Abstract
Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) represent two classes of important non-coding RNAs in eukaryotes. Although these non-coding RNAs have been implicated in organismal development and in various human diseases, surprisingly little is known about their transcriptional regulation. Recent advances in chromatin immunoprecipitation with next-generation DNA sequencing (ChIP-Seq) have provided methods of detecting transcription factor binding sites (TFBSs) with unprecedented sensitivity. In this study, we describe ChIPBase (http://deepbase.sysu.edu.cn/chipbase/), a novel database that we have developed to facilitate the comprehensive annotation and discovery of transcription factor binding maps and transcriptional regulatory relationships of lncRNAs and miRNAs from ChIP-Seq data. The current release of ChIPBase includes high-throughput sequencing data that were generated by 543 ChIP-Seq experiments in diverse tissues and cell lines from six organisms. By analysing millions of TFBSs, we identified tens of thousands of TF-lncRNA and TF-miRNA regulatory relationships. Furthermore, two web-based servers were developed to annotate and discover transcriptional regulatory relationships of lncRNAs and miRNAs from ChIP-Seq data. In addition, we developed two genome browsers, deepView and genomeView, to provide integrated views of multidimensional data. Moreover, our web implementation supports diverse query types and the exploration of TFs, lncRNAs, miRNAs, gene ontologies and pathways.
INTRODUCTION
It has become increasingly clear that eukaryotic genomes encode thousands of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) (1–4). Emerging evidence is revealing that lncRNAs and miRNAs serve as the nodes of signaling networks that regulate cancer, apoptosis, proliferation, differentiation and stem cell biology (1,2,5–8). However, the majority of studies that address these types of RNAs focus on defining the regulatory functions of lncRNAs and miRNAs, whereas few investigations are directed toward assessing how the lncRNA and miRNA genes themselves are transcriptionally regulated.
The major limitation in identifying the transcriptional regulatory relationships of lncRNAs and miRNAs has been the high false-positive rates of predictive algorithms for transcription factor binding sites (TFBSs) (9). Recently, chromatin immunoprecipitation with massively parallel DNA sequencing (ChIP-Seq) has provided a powerful way to identify TFBSs. The application of the ChIP-Seq technique has significantly reduced the rate of false-positive predictions of TFBSs (10–12). However, although ChIP-Seq technology can reliably identify TFBSs, few studies have used ChIP-Seq data to explore the transcriptional regulation of lncRNAs and miRNAs. For this reason, a high-quality integrated database that could facilitate the annotation and analysis of the transcriptional regulation of lncRNAs and miRNAs from ChIP-Seq data will be of great utility in the study of both the regulation of lncRNAs and miRNAs by TFs and the roles of this regulation in human diseases.
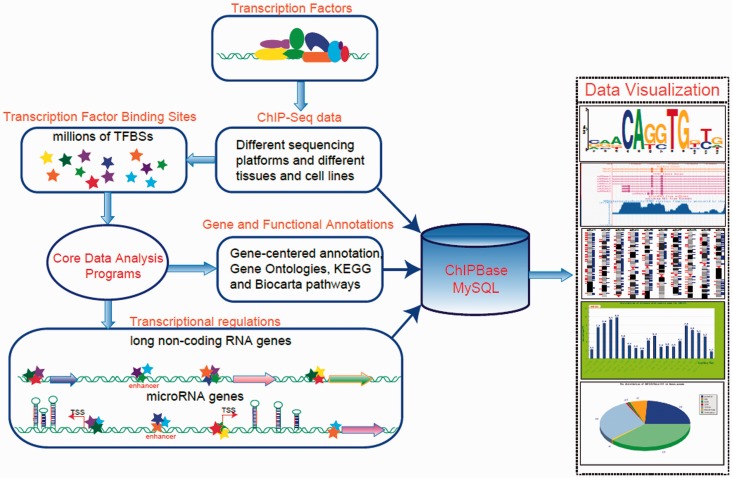
In this study, we developed the ChIPBase to facilitate the integrative and interactive display, as well as the comprehensive annotation and discovery, of TF-lncRNA and TF-miRNA interaction maps from ChIP-Seq data that were generated from diverse tissues and cell lines from six organisms: human, mouse, dog, chicken, Drosophila melanogaster and Caenorhabditis elegans (Figure 1). ChIPBase contains tens of thousands of TF-lncRNA and TF-miRNA regulatory relationships, as well as millions of TFBSs (Table 1). In addition, two novel web servers and two genome browsers were developed to comprehensively explore the relationships of TFs and ncRNAs from ChIP-Seq data.
