Background: DNA replication complex assembly is regulated to maintain genomic integrity.
Results: Proteolysis of a replication protein is dependent on a new ubiquitination pathway, which alters replication complex assembly.
Conclusion: A novel mechanism for degrading a replication protein is described.
Significance: This new pathway may contribute to regulation of DNA replication and genomic integrity.
Keywords: Cell Cycle, DNA Replication, E3 Ubiquitin Ligase, Protein Degradation, Ubiquitin
Abstract
The accurate replication of genetic information is critical to maintaining chromosomal integrity. Cdc6 functions in the assembly of pre-replicative complexes and is specifically required to load the Mcm2-7 replicative helicase complex at replication origins. Cdc6 is targeted for protein degradation by multiple mechanisms in Saccharomyces cerevisiae, although only a single pathway and E3 ubiquitin ligase for Cdc6 has been identified, the SCFCdc4 (Skp1/Cdc53/F-box protein) complex. Notably, Cdc6 is unstable during the G1 phase of the cell cycle, but the ubiquitination pathway has not been previously identified. Using a genetic approach, we identified two additional E3 ubiquitin ligase components required for Cdc6 degradation, the F-box protein Dia2 and the Hect domain E3 Tom1. Both Dia2 and Tom1 control Cdc6 turnover during G1 phase of the cell cycle and act separately from SCFCdc4. Ubiquitination of Cdc6 is significantly reduced in dia2Δ and tom1Δ cells. Tom1 and Dia2 each independently immunoprecipitate Cdc6, binding to a C-terminal region of the protein. Tom1 and Dia2 cannot compensate for each other in Cdc6 degradation. Cdc6 and Mcm4 chromatin association is aberrant in tom1Δ and dia2Δ cells in G1 phase. Together, these results present evidence for a novel degradation pathway that controls Cdc6 turnover in G1 that may regulate pre-replicative complex assembly.
Introduction
DNA replication is a highly coordinated process to precisely duplicate chromosomes during S phase of the cell cycle. Initiation of DNA synthesis requires assembly of pre-replicative complexes (pre-RCs)3 at replication origins (for review, see Ref. 1). Regulation of pre-RC assembly is critical to prevent over duplication of genetic material. Pre-RC assembly occurs during the G1 phase of the cell cycle, and reassembly is blocked for the remainder of the cycle (for review, see Ref. 2).
Cdc6 is an AAA+-ATPase required for pre-RC assembly (3–6). Cdc6 binds to the origin recognition complex along with another pre-RC protein, Cdt1, to recruit the Mcm2-7 replicative helicase complex to replication origins (7–11). Budding yeast Cdc6 is a highly unstable protein subject to multiple modes of degradation (12–14). In the best understood degradation pathway, cyclin-dependent kinase (Cdk) phosphorylation targets Cdc6 for ubiquitination and degradation via the ubiquitin ligase complex SCFCdc4 at the G1/S phase transition and during G2/M (13–17). However, Cdc6 is also unstable during G1, and the degradation pathway responsible has not been identified (14).
To identify other pathways that might target Cdc6 for ubiquitination and degradation, we assayed other E3 ubiquitin ligases that function in DNA replication or during S phase. Tom1 is a Hect domain E3 ubiquitin ligase that targets excess histone H3 for DNA replication checkpoint-dependent degradation (18). The human homolog of Tom1, Huwe1, targets human Cdc6 for degradation during the DNA damage checkpoint response (19, 20). This function appears to be conserved in Saccharomyces cerevisiae (19), although the contribution of Tom1 to Cdc6 degradation in an unperturbed cell cycle has not been investigated. The SCFDia2 ubiquitin ligase complex regulates DNA replication and is required for genomic stability (21–24). Potential targets of SCFDia2 include two proteins that travel with replication complexes at the replication fork, Mrc1 and Ctf4, although the physiological role of their degradation is unclear (25).
Here we show that both Tom1 and Dia2 contribute to Cdc6 degradation during G1. This work describes a new degradation pathway for Cdc6 and identifies a novel target for both the Tom1 and Dia2 ubiquitin ligase components.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
Yeast strains and oligonucleotides are described in supplemental Tables I and II. To generate the CDC6-3HA strain (DKY546), the 3HA-TRP1 fragment containing the 3′-end of CDC6 open reading frame and the 3′-UTR of CDC6 was amplified with primers DHK9 and 10 using pFA6a-3HA-TRP1 as a template (26) and integrated into strain DKY153 via homologous recombination. The HA-CDC6 TRP1 plasmid using a GAL1,10 promoter (pDHK1) was constructed via PCR amplification of CDC6 from genomic DNA with primers DHK6 and 7 and then inserted into SalI and BamHI sites of the p1216 plasmid. To generate the GST-CDC6 baculovirus construct (pZHW073), the CDC6 open reading frame was amplified with primers DHK25 and 26 using yeast genomic DNA and cloned into the pUNI-10 vector with NdeI and BamHI sites (pZHW072). Plasmid pZHW072 was then recombined with p1212 (27) using cre-lox recombination to generate plasmid pZHW073. To generate the FLAG-TOM1 HECT baculovirus construct (pDHK5), the Hect domain was amplified using yeast genomic DNA with primers DHK84 and 85 and cloned into the pUNI-10 vector with XhoI and BamHI sites to construct the plasmid pDHK4. Plasmid pDHK4 was then recombined with p1214 (27) using cre-lox recombination to generate the FLAG-TOM1 HECT baculovirus construct (pDHK5). The pUNI-10-CDC6ΔN, -DIA2 TPR, and -DIA2-ΔN214 plasmids (pDHK7, pZHW094, pZHW079) were constructed by amplifying the CDC6ΔN, DIA2 TPR, and DIA2-ΔN214 fragments using the plasmids pDHK6, pACK140, and pACK137 as a template with primers DHK104, 105, ZHW57, 54, 58, and AK43. The amplified fragments were then cloned into the XhoI and NotI, the NdeI and SalI, and the NdeI and BamHI sites of the pUNI-10 plasmid, respectively. The GST-CDC6ΔN, FLAG-DIA2 TPR, and FLAG-DIA2-ΔN214 baculovirus constructs (pDHK8, pZHW095 and pZHW080) were also generated via cre-lox recombination using p1212 and p1214 (27). To construct the cdc6ΔN-3HA strain (DKY875), the CDC6–3HA-TRP1 fragment was cloned into pBluescript SK+ plasmid with XhoI and NotI sites to generate plasmid (pDHK2). The N-terminal 2–47 amino acid was then deleted by amplifying the CDC6ΔN fragment using the plasmid pDHK2 as a template with primers DHK46, 82, 81, and 48 and cloned into the XhoI and NotI sites of the plasmid pDHK2 to construct the plasmid CDC6ΔN-3HA (pDHK6). The CDC6ΔN-3HA fragment cut with the XhoI and NotI restriction enzymes was integrated into strain DKY 153 via homologous recombination. To generate the galactose-inducible HA-tagged TOM1 HECT domain plasmid (pDHK15), the HECT domain was amplified via PCR with primers DHK122 and 123 and then inserted into BamHI and NotI sites of the p1220 plasmid.
