ABSTRACT
Pneumococcal pili have been shown to influence pneumococcal colonization, disease development, and the inflammatory response in mice. The role of the pilus-associated RrgA adhesin in pneumococcal interactions with murine and human macrophages was investigated. Expression of pili with RrgA enhanced the uptake of pneumococci by murine and human macrophages that was abolished by antibodies to complement receptor 3 (CR3) and not seen in CR3-deficient macrophages. Recombinant RrgA, but not pilus subunit RrgC, promoted CR3-mediated phagocytosis of coated beads by murine and human macrophages. Flow cytometry showed that purified CR3 binds pneumococcal cells expressing RrgA, and purified RrgA was shown to interact with CR3 and its I domain. In vivo, RrgA facilitated spread of pneumococci from the upper airways and peritoneal cavity to the bloodstream. Earlier onset of septicemia and more rapidly progressing disease was observed in wild-type mice compared to CR3-deficient mice challenged intranasally or intraperitoneally with pneumococci. Motility assays and time-lapse video microscopy showed that pneumococcal stimulation of macrophage motility required RrgA and CR3. These findings, together with the observed RrgA-dependent increase of intracellular survivors up to 10 h following macrophage infection, suggest that RrgA-CR3-mediated phagocytosis promotes systemic pneumococcal spread from local sites.
IMPORTANCE
Streptococcus pneumoniae is a major contributor to morbidity and mortality in infectious diseases globally. Symptomatology is mainly due to pneumococcal interactions with host cells leading to an inflammatory response. However, we still need more knowledge on how pneumococci talk to immune cells and the importance of this interaction. Recently, a novel structure was identified on the pneumococcal surface, an adhesive pilus found in about 30% of clinical pneumococcal isolates. The pilus has been suggested to be important for successful spread of antibiotic-resistant pneumococcal clones globally. Here we sought to identify mechanisms for how the pneumococcal pilin subunit RrgA contributes to disease development by interacting with host immune cells. Our data suggest a new way for how pneumococci may cross talk with phagocytic cells and affect disease progression. An increased understanding of these processes may lead to better strategies for how to treat these common infections.
Introduction
Streptococcus pneumoniae, commonly referred to as pneumococcus, is a major human pathogen contributing greatly to morbidity and mortality worldwide, especially in children and the elderly. Pneumococcal infections cause approximately as many deaths as tuberculosis and include diseases ranging from milder respiratory tract infections to more severe diseases such as pneumonia, meningitis, and septicemia. Although colonization of the nasopharynx precedes disease, pneumococci are frequently found colonizing the nasopharynges of healthy preschool children. The underlying mechanisms explaining how pneumococci, from a local site, pass mucosal barriers to cause systemic disease remains to be further elucidated.
S. pneumoniae strain T4 (TIGR4) expresses a pilus-like structure, encoded by the rlrA pilus islet 1, and shown to contribute to virulence in animal models (1). The rlrA pilus islet 1 is present in approximately 30% of all pneumococcal isolates (2, 3), depending on the clonal type, and consists of genes encoding three different pilus subunit proteins, RrgA, RrgB, and RrgC, which are covalently linked by three pilus-specific sortases. RrgB is the major stalk protein of the pilus, and in the absence of a pilus shaft, monomeric RrgA is located on the surface and is sortase-linked to the cell wall. However, clinical isolates producing RrgA in the absence of pili have not yet been reported. The pneumococcal pilus and specifically the RrgA protein promote adhesion to lung epithelial cells in vitro and virulence in vivo in murine models (1, 4). The crystal structure of RrgA was recently solved (5). It was demonstrated that the 893-residue-long adhesin formed an elongated structure composed of four domains of which the major domain, the D3 domain, adopts an integrin I collagen recognition domain suggested to interact with extracellular matrix (ECM) proteins. Indeed, purified RrgA has been shown to bind fibronectin, laminin, and collagen I, but not to vitronectin (6).
The innate immune system involves effectors and immune cells and constitutes the first line of defense against invading pathogens. In the lungs, phagocytosis mediated by resident macrophages plays a central role in clearance of pneumococci early in infection, and bacterium-induced Toll-like receptor 9 (TLR9)-NF-κB signaling has been suggested to enhance the phagocytic capacity of alveolar macrophages (AMs) (7, 8). It has also been reported that influenza virus sensitization to pneumococcal infection might operate via an interferon-induced inhibition of bacterial clearance, mediated by AMs in the lungs (9).
Numerous surface receptors and associated signal transduction pathways are involved in the phagocytic machinery, leading to bacterial killing and later to the induction of an adaptive immune response. The complement system acts as a part of the innate immune response by opsonizing microbes in a specific manner. Complement receptors (CRs) on the surfaces of phagocytes recognize and internalize the opsonized pathogens. Opsonization of bacteria by immunoglobulins leads to similar enhanced uptake of pathogens by Fcγ receptors. Also, opsonin-independent phagocytosis, where ligands on the surfaces of the microorganisms are directly recognized by receptors on the plasma membranes of phagocytes, has been reported. Scavenger receptors, like macrophage receptor with a collagenous structure (MARCO), promote phagocytosis of bacteria nonopsonically and have been shown to protect against pneumococcal infections (10–12). There are also receptors that can be involved in either pathway, such as complement receptor 3 (CR3, CD11b/CD18, Mac-1) (13). CR3 is expressed on polymorphonuclear leukocytes, monocytes/macrophages, and activated lymphocytes and mediates both opsonin-dependent and -independent phagocytosis. It recognizes multiple microbial adhesins by direct protein-protein interactions (14, 15). CR3 binds to a variety of molecules in the host, such as intercellular adhesion molecule 1 (ICAM-1) (16), fibrinogen (17), and heparin (18). Binding of CR3 induces different functions such as leukocyte extravasation and migration. Activation of CR3 also upregulates other key adhesion and defense receptors on leukocytes (19, 20).
Here, we sought to determine whether an interaction can be found between pneumococcal pili and phagocytes, whether the pilus-associated adhesin RrgA is required for this process, and if such an interaction translates into in vivo effects using mouse infection models.
RESULTS
RrgA on pneumococcal pilus 1 promotes nonopsonic complement receptor 3 (CR3)-dependent uptake of S. pneumoniae by murine and human macrophages.
To examine whether expression of RrgA affects phagocytosis, pneumococcal strain T4 (TIGR4) expressing RrgA containing pili and mutant derivatives of T4 were incubated on monolayers of murine bone marrow-derived macrophages (BMDMs). T4 is an encapsulated and piliated serotype 4 pneumococcal strain originally isolated from a patient with invasive disease. Its isogenic mutant, the T4ΔrrgA strain, lacks RrgA but expresses a RrgB- and RrgC-positive pilus (4). We also created a complemented mutant of T4ΔrrgA, T4ΔrrgA∇(lacE::rrgA) in which rrgA was reinserted into the pneumococcal chromosome in trans.
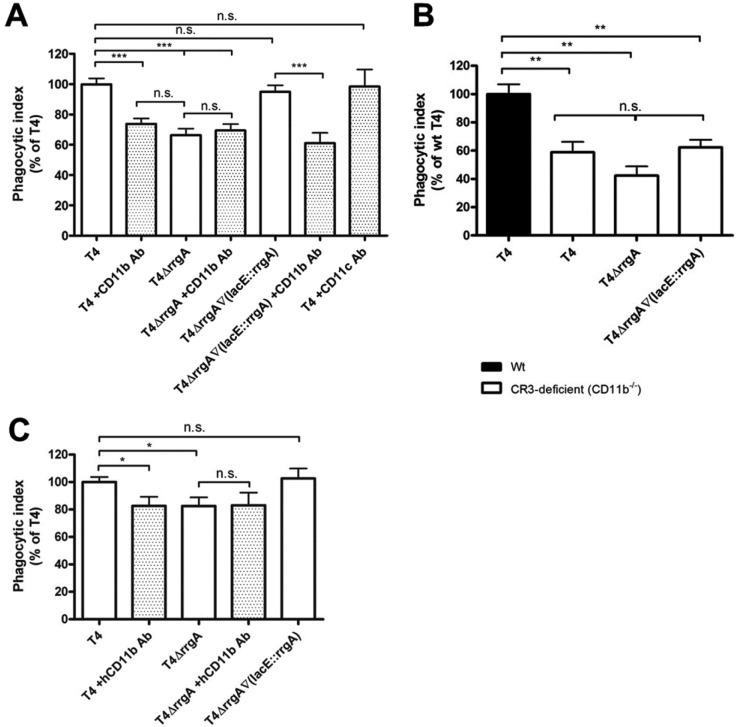
After 1 h of phagocytosis, the strains expressing pilus-associated RrgA [T4 and T4ΔrrgA∇(lacE::rrgA) strains] were internalized to a significantly higher degree than the rrgA deficient mutant, the T4ΔrrgA strain (Fig. 1A) (P = 0.0005). For a control, we used cytochalasin D to block actin cytoskeleton rearrangement in cells. With cytochalasin D treatment, the uptake of pneumococci was almost completely abolished, and there was no difference in adherence of the three pneumococcal strains to the cells (data not shown).