Figure 1.
A system-level overview of the core framework of ChIPBase. All of the results that are generated by ChIPBase are deposited in MySQL relational databases and displayed in the visual browser and web page.
Table 1.
The data that are incorporated into ChIPBase
| Species | Experiments | TFBSs | TFBS cluster | TF-miRNA | TF-lncRNA |
|---|---|---|---|---|---|
| Human | 329 | 6 134 098 | 1 525 778 | 41 809 | 640 741 |
| Mouse | 119 | 2 042 652 | 478 331 | 7306 | 109 383 |
| Dog | 2 | 65 674 | 52 368 | 149 | / |
| Chicken | 3 | 40 635 | 37 108 | 391 | / |
| D. melanogaster | 53 | 160 202 | 29 565 | 1788 | 98 710 |
| C. elegans | 37 | 216 992 | 42 666 | 1790 | / |
These statistics indicate the numbers of sequencing experiments (ChIP-Seq), TFBSs, TFBS clusters (transcription factor binding clusters), TF-lncRNA interactions and TF-miRNA interactions that are incorporated into ChIPBase. These data are from six organisms, namely, human, mouse, dog, chicken, D. melanogaster and C. elegans. Known lncRNAs for dog, chicken and C. elegans are not available, and therefore the TF-lncRNA interactions for these species are denoted as ‘/’ in the table.
MATERIALS AND METHODS
A total of 543 ChIP-Seq peak data sets for 252 different transcription factors were compiled from multiple related studies and downloaded from the NCBI GEO database (13), the ENCODE (14) and modENCODE (15,16) databases, or the Supplementary Data of the original research articles (Supplementary Table S1). We have also manually curated metadata (such as TF name, refSeq accession number, gene symbols and detailed descriptions and expression patterns of TFs) to ensure annotation consistency. These peak data sets were converted to latest genome version using liftOver tool from the UCSC genome browser website (17), and peaks whose genomic regions could not be transformed into latest version of the genome were discarded. In addition, some data sets in BedGraph format were downloaded to construct peak tracks and displayed in our deepView browser to allow users to check TFBSs. In each species, TFBSs from different transcription factors and many different cell lines were combined and sorted according to their genomic positions. And then, the overlapping TFBSs were grouped into clusters and were imported into database; each cluster included at least one TFBS. Known transcription factor binding matrices were downloaded from the JASPAR (18), Transfac (19), Cistrome (20) and UniPROBE (21) databases.
All of the known lncRNAs or large intergenic non-coding RNAs were downloaded from the Supplementary Data of the six original research articles that addressed these RNAs (22–27) or extracted from Ensembl (28), refSeq (17) and UCSC Bioinformatics website (17). Known functional lncRNAs were downloaded from lncRNAdb database (29). All of the known miRNAs were downloaded from miRBase [release 17.0, (30)]. miRNA targets were downloaded from starBase database (31). All of the refSeq genes were downloaded from the UCSC bioinformatics websites (17). Other known non-coding RNAs were downloaded from the Ensembl database (28) or the UCSC websites (17) or were obtained from the relevant literature. The human (UCSC hg19), mouse (UCSC mm9, NCBI Build 37), dog (UCSC canFam2), chicken (UCSC galGal3), D. melanogaster (UCSC dm3) and C. elegans (WS190) genome sequences were downloaded from the UCSC bioinformatics websites (17).