Reverse Transcription PCR (RT-PCR)
Cultures were grown to mid-log phase (2 × 107 cells/ml) at 30 °C, and total RNA was isolated with PURE LINK micro- to midi-Kit (Invitrogen) using the manufacturer's protocol. After DNase I treatment, 3 μg of total RNA was reverse-transcribed using Superscript II (Invitrogen) with oligo(dT)50 primer. The cDNA was amplified with primers DHK11 and 12 or ACT1–5 and ACT1–3.
In Vitro Binding Assays
1 mg of total cell lysate isolated from baculovirus-infected Hi5 insect cells was immunoprecipitated with anti-FLAG M2 monoclonal (Sigma), anti-Myc 9E10 monoclonal (Covance), or anti-GST polyclonal (Santa Cruz) antibodies and immunoblotted with anti-FLAG M2, anti-Myc 9E10, and anti-GST (Santa Cruz) antibodies.
Stability Assays
Cells at 1 × 107 cells/ml were arrested with 40 μg/ml α factor for 3 h. Cycloheximide was added at 100 μg/ml. Cell pellets were washed and lysed by vortexing with glass beads in 20% trichloroacetic acid (TCA) for 3 min. Lysed cell pellets were centrifuged at 3,000 rpm for 10 min and resuspended in Laemmli buffer. Precipitated proteins were neutralized with 1 m Tris and boiled for 5 min. Protein concentration was quantified using the RC/DC protein assay kit (Bio-Rad). 20 μg of total protein was run on 8% SDS-PAGE. Protein abundance was measured using the ImageJ software and normalized against a loading control.
Immunoprecipitation
Cells were cultured to mid-log phase (2 × 107 cells/ml) and collected by centrifuging at 4,000 rpm for 2 min. Total cell lysate was isolated by vortexing the cells with glass beads in radioimmune precipitation assay buffer (1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 20 mm Tris, pH 7.5, 10% glycerol) with protease inhibitor mixture (Roche Applied Science) for 40 min at 4 °C. Protein concentration was determined using the Bio-Rad protein assay. 1 mg of total lysate was incubated with affinity matrix anti-HA.11 (Covance) and protein A/G-agarose (Santa Cruz) for 3 h at 4 °C. Samples were washed with radioimmune precipitation assay buffer four times and boiled in Laemmli buffer for 5 min.
In Vivo and in Vitro Ubiquitination Assays
Cells were grown to mid-log phase and treated with dimethyl sulfoxide or 50 μm MG132 for 2 h. 2 mg of total protein was used for immunoprecipitation with anti-HA.11 monoclonal antibodies (Covance). Proteins were analyzed by 8% SDS-PAGE followed by an immunoblot assay with anti-P4D1 monoclonal ubiquitin and anti-HA.11 antibodies (Covance). ImageJ software was used to quantify ubiquitinated Cdc6. For in vitro ubiquitination, 20 μg of crude yeast extracts isolated from wild type, tom1Δ, and dia2Δ strains were incubated with 40 μg GST-Cdc6 protein bound to the glutathione-Sepharose 4B beads (GE Healthcare) at 30 °C for 45 min. Samples were run on 6% SDS-PAGE and immunoblotted with anti-GST polyclonal antibodies (Santa Cruz).
Chromatin Fractionation
Chromatin fractionation assay was performed as described (28). Protein concentration was quantified using the RC/DC protein assay kit. 30 μg of whole cell extract and crude chromatin fraction were resolved on 8 and 15% SDS-PAGE, respectively, and analyzed by immunoblotting with anti-HA.11 (Covance), anti-3-phosphoglycerate kinase 22C5 (anti-Pgk1, Invitrogen), and anti-histone H3 (Abcam) antibodies. The ratio of chromatin-bound Cdc6 to histone H3 was measured using the ImageJ software.
Chromatin Immunoprecipitation
Total yeast cell lysates were prepared for chromatin immunoprecipitation as described previously (29). PCR was performed using primers for early firing origins ARS1 and ARS305 and nonorigin region ACF2.
RESULTS
Tom1 and Dia2 Regulate Cdc6 Degradation
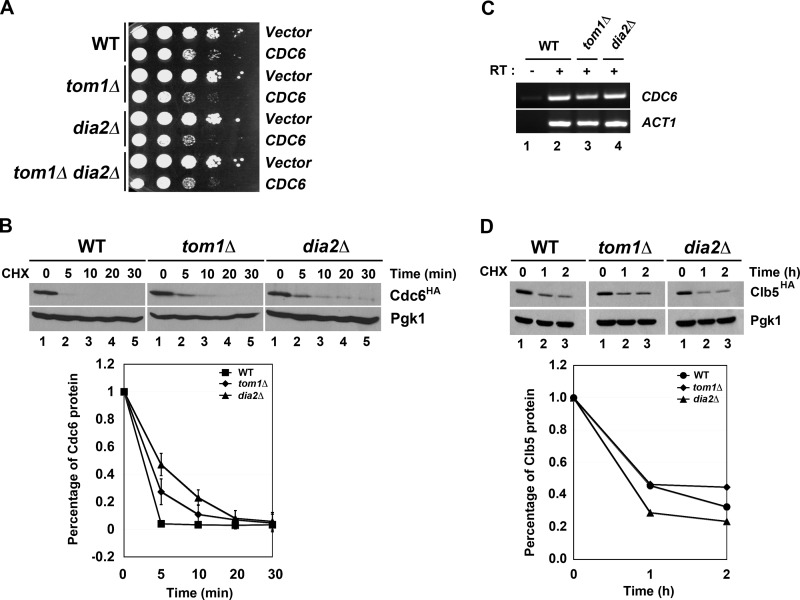
To determine whether other ubiquitin ligases might have a role in Cdc6 protein turnover, we examined whether overexpression of CDC6 caused a growth defect in ligase mutants that had known roles in DNA replication or the cell cycle. Two ligases that fit this profile include the F-box protein Dia2 and the Hect domain E3 Tom1 (20–23). Previous work has shown that Tom1 can target Cdc6 for degradation in response to DNA damage (20), but a role for Tom1 in Cdc6 turnover in an unperturbed cell cycle has not been examined. Overexpression of CDC6 causes a growth defect in wild type yeast cells (30, 31), but we observed an exacerbated growth phenotype in both dia2Δ and tom1Δ cells (Fig. 1A). Intriguingly, the CDC6 overexpression phenotype in the tom1Δ dia2Δ double mutant mimicked the phenotype of the single mutants.
