Background: Mediator is a general coactivator complex for regulated RNA polymerase II transcription.
Results: Mediator is phosphorylated on most subunits, and phosphorylation can contribute to function.
Conclusion: Post-translational modifications of a transcription coactivator can contribute to its function in gene regulation.
Significance: Transcription coactivators can be strongly modified post-translationally, and this can contribute to function.
Keywords: Coactivator Transcription, Gene Regulation, Gene Transcription, Mass Spectrometry (MS), Phosphorylation
Abstract
The multiprotein complex Mediator is a coactivator of RNA polymerase (Pol) II transcription that is required for the regulated expression of protein-coding genes. Mediator serves as an end point of signaling pathways and regulates Pol II transcription, but the mechanisms it uses are not well understood. Here, we used mass spectrometry and dynamic transcriptome analysis to investigate a functional role of Mediator phosphorylation in gene expression. Affinity purification and mass spectrometry revealed that Mediator from the yeast Saccharomyces cerevisiae is phosphorylated at multiple sites of 17 of its 25 subunits. Mediator phosphorylation levels change upon an external stimulus set by exposure of cells to high salt concentrations. Phosphorylated sites in the Mediator tail subunit Med15 are required for suppression of stress-induced changes in gene expression under non-stress conditions. Thus dynamic and differential Mediator phosphorylation contributes to gene regulation in eukaryotic cells.
Introduction
Regulation of transcriptional activity is dependent on gene-specific transcription factors that respond to environmental signals. In eukaryotic cells, these transcription factors require co-activator complexes to transmit signals to the RNA polymerase (Pol)8 II machinery. Among these co-activators, the Mediator complex plays a key role by interacting with transcription factors and the Pol II machinery. The ability of the Mediator to form an interface between gene-specific regulators and the Pol II initiation complex led to the assumption that Mediator functions as an adaptor that is recruited to promoters by gene-specific factors (1). Mediator complexes have been identified in yeast (2, 3), plants (4), and metazoans (1). Mediator dysfunction leads to a variety of diseases, including cardiovascular diseases (5, 6), metabolic disorders (5), mental retardation (5), and cancer (1, 5). Mediator from Saccharomyces cerevisiae consists of 25 subunits that are organized in the head, middle, tail, and kinase modules (7, 8). Whereas the head module interacts with the Pol II machinery, the middle and the tail modules bind regulatory factors.
Mediator is a target of cellular signaling pathways, but it is poorly understood how it integrates regulatory signals and how it transfers the output to the Pol II machinery. In human Mediator, subunit Med1 is targeted by the thyroid hormone receptor (9) and is phosphorylated by ERK kinase during thyroid signaling (10). The human Mediator subunit Med23 is an end point of the insulin-signaling pathway, which induces MAP kinase-dependent activation of Elk1 (11, 12). In yeast Mediator, subunit Med15 is targeted by Oaf1, a transcription activator involved in sensing fatty acid levels (13). Subunit Med15 also interacts with Pdr1, a factor involved in multidrug resistance (14, 15). The yeast Mediator subunits Med2, Med4, and Med13 are phosphorylated, and these phosphorylation events play a role in transcription, Kin28-dependent processes, and the Ras/PKA pathway, respectively (16–19). However, Mediator phosphorylation and its possible contribution to Mediator function have not been investigated in a systematic way.
A paradigm for a conserved signaling pathway is the response of yeast cells to high salt concentrations. Osmotic stress activates the conserved MAP kinase cascade, which leads to cell cycle arrest (20, 21), affects interaction between proteins and chromatin (22), and induces transcription of stress-responsive genes (23–26). Osmotic stress response includes three phases (27–29). During the initial shock phase, transcription activity is severely reduced. Stress-induced genes are heavily transcribed within 12–24 min (induction phase), and cells then fully adapt to growth in high-salt conditions (recovery phase). Response to osmotic stress goes along with significant changes in the phosphoproteome (30).
Due to recent technological and methodological advances, mass spectrometry (MS)-based proteomics has established itself as a powerful and versatile approach for global and quantitative investigation of many aspects of biology (31). Stable isotope labeling of amino acids in culture (SILAC) (32) is one of the most popular quantitative proteomics methods with a wide range of biological applications (33). Although mass spectrometry is well suited for the study of nearly all post-translational modifications (34), it has proven particularly successful in characterizing phosphorylation dynamics (35). Due to the availability of efficient techniques for phosphopeptide enrichment, tens of thousands of phosphorylation sites can be identified in a single experiment (36–39).
Here, we used affinity purification and MS to show that yeast Mediator is phosphorylated at 17 of its 25 subunits in vivo. We then used SILAC to show that the phosphorylation level at some of the identified sites changes during the osmotic stress response. Finally, we used mutagenesis in combination with dynamic transcriptome analysis (DTA) and comparative DTA (cDTA) to show that phosphorylated amino acid residues on the Mediator subunit Med15 are involved in suppression of stress-induced changes in gene expression under non-stress conditions. These results demonstrate that Mediator is extensively modified post-translationally and that Mediator phosphorylation contributes to transcription regulation of genes responding to environmental changes.
MATERIALS AND METHODS
Yeast Strains and Growth
For purification of endogenous Mediator, we introduced a tandem affinity purification (TAP) tag at the N terminus of Med17 in S. cerevisiae strain RS453 with mating type α (40). For SILAC, we modified this strain by disruption of the lys1 gene with KanMX6 to introduce lysine auxotrophy. Cells were grown in YPD or SILAC medium (6.7 g/liter Formedia yeast nitrogen base without amino acids, 2% glucose, 200 mg/liter adenine, 200 mg/liter tyrosine, 10 mg/liter histidine, 10 mg/liter methionine, 60 mg/liter phenylalanine, 40 mg/liter tryptophan, 20 mg/liter uracil, 60 mg/ml leucine, arginine 20 mg/liter) at 30 °C to late log-phase. For stable isotope labeling, SILAC medium contained 30 mg/liter 13C6-, 15N2-lysine (heavy lysine, Cambridge Isotopes) for the salt stress sample, or 30 mg/ml 12C6-, 14N2-lysine (light lysine, Sigma Aldrich) for the control. Salt stress was introduced by adding crystalline sodium chloride to the heavy lysine-containing medium to a final concentration of 0.5 m. After incubation for 20 min, equal amounts the two populations were mixed.