Pre-miRNAs were grouped into transcriptional units. TFs might not almost exclusively bind at proximal promoters of lncRNAs or miRNAs. For protein-coding genes, more than half of the observed binding events are distal events (32). Moreover, the distances between transcription initiation sites (TSSs) and miRNA genes dramatically vary, ranging from a hundred bases to thousands of bases upstream (33,34). To incorporate proximal and distal binding events (32), for intergenic miRNAs, the 30-kb region upstream and the 10-kb region downstream of the TSS of the first pre-miRNA in the same transcriptional unit were chosen as the regulatory domain (32) of the examined miRNAs. For intronic miRNAs, the 30-kb region upstream and the 10-kb region downstream of TSS of the host genes contained miRNAs were chose as the regulatory domain of the examined miRNAs. The same strategy was used for lncRNAs; we chose a 30-kb region upstream and a 10-kb region downstream of the TSS of each lncRNA as the regulatory domain (32) of each lncRNA. Five-kb upstream region and 1-kb downstream region of each lncRNA and miRNA were chosen as promoter region (32). In each species, regulatory domains/regions of each lncRNA/miRNA were intersected with TFBSs of each data set to identify TFs that regulated the examined ncRNAs. And then, TFBSs overlapping with regulatory domains and corresponding lncRNAs were imported into MySql database.
DATABASE CONTENT
The genome-wide binding maps and high-occupancy target regions of transcription factors
We integrated ∼8.7 million TFBSs from 543 ChIP-Seq experiments in various tissues or cell lines to provide comprehensive genome-wide transcription factor binding profiles. To provide more useful information, we generated extensive annotations and analyses for transcription factors and TFBSs. For each TFBS, we identified the nearest/target gene and the distance between the site and the gene, as well as the expression pattern of the TF and its target gene in various tissues or cell lines (Supplementary Figure S1). For each ChIP-Seq experiment, we identified the distribution of TFBSs in the body of the gene and the distribution of the distances of the TFBSs that are associated with the TSSs of the nearest genes, and we provided descriptions of the ChIP-Seq experiments and the expression patterns of the TFs (Supplementary Figure S2). In addition, we offered the chipGO and chipKEGG tools to explore the features of the lists of TF-target interactions that are derived from the ChIP-Seq data (Figure 1 and Supplementary Figure S3).
To directly investigate potential high-occupancy target (HOT) regions on a genome-wide scale, we grouped ∼8.7 million TFBSs into ∼2 million clusters (Table 1). Each cluster contains between 1 and 74 transcription factors. We designated the genomic locations that were bound by many TFs as HOT regions. For instance, we identified 26 664 HOT regions that were bound by ≥15 factors in the human genome. In addition, we generated distribution maps of the numbers of transcription factors in the clusters. The maps are presented in the form of cluster peaks, which are displayed in our deepView genome browser (Figure 1 and Supplementary Figure S4). This display method allows us easily to determine HOT regions of TFs. We also identified the nearest/target genes of these clusters and created a web interface to display this information (Figure 1 and Supplementary Figure S4).
The annotation and identification of TF-lncRNA and TF-miRNA regulatory relationships
To investigate TF–lncRNA and TF–miRNA regulatory relationships, the regulatory domains (see the Materials and Methods section for a detailed description) of lncRNAs and miRNAs were intersected with all TFBSs from diverse tissues and cell lines. In total, we identified ∼848 834 TF-lncRNA regulatory relationships between 221 TFs and 38 293 lncRNA transcripts, as well as 53 233 TF-miRNA regulatory relationships between 249 TFs and 2294 miRNA clusters (Table 1). Because of its integration of the large number of high-resolution ChIP-Seq data from diverse tissues and cell lines, this analysis provides an enhanced resolution of these regulatory relationships. Moreover, to enable us to explore the interplay between miRNA transcriptional and posttranscriptional regulation, we integrated the targets of miRNAs from our starBase database (31) into the TF-miRNA networks. Cytoscape (web version) (35) were used to display and draw the TF-miRNA and miRNA-target networks.