FIGURE 1.
Tom1 and Dia2 control Cdc6 proteolysis. A, overexpression of Cdc6 results in an exacerbated growth defect in tom1Δ, dia2Δ, and tom1Δ dia2Δ strains. The 10-fold serial dilutions of wild type, tom1Δ, dia2Δ, and tom1Δ dia2Δ cells carrying empty vector or CDC6 under the control of GAL1,10 promoter were spotted onto minimal plates with 2% galactose. Plates were incubated at room temperature for 2–3 days. B, Cdc6 is partially stabilized in tom1Δ and dia2Δ mutants. Wild type, tom1Δ, and dia2Δ strains were grown to mid-log phase at 30 °C. After cycloheximide (CHX) treatment (100 μg/ml), samples were taken at the indicated times and immunoblotted with anti-HA and -Pgk1 antibodies. Pgk1 was used as a loading control. C, Cdc6 mRNA level is not changed in both tom1Δ and dia2Δ mutants. RT-PCR assay was conducted to examine the level of Cdc6 transcript in wild type, dia2Δ, and tom1Δ strains. ACT1 was used as a loading control. RT, reverse transcriptase. D, Clb5 stability is not affected in tom1Δ and dia2Δ mutants. Wild type, tom1Δ, and dia2Δ cells were grown to mid-log phase in minimal medium supplemented with 2% raffinose. Clb5 transcription was induced by addition of galactose for 1 h and repressed by adding glucose. 100 μg/ml cycloheximide was added to block protein translation.
We then performed a protein stability assay using HA-tagged Cdc6 expressed from the endogenous locus in wild type, tom1Δ, and dia2Δ cells. Log phase cultures were treated with cycloheximide to inhibit protein synthesis and Cdc6 protein levels examined over time. As shown in Fig. 1B, Cdc6 was partially stabilized in both dia2Δ and tom1Δ cells. No changes in CDC6 mRNA abundance were detected in these strains (Fig. 1C), consistent with the possibility that Dia2 and Tom1 might control Cdc6 ubiquitination. Another unstable cell cycle protein, Clb5, was not stabilized in dia2Δ or tom1Δ cells, indicating that the results with Cdc6 were not due to nonspecific defects in protein degradation (Fig. 1D).
Cdc6 Binds Dia2 and Tom1
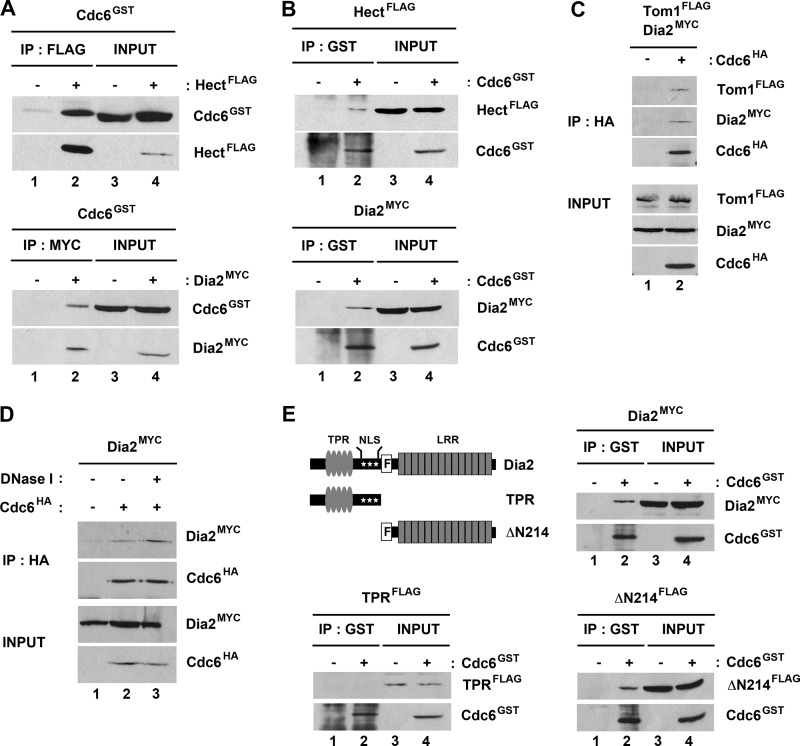
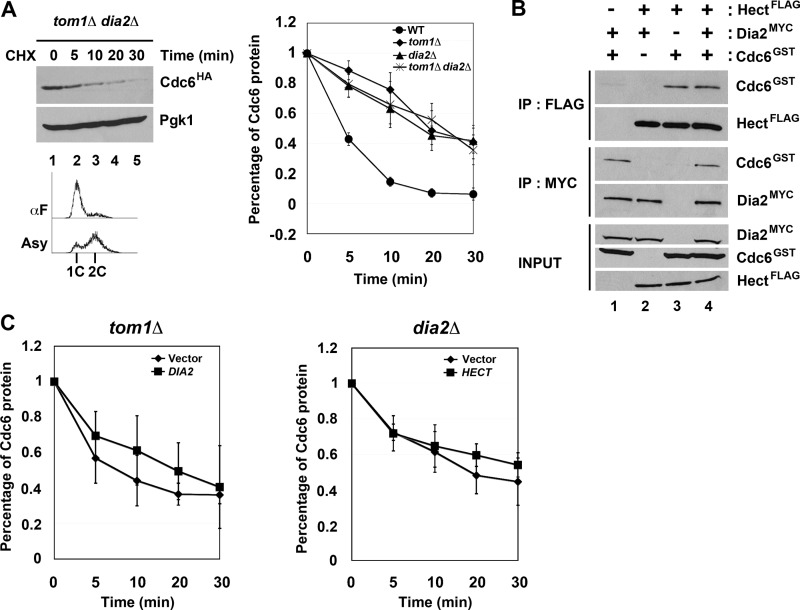
We examined whether Dia2 and Tom1 interact with Cdc6 using two different approaches. First, we tested whether GST-Cdc6 co-purified with either Myc-tagged Dia2 or the FLAG-tagged Hect domain of Tom1 when expressed in baculovirus-infected insect cells (Fig. 2). When either the FLAG-tagged Hect domain of Tom1 or Myc-tagged Dia2 was immunoprecipitated, we observed substantial co-purification of GST-Cdc6 (Fig. 2A). We also observed reciprocal co-purification when GST-Cdc6 was purified from the same cells (Fig. 2B). Next we tested whether endogenously expressed HA-tagged Cdc6 can co-immunoprecipitate endogenously expressed Myc-tagged Dia2 or FLAG-tagged Tom1 (Fig. 2C). Immunoprecipitation of Cdc6 resulted in Tom1 and Dia2 co-purification, indicating that these proteins interact under physiological conditions. GST-tagged Cdc6 purified from baculovirus-infected insect cells also interacted with endogenously expressed FLAG-tagged Tom1 and Myc-tagged Dia2 in yeast extracts (data not shown). Because Dia2 is a chromatin-bound protein, we considered the possibility that Dia2 interacted with Cdc6 indirectly by virtue of both proteins binding to chromatin, but we still observed co-precipitation when lysates were treated with DNase I (Fig. 2D). We conclude that both Tom1 and Dia2 form a complex with Cdc6, although we cannot determine whether all three proteins assemble into the same complex from these results.