Mediator Purification
Cells were lysed by beat beating (500 rpm, 4 min, 2 cycles) using glass beads (0.5-mm diameter). Purification of endogenous Mediator using Protein A/IgG (GE Healthcare) affinity precipitation was performed as described (40), except that all buffers contained 1 mm DTT, protease inhibitors (0.5 m phenylmethylsulfonyl fluoride, 2 mm benzamidine, 1 μm leupeptin, 2 μm pepstatin), and phosphatase inhibitors (40 mm p-nitrophenylphosphate, 2 mm sodium-pyrophosphate, 2 mm sodium orthovanadate, 50 mm sodium fluoride). IgG-bound proteins were washed with high salt buffer (250 mm sodium chloride, 50 mm Tris/HCl, pH 7.5, MgCl2, 0.15% m/w Nonidet P-40, 1 mm DTT) and low-salt buffer (100 mm sodium chloride 50 mm Tris/HCl, pH 7.5, MgCl2, 1 mm DTT). Proteins were cleaved from IgG beads by tobacco edge virus (TEV) protease for 60 min at 18 °C. Eluted proteins were precipitated by adding four volumes of methanol, one volume of chloroform, and three volumes of protease-free double-distilled water. After centrifugation, the pellet was washed with four volumes of methanol and centrifuged again. Remaining methanol was discarded, and the protein precipitate was dried. A sample was analyzed by 4–12% SDS-PAGE (NUPAGE, Invitrogen).
Mass Spectrometry
The protein pellet was dissolved and denatured in 8 m urea (Roth), reduced with 1 mm DTT for 45 min and alkylated with 5 mm iodoacetamide for 45 min. Proteins were digested in solution either with endoproteinase trypsin or Lys-C (Wako) overnight at room temperature. Phosphopeptide enrichment was performed using titanium dioxide (TiO2) beads (41). LC-MS/MS analysis was performed on an LTQ-Orbitrap instrument (Thermo Fisher) connected to a nanoflow HPLC system (Agilent 1100 or Proxeon EASY-nLC system) through a Proxeon nanoelectrospray ion source. Peptides were separated on an in-house packed 75 μm reversed-phase C18 column. Survey scans were acquired in the Orbitrap analyzer, and the 10 most intense precursor ions were subjected to collision-induced fragmentation and acquisition in the ion trap. The instrument was operated with the “lock mass option” (42), and multistage activation was enabled to improve phosphopeptide fragmentation. The MS raw data files were processed with MaxQuant (version 1.1.1.18) (43, 44). The MS/MS spectra were searched against the concatenated target decoy ORF protein database, concatenated with reversed versions of all sequences and combined with the most commonly observed contaminants. Enzyme specificity was set to LysC and up to three missed cleavages, and three labeled lysine residues were allowed. Cysteine carbamidomethylation was considered as a fixed modification, and methionine oxidation, protein N-acetylation, and phosphorylation on serine, threonine, and tyrosine residues were set as variable modifications.
Mutant Yeast Strains
D7P and D30P mutant strains were generated in three steps. First, to replace selected phosphorylated serine and threonine residues in the Med15/YOL051w coding sequence by alanine (supplemental Tables S3 and S4), we synthesized (GeneART) a DNA construct containing the 5′-UTR (583 bp upstream). The modified coding sequence of Med15/YOL051w (D30P mutant sequence), and the 3′-UTR (624 bp downstream) were inserted in vector prs-315 (AMP/Leu) (45) under the control of the Cen6/ARSH4 promoter with the use of NotI/SalI restriction sites. Second, a Med15/YOL051w::URA3 strain was generated by integration of a Ura3 cassette harboring overlapping sequences to Med15/YOR051w 5′- and 3′-UTR regions into BY4741 strain (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, Euroscarf) by homologous recombination. Med15/YOL051w::URA3 strain was selected twice on URA minus medium and confirmed by sequencing. Third, the URA3 cassette was removed by D7P or D30P mutant sequence using homologous recombination and selected twice on 5′-fluorotic acid containing medium. The exact genetic position and completeness of the genomic D7P or D30P mutant insertion were analyzed by sequencing. For mutant D7P, all steps were as above except that we synthesized (Invitrogen) a DNA construct containing the Med15 positions (2114 and 3333 bp) with the modified coding sequence of Med15/YOL051w (D7P mutant sequence), in vector prs-315 (AMP/Leu), and that we generated a Med15/YOL051w::URA3 strain by integration of a Ura3 cassette harboring overlapping sequences to Med15/YOR051w between the relative positions 2139 and 3243 bp into BY4741 strain (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, Euroscarf).
DTA
For DTA, the D30P mutant and wild-type strains (BY4741, MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, Euroscarf) were transformed with plasmid YEpEBI311 (2 micron, Leu2) carrying the human equilibrative nucleoside transporter hEnt1. Cells were grown in SILAC medium without leucine. Cells were grown to mid-log-phase of 0.8 (corresponding to 1.75 × 107 cells). For control samples, 4-thiouridine (Sigma) was added to the medium and adjusted to a final concentration of 500 μm. Cells were further incubated for 6 min (labeling time) and harvested by centrifugation at 4000 rpm for 1 min. For stress samples, sodium chloride was added to the sample to a final concentration of 0.8 m. 4-Thiouridine was added 18 min after salt addition, and cells were harvested after a 6-min labeling time as above. DTA was performed with Affymetrix arrays as described previously (27).
cDTA
For cDTA, we used the published protocol (46) with the following exceptions. The D7P mutant strain, Δmed15 and wild-type strain (BY4741, MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, Euroscarf) were transformed with plasmid YEpEBI311 (2 micron, Leu2) carrying the human equilibrative nucleoside transporter hEnt1. Cells were grown in SILAC medium without leucine to mid-log-phase of 0.8 (corresponding to 1.75 × 107 cells) and labeled with 4-thiouridine for 6 min. Cells were centrifuged at 2465 × g at 30 °C for 1 min and resuspended in RNAlater solution (Ambion/Applied Biosystems). The cell concentration was determined by Cellometer N10 (Nexus) before flash-freezing in N2 (liquid). Schizosaccaromyces pombe cells were grown in YES medium overnight, diluted to A600 = 0.1 and grown to A600 = 0.8. 4-thiouridine (Sigma) was added to a final concentration of 500 μm, and cells were incubated for 6 min. Cells were centrifuged at 2465 × g at 30 °C for 1 min and resuspended in RNAlater solution (Ambion/Applied Biosystems) before flash-freezing in liquid nitrogen. The S. pombe cells were taken from a general stock to eliminate errors by experimental variations. S. pombe cells were counted by Cellometer N10 (Nexus) and mixed with S. cerevisiae cells in a 1:3 ratio, resulting in 4 × 108 cells in total. Control and stress samples were treated as described above. Total RNA extraction, labeled RNA purification, sample hybridization, and microarray scanning were as described (27, 46).