WEB INTERFACE
The use of the deepView and genomeView genome browsers for the comparative analysis of TFBSs and the TF–lncRNA or TF–miRNA regulatory relationships
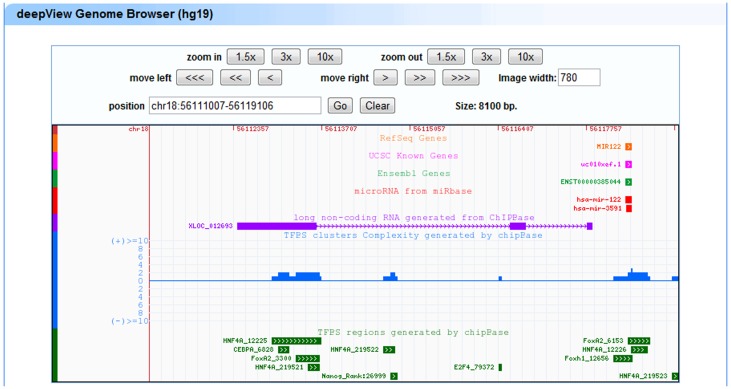
The large quantity of TFBSs and high-throughput ChIP-Seq data has increased the demand for visual tools that allow for the rapid visual correlation of different types of information. To enable the user to browse seamlessly along the genome and to zoom effortlessly in a very large set of ChIP-Seq data, our improved deepView genome browser (31,36) was developed. This browser provides an integrated view of TFBSs, lncRNAs, miRNAs, protein-coding genes, TF cluster peaks and TF clusters (Figure 2). In the deepView genome browser, the ‘zoom out’ or ‘zoom in’ button can be used to extend or shrink the width of the displayed coordinate range. A click on a track item (e.g. a miRNA, lncRNA or TFBS) of interest launches a multiple-alignment trace viewer that displays all of the traces that are relevant to the item in question or links to external resources, such as NCBI, UCSC and miRBase, that can be used to obtain more comprehensive information.
Figure 2.
Illustrative screen shots from the deepView browser. The deepView browser provides TFBSs that have been identified from ChIP-Seq data, predicted TFBSs, lncRNAs, miRNAs, protein-coding genes and TFBS clusters.
To provide the whole-genome-scale visualization of large-scale TFBSs, miRNAs and lncRNAs, a new genome browser, genomeView, was developed in this study (Figure 3). The user of this browser can view data for a single ChIP-Seq experiment across the entire genome in the context of miRNAs or lncRNAs. TFBSs and miRNAs or lncRNAs are displayed for each location in the genome as a profile over the chromosome ideogram. This feature allows the user to quickly observe genome-scale patterns in the regulatory data and identify regions of interest for further visualization in our deepView genome browser.
Figure 3.
Snapshot of the genomeView browser. The genomeView browser provides the whole-genome-scale visualization of large-scale TFBSs, miRNAs and lncRNAs.
The web-based exploration of TF-lncRNA and TF-miRNA regulatory relationships
We provide two web interfaces, LncRNA and MicroRNA, which may be used to display the TF-lncRNA and TF-miRNA interaction relationships, respectively (Supplementary Figures S5 and S6). Users can browse the relationships by entering a lncRNA name. When a user starts typing a lncRNA name in the search box, suggested lncRNA names are displayed in the list box. The user can then either choose a lncRNA from the list box or finish typing a full gene name. The user can also select a TF and search for lncRNAs that are regulated by the selected TF. If users do not enter lncRNA name and TF name, webpage will output all the TF-lncRNA interactions. Users can download these interactions to construct more complex networks composed by dozens to hundreds of lncRNAs. The results of the search are listed as the TF-lncRNA table. For the lncRNA interface, the numbers of TFBSs for each lncRNA in are indicated in a table. The users can click on a number within the table to launch a detailed page that provides further information about the TF-lncRNA interaction in question. The user can also click on the title of the table to sort TF-lncRNA interactions according to various features, such as the number of TFBSs, the lncRNA names or the TF names. The detailed information for a TF-lncRNA interaction includes a description of the TF gene and its distance to the start site of the lncRNA (Supplementary Figure S5). The ‘references’ section enables the retrieval of the primary articles yielding the annotation data. Click the article title link to visit the NCBI PUBMED website.
The microRNA interface is organized similarly to the LncRNA interface. The user can select a miRNA and a TF gene from a drop-down menu to explore TF-miRNA interactions. The numbers of upstream and downstream TFBSs, the genomic coordinates and the distance to the start site of the miRNA are all presented in a table (Supplementary Figure S6A and B). In addition, we have constructed a webpage, Networks, to simply display TF-miRNA, miRNA-target and TF-target interactions (37) using cytoscape (web version) (35) by integrating our starBase database. Users can select different miRNA target regulated by examined miRNA to construct regulatory networks (Supplementary Figure S6C). For example, we recapitulated the published c-Myc, E2F1 and miR-20 network (38) by selecting hsa-miR-20a and miRNA-target gene E2F1 in Networks webpage (Supplementary Figure S6D).