FIGURE 2.
Tom1 and Dia2 interact with Cdc6. A and B, Tom1 Hect domain and Dia2 interact with Cdc6. Hi5 insect cells were co-infected with GST-Cdc6 and FLAG-Hect or Myc-Dia2 baculoviruses. FLAG-Hect, Myc-Dia2, or GST-Cdc6 protein was immunoprecipitated (IP) with anti-FLAG, -Myc, or -GST antibodies and analyzed by immunoblotting. C, endogenously expressed Tom1 and Dia2 co-precipitate with Cdc6. Total cell lysates from strains expressing FLAG-Tom1 and Myc-Dia2 or FLAG-Tom1, Myc-Dia2, and HA-Cdc6 were immunoprecipitated with anti-HA.11 affinity matrix and immunoblotted with anti-FLAG, -Myc, and -HA antibodies. D, Dia2 binding to Cdc6 is resistant to DNase I treatment. Cell lysate from the strain expressing Myc-Dia2 and HA-Cdc6 strain was treated with 20 units of DNase I (Promega) for 45 min on ice. Cdc6 was immunoprecipitated with anti-HA antibodies and immunoblotted with anti-HA and -Myc antibodies. E, the leucine-rich repeat domain of Dia2 is required for its interaction with Cdc6. Total cell lysates from GST-Cdc6 and Myc-Dia2, FLAG-TPR, or FLAG-ΔN214 baculovirus-infected Hi5 insect cells were immunoprecipitated with anti-GST antibodies and analyzed by immunoblotting with anti-GST, -Myc, or -FLAG antibodies.
Tom1 is a large protein with multiple domains, including the conserved Hect domain found in the C terminus of the protein that contains the catalytic cysteine (32). Our results with insect cell-expressed protein indicate that the Tom1 Hect domain interacts with Cdc6 (Fig. 2, A and B). Dia2 is an F-box protein that contains TPR repeats in the N terminus and an LRR domain in the C terminus (22, 23). Because F-box proteins typically associate with substrate proteins via their C-terminal repeat domains, we asked whether a truncated Dia2 protein lacking the N terminus still bound Cdc6. We co-expressed ΔN214 Dia2, which lacks the N-terminal TPR repeats but still contains the F-box domain and LRR region, with GST-Cdc6 in insect cells. GST-Cdc6 co-purified with ΔN214 Dia2 under these conditions (Fig. 2E). Together, these results suggest that the N termini of Tom1 and Dia2 are dispensable for binding Cdc6.
Tom1 and Dia2 Control Cdc6 Degradation in G1 Phase
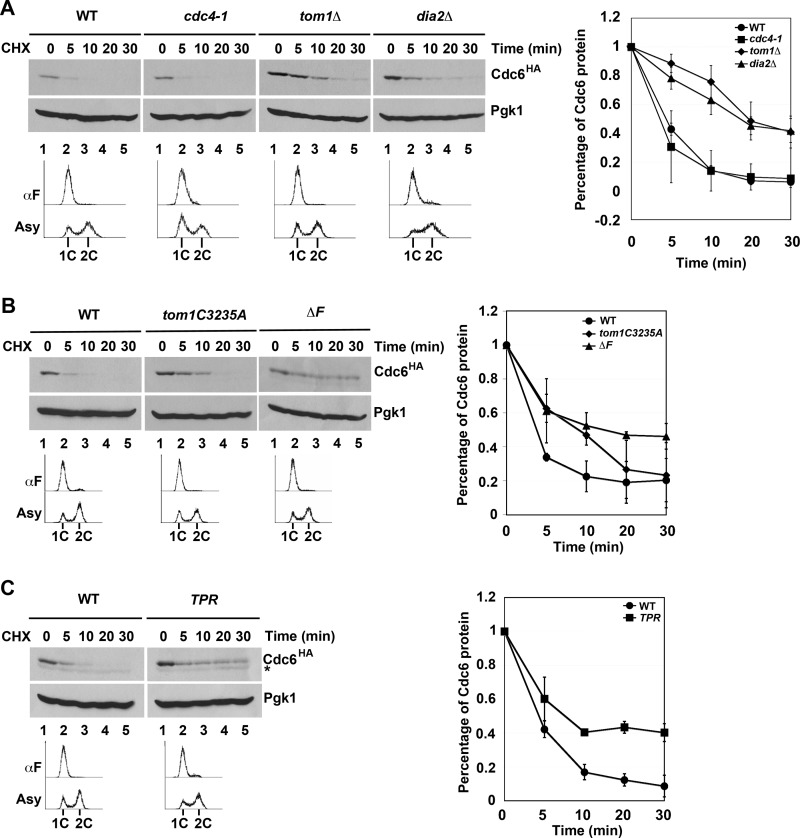
Previous work from the Diffley laboratory has shown that there are three modes of Cdc6 degradation (14). Mode 1 functions prior to Start during G1 and is Cdk-independent, whereas Modes 2 and 3 are Cdk- and Cdc4-dependent and function in early S and G2/M, respectively (14). The identity of the E3 ligase in Mode 1 was not determined, although genetic evidence suggested that neither the APC nor SCF complexes were responsible (14). We tested whether Tom1 or Dia2 might contribute to the turnover of Cdc6 in G1 using the cycloheximide stability assay in cells arrested with αF. As has been previously shown (14), Cdc6 is still unstable in cdc4-1 cells under these conditions (Fig. 3A). Strikingly, we found that Cdc6 was stabilized in both tom1Δ and dia2Δ strains (Fig. 3A). The observation that Dia2 is involved in Cdc6 turnover in G1 was surprising, as Cdc6 is not stabilized in scf mutants in G1 (14).
FIGURE 3.