FIG 1 .
The pneumococcal pilus subunit RrgA enhances uptake by murine and human macrophages through interaction with CD11b (CR3). (A and B) The bacteria were labeled with FITC and incubated with monolayers of bone marrow-derived macrophages (BMDMs) for 1 h at an MOI of ca. 60 bacteria per cell, and the fluorescence intensity was measured by FACS. The phagocytic index for each sample was calculated by multiplying the percentage of internalized cells by the mean fluorescence intensity and dividing this by the mean of the wild-type (T4) sample multiplied with the percentage of FITC-positive cells. (A) Phagocytosis assays were performed with murine BMDMs. The mutant lacking RrgA (T4ΔrrgA) was taken up to a significantly lower extent than the bacteria expressing RrgA (T4 and T4ΔrrgA∇lacE::rrgA strains). BMDMs were pretreated with antibodies (Ab) against mouse CD11b. This inhibited the uptake of the T4 and T4ΔrrgA∇(lacE::rrgA) strains, which under these conditions reached levels similar to those of the uptake of the T4ΔrrgA mutant. The antibody treatment did not, however, affect the uptake of the T4ΔrrgA mutant. Antibodies directed against mouse CD11c did not influence the uptake of T4. Each condition was done in triplicate and was repeated at least three times. Values that were significantly different (P < 0.001) are indicated by a bar and three asterisks. Values that were not significantly different are indicated by a bar and n.s. (for not significant). (B) CR3-deficient (CD11b−/−) BMDMs exhibited lower levels of uptake of T4 than macrophages from wild-type mice (Wt). The decrease was comparable to the levels of uptake of the T4ΔrrgA mutant by wild-type macrophages. There were no statistically significant differences in the levels of uptake of the different pneumococcal strains by the CR3-deficient macrophages. Each experiment was done in triplicate and was repeated at least three times. Values that were significantly different (P < 0.01) are indicated by a bar and two asterisks. Values that were not significantly different are indicated by a bar and n.s. (for not significant). (C) In concordance with the findings in murine macrophages, a mutant lacking RrgA (T4ΔrrgA) was taken up by human phorbol myristate acetate (PMA)-differentiated THP-1 cells to a significantly lower extent than bacteria expressing RrgA. Furthermore, a specific antibody against human CD11b inhibited the uptake of the T4 strain but did not significantly affect the uptake of the T4ΔrrgA mutant. Values are means plus standard errors of the means (SEM) (error bars). Values that were significantly different (P < 0.05) are indicated by a bar and one asterisk. Values that were not significantly different are indicated by a bar and n.s. (for not significant).
To identify a putative receptor on BMDMs, we considered CR3, also named integrin CD11b/CD18 or Mac-1. Pretreatment of BMDMs with anti-CD11b antibodies decreased the uptake of pneumococcal strain T4 and the T4ΔrrgA∇(lacE::rrgA) strain to the level of uptake of the T4ΔrrgA mutant (P < 0.001) (Fig. 1A). Antibody treatment had no effect on the uptake of the T4ΔrrgA mutant. We also treated murine BMDMs with anti-CD11c antibodies and found that they did not influence uptake of T4, thereby confirming the antibody specificity in inhibition (Fig. 1A).
To further confirm a role for CR3 in pneumococcal phagocytosis, we performed phagocytosis assays with BMDMs from CD11b−/− mice, which lack functional CR3. These macrophages showed a 40% reduction in the uptake of strain T4 expressing pili with RrgA (Fig. 1B) (P < 0.0012), which is similar to the level of uptake of the RrgA-deficient bacteria by wild-type macrophages. When comparing uptake of the different pneumococcal strains in CD11b−/− macrophages, no statistically significant differences were observed (Fig. 1B). Targeting CD11b by antibodies or its complete absence only partially inhibited BMDM phagocytosis, testifying to the role of other phagocytic receptors in pneumococcal uptake by macrophages.
Next we used human macrophages (THP-1 cells) in phagocytosis assays of the T4, T4ΔrrgA, and T4ΔrrgA∇(lacE::rrgA) strains (Fig. 1C). A moderate but significant decrease of T4ΔrrgA phagocytosis compared to the wild type and the RrgA-complemented mutant was found. A monoclonal antibody specific for human CD11b decreased THP-1 phagocytosis only for strain T4 expressing wild-type pili but had no effect on the uptake of the T4ΔrrgA mutant (Fig. 1C).
Recombinant RrgA, but not RrgC, promotes CR3-mediated phagocytosis of coated beads by murine and human macrophages.
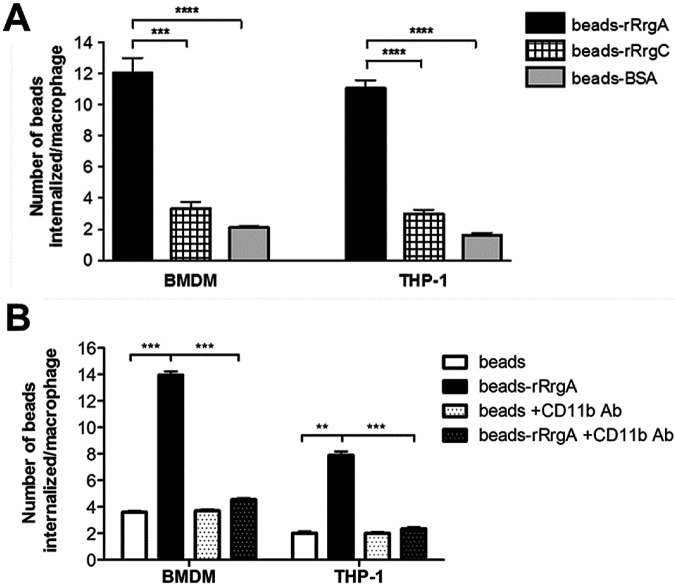
To directly demonstrate a role for the RrgA adhesin in phagocytosis, we conjugated full-length recombinant RrgA (rRrgA) to fluorescent microspheres and monitored uptake by murine BMDMs and by human THP-1 cells (Fig. 2A). For a relevant control, we made use of rRrgC, an ancillary pilus 1 component believed to anchor the completed pilus to the peptidoglycan cell wall. In both BMDMs and THP-1 cells, phagocytosis was three and four times higher for beads coated with rRrgA than for beads coated with rRrgC and bovine serum albumin (BSA), respectively. Antibodies specific to murine and human CD11b completely inhibited phagocytosis of rRrgA-coated beads by murine BMDMs and human THP-1 cells, respectively (Fig. 2B).
FIG 2 .
rRrgA promotes uptake of latex beads into macrophages. (A) BMDMs and THP-1 cells were challenged with latex beads covered with rRrgA (beads-rRrgA) or with rRrgC (beads-rRrgC) or with bovine serum albumin (BSA) (beads-BSA). We compared the number of coated beads that were taken up by both murine and human macrophages using BMDMs from mice as well as human PMA-differentiated THP-1 cells. The number of phagocytosed beads increased significantly when the beads were coated with rRrgA. (B) Pretreatment with mouse or human CD11b-specific antibodies significantly blocked the CD11b-dependent uptake of beads coated with rRrgA (beads-rRrgA +CD11b Ab) without influencing the number of internalized uncoated beads (beads +CD11b Ab) in BMDMs and THP-1 cells, respectively. Each treatment was done in duplicate four times. For each sample, the number of internalized beads was counted in a minimum of 10 visual fields containing at least 10 cells. Values are means plus SEM (error bars). Values that are significantly different by the Mann-Whitney U test are indicated by bars and asterisks as follows: **, P < 0.01; ***, P < 0.0002; ****, P < 0.0001.
Purified CR3 binds directly to pneumococcal cells expressing pilus-associated RrgA and to recombinant RrgA.