The interface of the transcriptional regulatory for other ncRNAs (such as snoRNAs, tRNAs, snRNAs, etc.) is also provided and organized similarly to the lncRNA and miRNA interfaces. Users can explore their regulatory interactions by similar ways.
The web-based annotation of transcription factor binding regions
We also provide the annotatedTool program, which offers a simple and user-friendly interface to annotate transcription factor binding regions (TFBRs). The user is required to select an intended organism and annotated TSSs of known protein-coding genes, lncRNAs or miRNAs and then upload TFBRs in the browser extensible (BED) format. After the user has completed the data submission, a typical iteration of the annotatedTool program may require several minutes to finish. The output of this program consists of three parts: the distribution of distances between the center of the TFBR and the TSS, information about the nearest gene and a link to the deepView genome browser, which allows the user to view various features of each target region (Supplementary Figure S7).
EXAMPLE APPLICATIONS
In the following section, we will present several example applications of ChIPBase.
hsa-miR-122, a target of liver-enriched transcription factors
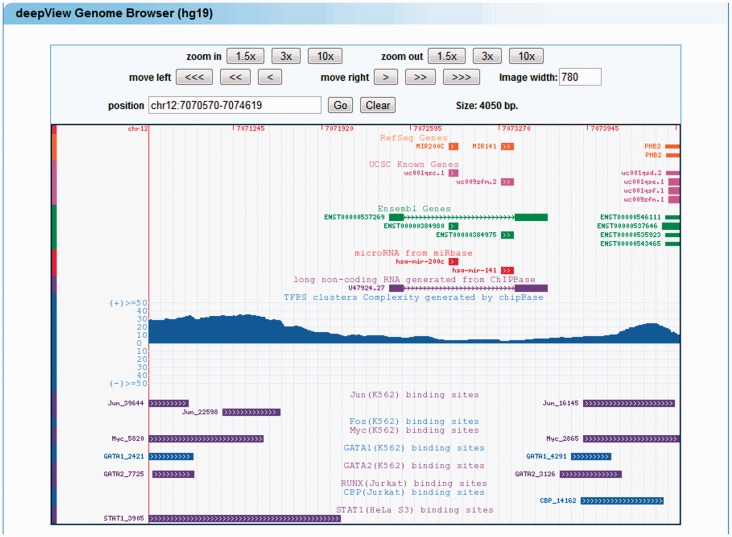
Let us assume that we are interested in liver-specific TFs as transcriptional regulators of miR-122, which is also expressed in the liver. We select three liver-enriched TFs (HNF4A, CEBPA and HNF3B/FoxA2) and the miR-122 gene in the microRNA webpage. The results page summarizes all of the query results: (i) there are five ChIP-Seq experiments for these three TFs (Supplementary Figure S8A and B); (ii) there are HNF4A ChIP-Seq data from three different experiments; and (iii) HNF4A and HNF3B/FoxA2 have multiple binding sites in the regulatory domain of miR-122 (Supplementary Figure S8A and B). We navigate to the corresponding deepView genome browser by locating the miR-122 regulatory domain, which opens up a genome browser view that effectively recapitulates the published TFBSs (39) (Figure 4).
Figure 4.
Liver-enriched transcription factors regulate the expression of hsa-miR-122. The liver-enriched transcription factors (HNF4A, CEBPA and HNF3B/FoxA2) bind to the promoter regions of the primary transcript (XLOC_012693) of hsa-miR-122.