Sensitivity Screen
We tested the D7P and D30P mutant for sensitivity to high salt conditions and the Δmed15 deletion mutant for different stress conditions. Wild-type D7P and D30P mutant, Δmed15 deletion mutant cells (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YOR051w::KanMX (Euroscarf)), and Δhog1 deletion mutant (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0; YLR113w::KanMX (Euroscarf)) were grown in liquid YPD medium (1% yeast extract, 2% bacto-peptone, 2% glucose, 2% agar, 200 mg/liter adenine) to mid logarithmic phase. The cells were pelleted by centrifugation and washed twice with sterile, double distilled water. The first cell suspension was adjusted to an optical density of 1.0 at 600 nm and diluted to 10−1, 10−2, 10−3, and 10−4. The resulting cell suspensions were spotted on YPD/agar solid medium and incubated either at 25, 30, or 37 °C for 3 or 5 days. Sensitivity to high salt concentrations was tested by the same procedure except that YPD/agar solid medium contained either 0, 0.4, or 1.2 m sodium chloride (Δmed15 deletion mutant) or 0 and 1.2 m (D7P and D30P mutant), and cells were incubated at 30 °C for 5 days.
RESULTS
Mediator Is Phosphorylated on Many Sites in Vivo
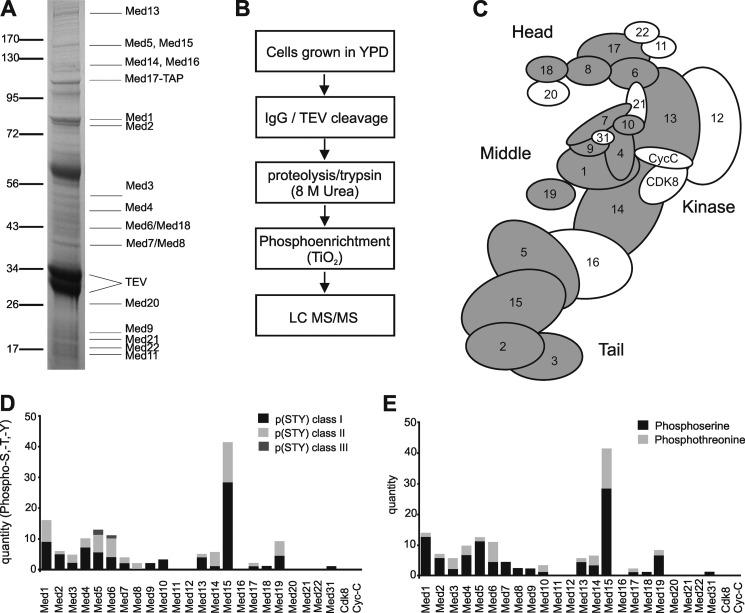
To obtain endogenous Mediator, we generated a yeast strain carrying an N-terminal TAP-tagged version of Med17. Cells were grown in YPD medium to late-logarithmic phase, and Mediator was affinity-purified in the presence of phosphatase inhibitors (Fig. 1A; see “Material and Methods”) (40, 47). Pure Mediator was digested with trypsin to generate peptides that were subjected to a phospho-enrichment procedure (41). Phosphopeptides were analyzed by liquid chromatography-tandem mass-spectrometry (LC-MS/MS) on a high-resolution linear ion trap Orbitrap instrument (Fig. 1B; see “Material and Methods”).
FIGURE 1.
Mediator is highly phosphorylated in vivo. A, SDS-PAGE analysis of endogenous Mediator proteins that were co-purified with TAP-Med17 from wild-type S. cerevisiae. Protein bands were stained with Coomassie Blue and identified by mass spectrometry (TEV, TEV protease bands). B, diagram illustrating the workflow for analysis of Mediator phosphosites by high-performance LQ MS/MS. C, schematic view of Mediator with Med subunits labeled with numbers and highlighted in gray if they were found to be phosphorylated under normal growth C conditions. The four modules of Mediator are also indicated. D, bar chart showing the number of phosphosites classified into p(STY) classes I (black), II (light gray), and III (dark gray). E, bar chart showing the number of phosphorylated serine (black), threonine (light gray), and tyrosine (dark gray) residues in Mediator proteins.
The phosphopeptides contained 125 unique phosphorylated sites that were heterogeneously distributed over 17 different Mediator subunits (Fig. 1C). Phosphosites were concentrated in the Mediator middle (36%) and tail (48%) modules. Of the 125 phosphosites, 37 have previously been reported (16, 17, 30, 48–54), whereas 88 were novel (supplemental Table S1). Because phosphosite localization can be challenging when a peptide contains multiple serine, threonine, or tyrosine residues, we assigned each phosphosite to one of three localization classes. Phosphosites were categorized into p(STY)-class I, II, or III, if the localization probability value for the phospho group was above 0.75, between 0.25 and 0.75, or below 0.25, respectively (35, 55). Of the 125 phosphosites, 82, 38, and 5 were ascribed to class I, II, and III, respectively (Fig. 1, D and E). These results reveal that Mediator is phosphorylated on many sites during exponential cell growth in vivo (Fig. 2, A–C; supplemental Table S1).
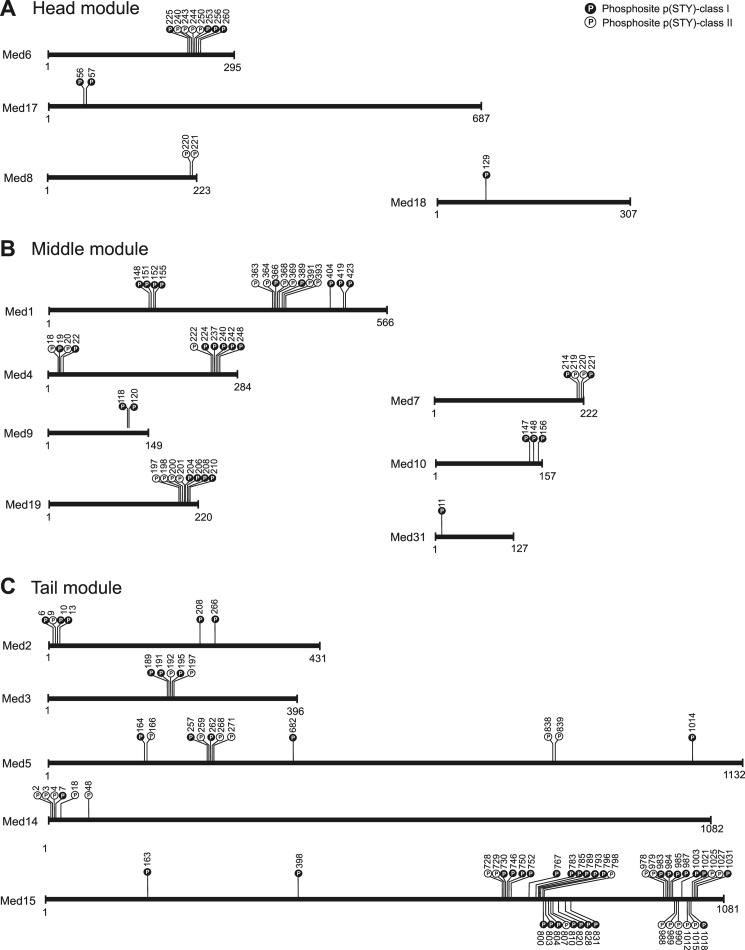
FIGURE 2.