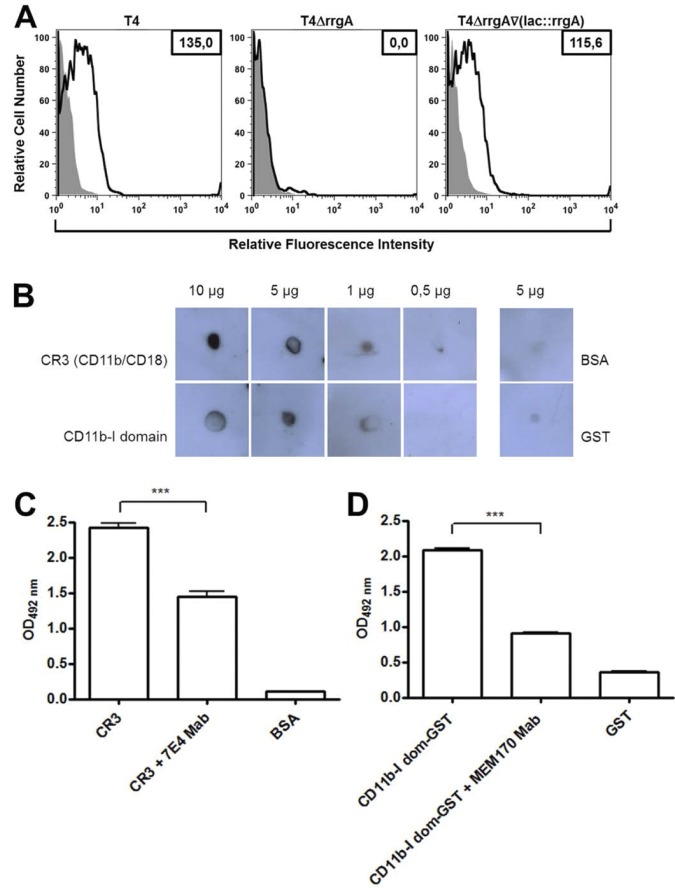
The RrgA adhesin has been demonstrated to bind to a number of extracellular matrix proteins, including collagen I, fibronectin, and laminin in contrast to the other two pilus 1 constituents, RrgB and RrgC (6). To find evidence for a direct interaction between RrgA-expressing pneumococci and CR3, we performed flow cytometry analyses of purified human CR3 (CD11b/CD18) binding to S. pneumoniae T4, T4ΔrrgA, and T4ΔrrgA∇(lacE::rrgA) strains, making use of anti-CD11b monoclonal antibody (MAb) (peridinin chlorophyll protein [PerCp]-Cy5.5 labeled). CR3 bound efficiently to the two RrgA-expressing strains, whereas there was no binding to the T4ΔrrgA strain (Fig. 3A). The addition of extracellular matrix proteins (collagen I, fibronectin, laminin, or vitronectin) did not inhibit or enhance CR3 binding to pneumococcal cells expressing RrgA (see Fig. S1 in the supplemental material), suggesting a direct interaction between CR3 and RrgA.
FIG 3 .
Pneumococcal cell surface-associated and purified RrgA binds to purified human CR3 as well as to the CD11b-I domain. (A) Flow cytometry analysis of CR3 binding to RrgA protein associated with the S. pneumoniae cell surface. T4 strain and its mutants (108 live bacteria) were incubated with 10 µg/ml of purified CR3 (histograms outlined by thick black lines) or PBS (light gray filled histograms), and CR3 binding was detected with an anti-CD11b MAb (PerCp-Cy5.5 labeled) at a final concentration of 10 µg/ml. CR3 binding is observed by a shift to the right in the population of bacteria incubated with the purified CR3 protein and expressing RrgA on their surface, i.e., T4 and T4ΔrrgA∇(lacE::rrgA) cells, compared with cells incubated with PBS (control) (gray) or with CR3 but not expressing RrgA, i.e., T4ΔrrgA cells. Binding was calculated by subtracting the mean fluorescence intensity of bacteria incubated with purified CR3 protein from that of the bacteria incubated with PBS (ΔMFI). The average ΔMFI values from three independent experiments are shown as numbers in the boxes in the top right-hand corner. (B) Human CR3 or CD11b I domain-GST was blotted in different concentrations onto a nitrocellulose filter, blocked, and subsequently incubated with 2 µg of rRrgA. Dose-dependent binding was detected using an antibody directed against RrgA. BSA or GST was used as a negative control. (C and D) rRrgA binding to purified CR3 (C) and to CD11b-I domain (dom) (D) was significantly decreased when specific antibody against CD18 (7E4 Mab) and CD11b (MEM170 Mab) were used, respectively, to inhibit the binding in an ELISA. Values are means plus standard deviations (SD) (error bars). Values that are significantly different (P < 0.0001) are indicated by a bar and three asterisks. OD492 nm, optical density at 492 nm.
Next we studied the interaction between purified CR3 and rRrgA using a far-Western dot blotting technique in which purified human CR3 was blotted onto nitrocellulose membranes in concentrations of 0.5, 1, 5, and 10 µg (Fig. 3B). Since most known interactions between CR3 and its ligands occur via the CD11b integrin domain, purified CD11b-I was also used at equivalent concentrations. Five micrograms of BSA was used as a negative control. For a probe, we used 2 µg of full-length recombinant RrgA and monitored binding with a specific rabbit antibody against RrgA (anti-RrgA). rRrgA was found to bind in a dose-dependent manner to both CR3 and CD11b-I (Fig. 3B). Furthermore, binding to purified CR3 and CD11b-1 decreased when a specific antibody against CD18 and CD11b was used in an ELISA (Fig. 3C and D). Also, rRrgA binding to CR3 and CD11b-I was dependent on the concentration of the RrgA adhesin (see Fig. S2A and S2B in the supplemental material).
Absence of RrgA and/or murine expression of CR3 decreased the number of bacteria in the bloodstream early after intranasal and intraperitoneal challenge.
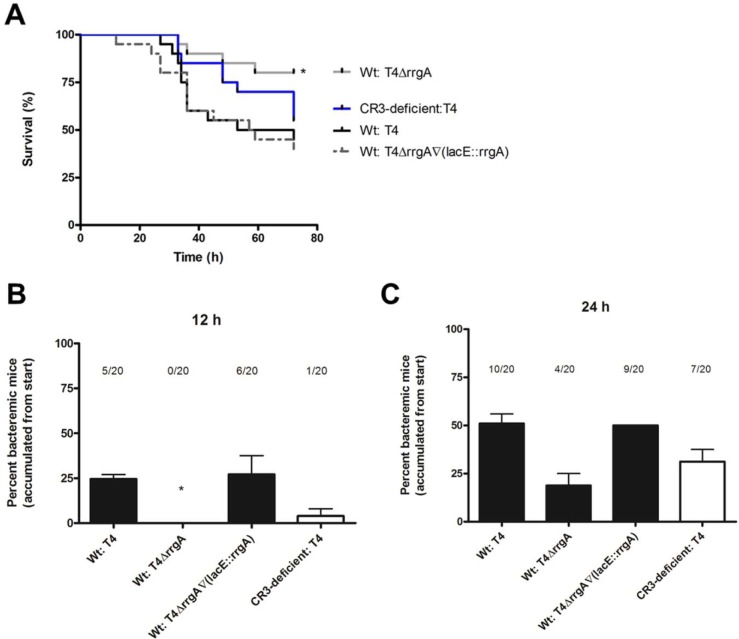
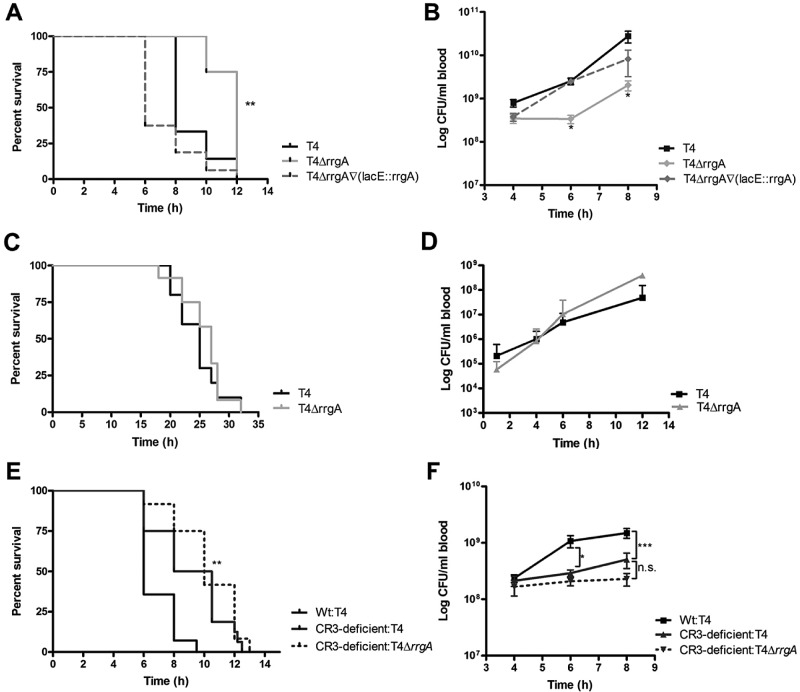
We next investigated whether pneumococcal expression of RrgA and host expression of CR3 affects virulence and dissemination of pneumococci from a local site to the bloodstream. As intranasal (i.n.) inoculation of pneumococci in mice mimics the natural route of infection in humans, we first investigated whether RrgA and CR3 influence disease kinetics in an infection model of pneumococcal pneumonia. We inoculated 5 × 106 CFU i.n. into C57BL/6 mice and monitored the animals up to 72 h postinfection (Fig. 4A to C). Wild-type mice infected with the T4 or T4ΔrrgA∇(lacE::rrgA) strain showed increased mortality compared to wild-type mice infected with the T4ΔrrgA strain (Fig. 4A) (P = 0.0187 in the survival analysis with the Kaplan-Meier log rank test). Furthermore, the two strains expressing RrgA could be detected in blood samples from wild-type mice as early as 12 h postinfection, whereas no T4ΔrrgA bacteria were recovered from blood samples from any mice at this early time point (Fig. 4B). CD11b−/− mice inoculated i.n. with the T4 strain succumbed later to infection than wild-type mice challenged with the same strain (Fig. 4A), and CR3-deficient mice also exhibited a delay in the onset of bacteremia (Fig. 4B and C).