lncRNAs as targets of embryonic stem cell transcription factors
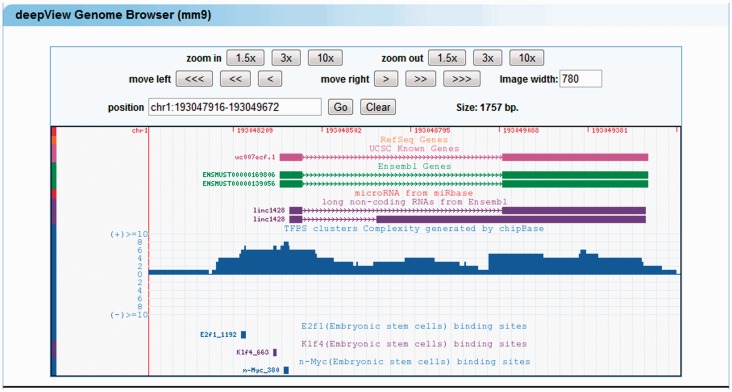
To relate a lncRNA gene to the core transcriptional circuitry of embryonic stem (ES) cells, we select nine pluripotency-associated transcription factors (including Oct4, Sox2, Nanog, c-Myc, n-Myc, Klf4, Zfx, E2F1 and Smad1) in the mouse genome at the lncRNA webpage. The results page summarizes the pluripotency-associated transcription factors that bind in the regulatory domains of lncRNAs. A click on linc1428, a known ES-cells-associated lncRNA, launches a deepView genome browser view that also recapitulates the published TFBSs of E2F1, n-Myc and Klf4 (25) (Figure 5).
Figure 5.
linc1428 as a target of ES cell transcription factors. The ES cell transcription factors (E2F1, n-Myc and Klf4) bind to promoter regions of the linc1428 lncRNA.
DISCUSSIONS AND CONCLUSIONS
Ultra-high-throughput next-generation sequencing technology has recently been developed for mapping TFBSs (10,11). In this study, we performed a large-scale integration of public TFBSs that have been generated by high-throughput ChIP-Seq technology and provide the most comprehensive TF data set for various cell types that are available at the present time. We also provide comprehensive transcriptional regulatory maps of lncRNAs and miRNAs by connecting TFs to these non-coding genes.
The transcriptional regulation of the majority of miRNAs and almost all of the discovered lncRNAs is currently unknown. Recent studies have revealed that the deregulation of miRNAs and lncRNAs is correlated with various human cancers and diseases (6,7), and that this deregulation is often due to the aberrant expression of TFs (39). In the current study, we developed the ChIPBase database to decode the transcriptional regulation of lncRNA and miRNA genes from ChIP-Seq data. We can use ChIPBase to recapitulate the known transcriptional regulatory relationships of miRNAs and lncRNAs. For example, ChIPBase can be used to identify that the liver-specific miR-122 is regulated by three liver-enriched TFs (39), and that the linc2048 lncRNA is regulated by embryonic stem cell (ESC)-associated transcription factors (25). In addition, the integration of a large quantity of ChIP-Seq data from diverse tissues and cell lines allows us to provide enhanced resolution and novel findings.
In comparison with other sources, for elucidating the transcriptional regulation of lncRNAs and miRNAs, or storing and analyzing ChIP-Seq data, the distinctive features in our ChIPBase database are as follows. (i) Our ChIPBase database is the first database that provides the transcriptional regulation maps for lncRNA genes. (ii) The other databases that are related to transcriptional regulation for miRNAs, including transmiR (40) and CircuitsDB (41), only collect computationally predicted or experimentally supported TF-miRNA interactions. By contrast, ChIPBase provides the comprehensive TF-miRNA regulatory relationships that have been identified from high-throughput ChIP-Seq data. The entries in TransmiR database contain only the name of TFs and their corresponding target miRNAs. The detail information of TFBSs and TFs, however, is not included. Also, the TransmiR database may not contain the comprehensive target miRNAs of the corresponding TFs. We used two TFs, E2F1 and MYC, whose target miRNAs are the most comprehensive in TransmiR to perform comparison between TransmiR database and our ChIPBase database. When considered only the relationships of TFs and target miRNAs, ChIPBase could identify 77% (20/26) E2F1–miRNA and 75% (21/28) MYC–miRNA relationships. These data indicated that ChIPBase could recover majority of TF-miRNA interactions documented in TransmiR. Moreover, our database also identifies tens of novel E2F1–miRNA and MYC–miRNA relationships that were not included in transmiR data. In addition, TransmiR does not contain HN4FA-miR-122 and CEBPA-miR-122 interactions described in our Example Applications sections. (iii) hmChIP (42) is a database of genome-wide ChIP data in human (hg18 version) and mouse (mm8 