Tom1 and Dia2 control Cdc6 proteolysis in G1. A, Cdc6 is stabilized in both tom1Δ and dia2Δ mutants in G1. The indicated strains were arrested with α factor (αF) for 3 h and shifted to 37 °C for 30 min prior to performing stability assays as in Fig. 1B. Three independent experiments were used for quantification. Error bars indicate S.D. B, the catalytic activity of Tom1 Hect domain and the F-box domain of Dia2 is required for Cdc6 turnover. Wild type, tom1C3235A, dia2ΔF strains arrested with α factor were used in protein stability assays. Samples were collected at indicated times after cycloheximide (CHX) addition. Pgk1 was used as a loading control. Three independent experiments were used for quantification. C, the TPR mutant stabilizes Cdc6. Samples from wild type and TPR strains were prepared as in A. The asterisk indicates a nonspecific band. Error bars indicate S.D.
The most straightforward explanation for our results is that Cdc6 may be a direct ubiquitination target of both Tom1 and Dia2. To test this hypothesis, we examined whether the ubiquitin ligase activity of Tom1 or Dia2 was required for Cdc6 protein turnover. Tom1 is a member of the Hect domain E3 ubiquitin ligase family, which uses a catalytic cysteine in the transfer of ubiquitin to substrate proteins (32). Cdc6 was stabilized in G1 in a Tom1 mutant that had this cysteine replaced with alanine (Fig. 3B). Likewise, Dia2 contains an F-box domain required for assembly with the other SCF components to form a functional E3 ligase complex (22, 23). Cdc6 is stabilized in a dia2 ΔF-box mutant in cells arrested in G1 (Fig. 3B). This result suggests a requirement for the F-box domain of Dia2, but it is at odds with previous work indicating that scf mutants do not stabilize Cdc6 in G1 (14). We verified that skp1-11, cdc53-1, and cdc34-2 mutants do not stabilize Cdc6 in G1-arrested cells (data not shown). However, when we examined Cdc6 turnover in a Dia2 mutant lacking both the F-box domain and the C-terminal LRR region (TPR mutant), we also observed Cdc6 stabilization (Fig. 3C). Our results suggest that Dia2 contributes to Cdc6 turnover in an F-box-dependent manner but that this function is likely independent of a traditional SCF complex.
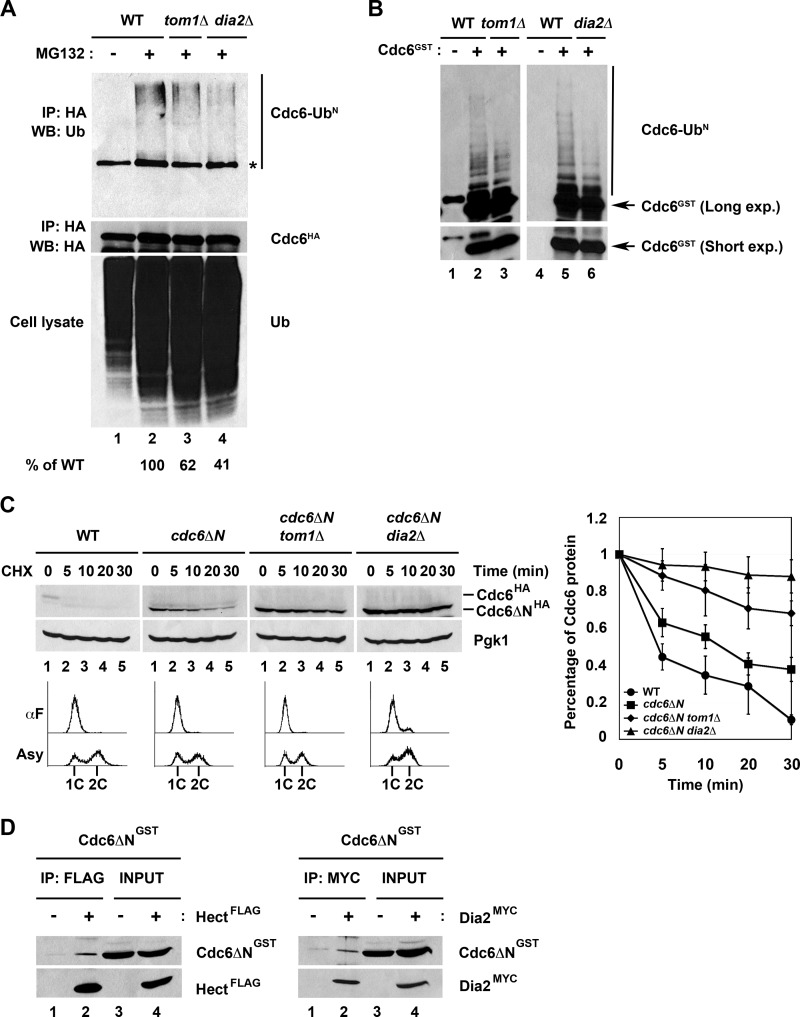
We next asked whether Tom1 and Dia2 are required for Cdc6 ubiquitination. We immunoprecipitated HA-tagged Cdc6 from wild type, tom1Δ, and dia2Δ cells that had been incubated with the proteasome inhibitor MG132. We probed the immunoprecipitates with anti-ubiquitin antibodies. Under these conditions, we observed modified forms of Cdc6 in proteasome-inhibited cells, but these forms were reduced to ∼60% of wild type in tom1Δ cells and to 40% of wild type in dia2Δ cells (Fig. 4A). In addition, we developed an in vitro ubiquitination assay using GST-Cdc6 purified from baculovirus-infected insect cells. When GST-Cdc6 was incubated with crude yeast extracts, we observed Cdc6 ubiquitin conjugates (Fig. 4B). We performed this assay using crude extracts from tom1Δ and dia2Δ strains and observed decreased ubiquitination (Fig. 4B). Together, our results indicate that Cdc6 ubiquitination and protein stability are dependent on both Tom1 and Dia2.
FIGURE 4.
Tom1 and Dia2 target Cdc6 for ubiquitin-dependent degradation and recognize a domain in Cdc6 C-terminal to amino acid 47. A, Cdc6 ubiquitination is dependent on Tom1 and Dia2. Wild type, tom1Δ, and dia2Δ strains were cultured to mid-log phase and treated with dimethyl sulfoxide or 50 μm MG132 for 2 h. Cdc6 protein was immunoprecipitated (IP) with anti-HA antibodies and visualized by immunoblot assays (WB) with anti-ubiquitin antibodies. The asterisk indicates a nonspecific band. Percentage of ubiquitinated Cdc6 was quantified using ImageJ software. B, in vitro ubiquitination of Cdc6 is shown. GST-Cdc6 protein expressed from baculovirus-infected insect cells was purified using glutathione-Sepharose 4B beads. GST-Cdc6 protein was incubated with crude yeast extracts purified from wild type, tom1Δ, or dia2Δ strains at 30 °C for 45 min. Samples were run on 6% SDS-PAGE and immunoblotted with anti-GST antibodies. C, Cdc6ΔN stabilization was increased in tom1Δ and dia2Δ mutants in G1. Wild type, cdc6ΔN, cdc6ΔN tom1Δ, and cdc6ΔN dia2Δ strains arrested with α factor (αF) were used for stability assays. Three independent experiments were used for quantification. D, Cdc6ΔN binds to Tom1 Hect domain and Dia2. Total lysates isolated from Hi5 insect cells infected with GST-Cdc6ΔN, FLAG-Hect, and Myc-Dia2 baculoviruses were incubated together as shown in the figure. FLAG-Hect or Myc-Dia2 was immunoprecipitated with anti-FLAG or anti-Myc antibodies and analyzed by immunoblotting.