FIG 4 .
RrgA induces faster onset of bacteremia and decreased survival after intranasal challenge of mice. Mice were inoculated i.n. with ca. 5 × 106 CFU per mouse. Blood samples were taken at various time points, and survival was assessed. (A) Wild-type mice infected with the T4ΔrrgA mutant and CR3-deficient (CD11b−/−) mice survived to a higher extent (80 and 55%, respectively) than wild-type (Wt) mice infected with strains expressing RrgA, i.e., T4 and T4ΔrrgA∇(lacE::rrgA) strains, which had survival rates of 45 and 40%, respectively. Survival was analyzed using the Kaplan-Meier log rank test. The asterisk refers to wild-type mice infected with the T4∆rrgA strain compared to the T4 strain. (B and C) Percentages of bacteremic mice from the start of the experiment. Wild-type mice inoculated with RrgA+ bacteria were bacteremic 12 h postinfection. Bacteria appeared later in blood samples from wild-type mice infected intranasally with the T4ΔrrgA mutant and CR3-deficient (CD11b−/−) mice infected with the T4 strain. There were 10 mice per group in each of the i.n. infection experiments, and the experiments were repeated three times. The value for wild-type mice infected with the T4ΔrrgA mutant was significantly different (P < 0.05) from the value for wild-type mice infected with T4 strain as indicated by the asterisk.
When we challenged C57BL/6 mice intraperitoneally (i.p.) with the T4 strain and the different isogenic mutants, we observed that mice infected with the T4ΔrrgA mutant lacking RrgA survived significantly longer than mice infected with either the T4 or T4ΔrrgA∇(lacE::rrgA) strain both expressing RrgA (Fig. 5A) (P = 0.0025 in the survival analysis with the Kaplan-Meier log rank test) (4). The prolonged survival of mice infected i.p. with the T4ΔrrgA mutant was accompanied by lower levels of bacteria in the bloodstream (Fig. 5B).
FIG 5 .
Pneumococcal expression of RrgA and mouse expression of CR3 promotes dissemination of bacteria from the peritoneal cavity to the bloodstream. Mice were infected i.p. or i.v. with ca. 5 × 106 or 5 × 105 CFU per mouse, respectively. Blood samples were taken at various time points. (A) Wild type (Wt) mice infected i.p. with the strain lacking RrgA (T4ΔrrgA) survived significantly longer than mice infected with strains expressing RrgA, i.e., T4 and T4ΔrrgA∇(lacE::rrgA) strains (P < 0.01). (B) The levels of bacteria lacking RrgA were lower in the bloodstream of wild-type mice. (C and D) Wild-type mice infected i.v. exhibited similar survival curves regardless of which strain they were infected with (C), and there were no differences in the growth of the bacteria in the blood of wild-type mice (D). (E) CR3-deficient (CD11b−/−) mice infected i.p. with either T4 or T4ΔrrgA survived significantly longer than wild-type mice. (F) T4 and T4ΔrrgA strains appeared later and in lower numbers in the bloodstream of CR3-deficient mice compared to wild-type mice. There were no statistically significant differences between the different groups of CR3-deficient mice compared to wild-type mice infected with the T4 strain. There were 10 and 5 mice per group in each of the i.p. and i.v. infection experiments, respectively, and the experiments were repeated three times. Values that are significantly different are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant.
To rule out the possibility that the prolonged survival of mice infected with the T4ΔrrgA mutant either i.n. or i.p. was due to slower growth of the bacteria in blood, we infected mice intravenously (i.v.) with the T4 strain and the T4ΔrrgA mutant. Using the i.v. infection route, there were no differences in the survival rate or bacterial growth in blood samples from mice infected with the two strains (Fig. 5C and D).
Finally, we investigated whether a lack of CR3 expression affected the survival of mice after i.p. inoculation with the T4 or T4ΔrrgA strain. When we performed i.p. experiments, we observed that there was a delay in the onset of symptoms and a significantly longer overall survival time of CR3-deficient (CD11b−/−) mice compared to wild-type mice (P = 0.0051 in the survival analysis with the Kaplan-Meier log rank test) (Fig. 5E). CD11b−/− mice had lower numbers of CFUs in blood when infected with both T4 and T4ΔrrgA strains compared to wild-type mice infected with T4, arguing that the dissemination of RrgA-expressing pneumococci is impaired in CD11b−/− mice (Fig. 5F).
Bacterial expression of the pilus-associated RrgA adhesin results in higher numbers of intracellular survivors at late time points after phagocytosis.
The results presented so far suggest that the RrgA adhesin, an ancillary protein of the pneumococcal 1 pilus, enhances phagocytosis through a direct interaction with the CR3 receptor expressed on murine and human macrophages. We expected that enhanced phagocytosis in vitro would translate into decreased virulence in vivo. In contrast, expression of RrgA in the bacteria together with CR3 in the host promoted bacterial virulence in mice and an early appearance of bacteria in the bloodstream after local infection.
To begin explaining these results, we first monitored the fate of RrgA-expressing and non-RrgA-expressing pneumococci after phagocytosis. The uptake of bacteria by macrophages leads to formation of phagosomes, where bacteria confront intracellular killing mechanisms. We monitored intracellular survival by performing gentamicin protection assays where T4, T4ΔrrgA, and T4ΔrrgA∇(lacE::rrgA) strains were incubated with macrophages (BMDMs). After antibiotic killing of extracellular bacteria, the number of internalized viable pneumococci was determined by quantitative plating at various time points. There were significantly fewer CFUs of the T4ΔrrgA mutant inside the macrophages at the beginning of the experiment (Fig. 6) (P = 0.0011), which further supported the differences in uptake observed in the fluorescence-activated cell sorting (FACS)-based phagocytosis assay (Fig. 1). The rate of bacterial killing was similar for all pneumococcal strains (Fig. 6). Interestingly, the increased uptake of RrgA-expressing bacteria and the unaffected intracellular killing rate resulted in significantly higher numbers of survivors for the T4 and T4ΔrrgA∇(lacE::rrgA) strains compared to the T4ΔrrgA strain even 10 h after infection (P < 0.05).
FIG 6 .
