo, the corresponding WM, a figure containing its position-dependent site frequencies in high-CpG and low-CpG promoters and a sorted table listing all promoters that have predicted TFBSs for the motif.
Genome browser interface
In the current version of SwissRegulon, we use the updated version of Generic Genome Browser (25) (version 2.45) as an interactive front end to the database. The new version of GBrowse operates much faster than previous versions and allows much more easy navigation across the genome. The page layout is similar to the previous version but includes a few changes. There is a toolbar on the top of the page giving access to various operations like exporting current track data in different formats, sharing data and getting help on GBrowse. Underneath this is a tab bar giving access to different panels: the browser itself, a panel for selecting which tracks to display, a snapshot panel for managing bookmarked regions, a panel that allows users to upload his/her own tracks and a panel for setting preferences.
Below the tab bar is a short ‘Landmark or region’ form, where users can either type a search term (e.g. a gene name) or explicitly specify a pair of genomic coordinates. The genomic region of interest is shown hierarchically in three panels: ‘Overview’, ‘Region’ and ‘Details’. The ‘Details’ panel shows the chosen tracks in the genomic region of interest. Each track can be easily turned on/off or customized by clicking on it. All displayed features are clickable and link to more detailed information about the feature.
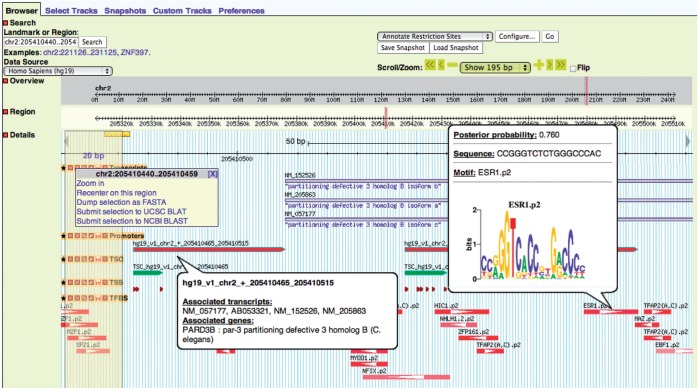
GBrowse offers a number of convenient ways for quick navigation. There are self-explanatory ‘Scroll/Zoom’ buttons and a drop-down menu for zooming to predefined resolutions. In addition, the ‘Overview’ and ‘Region’ panels have rulers that allow the user to select a region to zoom and jump to the selected region. Selecting a region in the ‘Details’ panel brings a menu with a few available operations. Clicking to a ruler in any panel re-centers the view to the selected location. Placing the mouse pointer on any of the features in the ‘Details’ panel brings up more detailed information about the feature, which may include a preview of the page that clicking on the feature links to. Figure 2 shows an example browser window that illustrates several of these features.
Figure 2.
The main browser panel showing a region around the promoter of the PARD3B gene in human. In this example, five tracks are shown: transcripts, promoters, TSC, TSS and TFBSs. The figure demonstrates selection of a region with the mouse and the associated drop-down menu with options. It also illustrates the pop-up windows with information about the promoter and one of the TFBSs, which will appear when placing the mouse pointer on these features.
Data representation within the genome browser
Annotated regulatory sites are displayed as boxes with an arrow inside (Figure 2), which indicates the strand of the site, and is labeled by the name of the TF(s) recognizing the site (when known). The color of the box indicates the site’s posterior probability, i.e. a more intense color indicates higher probability. The pop-up window that appears when placing the mouse pointer on a site contains the motif name, its posterior probability, the site sequence and a motif logo (if available). Clicking on a regulatory site opens a new window with detailed information about the site as described earlier.
Promoters are displayed as an arrow indicating the strand of the promoter (Figure 2) and are labeled with a unique identifier. A promoters’ pop-up window shows lists of genes and transcripts associated with the promoter. Clicking on the promoter opens a new window with detailed information on the promoter including its identifier, coordinates, strand, associated transcripts with corresponding gene names and information about each associated gene.
TSCs are displayed as arrows indicating their strand and a unique identifier. Their pop-up windows show the associated promoter and the position of the most highly transcribed TSS within the cluster. Finally, our annotation data is overlaid with transcript annotation from UCSC (human and mouse) and Genbank annotation for other organisms.
Downloads
All data are available for download. Genome annotations (promoters, TFBSs, TSCs and TSSs) are provided in GFF format (http://www.sequenceontology.org/resources/gff3.html). The WM annotation includes a file with WMs in the standard TRANSFAC format, and a plain text file containing motif-to-gene associations. The nucleosome occupancy data is provided in wig format (http://genome.ucsc.edu/goldenPath/help/wiggle.html). The description of the different formats can be found in the ‘Documentation’ section of the SwissRegulon web site. The ‘Software’ section of the web site provides downloads for motif and binding site prediction software, e.g. MotEvo (11) and PhyloGibbs (5). The latter also has a web interface, which can also be accessed through the SwissRegulon web site. Finally, there is also a link to a web-server for our Integrated Motif Activity Response Analysis (ISMARA). ISMARA allows users to automatically analyze their gene expression (microarray or RNA-seq) or ChIP-seq data in terms of our genome-wide predicted binding sites, with the aim of identifying the key TFs, their activities and their targets, in a given system of interest.