version). It just provides the Protein–DNA binding intensities from individual samples for user-provided genomic regions. Currently, hmChIP does not explore the transcriptional regulatory of lncRNAs or miRNAs, even for protein-coding genes. By contrast, our ChIPBase provides comprehensive transcriptional regulatory relationships of lncRNAs, miRNAs and other ncRNAs, as well as comprehensive annotation of TFBSs from 543 ChIP-Seq data sets from six organisms. (iv) To enable the user to browse seamlessly along the genome and to zoom effortlessly in a very large set of ChIP-Seq data, our improved deepView genome browser was developed to provide an integrated view of TFBSs that have been identified from ChIP-Seq data, predicted TFBSs, ncRNAs, protein-coding genes and TFBS clusters (Figures 2 and 3). (v) We developed two web tools, annotatedTool and genomeViewer to annotate and discover the transcriptional regulatory relationships of lncRNAs and miRNAs from ChIP-Seq data (Figure 1). (vi) We constructed genome-wide transcription factor binding profiles from ChIP-Seq data. Combinatorial transcription factor interactions that control the transcriptional regulations of lncRNAs and miRNAs were easily identified by searching for appropriate profiles in the genome browser (Figure 2). (vii) ChIPBase also provides the gene ontology annotation, biological pathways and expression patterns of transcription factor binding targets (Figure 1). This supplementary information may provide valuable insights into the function of each TF, lncRNA and miRNA. Finally, the data and the integrative, interactive and versatile displays that are provided by the ChIPBase database will aid future experimental and computational studies in their attempts to elucidate the regulation of lncRNAs and miRNAs by TFs and assess the roles of these regulatory relationships in human diseases.
FUTURE DIRECTIONS
As a means of comprehensively integrating ChIP-Seq data, ChIPBase is expected to provide considerable resources to assist researchers that are investigating the TF-lncRNA and TF-miRNA regulatory networks and examining the biological functions of the genes and ncRNAs with expression levels that are controlled by transcription factors. As ChIP-Seq technology is applied to a broader set of species, cell lines, tissues and conditions, ChIPBase will continue to be developed and refined toward the achievement of the following goals: (i) the better integration and cross-comparison of diverse ChIP-Seq data sets and data resources. (ii) the correlation of these diverse ChIP-Seq data with lncRNAs and miRNAs. (iii) we will continue to extend the amount of storage space and improve the performance of our computer servers for storing and analysing these new data, and improve the database to accept upload of new data by the users. In addition, we intend to integrate the epigenomic data that are generated by ChIP-Seq technology into ChIPBase to improve our understanding of eukaryotic regulatory networks.
AVAILABILITY
ChIPBase is freely available at http://deepbase.sysu.edu.cn/chipbase/. The ChIPBase data files can be freely downloaded and used in accordance with the GNU Public License.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1, Supplementary Figures 1–8 and Supplementary References [15,16,43–98].
FUNDING
Ministry of Science and Technology of China, National Basic Research Program [No. 2011CB811300]; National Natural Science Foundation of China [No. 31230042, 30900820, 81070589]; funds from Guangdong Province [No. S2012010010510]; The project of Science and Technology New Star in ZhuJiang Guangzhou city [No. 2012J2200025]; Fundamental Research Funds for the Central Universities [No. 2011330003161070]; China Postdoctoral Science Foundation [No. 200902348]; Guangdong Province Key Laboratory of Computational Science and the Guangdong Province Computational Science Innovative Research Team (in part). Funding for open access charge: Ministry of Science and Technology of China, National Basic Research Program [No. 2011CB811300].
Conflict of interest statement. None declared.