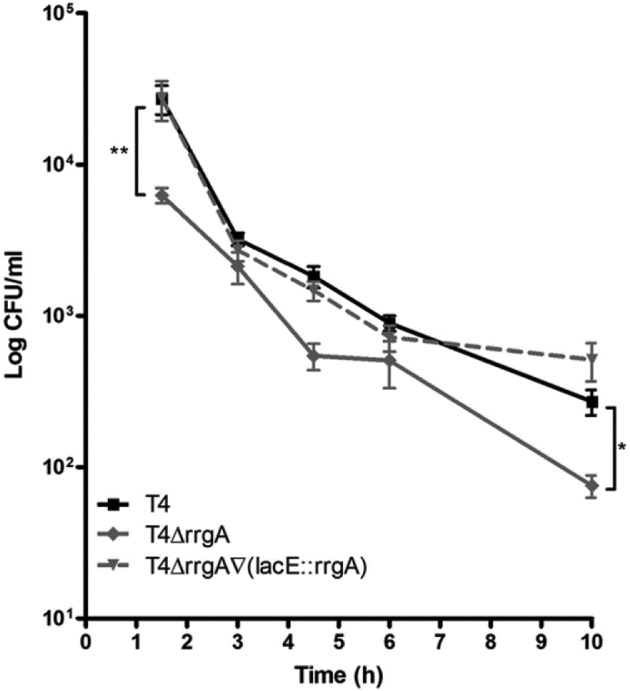
Killing by macrophages is not affected by the presence of RrgA, but intracellular bacteria are found up to 10 h postchallenge. Bacteria were incubated onto monolayers of BMDMs for 1 h. Extracellular bacteria were killed by the addition of gentamicin to the assay medium. At various time points, cells were lysed with 2% saponin, and viable bacteria were determined by plating serial dilutions. The rate of bacterial death was similar for all strains, but bacteria expressing RrgA, i.e., T4 and T4ΔrrgA∇(lacE::rrgA) strains, were taken up to a greater extent, which led to higher numbers of intracellular bacteria after 10 h. Each treatment was done in triplicate and was repeated at least three times. Values that are significantly different are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01.
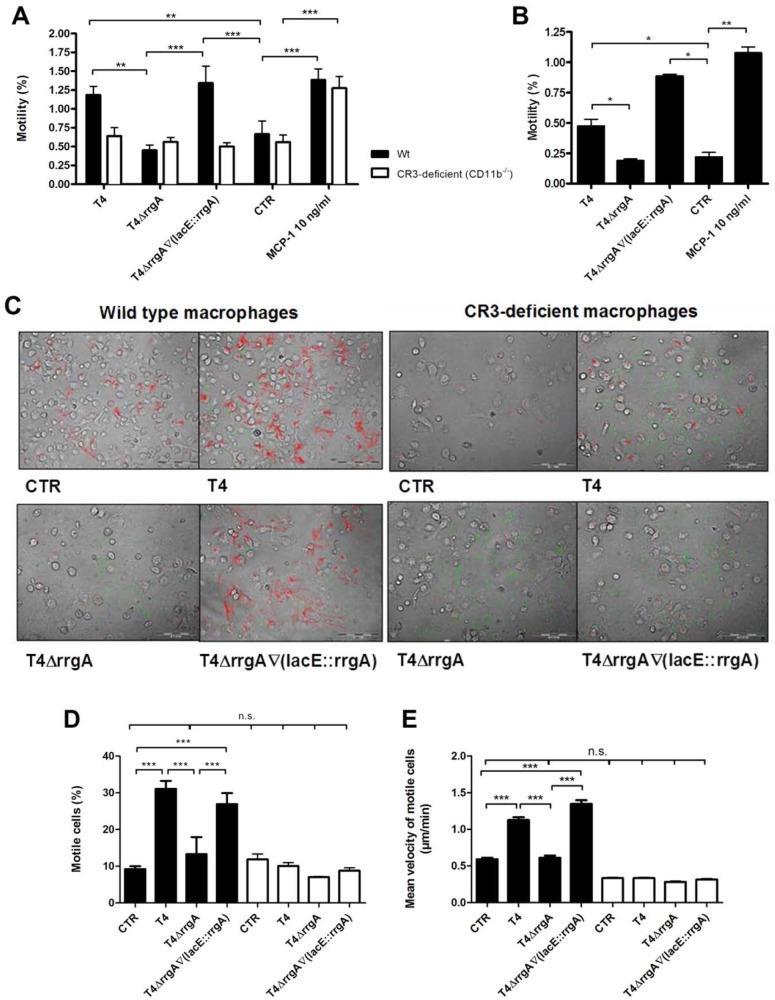
The interaction between RrgA and CR3 leads to increased motility and migratory behavior of murine BMDMs and human THP-1 cells.
CR3 is a highly versatile pattern recognition receptor that when ligated activates leukocytes via signaling complexes and actin reorganization. This activation not only mediates phagocytosis but also promotes leukocyte transmigration. The motility/migration of immune cells affects infection, inflammation, and wound healing, and pathogens may utilize this cellular function in order to disseminate within the host (21–24). Therefore, we investigated whether the identified interaction between RrgA and CR3 affected the motility/migration of murine BMDMs (Fig. 7A) and human THP-1 cells (Fig. 7B). Motility/migration assays were performed using a Transwell system, in which we determined that strain T4 expressing RrgA increased the motility of BMDMs and THP-1 cells approximately twofold compared to untreated control cells (Fig. 7A, P < 0.01, and Fig. 7B, P < 0.05). Interestingly, the T4ΔrrgA strain, lacking RrgA, did not induce the motility/migration of macrophages, while induction of motility/migration was restored in the complemented strain, T4ΔrrgA∇(lacE::rrgA) strain. Both CR3-expressing and CR3-deficient (CD11b−/−) macrophages migrated across the Transwell chamber when macrophage chemoattractant 1 (MCP-1) was present in the bottom chamber. However, the two RrgA-expressing strains failed to induce motility/migration in CR3-deficient (CD11b−/−) BMDMs (Fig. 7A), supporting the role for this receptor in macrophage motility/migration upon interaction with pneumococcal RrgA. Finally, when BMDMs were preincubated with antibodies against CD11b, there were no differences in motility/migration between untreated control cells and cells challenged with RrgA+ or RrgA− bacteria (data not shown).
FIG 7 .
The interaction between RrgA and CR3 leads to enhanced motility of murine and human macrophages. Wild-type (Wt) and CR3-deficient (CD11b−/−) BMDMs were challenged with pneumococci. (A) Macrophage motility was measured using a Transwell system. The percentages of cells that migrated were calculated by dividing the output by the input. In wild-type BMDMs, the T4 strain increased the motility of macrophages approximately twofold compared to the untreated control (CTR), but the T4ΔrrgA mutant failed to do so. The T4ΔrrgA∇(lacE::rrgA) strain induced motility to levels similar to those of the T4 strain. No statistically significant difference was found between cells infected with T4. All strains failed to induce motility in CR3-deficient macrophages, but the addition of macrophage chemoattractant protein 1 (MCP-1) in the bottom chamber led to increased migration of wild-type and CR3-deficient cells. (B) The presence of RrgA also enhanced the motility of human macrophages (THP-1 cells). (C to E) Motility was monitored using time-lapse video microscopy for up to 6 h. (C) Representative pictures illustrating all cell movements (red), FITC-labeled pneumococci (green), and BMDMs (gray). (D) Wild-type BMDMs infected with strains expressing RrgA were more motile than those infected with the T4ΔrrgA mutant. (E) T4 and T4ΔrrgA∇(lacE::rrgA) strains, but not the T4ΔrrgA strain, moved faster than untreated controls. (D and E) CR3-deficient macrophages did not display either induction of motility or an increase in velocity when challenged with any of the pneumococcal strains. Values that are significantly different are indicated by bars and asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Values that were not significantly different are indicated by a bar and n.s. (for not significant).
To further confirm that the motile/migratory behavior of BMDMs is induced by RrgA and CR3, we performed additional motility assays using time-lapse video microscopy. Single-cell tracking revealed that the motility/migration of BMDMs was induced by pneumococci expressing RrgA [T4 and T4ΔrrgA∇(lacE::rrgA) strains], but not by the T4ΔrrgA strain (P < 0.001 [Fig. 7C and D]). CR3-deficient BMDMs were not affected by the addition of any of the pneumococcal strains; their motility/migration remained at control levels (Fig. 7C and D). Also, the velocity of the cells was measured using this method, which revealed that cells infected with T4 and T4ΔrrgA∇(lacE::rrgA) strains moved faster than uninfected control cells (Fig. 7D) (P = 0.00015). In contrast, BMDMs infected with the T4ΔrrgA mutant did not move faster than the control cells. The velocity of the CR3-deficient BMDMs did not increase after bacterial challenge and remained at control levels throughout the experiment.
DISCUSSION
We show for the first time that expression of pili on the pneumococcal surface leads to enhanced phagocytosis by murine BMDMs and human macrophages (THP-1 cells). Furthermore, we provide evidence that the pilus-associated adhesin RrgA, rather than the pilus-forming RrgB or pilus-associated RrgC subunits, promotes phagocytosis (25). Moreover, we show that a specific antibody against murine CD11b decreased phagocytosis by BMDMs and THP-1 cells of pneumococci expressing RrgA to a similar level of phagocytosis as that of RrgA− bacteria, suggesting that RrgA promotes phagocytosis via CR3. Finally, interaction between RrgA and CR3 appears to be direct and mediated by the CD11b integrin domain of CR3, thereby representing the first identified cellular receptor for a Gram-positive pilus. Even though RrgA has been shown to interact with a number of extracellular matrix proteins, purified CR3 binding to RrgA-expressing pneumococci was not affected by the addition of extracellular matrix proteins, like collagen 1, known to interact with either of these two proteins (6).