Distribution of phosphorylation sites in Mediator subunits. A–C, schematic illustration of phosphosite positions on the primary structure of Mediator subunits belonging to the head (A), middle (B), and tail (C) modules. Phosphosites classified in p(STY)-class I (high localization probability) are highlighted in black and p(STY)-class II (medium localization probability) in white, respectively.
Mediator Phosphorylation Changes During Stress
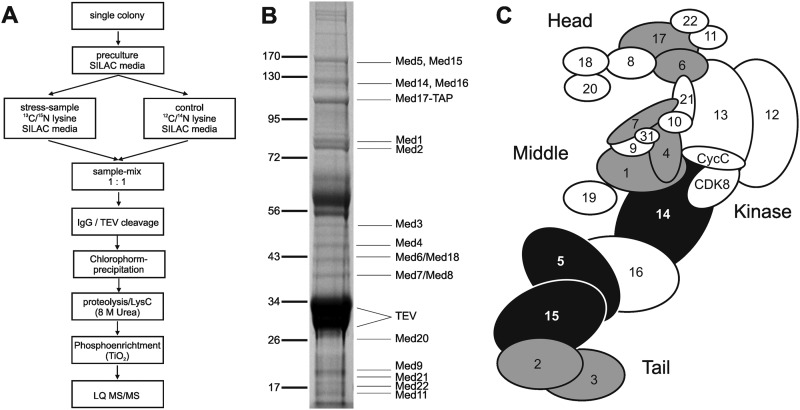
We next tested whether the phosphorylation status of Mediator changes in response to an external stimulus. We used SILAC to monitor the response of yeast cells to osmotic stress. Yeast cells auxotrophic for lysine were grown either in light or heavy (13C6/15N2) lysine-containing SILAC medium to mid-logarithmic phase (see “Material and Methods”). Sodium chloride was added to a concentration of 0.5 m to the culture grown on heavy lysine, whereas the culture grown on light lysine was left untreated. Cells were harvested 20 min after salt addition, when expression of stress mRNAs is highly induced (27, 29). Equal amounts of both cultures were combined, and Mediator was isolated. Proteins were digested with endopeptidase LysC to ensure that peptides contained at least one lysine (Fig. 3, A and B) (56). The obtained peptides cover 53% of the amino acid sequence of Mediator core proteins (supplemental Fig. S3A). Levels of 20 Mediator subunits were unchanged providing an accurate basis for data normalization (supplemental Fig. S3B).
FIGURE 3.
Mediator phosphorylation changes during osmotic stress. A, schematic diagram of SILAC experiment workflow. B, SDS-PAGE analysis of endogenous yeast Mediator preparation (SILAC) that was used as input for mass spectrometry analysis. Copurified proteins were stained with Coomassie Blue (TEV, TEV protease bands). C, schematic view of Mediator with subunits that were detected to be phosphorylated under osmotic stress conditions. Subunits with unaltered phosphorylated sites are in gray, and subunits with phosphorylated sites that show altered levels of phosphorylation are in black.
Phosphopeptides were enriched and analyzed by MS (see “Material and Methods”). We detected peptides from 21 Mediator subunits. These peptides contained 29 unique phosphosites, which derived from subunits Med1, 2, 3, 4, 5, 6, 7, 14, 15, and 17. A replicate experiment revealed 23 additional sites, resulting in a total of 52 unique phosphosites (Fig. 3C; supplemental Table S2). Of these, 15 sites in Med5 and Med15 showed significantly decreased phosphorylation with a ratio of heavy to light lysine-containing peptide intensities between 0.2 and 0.6, whereas one site in Med14 showed significantly increased phosphorylation (Table 1). All of these dynamically phosphorylated sites were located in predicted loops of the Mediator tail module (supplemental material section 2; supplemental Fig. S4), which interacts with transcription regulators (14, 57).
TABLE 1.
Mediator sites that change phosphorylation level during salt stress
The post translational modification (PTM) score assigns localization probabilities for combinations of serine, threonine and tyrosine phosphorylation based on the most intense fragment ions (44).
| Subunit | Amino acid | Position | Sequence window | pSTY-class | Localization p value | PTM score | Ratio H/L normalized by proteins |
|---|---|---|---|---|---|---|---|
| Med5 | S | 257 | TNEFVGSPSLTSP | II | 0.504487 | 35.82 | 0.60258 |
| Med5 | S | 259 | EFVGSPSLTSPQY | II | 0.349267 | 35.82 | 0.60258 |
| Med5 | T | 261 | VGSPSLTSPQYIP | II | 0.351505 | 35.82 | 0.60258 |
| Med5 | S | 262 | GSPSLTSPQYIPS | II | 0.728166 | 35.82 | 0.60258 |
| Med5 | S | 268 | SPQYIPSPLSSTK | III | 0.188482 | 35.82 | 0.60258 |
| Med5 | S | 271 | YIPSPLSSTKPPG | III | 0.190959 | 35.82 | 0.60258 |
| Med5 | S | 272 | IPSPLSSTKPPGS | II | 0.306 | 35.82 | 0.60258 |
| Med5 | T | 273 | PSPLSSTKPPGSV | II | 0.306287 | 35.82 | 0.60258 |
| Med14 | T | 1036 | DTKRLGTPESVKP | I | 0.999212 | 40.65 | 1.4734 |
| Med15 | S | 746 | TPKVPVSAAATPS | I | 1 | 139.73 | 0.59134 |
| Med15 | T | 750 | PVSAAATPSLNKT | I | 0.75299 | 139.73 | 0.59134 |
| Med15 | S | 796 | QQPTPRSASNTAK | II | 0.431756 | 62.19 | 0.28888 |
| Med15 | S | 798 | PTPRSASNTAKST | II | 0.431756 | 62.19 | 0.28888 |
| Med15 | T | 769 | GRTKSNTIPVTSI | I | 0.999999 | 121.83 | 0.23904 |
| Med15 | S | 767 | VNGRTKSNTIPVT | I | 1 | 121.83 | 0.23904 |
| Med15 | T | 800 | PRSASNTAKSTPN | II | 0.328631 | 62.18 | 0.28888 |
Med15 Phosphosites Contribute to Suppression of Stress-induced Changes in Gene Expression under Non-stress Conditions
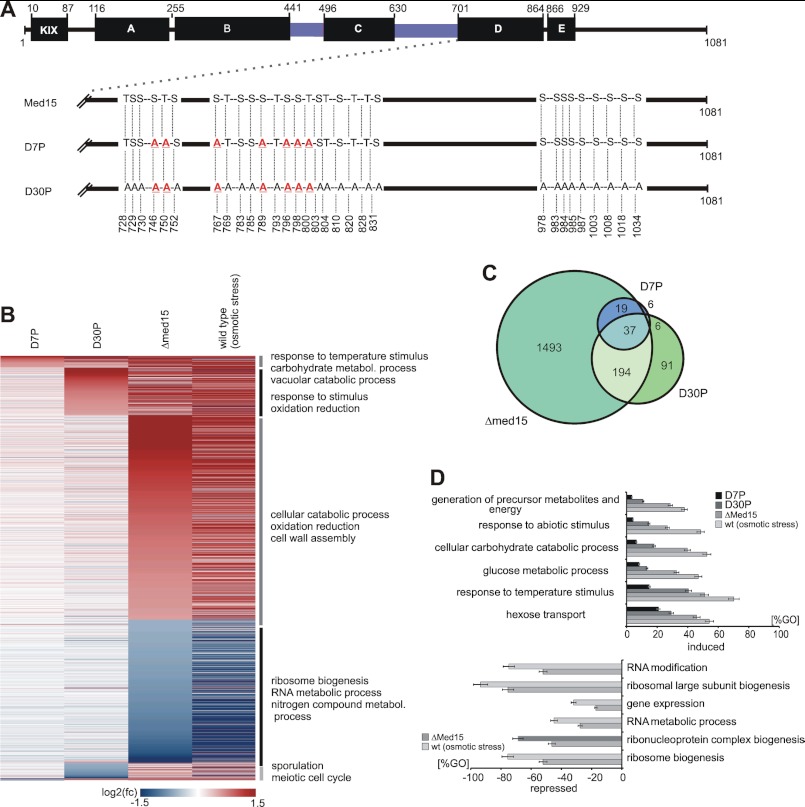