REFERENCES
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat . Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum . Mol. Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 10.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robasky K, Bulyk ML. UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2011;39:D124–D128. doi: 10.1093/nar/gkq992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP. Identification and properties of 1119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol. Evol. 2012;4:427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai X, Schmitz U, Gupta SK, Bhattacharya A, Kunz M, Wolkenhauer O, Vera J. Computational analysis of target hub gene repression regulated by multiple and cooperative miRNAs. Nucleic Acids Res. 2012;40:8818–8834. doi: 10.1093/nar/gks657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friard O, Re A, Taverna D, De Bortoli M, Cora D. CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinformatics. 2010;11:435. doi: 10.1186/1471-2105-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L, Wu G, Ji H. hmChIP: a database and web server for exploring publicly available human and mouse ChIP-seq and ChIP-chip data. Bioinformatics. 2011;27:1447–1448. doi: 10.1093/bioinformatics/btr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of beta-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38:5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin LA, Biggin MD, Eisen MB. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. Plos Biol. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cicatiello L, Mutarelli M, Grober OMV, Paris O, Ferraro L, Ravo M, Tarallo R, Luo SJ, Schroth GP, Seifert M, et al. Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am. J. Pathol. 2010;176:2113–2130. doi: 10.2353/ajpath.2010.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J. Biol. Chem. 2010;285:1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grober OM, Mutarelli M, Giurato G, Ravo M, Cicatiello L, De Filippo MR, Ferraro L, Nassa G, Papa MF, Paris O, et al. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics. 2011;12:36. doi: 10.1186/1471-2164-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heng JCD, Feng B, Han JY, Jiang JM, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. Plos Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu M, Yu JD, Taylor JMG, Chinnaiyan AM, Qin ZHS. On the detection and refinement of transcription factor binding sites using ChIP-Seq data. Nucleic Acids Res. 2010;38:2154–2167. doi: 10.1093/nar/gkp1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang JS, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 2009;23:208–222. doi: 10.1101/gad.1750709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de la Calle-Mustienes E, Smeenk L, Rinne T, Parsaulian L, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. Plos Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu XY, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 63.Lee BK, Bhinge AA, Iyer VR. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011;39:3558–3573. doi: 10.1093/nar/gkq1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lo KA, Bauchmann MK, Baumann AP, Donahue CJ, Thiede MA, Hayes LS, des Etages SA, Fraenkel E. Genome-wide profiling of H3K56 acetylation and transcription factor binding sites in human adipocytes. PLoS One. 2011;6:e19778. doi: 10.1371/journal.pone.0019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacIsaac KD, Lo KA, Gordon W, Motola S, Mazor T, Fraenkel E. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput. Biol. 2010;6:e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin D, Pantoja C, Minan AF, Valdes-Quezada C, Molto E, Matesanz F, Bogdanovic O, de la Calle-Mustienes E, Dominguez O, Taher L, et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes (vol 18, pg 708, 2011) Nat. Struct. Mol. Biol. 2011;18:1084–1084. doi: 10.1038/nsmb.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda-Carboni GA, Guemes M, Bailey S, Anaya E, Corselli M, Peault B, Krum SA. GATA4 regulates estrogen receptor-alpha-mediated osteoblast transcription. Mol. Endocrinol. 2011;25:1126–1136. doi: 10.1210/me.2010-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One. 2011;6:e19470. doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 72.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl Acad. Sci. USA. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao YJ, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 75.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZDD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. Plos Genet. 2011;7:e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol. Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol. Cell. Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 83.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verzi MP, Shin H, Ho LL, Liu XS, Shivdasani RA. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 2011;31:2026–2039. doi: 10.1128/MCB.01250-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vivar OI, Zhao XY, Saunier EF, Griffin C, Mayba OS, Tagliaferri M, Cohen I, Speed TP, Leitman DC. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J. Biol. Chem. 2010;285:22059–22066. doi: 10.1074/jbc.M110.114116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T, Balzereit D, Wruck W, Soldatov A, Vingron M, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem. 2011;286:23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo . EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 93.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu HB, Johnson R, Kunarso G, Stanton LW. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 2011;21:1284–1293. doi: 10.1101/gr.114488.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu JD, Yu JJ, Mani RS, Cao Q, Brenner CJ, Cao XH, Wang XJ, Wu LT, Li J, Hu M, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu M, Riva L, Xie HF, Schindler Y, Moran TB, Cheng Y, Yu DN, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao L, Glazov EA, Pattabiraman DR, Al-Owaidi F, Zhang P, Brown MA, Leo PJ, Gonda TJ. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. Plos Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]