Deletion of an N-terminal region in Cdc6 has been shown to lead to a stabilized protein (13). The region encompassing amino acids 2–47 contains four CDK consensus sites that are important for SCFCdc4-mediated degradation of Cdc6 at the G1 to S phase transition. However, this N-terminal truncation mutant of Cdc6 (Cdc6ΔN) is still stabilized in G1 cells, even though Cdc4 does not control Cdc6 proteolysis in this phase (13, 14). Thus, we sought to determine whether the Tom1 or Dia2 was responsible for stabilization of this mutant in G1. We generated strains that endogenously expressed HA-tagged Cdc6ΔN in wild type, tom1Δ, and dia2Δ strains and performed protein stability assays. As previously reported, Cdc6ΔN is partially stabilized in G1-arrested cells (13). Surprisingly, Cdc6ΔN stability is increased in tom1Δ and dia2Δ cells, indicating that Tom1 and Dia2 likely interact with a different region of the Cdc6 protein (Fig. 4C). To test whether Tom1 and Dia2 associate with a Cdc6 protein lacking the N terminus, we co-incubated GST-tagged Cdc6ΔN, FLAG-tagged Tom1 Hect domain, and Myc-tagged Dia2 expressed from baculovirus-infected insect cells and performed immunoprecipitation assays. When either the Tom1 Hect domain or Dia2 was immunoprecipitated, Cdc6ΔN co-purified (Fig. 4D), indicating that these proteins are able to form a complex. We conclude that Dia2 and Tom1 recognize a domain in Cdc6 downstream of amino acid 47.
Our results suggested that Tom1 and Dia2 are required for Cdc6 ubiquitination and degradation, but it was not clear whether Tom1 and Dia2 work together or independently, although the CDC6 overexpression results from Fig. 1 suggested that Tom1 and Dia2 did not act in redundant, separate pathways. To investigate this further, we examined Cdc6 protein stability in a tom1Δ dia2Δ double mutant arrested in G1 with α factor. When we plotted the rate of turnover in the double mutant compared with wild type and the single mutants, the rate of turnover in the double mutant was indistinguishable from the turnover in tom1Δ cells (Fig. 5A). This is consistent with our previous results that suggested Tom1 and Dia2 are not redundant.
FIGURE 5.
Tom1 and Dia2 cannot substitute for each other in Cdc6 degradation. A, Cdc6 turnover rate in tom1Δ dia2Δ mutant is indistinguishable from the tom1Δ or dia2Δ mutant. The results of quantification of wild type, tom1Δ, dia2Δ, and tom1Δ dia2Δ mutants are shown in the graph. B, Tom1 Hect domain and Dia2 bind Cdc6 independently of each other. Insect cell lysates expressing the indicated baculoviruses were incubated with anti-FLAG or anti-Myc antibodies and analyzed by immunoblotting with anti-FLAG, -Myc, or -GST antibodies. C, overexpression of the alternate ubiquitin ligase does not rescue the Cdc6 degradation defect in tom1Δ and dia2Δ mutants. Left, tom1Δ cells carrying empty vector or DIA2 under the control of GAL1,10 promoter were grown to mid-log phase and arrested with α factor for 3 h in minimal medium supplemented with 2% raffinose. Expression of DIA2 was induced with the addition of 2% galactose for 30 min. Stability assays were performed as in Fig. 3. Flow cytometry was used to monitor the α factor (αF) arrest. Right, the same experiment was performed with dia2Δ cells expressing the Tom1 Hect domain. Error bars indicate S.D. IP, immunoprecipitation; CHX, cycloheximide.
We also tested whether Tom1 and Dia2 bind Cdc6 in a cooperative or competitive manner. Our binding assays using baculovirus-expressed protein shown in Fig. 2 suggested that Tom1 and Dia2 can bind Cdc6 independently, as co-purification is observed when Cdc6 and either ubiquitin ligase is co-expressed. In addition, endogenous Cdc6 still immunoprecipitated with either Tom1 or Dia2 in cells lacking the other ligase (data not shown). To test whether Tom1 and Dia2 might affect each other's interaction with Cdc6, we compared the efficiency of co-purification with Cdc6 when both Tom1 and Dia2 are present (Fig. 5B). We incubated equal amounts of GST-Cdc6 with Myc-tagged Dia2, the FLAG-tagged Hect domain of Tom1, or a mixture of Myc-Dia2 and FLAG-Hect domain of Tom1 and then performed immunoprecipitations. Interestingly, we did not observe any enhancement or inhibition of Cdc6 co-purification with either Dia2 or Tom1 when all three proteins were present. We also examined whether Tom1 and Dia2 can compensate for each other's function when overexpressed. We examined Cdc6 protein turnover in tom1Δ cells overexpressing DIA2 and in dia2Δ cells overexpressing the Tom1 Hect domain. In each case, the degradation of the Cdc6 protein was not significantly altered by overexpression of the other ubiquitin ligase (Fig. 5C). Together, these results suggest that Tom1 and Dia2 bind Cdc6 independently of each other and that Tom1 and Dia2 cannot compensate for each other in Cdc6 protein turnover.