The above analysis identified 30 phosphorylated sites that cluster in the Med15 C-terminal region (Fig. 2C; supplemental Fig. S4). Seven of these sites are dynamically changed in response to osmotic stress. To test whether the phosphorylated sites are functionally involved in the regulation of transcription, we created two mutant strains that carry point mutations on selected sites (Fig. 4A; supplemental Tables S3 and S4). The first mutant, referred to as D7P, carries alanine mutations on the seven dynamically phosphorylated sites, whereas in the second mutant, D30P, all 30 phosphorylated sites clustering near the C terminus of Med15 were mutated to alanine (Fig. 4A). Mutation of phosphorylated serine and threonine residues to alanine is functionally similar to the dephosphorylated state (58). The clustering of 30 phosphosites in the Med15 C terminus suggested that combinatorial phosphorylation on multiple sites affect biological outcome (59, 60) as was shown for cell cycle control (61) and for transcription regulation (62).
FIGURE 4.
Effect of mutated phosphosites on global gene expression under normal growth conditions. A, functional organization of ScMed15. Med15 is organized in several regions as described previously (KIX-domain; A–E; black bars, which interact to different transcription factors (14, 82), Msn2 (77), Gal4 (84), Gcn4 (82, 85), Pdr1 (13, 15), Oaf1 (13)). Q-rich regions are colored blue. Mutant sequence features are shown below. The D7P mutant harbors genomic point mutations of the seven serine and threonine positions to alanine, which are dynamically phosphorylated during response to high salt concentrations. The D30P mutant harbors the complete set of all 30 phosphosites (identified during this study), which mimic the dephosphorylated state of the Med15 C terminus. B, heat map illustrating the effect of the mutated phosphosites on global gene expression (labeled mRNA fraction, 6 min labeling time). The genes are arranged by the highest induction or repression fold changes in order of D7P, D30P, and Δmed15. The horizontal lines represent genes, which are differentially expressed at least 1.5-fold in at least one of the data sets. Red indicates genes, which are induced compared with the wild-type control. Blue indicates repressed genes. Genes were ranked by highest/lowest fold-change in order of D7P, D30P, and Δmed15. C, Venn diagram illustrating the overlap between the data sets of D7P, D30P, and Δmed15. D, diagram representing enriched GO terms ranked by Fisher's exact test. Upper diagram, the bars represent the percentage of genes induced in the D7P (black), D30P (dark gray), and Δmed15 (middle gray) strains relative to all genes in the GO term. Lower diagram, the bars represent the percentage of genes repressed in the D7P (black), D30P (dark gray), and Δmed15 (light gray) mutant relative to all genes in the GO term.
We performed DTA for D30P mutant and wild-type cells under normal conditions. DTA uses metabolic RNA labeling to globally monitor mRNA synthesis and decay rates with high sensitivity (27). We carried out metabolic RNA labeling by addition of 4-thiouridine for 6 min, extracted RNA, separated labeled RNA from pre-existing RNA, and analyzed the labeled RNA to generate expression profiles on Affymetrix microarrays (27). We performed cDTA under identical experimental conditions for the D7P mutant, a strain lacking subunit Med15 (Δmed15) and the wild-type strain under normal conditions. cDTA allows for a direct comparison of synthesis and decay rates in different yeast strains (46).
Mutation of the seven dynamically phosphorylated sites in the D7P strain significantly altered the expression of 64 genes. We found 53 genes induced, suggesting a mild negative effect of the dynamic phosphosites on transcription. We searched for significantly enriched GO terms, which are ranked by Fisher's exact test. Among the subset of 53 induced genes, we identified a significant enrichment of genes involved in response to temperature stimulus, autophagy, and carbohydrate metabolism. We found stress markers induced by mutation of dynamically phosphorylated sites under physiological conditions. For example, we detected Hsp12, a responder to heat shock and oxidative and osmotic stress (63, 64), Ddr2, which is induced under environmental stress conditions (65–67), and the glycogen synthases Gsy1 and Gsy2, which are also induced under several environmental stress conditions (68–70). These observations indicate that the dynamic phosphosites contribute to the repression of genes involved in response to stress conditions under non-stress conditions (supplemental Table S10; Fig. 4B).
The expression profile of the strain carrying thirty mutated phosphosites in the Med15 C-terminal region (D30P) showed significantly altered transcription for 326 genes under normal growth conditions. The loss of function caused by the D30P mutation led to the induction of 240 genes, indicating that native phosphorylation of these sites has a negative effect on transcription. In comparison with the genes induced by the mutated dynamic phosphosites (D7P), the stable phosphosites act on additional pathways. A GO term analysis revealed the induction of genes involved in response to stimulus, vacuolar protein catabolic process, autophagy, and oxidation and reduction (supplemental Tables S8 and S9; Fig. 4B). These GO terms reflect the cellular adaption to environmental changes (71–73). Taken together, the overall transcriptome changes observed in the D30P mutant strain are similar to those observed in the D7P strain, albeit more pronounced.