FUTURE DEVELOPMENTS
For the coming years, the key updates and extensions that we intend to implement are the following. First, an important model organism that is currently missing from SwissRegulon is the fruit fly Drosophila melanogaster. Our curation of Fly regulatory motifs and genome-wide predictions are already in an advanced stage of completion, and we expect to be able to offer genome-wide TFBS annotations for D. melanogaster in the near future. We are also in the course of updating our regulatory site predictions for E. coli, including a newly curated set of WMs, and expect to be able to provide these fairly soon.
A key limitation of SwissRegulon’s TFBS annotations is that, in multicellular eukaryotes, the predictions are limited to promoter regions. Although these regions likely contain a significant fraction of relevant regulatory sites, it is well known that many important regulatory sites are contained in distal cis-regulatory modules (or enhancers) (26). Recent developments in high-throughput mapping and analysis of chromatin state along the genome have uncovered that distal regulatory regions can be recognized by their DNase I sensitivity (27), methylation status (28) and particular combinations of histone modifications (29), allowing a more systematic mapping of distal cis-regulatory modules. Based on such information, we are currently curating a number of sets of distal regulatory regions and expect to be able to provide TFBS predictions for these sets in the near future.
SwissRegulon currently provides an overview page for each regulatory motif that, in particular, provides a sorted list of all promoters/genes targeted by the motif. We intend to develop similar pages for each individual promoter/gene. These pages will thus contain an easy overview of all ‘regulatory inputs’ that are predicted for a given promoter or gene of interest.
Another crucial factor limiting the completeness of genome-wide TFBS predictions is the fact that, for many TFs, the sequence specificity is unknown. However, with the dramatically decreasing sequencing costs, and the more easily accessible protocols for ChIP-seq analysis, the number of available ChIP-seq data-sets is increasing rapidly. We have developed an automatic pipeline for processing ChIP-seq data, identifying high-quality binding peaks, and using motif inference programs such as PhyloGibbs to infer regulatory motifs from such data sets. In the near future, we intend to use this automated pipeline to significantly expand the number of TFs for which regulatory motifs are available.
Finally, our new search function has proven itself as useful tool for quick access to the information but currently only contains information from the annotations of human and mouse, and we intend to extent it in the near future to include all eukaryotic and prokaryotic species that are in the database.
FUNDING
Swiss Institute of Bioinformatics (to M.P.); Swiss National Science Foundation [31003A-118318, in part]; SystemsX.ch through the CellPlasticity project. Funding for open access charge: University of Basel.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulyk ML. Protein binding microarrays for the characterization of DNA-protein interactions. Adv. Biochem. Eng. Biotechnol. 2007;104:65–85. doi: 10.1007/10_025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Nimwegen E. Finding regulatory elements and regulatory motifs: a general probabilistic framework. BMC Bioinformatics. 2007;8(Suppl. 6):S4. doi: 10.1186/1471-2105-8-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Nimwegen E, Zavolan M, Rajewsky N, Siggia ED. Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics. Proc. Natl Acad. Sci. USA. 2002;99:7323–7328. doi: 10.1073/pnas.112690399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddharthan R, Siggia ED, van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molina N, van Nimwegen E. Universal patterns of purifying selection at noncoding positions in bacteria. Genome Res. 2008;18:148–160. doi: 10.1101/gr.6759507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balwierz PJ, Carninci P, Daub CO, Kawai J, Hayashizaki Y, Van Belle W, Beisel C, van Nimwegen E. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 2009;10:R79. doi: 10.1186/gb-2009-10-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erb I, van Nimwegen E. Transcription factor binding site positioning in yeast: proximal promoter motifs characterize TATA-less promoters. PLoS One. 2011;6:e24279. doi: 10.1371/journal.pone.0024279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold P, Erb I, Pachkov M, Molina N, van Nimwegen E. MotEvo: integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics. 2012;28:487–494. doi: 10.1093/bioinformatics/btr695. [DOI] [PubMed] [Google Scholar]
- 12.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, van Nimwegen E, Rajewsky N, Siegal ML. Correlating gene expression variation with cis-regulatory polymorphism in Saccharomyces cerevisiae. Genome Biol. Evol. 2010;2:697–707. doi: 10.1093/gbe/evq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 16.Severin J, Waterhouse AM, Kawaji H, Lassmann T, van Nimwegen E, Balwierz PJ, de Hoon MJ, Hume DA, Carninci P, Hayashizaki Y, et al. FANTOM4 EdgeExpressDB: an integrated database of promoters, genes, microRNAs, expression dynamics and regulatory interactions. Genome Biol. 2011;10:R39. doi: 10.1186/gb-2009-10-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaji H, Severin J, Lizio M, Forrest AR, van Nimwegen E, Rehli M, Schroder K, Irvine K, Suzuki H, Carninci P, et al. Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res. 2011;39:D856–D860. doi: 10.1093/nar/gkq1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC genome browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notredame C. Computing multiple sequence/structure alignments with the T-coffee package. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi0308s29. Chapter 3, Unit 3.8.1–25. [DOI] [PubMed] [Google Scholar]
- 22.Moses AM, Chiang DY, Pollard DA, Iyer VN, Eisen MB. Monkey: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmack CS, McCue LA, Newberg LA, Lawrence CE. PhyloScan: identification of transcription factor binding sites using cross-species evidence. Algorithms Mol. Biol. 2007;2:1. doi: 10.1186/1748-7188-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha S, van Nimwegen E, Siggia ED. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19:i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- 25.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davidson EH. Genomic Regulatory Systems. San Diego: Academic press; 2001. [Google Scholar]
- 27.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 29.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]