Cdc6 Chromatin Association in Ubiquitin Ligase Mutants
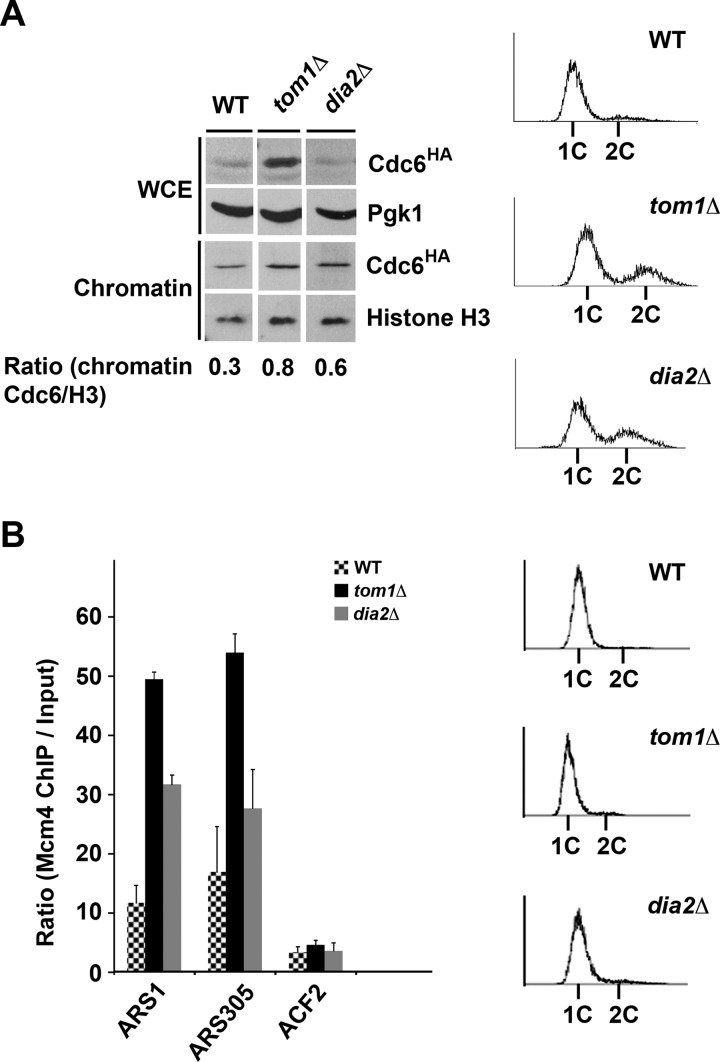
Cdc6 associates with chromatin in late M and early G1 to facilitate origin licensing and pre-RC formation (3, 4, 7). We examined the chromatin association of Cdc6 during G1 in tom1Δ and dia2Δ cells. We used wild type cells as a negative control. Cells were arrested in G1 with α factor, and then samples were collected for chromatin fractionation. The abundance of Cdc6 is significantly increased in the whole cell extract from tom1Δ cells relative to wild type, consistent with impaired degradation in this strain (Fig. 6A). The chromatin association of Cdc6 in the tom1Δ and dia2Δ strains was significantly higher than wild type. Overall, these results suggest that a greater fraction of Cdc6 associates with chromatin in late G1 in tom1Δ and dia2Δ cells, as ubiquitination of Cdc6 is impaired.
FIGURE 6.
Failure to degrade chromatin-bound Cdc6 by a Tom1- and Dia2-dependent pathway enhances the association of Mcm4 with early origins. A, chromatin-bound Cdc6 is increased in tom1Δ and dia2Δ cells. The indicated strains were arrested with α factor for 3 h, and samples were prepared for chromatin fractionation. Samples were analyzed by immunoblot assays with anti-HA, -Pgk1, and -histone H3 antibodies. Flow cytometry was used to monitor cell cycle arrest. WCE, whole cell extract. The ratio of chromatin-bound Cdc6 to histone H3 was measured using ImageJ software. B, Mcm4 origin association is increased in tom1Δ and dia2Δ cells. The indicated strains were arrested as in A. Samples were prepared for ChIP assays with HA-tagged Mcm4. ARS1 and ARS305 are early firing origins. ACF2 is nonorigin control. The ratio of Mcm4 chromatin IP to input is shown in the graph. Three replicates of each experiment were quantified using ImageJ. Error bars indicate S.D.
To examine whether the aberrant Cdc6 chromatin association led to increased association of the Mcm2-7 complex with replication origins, we used chromatin immunoprecipitation. Wild type, tom1Δ, and dia2Δ cells expressing HA-tagged Mcm4 were arrested in G1, and Mcm4 chromatin association at two early firing origins (ARS1 and ARS305) and one nonorigin region (ACF2) was assayed (Fig. 6B). All strains exhibited only negligible Mcm4 binding at the nonorigin region ACF2. Mcm4 association at ARS1 and ARS305 was substantially higher than wild type in tom1Δ cells. In dia2Δ cells, Mcm4 was strongly associated with ARS1, but the effect at ARS305 was less obvious. These results suggest that increased Cdc6 abundance and chromatin association in both the tom1Δ and dia2Δ strains may lead to increased association of Mcm proteins at origins.
DISCUSSION
Altogether our data indicate that the Hect domain E3 ligase Tom1 and the F-box protein Dia2 are required to target Cdc6 for ubiquitin-mediated destruction during G1 phase of the cell cycle. Our results suggest that Tom1 and Dia2 bind Cdc6 independently of each other and cannot substitute for each other. We favor a model in which Tom1 and Dia2 function in the same pathway, as the degradation of Cdc6 is not enhanced in a tom1Δ dia2Δ double mutant.
One possible explanation is that Dia2 and Tom1 act in a sequential pathway to target Cdc6 for degradation, although there is no evidence to indicate which ligase acts first or whether there are intervening steps in the pathway. It seems that Dia2 functions outside the context of an SCF complex in this pathway, as we do not observe any defect in Cdc6 degradation in scf mutants, similar to previous studies (13, 14). A non-SCF role for Dia2 has not been previously described, but other F-box proteins have been shown to function outside of traditional SCF complexes (33–36). It is possible that additional factors are required to complex with Dia2 to contribute to the ubiquitination of Cdc6.
We recently determined that Tom1 also targets Dia2 for ubiquitin-mediated degradation during G1 (37). In principle, this pathway could affect Cdc6 degradation via Tom1. However, we did not find any evidence that Dia2 competes with Cdc6 for binding to Tom1. Moreover, if Dia2 competed with Cdc6, we would expect that Cdc6 degradation would be enhanced in dia2Δ cells, rather than inhibited. It is possible that Dia2 degradation is a by-product of Cdc6 degradation. Such a scenario would suggest that Dia2 is an accessory factor for Tom1, but in this case we might expect cooperative binding, which we did not observe. Future studies will be necessary to delineate the precise mechanistic roles of Dia2 and Tom1 in targeting Cdc6 for degradation.
Our results support the idea that Tom1 and Dia2 account for a fraction of the G1-specific degradation of Cdc6. The observation that they act separately from the Cdc4 pathway is consistent with Mode 1 degradation, but investigation of whether Cdk activity affects the activity of Tom1 or Dia2 will be necessary to resolve this question. One possibility is that Cdc6 is targeted for degradation after DNA damage, as Tom1 has been shown to have a role in such a pathway (19) and dia2Δ cells exhibit endogenous DNA damage (21–23). However, we think this is unlikely during G1 phase, as dia2Δ cells only accumulate DNA damage foci during S and G2/M (22, 23). Rather, we think it is likely that Tom1 and Dia2 represent a novel mode of Cdc6 degradation. Indeed, the observation that Cdc6ΔN stability is enhanced in G1 in the absence of Tom1 and Dia2 suggests that there may be still another degradation pathway for Cdc6 in G1.