We next compared the D7P and D30P profiles with the Δmed15 profile under normal growth conditions. Med15 deletion altered the expression of 1743 genes. The transcriptome changes observed in the Δmed15 mutant strain resemble those observed during a salt stress response, albeit it was less pronounced (Fig. 4C). We analyzed this subset of genes for enriched regulatory factors and ranked the factors by Fisher's exact test. Among the induced genes, targets of Sok2 and Rap1 were significantly enriched. We also found a general dependence on the general coactivator SAGA and transcription factor Msn2 (supplemental Fig. S1).
We next tested whether the observed D7P- and D30P-dependent induction of genes contributes to the cellular response to osmotic stress. We focused on the expression profile of the wild-type within the stress induction phase (18–24 min after addition of salt). The GO terms associated with carbohydrate metabolism and response to stimulus were enriched in the wild-type profile under osmotic stress conditions but also in the mutants D7P, D30P, and Δmed15 under non-stress conditions (Fig. 4D). We calculated the induction level of each GO term by the percentage of genes induced by D7P relative to the whole set of genes in the corresponding GO term. We found the highest induction by D7P mutations for the GO term response to temperature stimulus (15%) and for hexose transport (20%) (Fig. 4D). Taken together, both dynamically and stably phosphorylated sites in Med15 contribute to the repression of genes involved in the response to environmental changes under non-stress conditions.
Med15 Contributes to Activation of Genes Involved in Ribosome Biogenesis During Non-stress Conditions
The Δmed15 mutation caused a selective repression of a subset of genes, which are involved in ribosomal biogenesis. The highest ranked GO terms are ribosome biogenesis, ribonucleoprotein complex biogenesis, RNA metabolic process, gene expression, ribosomal large subunit biogenesis, and RNA modification (supplemental Tables S6 and S7). We performed model-based gene set analysis (MGSA) (74) analysis to identify a selective enrichment of transcription factor targets. Among the subset of repressed genes, we found a significant enrichment of genes regulated by Leu3 and Fhl1 (data not shown). Leu3 regulates expression of genes that are involved in branched amino acid synthesis, and it is functionally related to Gcn4 (75). Fhl1 regulates transcription of ribosomal protein genes and is regulated under different growth and stress conditions (76). Fhl1 function links gene expression of ribosomal protein genes to the TOR pathway (76).
We next compared the subset of repressed genes of the Δmed15 data set with the data set of response of wild-type cells to osmotic stress. The down-regulation of ribosome biogenesis genes in the Δmed15 mutant under normal conditions is to a large extent similar to the repression under osmotic stress conditions in wild-type cells (Fig. 4D). These observations indicate a functional role of Med15 in the expression of ribosome biogenesis genes and thus cell growth that is suppressed during salt stress (Fig. 4, B and D).
Mutated Dynamic Phosphosites Do Not Alter Osmotic Stress-induced Gene Expression
Recent work revealed that Med15 interacts with the general stress regulatory factors Msn2/Msn4 (77, 78) and that mutations of Med15 had a strong effect on TATA-containing and SAGA-regulated genes (79). Based on these findings, we searched the cDTA and DTA data sets for D7P, D30P, Δmed15, and the wild-type data set at 0.8 m NaCl (induction phase, 18–24 min) for enrichment of TATA-containing promoters (80) and Msn2/4 and Hog1 targets. Consistent with previous findings, we found general enrichment of TATA-containing genes in both induced and repressed genes (p value, 6.25 × 10−11). Furthermore, we observed a predominance of Msn2/4- and Hog1-regulated genes within the subset of induced genes (p value, 6.32 10−42) (supplemental Fig. S2).
Because the Δmed15 strain exhibits a slow growth phenotype sensitive to temperature changes and osmotic stress, we tested the D7P and D30P mutant strains for sensitivity to temperature and salt stress (see “Material and Methods,” supplemental material section 3.0, supplemental Fig. S5 and S6). Whereas the D7P mutant strain exhibited no significant growth phenotype, the D30P mutant exhibits a weak slow growth phenotype under temperature and salt stress conditions (supplemental Fig. S5 and S6). We performed a liquid growth assay to determine the doubling times under stress conditions (YPD; 0.8 m NaCl). Relative to wild-type and D7P cells (145 min), the doubling time slightly increased from D30P (193 min) to Δmed15 (205 min).
To investigate whether this phenotype is a result of impaired transcription regulation, we performed DTA (for D30P and wild-type) and comparative DTA (for D7P, Δmed15, and wild-type) under osmotic stress conditions. Cells were grown to mid-log phase and divided into control and stress samples. For the control samples, we carried out metabolic RNA labeling as described (27, 46). For the stress samples, we induced osmotic stress by addition of sodium chloride to a final concentration of 0.8 m, added 4-thiouridine (DTA) after 18 min and extracted RNA after 6 min of labeling (supplemental material section 4). The time frame of 18–24 min is consistent with the previous observation of the induction phase, when cells activate the gene expression program to antagonize osmotic pressure (27, 29) (see “Materials and Methods”). Upon stress, levels of 3516 newly transcribed (labeled) mRNAs exhibit at least a 1.5-fold change in at least one of the data sets. We compared the labeled RNA fraction of D7P, D30P, Δmed15, and wild-type responding to 0.8 m sodium chloride within the time frame of 18–24 min after salt addition. The response to osmotic stress changed ∼50% of newly synthesized RNAs (supplemental Fig. S2A).
To identify gene expression changes during the stress response caused by the mutated phosphosites, we calculated a linear model. We used all data sets as dependent variables and regressors according to their condition in a linear regression analysis. We found that mutations D7P, D30P, and Δmed15 do not have any additional effect on salt stress-induced expression changes (supplemental Fig. S2B). We conclude that Med15 phosphosites are involved in repression of genes under non-stress conditions but do not have additional, specific effects on gene expression under osmotic stress conditions.
DISCUSSION
Mediator functions by receiving signals from different pathways to generate an output for the general transcription machinery (1). However, a functional influence of post-translational modification of the Mediator has not been investigated systematically. By combining high-resolution MS, quantitative proteomics (SILAC), and dynamic transcriptome analysis, we demonstrate here an extensive phosphorylation of Mediator and a contribution of Mediator phosphorylation to gene regulation.
Earlier work has shown that Mediator is phosphorylated on subunits Med2, Med4, Med13, and Med14 and that Med2 phosphorylation can influence Mediator function (16–19). Previous proteomic analysis additionally revealed that subunits Med1, Med5, Med6, Med15, and Med17 are phosphorylated (30, 48–52). Here, we used affinity purification and high-resolution MS to map 125 sites on 17 Mediator subunits that are phosphorylated in vivo under normal growth conditions. Of these, 88 were novel, whereas 37 were reported previously (16–19, 30, 48–52, 81) and confirmed here. The majority of these phosphosites were located in the middle and tail modules. Quantitative proteomics using SILAC revealed that a subset of these sites has different levels of phosphorylation during the osmotic stress response. Phosphorylation levels generally decrease during osmotic stress, consistent with previous observations (30). In particular, we identified 30 phosphorylated sites near the Med15 C terminus, of which seven changed phosphorylation levels during stress.