Our data demonstrate that internalization of RrgA+ and RrgA− bacteria follows a similar route, leading to phagosome formation and intracellular killing. However, pneumococci expressing RrgA were phagocytosed to a greater extent and BMDMs harbored higher numbers of viable RrgA+ than RrgA− bacteria over a longer time period (at least 10 h). Pneumococci produce several components toxic to macrophages, such as pneumolysin and H2O2, and therefore, prolonged intracellular survival time might lead to death of infected macrophages prior to the complete eradication of ingested bacteria.
When pneumococci expressing RrgA were injected intranasally and intraperitoneally into wild-type mice, they reached the bloodstream faster and in higher numbers compared to RrgA− bacteria which, irrespective of infection route, also resulted in an earlier onset of severe disease. In contrast, RrgA-expressing or non-RrgA-expressing pneumococci did not differ in growth after i.v. challenge, and the time to onset of severe disease was also not affected by RrgA. This suggests that RrgA does not promote resistance to clearing mechanisms in the bloodstream.
Expression of RrgA and CR3 was required for rapid appearance of bacteria in the bloodstream when mice were challenged intranasally or intraperitoneally. This supports a role for an interaction between these proteins in vivo for pneumococcal dissemination.
It has been demonstrated that CR3 is upregulated on AMs in the lungs after infection with S. pneumoniae (26). AMs are central to the normal surveillance of pneumococci. They may, however, have a dual function in host defense, as it was recently demonstrated that AMs migrate from the lungs to the lung-draining lymph nodes early after infection with an S. pneumoniae 6B isolate (27). Also, the migration of activated macrophages from the peritoneal cavity to the lymph nodes and blood has been shown to depend on CR3 (28). Thus, activated macrophages carrying pneumococci might migrate in a CR3-dependent manner to regional lymph nodes, and from there, infected phagocytic cells or liberated live bacteria are thought to reach the bloodstream, thereby contributing to systemic spread from local sites. However, further in vivo studies are needed to demonstrate that pneumococci can be translocated into the bloodstream via infected immune cells. Also, it is important to stress that other pneumococcal virulence properties, such as PspC, might mediate a direct translocation of bacteria across biological barriers.
For a commensal pathogen like Streptococcus pneumoniae, where healthy carriage vastly dominates over disease, each interaction between microbe and host may have dual effects, sometimes favoring the host and other times the microbe. Here we demonstrate that the pilus-associated adhesin RrgA promotes phagocytosis by BMDMs in a process requiring CR3. This process is likely to enhance bacterial clearance. However, the simultaneous activation of motility and migratory behavior of macrophages may tilt the balance in favor of the ingested microbe. The bistability of pilus expression in a pneumococcal population (29, 30) and the fact that only a subset of pneumococcal strains harbor the pilus 1 islet are additional arguments for the dual role of pneumococcal pili in its interaction with the host.
MATERIALS AND METHODS
Bacterial strains and construction of mutants.
S. pneumoniae T4 (TIGR4) of serotype 4 belongs to a clone (ST205) with high invasive disease potential in Sweden (31–33). The insertion-deletion mutagenesis used for creating the rrgA mutant strain T4ΔrrgA, is described elsewhere (1, 34). Complementation in trans was achieved by inserting a second, intact copy of rrgA into the lacE lactose utilization operon, creating the T4ΔrrgA∇(lacE::rrgA) strain (4, 35). Mutants were checked by PCR, sequencing, and immunogenicity using antibodies to pilin subunits.
Murine BMDMs and human macrophage-like THP-1 cells.
Murine BMDMs were extracted (7, 36), and cells were plated in 24-well plates (1 × 106 cells per well) and incubated for 7 days at 37°C and 5% CO2. Human monocytic leukemia THP-1 cells (American Type Culture Collection [ATCC], Manassas, VA) were cultured in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCSi), 2 mM l-glutamine, and 10 mM HEPES. To induce differentiation, THP-1 cells (5 × 105 cells per well) were seeded onto a 24-well plate with 100 ng/ml of phorbol myristate acetate (PMA) (Sigma) for 30 h at 37°C and 5% CO2. Before use, cells were washed to remove nonadherent cells. Each condition was performed in triplicate, and the experiments were repeated at least three times.
Mouse challenge.
The mice were 6 to 10 weeks old and were matched by age and sex. The mice were inoculated intranasally, intraperitoneally, or intravenously with ca. 5 × 106 CFU per mouse. Bacterial growth in blood was monitored by taking blood samples from the tail at various time points and plating the samples in serial dilutions on blood agar plates. The health status of the mice was carefully monitored, and clinical scores were given.
In vitro phagocytosis assays.
Bacteria were resuspended in a solution containing 2.5 mg fluorescein isothiocyanate (FITC) (isomer I; Sigma), 1 ml dimethyl sulfoxide (DMSO) (Sigma), and 9 ml FITC buffer (0.05 M Na2CO3 [Merck, Darmstadt, Germany] and 0.1 M NaCl [Merck] in double-distilled water) and incubated on ice for 1 h. After the bacteria were washed 3 times, macrophages were challenged with FITC-labeled pneumococci at a multiplicity of infection (MOI) of about 60 bacteria per cell, spun down at 1,500 rpm for 5 min, and incubated for 60 min at 37°C and 5% CO2. Serum-free RPMI 1640 (Gibco/Invitrogen) supplemented with 2 mM glutamine and 10 mM HEPES was used as assay medium. Following bacterial challenge, the cells were washed 3 times with cold phosphate-buffered saline (PBS). The cells were detached by 15- to 30-min incubation on ice with PBS supplemented with 5 mM EDTA at pH 8.0 and 4.0 mg ml−1 lidocaine (Sigma). The cells were transferred to 96-well V-bottom plates, washed in PBS, and resuspended in 2% paraformaldehyde (Sigma). Before data acquisition by FACS, extracellular fluorescence was quenched by adding trypan blue (Sigma) at pH 5.5. To inhibit phagocytosis, 10 µg ml−1 of cytochalasin D (Sigma) was added 15 min prior to bacterial challenge. For antibody blocking experiments, the following affinity-purified antibodies were used: anti-mouse CD11c (N418), anti-mouse CD11b (M1/70) (eBioscience Inc., San Diego, CA) and anti-human CD11b (VIM12) (Invitrogen). BMDMs were pretreated with selected anti-mouse antibody and THP-1 cells were pretreated with selected anti-human antibody at a concentration of 20 µg µl−1 for 30 min at 37°C and 5% CO2 prior to bacterial challenge. During FACS acquisition, 1 × 104 cells were counted in each sample. Each condition was added in triplicate, and the experiments were repeated at least three times. The phagocytic index for each sample was calculated by multiplying the percentage of FITC-positive cells with the mean fluorescence intensity (MFI), and dividing this by the mean MFI for the wild-type (T4) samples multiplied with the percentage of FITC-positive cells.
For the microbead phagocytosis assays, we used full-length wild-type rRrgA (amino acid [aa] 39 to aa 862) (1, 4, 37), full-length wild-type rRrgC (1, 4), and BSA conjugated to carboxylate-modified yellow-green fluorescent microsphere (fluorosphere carboxylate from Molecular Probes, Invitrogen) according to the manufacturer’s protocol. THP-1 and BMDM cells, differentiated onto chamber slides (Nunc, Rochester, NY), were treated with 10 µg ml−1 of latex beads alone or beads covered with rRrgA, rRrgC, or BSA and incubated for 3 h at 37°C and 5% CO2. The inhibition assay was performed as previously described using anti-mouse (M1/70) and anti-human (VIM12) anti-CD11b antibodies. Each treatment was repeated in duplicate. Each preparation was examined at ×100 magnification under oil immersion. At least 10 visual fields for each sample containing at least 10 or more cells were selected in a random fashion, and the number of internalized beads was counted manually, taking care to exclude beads that were only adherent to the cell surface. Differences in the number of beads were tested with the nonparametric Mann-Whitney U test.
Purification of CR3 (CD11b/CD18).
Human CD11b/CD18 integrin was purified from human blood buffy coat cell lysates by affinity chromatography on MEM170 monoclonal antibody-Sepharose and eluted at pH 11.5 in the presence of 2 mM MgCl2 and 1% octyl glucoside. Human blood buffy coats were from Finnish Red Cross Blood Transfusion Service, Helsinki, Finland. Integrin purity was checked by polyacrylamide gel electrophoresis in the presence of SDS (see Fig. S3 in the supplemental material).