The functional significance of degrading Cdc6 during G1 is not entirely clear. Previous work from the Diffley laboratory has suggested that a greater fraction of Cdc6 is chromatin-bound during G1 (13). It is possible that degradation of Cdc6 may be used to regulate pre-RC assembly on replication origins in G1. The increased accumulation of chromatin-bound Cdc6 and Mcm4 observed in dia2Δ and tom1Δ cells is consistent with this hypothesis. Alternatively, it is possible that Dia2 and Tom1 target soluble Cdc6 for degradation. Future studies will be necessary to distinguish between these possibilities.
In summary, we have identified two novel E3 ubiquitin ligase components, Tom1 and Dia2, which contribute to Cdc6 ubiquitination and degradation during the G1 phase of the cell cycle. In the absence of Tom1 and Dia2, Cdc6 and Mcm4 chromatin association is altered. Altogether, our results indicate that multiple ubiquitin ligases control Cdc6 degradation, perhaps to regulate pre-RC assembly.
Acknowledgments
We thank C. Brandl (University Western Ontario) for the tom1 C3235A strain and members of D. Clarke's laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM076663 (to D. M. K.).

This article contains supplemental Tables I and II.
- pre-RC
- pre-replicative complex
- Cdk
- cyclin-dependent kinase
- pgk
- phosphoglycerate kinase
- SCF
- Skp1/Cdc53/F-box protein.
REFERENCES
- 1. Sclafani R. A., Holzen T. M. (2007) Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41, 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blow J. J., Dutta A. (2005) Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang C., Weinreich M., Stillman B. (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81, 667–676 [DOI] [PubMed] [Google Scholar]
- 4. Cocker J. H., Piatti S., Santocanale C., Nasmyth K., Diffley J. F. (1996) An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379, 180–182 [DOI] [PubMed] [Google Scholar]
- 5. Perkins G., Diffley J. F. (1998) Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell 2, 23–32 [DOI] [PubMed] [Google Scholar]
- 6. Weinreich M., Liang C., Stillman B. (1999) The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl. Acad. Sc.i U.S.A. 96, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donovan S., Harwood J., Drury L. S., Diffley J. F. (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 94, 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanaka T., Knapp D., Nasmyth K. (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90, 649–660 [DOI] [PubMed] [Google Scholar]
- 9. Maiorano D., Moreau J., Méchali M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404, 622–625 [DOI] [PubMed] [Google Scholar]
- 10. Nishitani H., Lygerou Z., Nishimoto T., Nurse P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625–628 [DOI] [PubMed] [Google Scholar]
- 11. Devault A., Vallen E. A., Yuan T., Green S., Bensimon A., Schwob E. (2002) Identification of Tah11/Sid2 as the ortholog of the replication licensing factor Cdt1 in Saccharomyces cerevisiae. Curr. Biol. 12, 689–694 [DOI] [PubMed] [Google Scholar]
- 12. Piatti S., Lengauer C., Nasmyth K. (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S-phase and for preventing a reductional anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drury L. S., Perkins G., Diffley J. F. (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drury L. S., Perkins G., Diffley J. F. (2000) The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231–240 [DOI] [PubMed] [Google Scholar]
- 15. Elsasser S., Chi Y., Yang P., Campbell J. L. (1999) Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sánchez M., Calzada A., Bueno A. (1999) The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem. 274, 9092–9097 [DOI] [PubMed] [Google Scholar]
- 17. Perkins G., Drury L. S., Diffley J. F. (2001) Separate SCFCDC4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20, 4836–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh R. K., Kabbaj M. H., Paik J., Gunjan A. (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell. Biol. 11, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., Cook J. G. (2007) Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell 18, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall J. R., Lee H. O., Bunker B. D., Dorn E. S., Rogers G. C., Duronio R. J., Cook J. G. (2008) Cdt1 and Cdc6 are destabilized by rereplication-induced DNA damage. J. Biol. Chem. 283, 25356–25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124, 1069–1081 [DOI] [PubMed] [Google Scholar]
- 22. Koepp D. M., Kile A. C., Swaminathan S., Rodriguez-Rivera V. (2006) The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell 17, 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blake D., Luke B., Kanellis P., Jorgensen P., Goh T., Penfold S., Breitkreutz B. J., Durocher D., Peter M., Tyers M. (2006) The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 174, 1709–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morohashi H., Maculins T., Labib K. (2009) The amino-terminal TPR domain of Dia2 tethers SCFDia2 to the replisome progression complex. Curr. Biol. 19, 1943–1949 [DOI] [PubMed] [Google Scholar]
- 25. Mimura S., Komata M., Kishi T., Shirahige K., Kamura T. (2009) SCFDia2 regulates DNA replication forks during S-phase in budding yeast. EMBO J. 28, 3693–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 27. Liu Q., Li M. Z., Leibham D., Cortez D., Elledge S. J. (1998) The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8, 1300–1309 [DOI] [PubMed] [Google Scholar]
- 28. Liang C., Stillman B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11, 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93 [DOI] [PubMed] [Google Scholar]
- 30. Elsasser S., Lou F., Wang B., Campbell J. L., Jong A. (1996) Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell 7, 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bueno A., Russell P. (1992) Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 11, 2167–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saleh A., Collart M., Martens J. A., Genereaux J., Allard S., Cote J., Brandl C. J. (1998) TOM1p, a yeast Hect domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J. Mol. Biol. 282, 933–946 [DOI] [PubMed] [Google Scholar]
- 33. Yoshida Y., Murakami A., Iwai K., Tanaka K. (2007) A neural-specific F-box protein Fbs1 functions as a chaperone suppressing glycoprotein aggregation. J. Biol. Chem. 282, 7137–7144 [DOI] [PubMed] [Google Scholar]
- 34. Tafforeau L., Le Blastier S., Bamps S., Dewez M., Vandenhaute J., Hermand D. (2006) Repression of ergosterol level during oxidative stress by fission yeast F-box protein Pof14 independently of SCF. EMBO J. 25, 4547–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gearhart M. D., Corcoran C. M., Wamstad J. A., Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Escobar-Henriques M., Westermann B., Langer T. (2006) Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 173, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim D. H., Koepp D. M. (2012) The Hect E3 ubiquitin ligase Tom1 controls Dia2 degradation during the cell cycle. Mol. Biol. Cell 23, 4203–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]