To investigate whether these phosphosites contribute to Mediator function in gene regulation, we mutated a phosphorylation sites in Med15 and used DTA to investigate their contribution to gene regulation during the osmotic stress response. This revealed that the 30 clustered phosphosites in Med15 are involved in suppression of stress-induced alteration of transcription from genes responding to environmental changes. The seven dynamically phosphorylated sites were required for the repression of genes involved in temperature stimulus, autophagy, and carbohydrate metabolism under normal growth conditions. Under osmotic stress conditions, however, we found no significant contribution of the Med15 phosphosites to gene expression.
Taken together, our results show that Mediator is phosphorylated at multiple sites in vivo, that the level of phosphorylation can change during stress, and that a phosphorylated subunit region can contribute to transcription regulation by repressing stress genes under normal growth conditions. One possible explanation for our observations is that Med15 phosphorylation suppresses the association of Mediator with various activators that stimulate the expression of stress-induced genes. Consistent with this model, Med15 binds to transcription factors Msn2, Gcn4, Pdr1, Oaf1, Hsf1, and Ace1 (13, 14, 77, 82). Alternatively, transcriptional repressors that normally repress stress gene transcription would require certain Mediator phosphorylations for their function.
Acknowledgments
We thank L. Larivière, M. Seizl, T. Koschubs, S. Benkert, E. Czeko, R. Ringel, D. Martin, and other members of the Cramer laboratory for help and discussion.
This work was supported in part by the Mann and Cramer laboratories, which are supported by the Center for Integrated Protein Science Munich.

This article contains supplemental Methods, Tables S1–S10, Figs. S1–S6, additional references, and data.
Microarray data were deposited in ArrayExpress under accession code E-MTAB-1059.
- Pol
- RNA polymerase
- SILAC
- stable isotope labeling of amino acids in culture
- DTA
- dynamic transcriptome analysis
- cDTA
- comparative DTA
- TAP
- tandem affinity purification
- TEV
- tobacco edge virus
- SAGA
- Spt-Ada-Gcn5-Acetyl transferase.
REFERENCES
- 1. Malik S., Roeder R. G. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61, 1209–1215 [DOI] [PubMed] [Google Scholar]
- 3. Flanagan P. M., Kelleher R. J., 3rd, Sayre M. H., Tschochner H., Kornberg R. D. (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 [DOI] [PubMed] [Google Scholar]
- 4. Bäckström S., Elfving N., Wingsle G., Björklund S. (2007) Mol. Cell 26, 436–438 [DOI] [PubMed] [Google Scholar]
- 5. Spaeth J. M., Kim N. H., Boyer T. G. (2011) Mediator and human disease. Semin. Cell Dev. Biol. 22, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berti L., Mittler G., Przemeck G. K., Stelzer G., Günzler B., Amati F., Conti E., Dallapiccola B., Hrabé de Angelis M., Novelli G., Meisterernst M. (2001) Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a mediator subunit. Genomics 74, 320–332 [DOI] [PubMed] [Google Scholar]
- 7. Björklund S., Gustafsson C. M. (2005) The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30, 240–244 [DOI] [PubMed] [Google Scholar]
- 8. Cai G., Imasaki T., Takagi Y., Asturias F. J. (2009) Mediator structural conservation and implications for the regulation mechanism. Structure 17, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rachez C., Lemon B. D., Suldan Z., Bromleigh V., Gamble M., Näär A. M., Erdjument-Bromage H., Tempst P., Freedman L. P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398, 824–828 [DOI] [PubMed] [Google Scholar]
- 10. Belakavadi M., Pandey P. K., Vijayvargia R., Fondell J. D. (2008) MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol. Cell Biol. 28, 3932–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang G., Balamotis M. A., Stevens J. L., Yamaguchi Y., Handa H., Berk A. (2005) Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17, 683–694 [DOI] [PubMed] [Google Scholar]
- 12. Wang W., Huang L., Huang Y., Yin J. W., Berk A. J., Friedman J. M., Wang G. (2009) Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev. Cell 16, 764–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thakur J. K., Arthanari H., Yang F., Chau K. H., Wagner G., Näär A. M. (2009) Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284, 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jedidi I., Zhang F., Qiu H., Stahl S. J., Palmer I., Kaufman J. D., Nadaud P. S., Mukherjee S., Wingfield P. T., Jaroniec C. P., Hinnebusch A. G. (2010) Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX domain, to recruit mediator to target genes in vivo. J. Biol. Chem. 285, 2438–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thakur J. K., Arthanari H., Yang F., Pan S. J., Fan X., Breger J., Frueh D. P., Gulshan K., Li D. K., Mylonakis E., Struhl K., Moye-Rowley W. S., Cormack B. P., Wagner G., Näär A. M. (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 [DOI] [PubMed] [Google Scholar]
- 16. Hallberg M., Polozkov G. V., Hu G. Z., Beve J., Gustafsson C. M., Ronne H., Björklund S. (2004) Site-specific Srb10-dependent phosphorylation of the yeast Mediator subunit Med2 regulates gene expression from the 2-microm plasmid. Proc. Natl. Acad. Sci. U.S.A. 101, 3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guidi B. W., Bjornsdottir G., Hopkins D. C., Lacomis L., Erdjument-Bromage H., Tempst P., Myers L. C. (2004) Mutual targeting of mediator and the TFIIH kinase Kin28. J. Biol. Chem. 279, 29114–29120 [DOI] [PubMed] [Google Scholar]
- 18. Chang Y. W., Howard S. C., Herman P. K. (2004) The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell 15, 107–116 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y., Kung C., Fishburn J., Ansari A. Z., Shokat K. M., Hahn S. (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell Biol. 24, 1721–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexander M. R., Tyers M., Perret M., Craig B. M., Fang K. S., Gustin M. C. (2001) Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell 12, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Escoté X., Zapater M., Clotet J., Posas F. (2004) Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6, 997–1002 [DOI] [PubMed] [Google Scholar]
- 22. Proft M., Struhl K. (2004) MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 118, 351–361 [DOI] [PubMed] [Google Scholar]
- 23. Gasch A. P., Werner-Washburne M. (2002) The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2, 181–192 [DOI] [PubMed] [Google Scholar]
- 24. Macia J., Regot S., Peeters T., Conde N., Solé R., Posas F. (2009) Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci. Signal 2, ra13. [DOI] [PubMed] [Google Scholar]