Flow cytometry analysis of CR3 binding to pneumococci.
FACS analysis was performed to quantify the binding of CR3 to RrgA-expressing pneumococci. Bacteria were grown overnight on solid medium and then harvested into PBS. Bacteria (108 CFU) were incubated with 10 µg/ml of purified CR3 (CD11b/CD18) or with PBS as a negative control for 30 min at 37°C. After the bacteria were washed twice in PBS containing 0.1% BSA [PBS-BSA (0.1%)], binding was detected following incubation with an anti-CD11b MAb labeled with PerCp-Cy5.5 (BD BioScience) at a final concentration of 10 µg ml−1 for 1 h on ice. The cells were washed twice in PBS-BSA (0.1%) and once in PBS and then fixed in 2% paraformaldehyde for 20 min at room temperature. Finally, the bacteria were washed, and the fluorescence intensity was analyzed by a flow cytometer (CyAn ADP; Beckman Coulter). Bacteria were detected using log forward and log side scatter dot plots, and a gating region was set to exclude debris and larger aggregates of bacteria. A total of 1 × 104 bacteria (events) were analyzed for fluorescence using log scale amplifications. Comparison between the fluorescence intensity of the bacteria incubated with purified CR3 with that of bacteria incubated with PBS alone were illustrated by two-dimensional overlaid histograms representing fluorescence intensities calculated with voltage adjustment at FL4 linear channel. FACS results were also expressed as the change in the mean fluorescence intensity (ΔMFI), where ΔMFI was calculated by subtracting the mean fluorescence intensity of the bacteria incubated with purified CR3 from that of bacteria incubated with PBS alone.
Far-Western blots.
Human CR3 (CD11b/CD18) or CD11b-I domain-GST was blotted onto nitrocellulose membranes using the Manifold system in concentrations of 0.5, 1, 5, and 10 µg; 5 µg of BSA and 5 µg of glutathione S-transferase (GST) were used as negative controls. The membranes were blocked with PBS with 0.01% Tween 20 [PBS-Tween (0.01%)] and 5% milk for 30 min at room temperature. The membranes were washed briefly and incubated with 2 µg of rRrgA in the presence of PBS-Tween (0.01%) and 1% milk overnight at 4°C. The membranes were washed, primary antibody (Rb−anti-RrgA [1:1,000]) was added, and the membranes were incubated for 3 h at room temperature on a rocking platform. Finally, the membranes were incubated with secondary antibody (anti-Rb peroxidase [1:2,000]; Sigma) for 1 h. Experiments were repeated three times, and representative experiments are shown.
ELISA.
To assess direct binding of CR3 (CD11b/CD18) and CD11b I domain-GST to the rRrgA protein, microtiter plates were coated overnight at 4°C with 10 µg/ml of purified CR3 (or CD11b I domain-GST). Subsequent steps were conducted at room temperature (RT) for 1 h each. Nonspecific binding sites were blocked with PBS-BSA (3%), followed by the addition of rRrgA (different concentrations from 0 to 200 ng per well) in PBS-Tween (0.01%) (PBST). Bound rRrgA was detected with a specific antibody against rRrgA (Rb−anti-RrgA) and anti-rabbit serum conjugated to horseradish peroxidase (Sigma), both diluted 1:5,000 in PBST. Sigmafast-OPD (o-phenylenediamine dihydrochloride) (Sigma) was used for detection, and absorbance was measured at 492 nm. In inhibition assays, CR3 and CD11b I domain-GST were pretreated with 25 µg/ml of anti-CD18 7E4 MAb and anti-CD11b MEM170 MAb, respectively, for 30 min at RT.
In vitro gentamicin protection assay.
The gentamicin protection assay was performed as described elsewhere (7).
In vitro motility assays.
Differentiated BMDM cells were seeded in serum-free medium (RPMI 1640 supplemented with 2 mM glutamine and 10 mM HEPES) in small petri dishes (35 by 10 mm) at a density of 1 × 106 cells per dish. The cells were either left untreated (control) or challenged with pneumococci at an MOI of ca. 60 bacteria per cell, or recombinant mouse MCP-1 (10 ng ml−1; Invitrogen) was added to the bottom chamber (38). After 1.5 h, gentamicin was added to the medium (400 µg ml−1). After an additional 1.5 h, the cells were gently scraped off, counted (input), and transferred to Transwell inserts containing RPMI 1640, and supplemented with 2 mM glutamine, 10 mM HEPES, 100 µg ml−1 gentamicin, and 10% FCS (time point zero). Additionally, samples were taken from the medium to ensure that the extracellular bacteria were killed. The cells were incubated for 15 h before the inserts were removed, and the cells in the lower wells were scraped off and counted (output). The percentage of migrated cells was calculated by dividing the output by the input. Each condition was added in triplicate, and the experiments were repeated at least three times.
Time-lapse video microscopy.
Time-lapse video microscopy was performed by the method of Behnsen et al. (39).
Statistical analysis.
In vitro assays were analyzed using one-way analysis of variance (ANOVA) with Bonferroni’s posttest (mean fluorescence intensity, CFU numbers, and motile cells), unless otherwise stated. In vivo assays were analyzed using Kaplan-Meier log rank test (survival), Kruskal-Wallis test with Dunn’s posttest (CFU numbers), or chi-square test and Fisher’s exact test (proportion of bacteremic mice). Graph Pad Prism 4.0 software was used (Graph Pad Software, Inc.), and a P value of <0.05 was considered statistically significant.
SUPPLEMENTAL MATERIAL
CR3 binding to pneumococci expressing RrgA is not affected by the presence of extracellular matrix proteins (collagen I, fibronectin, laminin, or vitronectin). Flow cytometry analysis of CR3 binding to RrgA protein associated with the S. pneumoniae cell surface in the presence of extracellular matrix proteins (EMPs) was performed. The T4 strain and its mutants (108 live bacteria) were incubated with 10 µg/ml of the indicated EMP with 10 mg/ml of purified CR3 (histograms outlined by thick black lines) or without purified CR3 (light gray filled histograms). CR3 binding was detected with an anti-CD11b MAb (PerCp-Cy5.5 labeled) at a final concentration of 10 µg/ml and is observed by a shift to the right in the population of bacteria incubated with the purified CR3 protein and expressing RrgA on their surface, i.e., the T4 and T4ΔrrgA∇(lacE::rrgA) strains, compared with cells incubated with PBS (control) (filled gray) or with CR3 but not expressing RrgA, i.e., the T4ΔrrgA strain. Binding was calculated by subtracting the mean fluorescence intensity of bacteria incubated with purified CR3 protein plus the indicated EMP from that of the bacteria incubated with the same indicated EMP and PBS (ΔMFI). The average ΔMFI values from three independent experiments are shown as numbers in the box in the top right-hand corner of each graph. The addition of EMPs (collagen I, fibronectin, laminin, or vitronectin) did not inhibit or enhance CR3 binding to pneumococcal cells expressing RrgA. Download
The recombinant RrgA protein binds to purified human CR3 and to CD11b-I domain-GST in a concentration-dependent manner. (A and B) In ELISA experiments, increasing amounts of recombinant RrgA were incubated with coated CR3 (A) or CD11b-I domain-GST (B). Binding was detected by using a specific antibody against rRrgA (Rb−anti-RrgA) and an anti-rabbit IgG antibody conjugated to horseradish peroxidase (HRP). The binding of rRrgA to CD11b (A) and to the CD11b-I domain-GST (B) is dependent on the concentration of rRrgA. Download
Polyacrylamide gel of purified CR3 (CD11b/CD18 integrin). Purified CR3 was run on a 8% polyacrylamide gel in the presence of SDS and stained with Coomassie blue. The positions of the molecular mass markers are indicated to the left of the gel, and the positions of CD11b and CD18 chains are indicated by arrows to the right of the gel. Download
ACKNOWLEDGMENTS
This work was supported by grants from the Torsten and Ragnar Söderbergs foundation, the Swedish Research Council, the Swedish Royal Academy of Sciences, the Swedish foundation for Strategic Research, ALF grant from Stockholm County Council, EU commission (EIMID-IAPP and EIMID-ITN projects), IRTG Germany funded by DFG, Knut and Alice Wallenberg Foundation, the Academy of Finland, and the Finnish Medical Foundation.
We also thank Martin Rottenberg from the Karolinska Institutet, MTC, for providing rat anti-mouse F4/80 antibodies and Monica Moschioni at Novartis Vaccine for help and provision of sera and proteins.