- 25. Melamed D., Pnueli L., Arava Y. (2008) Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA 14, 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller C., Schwalb B., Maier K., Schulz D., Dümcke S., Zacher B., Mayer A., Sydow J., Marcinowski L., Dölken L., Martin D. E., Tresch A., Cramer P. (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Romero-Santacreu L., Moreno J., Pérez-Ortín J. E., Alepuz P. (2009) Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15, 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Molin C., Jauhiainen A., Warringer J., Nerman O., Sunnerhagen P. (2009) mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA 15, 600–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soufi B., Kelstrup C. D., Stoehr G., Fröhlich F., Walther T. C., Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. BioSyst. 5, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 31. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 32. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 33. Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 34. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 35. Macek B., Mann M., Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 36. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 37. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 39. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 41. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 42. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 43. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 44. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 45. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun M., Schwalb B., Schulz D., Pirkl N., Etzold S., Larivière L., Maier K. C., Seizl M., Tresch A., Cramer P. (2012) Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 22, 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 48. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteomics 7, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., Gygi S. P. (2007) Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 51. Chi A., Huttehhower C., Geer L. Y., Coon J. J., Syka J. E. P., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proct. Natl. Acad. Sci. USA 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 53. UniProt Consortium (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jain E., Bairoch A., Duvaud S., Phan I., Redaschi N., Suzek B. E., Martin M. J., McGarvey P., Gasteiger E. (2009) Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 56. Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 57. Park J. M., Kim H. S., Han S. J., Hwang M. S., Lee Y. C., Kim Y. J. (2000) In vivo requirement of activator-specific binding targets of mediator. Mol. Cell Biol. 20, 8709–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thorsness P. E., Koshland D. E. (1987) Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 262, 10422–10425 [PubMed] [Google Scholar]
- 59. Thomson M., Gunawardena J. (2009) Unlimited multistability in multisite phosphorylation systems. Nature 460, 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barik D., Baumann W. T., Paul M. R., Nova B., Tyson J. J. (2010) A model of yeast cell-cycle regulation based on multisite phosphorylation. Mol. Syst. Biol. 6, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kõivomägi M., Valk E., Venta R., Iofik A., Lepiku M., Balog E. R., Rubin S. M., Morgan D. O., Loog M. (2011) Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 480, 128–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Holmberg C. I., Tran S. E., Eriksson J. E., Sistonen L. (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem.. Sci. 27, 619–627 [DOI] [PubMed] [Google Scholar]
- 63. Welker S., Rudolph B., Frenzel E., Hagn F., Liebisch G., Schmitz G., Scheuring J., Kerth A., Blume A., Weinkauf S., Haslbeck M., Kessler H., Buchner J. (2010) Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 39, 507–520 [DOI] [PubMed] [Google Scholar]
- 64. Morano K. A., Grant C. M., Moye-Rowley W. S. (2012) The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kobayashi N., McClanahan T. K., Simon J. R., Treger J. M., McEntee K. (1996) Structure and functional analysis of the multistress response gene DDR2 from Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 229, 540–547 [DOI] [PubMed] [Google Scholar]
- 66. Treger J. M., Magee T. R., McEntee K. (1998) Functional analysis of the stress response element and its role in the multistress response of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 243, 13–19 [DOI] [PubMed] [Google Scholar]
- 67. Hirata Y., Andoh T., Asahara T., Kikuchi A. (2003) Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 14, 302–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Unnikrishnan I., Miller S., Meinke M., LaPorte D. C. (2003) Multiple positive and negative elements involved in the regulation of expression of GSY1 in Saccharomyces cerevisiae. J. Biol. Chem. 278, 26450–26457 [DOI] [PubMed] [Google Scholar]
- 69. Enjalbert B., Parrou J. L., Teste M. A., Francois J. (2004) Combinatorial control by the protein kinases PKA, PHO85, and SNF1 of transcriptional induction of the Saccharomyces cerevisiae GSY2 gene at the diauxic shift. Mol. Gen. Genomics 271, 697–708 [DOI] [PubMed] [Google Scholar]
- 70. Zähringer H., Thevelein J. M., Nwaka S. (2000) Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: variations of PKA effect during stress and growth. Mol. Microbiol. 35, 397–406 [DOI] [PubMed] [Google Scholar]
- 71. Rubinsztein D. C., Mariño G., Kroemer G. (2011) Autophagy and aging. Cell 146, 682–695 [DOI] [PubMed] [Google Scholar]
- 72. Singh R., Cuervo A. M. (2011) Autophagy in the cellular energetic balance. Cell Metabolism 13, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Murray D. B., Haynes K., Tomita M. (2011) Redox regulation in respiring Saccharomyces cerevisiae. Biochim. Biophys. Acta 1810, 945–958 [DOI] [PubMed] [Google Scholar]
- 74. Bauer S., Robinson P. N., Gagneur J. (2011) Model-based gene set analysis for Bioconductor. Bioinformatics 27, 1882–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hinnebusch A. G., Natarajan K. (2002) Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Martin D. E., Soulard A., Hall M. N. (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979 [DOI] [PubMed] [Google Scholar]
- 77. Lallet S., Garreau H., Garmendia-Torres C., Szestakowska D., Boy-Marcotte E., Quevillon-Chéruel S., Jacquet M. (2006) Role of Gal11, a component of the RNA polymerase II mediator in stress-induced hyperphosphorylation of Msn2 in Saccharomyces cerevisiae. Mol. Microbiol. 62, 438–452 [DOI] [PubMed] [Google Scholar]
- 78. Sadeh A., Baran D., Volokh M., Aharoni A. (2012) Conserved motifs in the Msn2-activating domain are important for Msn2-mediated yeast stress response. J. Cell Sci. 125, 3333–3342 [DOI] [PubMed] [Google Scholar]
- 79. Ansari S. A., Ganapathi M., Benschop J. J., Holstege F. C., Wade J. T., Morse R. H. (2012) Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 31, 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Seizl M., Hartmann H., Hoeg F., Kurth F., Martin D. E., Söding J., Cramer P. (2011) A conserved GA element in TATA-less RNA polymerase II promoters. PLoS One 6, e27595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T., van Leenen D., Holstege F. C. (2005) Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19, 511–522 [DOI] [PubMed] [Google Scholar]
- 82. Herbig E., Warfield L., Fish L., Fishburn J., Knutson B. A., Moorefield B., Pacheco D., Hahn S. (2010) Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol. Cell Biol. 30, 2376–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Larivière L., Seizl M., van Wageningen S., Röther S., van de Pasch L., Feldmann H., Strässer K., Hahn S., Holstege F. C., Cramer P. (2008) Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev. 22, 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jeong C. J., Yang S. H., Xie Y., Zhang L., Johnston S. A., Kodadek T. (2001) Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry 40, 9421–9427 [DOI] [PubMed] [Google Scholar]
- 85. Brzovic P. S., Heikaus C. C., Kisselev L., Vernon R., Herbig E., Pacheco D., Warfield L., Littlefield P., Baker D., Klevit R. E., Hahn S. (2011) The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]