Footnotes
Citation Orrskog S, et al. 2012. Pilus adhesin RrgA interacts with complement receptor 3, thereby affecting macrophage function and systemic pneumococcal disease. mBio 4(1):e00535-12. doi:10.1128/mBio.00535-12.
REFERENCES
- 1. Barocchi MA, et al. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103:2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguiar SI, Serrano I, Pinto FR, Melo-Cristino J, Ramirez M. 2008. The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 8:41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moschioni M, et al. 2008. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197:888–896 [DOI] [PubMed] [Google Scholar]
- 4. Nelson AL, et al. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Izoré T, et al. 2010. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18:106–115 [DOI] [PubMed] [Google Scholar]
- 6. Hilleringmann M, et al. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 4:e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albiger B, et al. 2007. Toll-like receptor 9 acts at an early stage in host defence against pneumococcal infection. Cell. Microbiol. 9:633–644 [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Barke RA, Charboneau R, Schwendener R, Roy S. 2008. Morphine induces defects in early response of alveolar macrophages to Streptococcus pneumoniae by modulating TLR9-NF-kappa B signaling. J. Immunol. 180:3594–3600 [DOI] [PubMed] [Google Scholar]
- 9. Sun K, Metzger DW. 2008. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 14:558–564 [DOI] [PubMed] [Google Scholar]
- 10. Arredouani M, et al. 2004. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. J. Exp. Med. 200:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arredouani MS, et al. 2006. The macrophage scavenger receptor SR-AI/II and lung defense against pneumococci and particles. Am. J. Respir. Cell Mol. Biol. 35:474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas CA, et al. 2000. Protection from lethal gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. J. Exp. Med. 191:147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Cabec V, Carréno S, Moisand A, Bordier C, Maridonneau-Parini I. 2002. Complement receptor 3 (CD11b/CD18) mediates type I and type II phagocytosis during nonopsonic and opsonic phagocytosis, respectively. J. Immunol. 169:2003–2009 [DOI] [PubMed] [Google Scholar]
- 14. Gbarah A, Gahmberg CG, Ofek I, Jacobi U, Sharon N. 1991. Identification of the leukocyte adhesion molecules CD11 and CD18 as receptors for type 1-fimbriated (mannose-specific) Escherichia coli. Infect. Immun. 59:4524–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishibashi Y, Claus S, Relman DA. 1994. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte CR3 (CD11b/CD18). J. Exp. Med. 180:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diamond MS, et al. 1990. ICAM-1 (CD54): a counter-receptor for Mac-1 (CD11b/CD18). J. Cell Biol. 111:3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wright SD, Reddy PA, Jong MT, Erickson BW. 1987. C3bi receptor (complement receptor type 3) recognizes a region of complement protein C3 containing the sequence Arg-Gly-Asp. Proc. Natl. Acad. Sci. U. S. A. 84:1965–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diamond MS, Alon R, Parkos CA, Quinn MT, Springer TA. 1995. Heparin is an adhesive ligand for the leukocyte integrin Mac-1 (CD11b/CD1). J. Cell Biol. 130:1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agramonte-Hevia J, González-Arenas A, Barrera D, Velasco-Velázquez M. 2002. Gram-negative bacteria and phagocytic cell interaction mediated by complement receptor 3. FEMS Immunol. Med. Microbiol. 34:255–266 [DOI] [PubMed] [Google Scholar]
- 20. Rosen H, Gordon S. 1987. Monoclonal antibody to the murine type 3 complement receptor inhibits adhesion of myelomonocytic cells in vitro and inflammatory cell recruitment in vivo. J. Exp. Med. 166:1685–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guidi-Rontani C. 2002. The alveolar macrophage: the Trojan horse of Bacillus anthracis. Trends Microbiol. 10:405–409 [DOI] [PubMed] [Google Scholar]
- 22. Kubica M, et al. 2008. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One 3:e1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen L, Pieters J. 2005. The Trojan horse: survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol. 15:269–276 [DOI] [PubMed] [Google Scholar]
- 24. Zlotkin A, et al. 2003. Trojan horse effect: phagocyte-mediated Streptococcus iniae infection of fish. Infect. Immun. 71:2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hilleringmann M, et al. 2009. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28:3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirby AC, Raynes JG, Kaye PM. 2006. CD11b regulates recruitment of alveolar macrophages but not pulmonary dendritic cells after pneumococcal challenge. J. Infect. Dis. 193:205–213 [DOI] [PubMed] [Google Scholar]
- 27. Kirby AC, Coles MC, Kaye PM. 2009. Alveolar macrophages transport pathogens to lung draining lymph nodes. J. Immunol. 183:1983–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao C., Lawrence DA, Strickland DK, Zhang L. 2005. A specific role of integrin Mac-1 in accelerated macrophage efflux to the lymphatics. Blood 106:3234–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Basset A, et al. 2011. Expression of the type 1 pneumococcal pilus is bistable and negatively regulated by the structural component RrgA. Infect. Immun. 79:2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Angelis G, et al. 2011. The Streptococcus pneumoniae pilus-1 displays a biphasic expression pattern. PLoS One 6:e21269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henriques-Normark B, et al. 2003. Clonal analysis of Streptococcus pneumoniae nonsusceptible to penicillin at day-care centers with index cases, in a region with low incidence of resistance: emergence of an invasive type 35B clone among carriers. Microb. Drug Resist. 9:337–344 [DOI] [PubMed] [Google Scholar]
- 32. Sandgren A, et al. 2005. Virulence in mice of pneumococcal clonal types with known invasive disease potential in humans. J. Infect. Dis. 192:791–800 [DOI] [PubMed] [Google Scholar]
- 33. Sjöström K, et al. 2006. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin. Infect. Dis. 42:451–459 [DOI] [PubMed] [Google Scholar]
- 34. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 35. Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Racoosin EL, Swanson JA. 1989. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J. Exp. Med. 170:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moschioni M, et al. 2010. The two variants of the Streptococcus pneumoniae pilus 1 RrgA adhesin retain the same function and elicit cross-protection in vivo. Infect. Immun. 78:5033–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deshmane SL, Kremlev S, Amini S, Sawaya BE. 2009. Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interferon Cytokine Res. 29:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Behnsen J, et al. 2007. Environmental dimensionality controls the interaction of phagocytes with the pathogenic fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 3:e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CR3 binding to pneumococci expressing RrgA is not affected by the presence of extracellular matrix proteins (collagen I, fibronectin, laminin, or vitronectin). Flow cytometry analysis of CR3 binding to RrgA protein associated with the S. pneumoniae cell surface in the presence of extracellular matrix proteins (EMPs) was performed. The T4 strain and its mutants (108 live bacteria) were incubated with 10 µg/ml of the indicated EMP with 10 mg/ml of purified CR3 (histograms outlined by thick black lines) or without purified CR3 (light gray filled histograms). CR3 binding was detected with an anti-CD11b MAb (PerCp-Cy5.5 labeled) at a final concentration of 10 µg/ml and is observed by a shift to the right in the population of bacteria incubated with the purified CR3 protein and expressing RrgA on their surface, i.e., the T4 and T4ΔrrgA∇(lacE::rrgA) strains, compared with cells incubated with PBS (control) (filled gray) or with CR3 but not expressing RrgA, i.e., the T4ΔrrgA strain. Binding was calculated by subtracting the mean fluorescence intensity of bacteria incubated with purified CR3 protein plus the indicated EMP from that of the bacteria incubated with the same indicated EMP and PBS (ΔMFI). The average ΔMFI values from three independent experiments are shown as numbers in the box in the top right-hand corner of each graph. The addition of EMPs (collagen I, fibronectin, laminin, or vitronectin) did not inhibit or enhance CR3 binding to pneumococcal cells expressing RrgA. Download
The recombinant RrgA protein binds to purified human CR3 and to CD11b-I domain-GST in a concentration-dependent manner. (A and B) In ELISA experiments, increasing amounts of recombinant RrgA were incubated with coated CR3 (A) or CD11b-I domain-GST (B). Binding was detected by using a specific antibody against rRrgA (Rb−anti-RrgA) and an anti-rabbit IgG antibody conjugated to horseradish peroxidase (HRP). The binding of rRrgA to CD11b (A) and to the CD11b-I domain-GST (B) is dependent on the concentration of rRrgA. Download
Polyacrylamide gel of purified CR3 (CD11b/CD18 integrin). Purified CR3 was run on a 8% polyacrylamide gel in the presence of SDS and stained with Coomassie blue. The positions of the molecular mass markers are indicated to the left of the gel, and the positions of CD11b and CD18 chains are indicated by arrows to the right of the gel. Download